Abstract
Fast evolving of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has caused the spreading of COVID-19 disease rapidly around the globe. The mutation, especially in the gene encoding spike protein has helped the virus adapt and evade human immune system, as well as affect the efficacy of the immunizations and treatments. SARS-CoV-2 variant carrying D614G amino acid change at the spike protein is the most dominant strain in the pandemic. Therefore, efficient detection of the SARS-CoV-2 variants including D614G mutation is critical to control the COVID-19 pandemic. Herein, we report a dual synthetic mismatches CRISPR/Cas12a (dsmCRISPR) method to detect the SARS-CoV-2 D614G mutation with high sensitivity and specificity. By targeting SARS-CoV-2 D614G mutation, synthetic mismatch crRNAs were designed from -3 to +3 position around the mutation site. To improve the sensitivity and specificity, a synthetic mismatch primer with a 3′-terminal base complementary to the D614G point mutation and a mismatch next to 3′-terminal base was used to specifically amplify the D614G mutation site with higher annealing temperature. Using synthetic mismatch crRNA-(-1), a higher ratio (13.45) of the fluorescence between G614 and D614 was observed. When combined with mismatch primer to amplify D614G mutation, the fluorescence ratio of G614/D164 template detected was increased by 73.53% to 23.12. This method can detect the SARS-CoV-2 D614G mutation nucleic acid with high sensitivity, which was validated with synthetic SARS-CoV-2 D614G RNA. Therefore, the dsmCRIPSR method has significant potential to serve as a sensitive and specific assay for SARS-CoV-2 D614G detection and could be further extended for the detection of other SARS-CoV-2 variants of interest.
Keywords: SARS-CoV-2, COVID-19, D614G, dsmCRISPR
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified in December 2019 as the causative agent of novel coronavirus disease (COVID-19) (WHO, 2020; Wu et al., 2020). As of June 27, 2021, there are 181,677,542 coronavirus cases and 3,935,351 deaths worldwide, according to the World Health Organization (WHO) (WHO, 2021). As a RNA virus, the genome of SARS-CoV-2 are prone to mutation during replication (Domingo et al., 1996). According to the sequencing results, same mutations recurred in the SARS-CoV-2 genome. The D614G mutation (aspartate change to glycine at position 614) in the spike (S) protein is the most prevalent variant resulting from an A-to-G nucleotide substitution at genomic position 234035. Despite D614G mutation was only detected in January 2020, Korber and colleagues reported that the mutation is circulating in many places in the world (Korber et al., 2020). D614G mutation is located at the S1 subunit of the S protein which mediates SARS-CoV-2 entry into cells. High viral RNA levels and pseudovirus titers were found in patients infected with the SARS-CoV-2 D614G variant (Korber et al., 2020). Hence, the D614G variant was speculated to be responsible for the high SARS-CoV-2 infectivity due to its high transmissibility (Korber et al., 2020; Li et al., 2020; Plante et al., 2020). Additionally, the mutations of L452R and E484Q within spike receptor-binding domain (RBD) were shown to have reduced susceptibility to monoclonal antibodies, including bamlanivimab, as well as convalescent plasma (Verghese et al., 2021), even though the exact mechanism is not well-established. As the pandemic is lasting for over one and half years, variants of SARS are replacing the initial wild type strain, which render the virus more transmissible and reduce vaccine efficacy, jeopardizing the current efforts in containing the disease.
Currently, the RNA-guide clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease (Cas)-based nucleic acid detection has been demonstrated to be sensitive, specific, and reliable (Chen et al., 2018; Li et al., 2019b). It was reported that Cas nucleases, including Cas12a, Cas12b, Cas13a, Cas14, exhibit nonspecific collateral cleavage activities, which is capable of unleashing indiscriminate nuclease activity upon RNA-guided DNA binding of nucleases (Gootenberg et al., 2017; Harrington et al., 2018; Li et al., 2019a; Li et al., 2018). The CRISPR/Cas12a system's trans-cleavage activity was first reported and patented in nucleic acid detection by Cheng in China (Cheng, 2017). Thereafter, various Cas12a-based biosensing platforms have been developed by researchers from different labs around the globe (Li et al., 2019b). With the social and economy impacts of COVID-19 globally, rapid and convenient methods for SARS-CoV-2 detection is in urgent need. Owing to its easy-to-implement and accurate characteristics, Cas12a-based virus detection system has been widely explored. Latterly, it has been combined with the reverse transcriptase loop-mediated isothermal amplification (RT-LAMP), recombinase polymerase amplification (RPA) and flow assay techniques for POC diagnostics (Broughton et al., 2020; Ding et al., 2020; Wang et al., 2021). However, the methods aforementioned are only suitable for detection of SARS-CoV-2 without single nucleotide mutation. Therefore, the development of a Cas12a-based detection of SARS-CoV-2 mutations with high sensitivity and specificity is urgently required.
In this study, we developed a dual synthetic mismatches CRIPSR/Cas12a-based method (dsmCRISPR) to detect SARS-CoV-2 D614G mutation. Firstly, the method employed a mismatched crRNA, -3 to +3 position around the D614G site, to discriminate the wild type (D614) and the mutant (G614) virus based on the target efficiency score calculated by CRISPR-DT. Secondly, we used a mismatched primer pair (including SNP site located at 3′ end of the forward primer) and the optimized PCR conditions to amplify the mutant rather than the wild type fragment selectively. To sum up, the dsmCRIPSR method provides a novel approach to detect the D614G mutation of SARS-CoV-2 with high sensitivity and specificity, which could be extended to the detection of any other SARS-CoV-2 variants of interest.
2. Materials & methods
2.1. Materials
Primers of RT-PCR, synthesized DNA, RNA and oligonucleotides for crRNA production were purchased from BGI (Shenzhen, China), and dissolved in DNase/RNase free distilled water (Life Technologies, Carlsbad, CA, USA). Then dsDNA and RNA templates were both diluted by a 10-fold gradient and the concentrations of DNA and RNA were ranged from 6 × 1012 to 6 × 100 copies/μL and 6 × 104 to 6 × 100 copies/μL, respectively. LbCas12a (Cpf1) was purchased from Huich Biotech (Shanghai, China). RNaseOUT Recombinant Ribonuclease Inhibitor was purchased from Life Technologies (Carlsbad, CA, USA). ssDNA reporter (/56-FAM/TTTTTTTTT/3BHQ1/) was synthesized from TranSheepBio (Shanghai, China).
2.2. Design of primers and crRNAs
The SARS-CoV-2 D614G (A to G) mutation target sequence was 90 bp Spike fragment with location from 23421 to 23473 in SARS-COV-2’s genome (GeneBank accession NC_045512.2). RT-PCR primers (Table 1 ) targeting SARS-CoV-2 D614G variant mutation were designed with Primer-BLAST tool available from the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The crRNAs with 20nt size were designed using online tool of CRISPR-DT (http://bioinfolab.miamioh.edu/CRIPSR-DT/).
Table 1.
Primers for G614 variant.
| Name | Sequences (5’-3’) |
|---|---|
| Forward | GTTGCTGTTCTTTATCAGCG |
| Reverse | AGAATAAACACGCCAAGTAGGAG |
2.3. Transcription of crRNAs
The transcription of crRNAs was prepared through annealing of complementary crRNA-R oligonucleotides with T7-crRNA-R (Table 2 ). The synthesis of crRNAs was performed at 37°C for 2 h using a T7 High Yield Transcription Kit (Thermo Scientific, San Jose, CA, USA). Then the transcription products of RNA were treated with DNase I and purified with RNA Clean & ConcentratorTM-5 (Zymo Research, Orange, CA, USA) according to the manufacturer's protocol and quantified with NanoDrop 2000 (Thermo Scientific, San Jose, CA, USA).
Table 2.
Oligonucleotides used for preparation of transcription templates in this study.
| Name | Sequences (5’-3’) |
|---|---|
| T7-crRNA-F | GAAATTAATACGACTCACTATAGGT |
| T7-crRNA-0-R | CTGTGCAGTTAACATCCTGAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTAATTTC |
| T7-crRNA-(-1)-R | CTGTGCAGTTAACACGCTGAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTAATTTC |
| T7-crRNA-(-2)-R | CTGTGCAGTTAACACCGTGAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTAATTTC |
| T7-crRNA-(-3)-R | CTGTGCAGTTAACGCCCAGAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTAATTTC |
| T7-crRNA-(+1)-R | CTGTGCAGTTAACGCCCTCAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTAATTTC |
| T7-crRNA-(+2)-R | CTGTGCAGTTAAGACCCTGAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTAATTTC |
| T7-crRNA-(+3)-R | CTGTGCAGTTATGACCCTGAATCTACACTTAGTAGAAATTACCTATAGTGAGTCGTATTAATTTC |
2.4. Cas12a-mediated DNA cleavage assay
Cas12a-mediated DNA cleavage assay contained 1 × NEB buffer 3.1, 0.05 μM Cas12a, 0.2 μM crRNA, 0.5 μM ssDNA reporter, 4U RNaseOUT Recombinant Ribonuclease Inhibitor with 6 × 109 copies of cleavage templates (Table 3 ) in 20 μL reaction volume. The reaction was carried out at 37°C for 30 min on Pro Flex PCR system (Thermo Fisher Scientific Inc., Waltham, MA, USA). The fluorescence emission was excited at 485 nm and detected at 525 nm using a SpectraMax iD5 Plate Reader (Molecular Devices, Sunnyvale, CA, USA) and reactions with no DNA/RNA serve as the background. The non-target sequence (NTS) control is SARS-CoV-2 N gene cloned into plasmid of pET28a-2019nCov-N, which was a kind gift from Dr. Feng Cong at Guangdong Laboratory Animals Monitoring Institute, Guangzhou, China. For visual detection, the images were photographed under UV light using a GelDoc XR Imaging System (Bio-Rad, Hercules, CA, USA).
Table 3.
Oligonucleotides used for preparation of cleavage templates in this study.
| Name | Sequences (5’-3’) |
|---|---|
| D614-F | CCAGGTTGCTGTTCTTTATCAGGATGTTAACTGCACAG AAGTCCCTGTTGCTAT |
| D614-R | ATAGCAACAGGGACTTCTGTGCAGTTAACATCCTGATAAAGAACAGCAACCTGG |
| G614-F | CCAGGTTGCTGTTCTTTATCAGGGTGTTAACTGCACAG AAGTCCCTGTTGCTAT |
| G614-R | ATAGCAACAGGGACTTCTGTGCAGTTAACACCCTGATAAAGAACAGCAACCTGG |
2.5. PCR condition of synthesized DNA template annealed in a temperature gradient
PCR was performed using Platinum Taq polymerase (Invitrogen, Carlsbad, CA, USA). The reaction system contained 1 × Invitrogen PCR buffer with 4 mM MgCl2, 200 μM of each dNTP, 0.5 U Platinum Taq polymerase, 0.4 μM of each primer (F and R), 1 μL of diluted synthesized DNA template (Table 4 ), and DNase/RNase-free water up to 20 μL. Thermal cycling procedure was 94°C for 2 min, 35 cycles at 94°C for 30 s, 50°C or indicated annealing temperature for 15 s and 72°C for 30 s. After amplification, 5 μL of PCR products were electrophoresed at 2% agarose gel under 90 V for 30 min and imaged in GelDoc XR Imaging System (Bio-Rad, Hercules, CA, USA).
Table 4.
Oligonucleotides synthesized as RT-PCR template.
| Name | Sequences (5’-3’) |
|---|---|
| D614-DNA | GTTGCTGTTCTTTATCAGGATGTTAACTGCACAGAAGTCCCTGTTGCTATTCATGCAGATCAACTTACTCCTACTTGGCGTGTTTATTCT |
| G614-DNA | GTTGCTGTTCTTTATCAGGGTGTTAACTGCACAGAAGTCCCTGTTGCTATTCATGCAGATCAACTTACTCCTACTTGGCGTGTT TATTCT |
| D614-RNA | GUUGCUGUUCUUUAUCAGGAUGUUAACUGCACAGAAGUCCCUGUUGCUAUUCAUGCAGAUCA ACUUA CUCCUACUUGGCGUGUUUAUUCU |
| G614-RNA | GUUGCUGUUCUUUAUCAGGGUGUUAACUGCACAGAAGUCCCUGUUGCUAUUCAUGCAGAUCA ACUUACUCCUACUUGGCGUGUUUAUUCU |
2.6. RT-PCR of synthesized RNA template
RT-PCR assay was performed using PrimeScript™ One Step RT-PCR Kit (Takara, Dalian, China). The reaction system contained 1 × Step Buffer, 1 μL PrimeScript 1 Step Enzyme Mix, 0.4 μM of each primer (F and R), 1 μL of diluted synthesized RNA template (Table 4), and RNase-free water up to 20 μL. Thermal cycling procedure was 50°C for 30 min, 94°C for 2 min, 35 cycles at 94°C for 30 s, 68°C for 15 s and 72°C for 30 s.
2.7. Total cell RNA extraction
To address the potential off-target issue and cross reaction with the human genome, the HEK293T human cell line (ATCC; Manassa, Va, USA) was cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, NY, USA), 100 units/mL penicillin and 100 mg/mL streptomycin at 37°C and 5% CO2. 5% CO2. Total cell RNA was extracted from HEK293T cells as previously described (Ma et al., 2020).
2.8. Statistical analysis
Statistical significances were conducted by unpaired two-tailed Student's t test using GraphPad Prism 8.0.2 software (San Diego, CA, USA). One-way ANOVA followed by Tukey's test was used when three or more groups were compared. Data are represented as mean ± SD (* represents P < 0.05, **represents P < 0.01, and ***represents P < 0.001).
3. Results
3.1. Design specific crRNA for SARS-CoV-2 D614G mutation
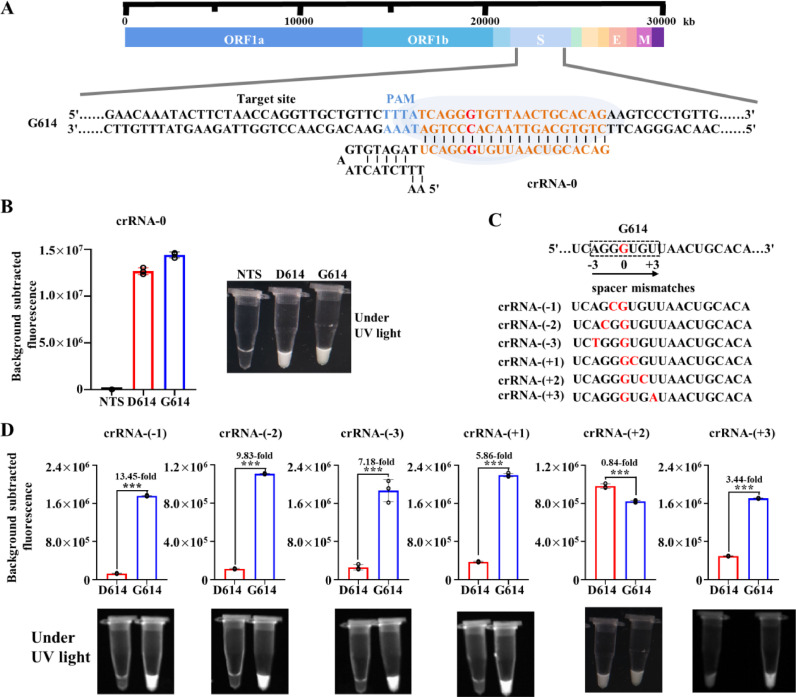
To obtain a high sensitivity of detection, we optimized the crRNA and designed a SARS-CoV-2 D614G mutation specific crRNA targeting the S gene (crRNA-0) (Fig. 1 A). There was only one single nucleotide difference of crRNA-0 at 23403 (A>G) in SARS-CoV-2’s genome (GeneBank accession NC_045512.2) between SARS-CoV-2 D614 and SARS-CoV-2 G614 (Fig. 1A). The results showed that crRNA-0 could not distinguish the D614 and G614 of SARS-CoV-2 (Fig. 1B). To get a better distinction, we introduced additional mismatches of the crRNAs from -3 to +3 around the D614G mutation site with high target efficiency calculated by the CRISPR-DT tool (Fig. 1C). The results showed that all crRNAs, containing synthetic mismatches at different positions, can discriminate D614 from G614 except crRNA-(+2) (Fig. 1D). A higher ratio (13.45) of the fluorescence between G614 and D614 was recorded in the crRNA-(-1) group than other groups. Thus, crRNA-(-1) was selected for further investigation.
Fig. 1.
crRNAs design and specificity of Cas12-based detection of SARS-CoV-2 D614G mutation. (A) SARS-CoV-2 genome map with the detailed sequence information of the designed crRNA targeting the D614G mutation. (B) Fluorescence signals of CRISPR/Cas12a-based detection of D614G mutation with crRNA-0. The presented images of visual detection are shown. NTS, non-template control reaction (n=3). (C) Schematic of the process for design of D614G specific spacers with synthetic mismatches. Synthetic mismatches are placed in -3 to +3 position of the SNP (A23403G) site of D614G mutation. (D) Fluorescence signals of CRISPR/Cas12a-based detection of D614G mutation with crRNAs design in (C). The presented images of visual detection were captured under UV light (n=3).
3.2. Sensitivity evaluation Cas12a-based detection of SARS-CoV-2 D614G mutation
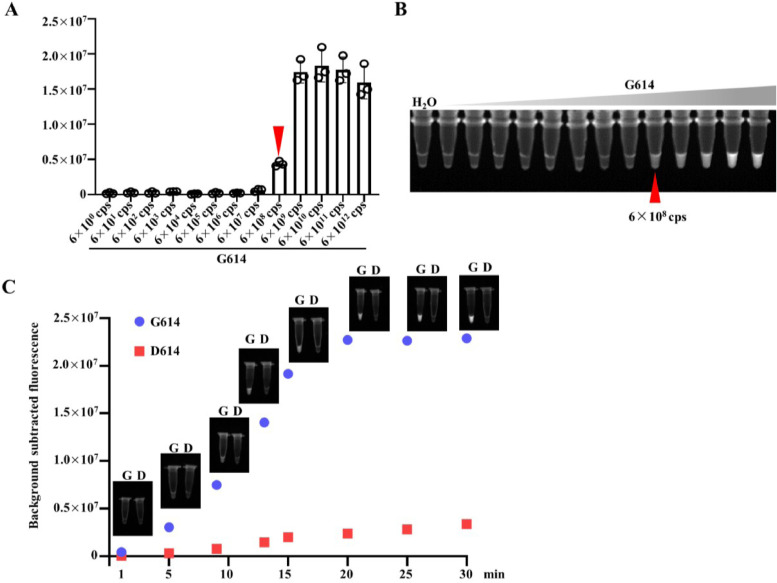
To determine the sensitivity of the Cas12-based visual detection, the synthetic DNA templates were prepared by using a 10-fold serial dilution in sterile water that ranged from 6 × 1012 to 6 × 100 copies in each reaction. As shown in Fig. 2 A and B, the detection limit is 6 × 108 copies. The results of time-course showed that the fluorescence signals increased overtime and reached the plateau in 20 min (Fig. 2C). After 30 min reaction, there was a slightly increase in fluorescence signals generated in D614 template (Fig. 2C). Therefore, 30 min was used as the Cas12a cleavage-time in further study.
Fig. 2.
Sensitivity evaluation Cas12a-based detection of SARS-CoV-2 D614G mutation. (A) Fluorescence signals of 10-fold gradually diluted DNA templates of G614 (from 6 × 1012 cps to 6 × 100 cps) detected by Cas12a-based detection (n=3). (B) The presented images of visual detection in (A) are shown. (C) Visual detection results of D614 (6 × 108 cps) and G614 (6 × 108 cps) for 1, 5, 9, 13, 15, 20, 25, 30 min, respectively. The presented images are shown here (n=3).
3.3. Optimization of Cas12a-based detection of SARS-CoV-2 D614G mutation
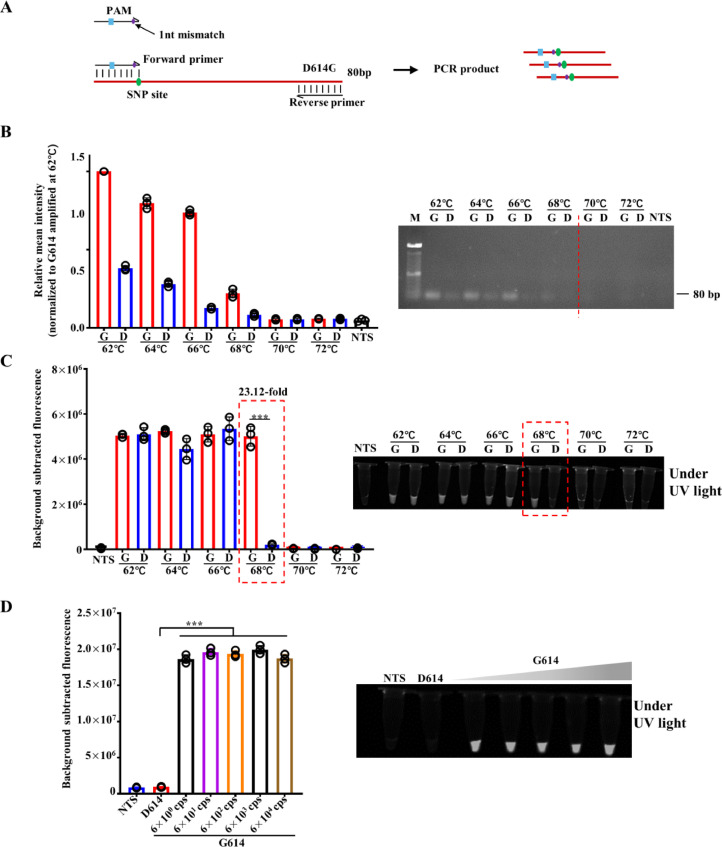
The protospacer adjacent motif (PAM) is required for the Cas12-based nucleic acid detection system. It could be introduced into the forward primer if it is not exist in the target sequence (Fig. 3 A) (Li et al., 2018). To further improve the sensitivity and specificity of the method, the 3′ end of the forward primer was designed at the SNP (A23403G) site of the D614G mutation. Therefore, the forward primer was loaded with one synthetic mismatch for the mutant template G614, while two for the wild type D614 at the 3′-terminal base (Fig. 3A). A single base mismatch at the 3′ end of the primer reduces PCR efficiency, and a double mismatched primer produces a more dramatic compromise of the PCR yield (Kwok et al., 1990). We next optimized the annealing temperature to selectively amplify the mutant G614 template rather than the wild type D614 template. As shown in Fig. 2B, both templates were amplified when the annealing temperature was below 68°C, whereas the PCR products were not detected when the annealing temperature was higher than 70°C. Notably, it was 68°C of the annealing temperature that gave the most significant difference in the PCR products of the G614 and D614 templets (Fig. 3C). The products were then mixed with Cas12a and incubated at 37°C for 30 min for fluorescent and visual detection, respectively. Consistent with the electrophoresis results, G614 could be discriminated from D614 with a higher G614/D614 ratio of 23.12 rather than 13.45 if only the synthetic mismatch crRNA were used (Fig. 3D). Furthermore, when the synthetic DNA templates of SARS-CoV-2 D614G mutation were 10-fold diluted from 6 × 104 to 6 × 100 copies/μL, the dsmCRISPR system successfully distinguished G614 from D614 even when the templates were as low as 6 × 100 copies (Fig. 3D).
Fig. 3.
Validation of primer mismatch amplification of SARS-CoV-2 D614G by PCR and Cas12-based detection. (A) Schematic of the process for design of D614G specific primer with synthetic mismatch. The 3′ end of the forward primer is located at the SNP (A23403G) site of D614G mutation with a synthetic mismatch which is the same as that of crRNA-1. (B) Amplification of synthetic templates of D614 (6 × 104 cps) and G614 (6 × 104 cps) by mismatch primer designed in (A) annealed in a temperature gradient, respectively. After amplification, the products were analyzed by electrophoresis in 2% agarose gel and quantified. M represents 100 bp DNA ladder. D represents D614. G represents G614. The relative mean intensity was normalized to G614 amplified at 62°C. (C) Fluorescence signals of CRISPR/Cas12a-based detection of amplification products of (B) with crRNA-(-1). The presented images of visual detection were captured under UV light (n=3). (D) Fluorescence signals of D614 (6 × 104 cps) and 10-fold gradually diluted DNA templates of G614 (from 6 × 104 cps to 6 × 100 cps) detected by dsmCRISPR (n=3). NTS, non-target sequence. The presented images of visual detection were captured under UV light.
3.4. Detection of the synthetic SARS-CoV-2 D614G RNA
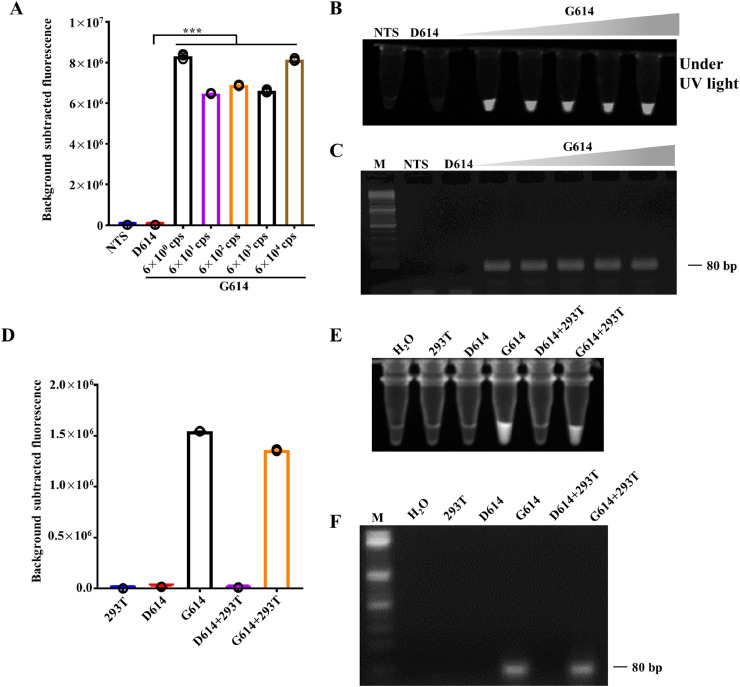
To verify the specificity of the synthetic mismatched primers for the G614 RNA template, we performed reverse transcription polymerase chain reaction (RT-PCR) to amplify the synthetic RNA sequence of SARS-CoV-2 D614G mutation. The RNA templates were 10-fold diluted to obtain 6 × 104 to 6 × 100 copies/μL. After RT-PCR amplification, the products were mixed with Cas12a proteins and incubated at 37°C for 30 min for fluorescent and visual detection, respectively. The results showed that the PCR products were detectable by this assay (Fig. 4 A-C). In order to validate the dsmCRIPSR method in clinical scenarios, D614 or G614 RNA templates were mixed with RNA samples isolated from HEK293T cells. After RT-PCR amplification, only samples containing G614 template were able to be amplified (Fig. 4F). Then, the products were mixed with Cas12a proteins and incubated at 37°C for 30 min for fluorescent and visual detection, respectively. The results showed that the dsmCRISPR method remained sensitivity and specificity under these conditions (Fig. 4D, E). Hence, when combined with RT-PCR, the dsmCRISPR system can detect the nucleic acids with template number as low as few copies, thus providing a highly sensitive and specific method to detect the SARS-CoV-2 D614G mutation.
Fig. 4.
Synthetic RNA of SARS-CoV-2 D614G detected by dsmCRISPR. (A) Fluorescence signals of D614 (6 × 104 cps) and 10-fold gradually diluted RNA templates of G614 (from 6 × 104 cps to 6 × 100 cps) detected by dsmCRISPR (n=3). NTS, non-target sequence. (B) The presented images of visual detection in (A) are shown. (C) After amplification, the products were analyzed by electrophoresis in 2% agarose gel. M represents 100 bp DNA ladder. (D) SARS-CoV-2 RNA fragments containing RNA G614 or D614 were mixed with or without total RNAs of HEK293T, then detected by dsmCRISPR NTS, non-target sequence. (E) The presented images of visual detection in (D) are shown. (F) After amplification, the products were analyzed by electrophoresis in 2% agarose gel. M represents 100 bp DNA ladder.
4. Discussion
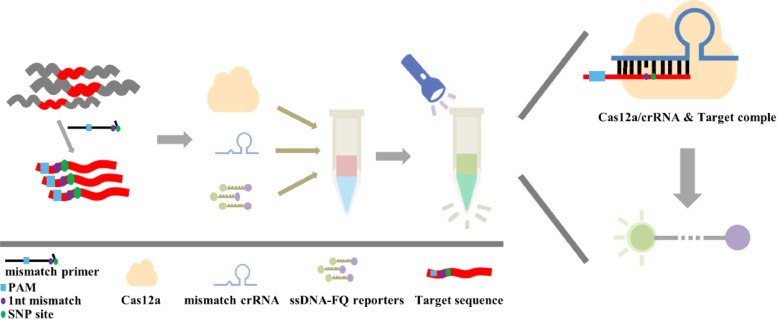
Cas12a-based detection of the unmutated genomic RNA of SARS-CoV-2 is recently reported to be a rapid, sensitive and reliable method potential for large-scale POC diagnostics (Verma et al., 2020). Research has demonstrated that Cas12a can be used to distinguish a single nucleotide mutation (Li et al., 2018). Therefore, in this study, a Cas12a-based detection system was established to detect SARS-CoV-2 D614G mutation (Fig. 5 ).
Fig. 5.
Scheme of dual synthetic mismatches CRIPSR (dsmCRISPR) detection method.
However, the crRNA completely complementary to G614 (crRNA-0) could not discriminate G614 from D614 (Fig. 1B). Research has previously shown that the sensitivity of Cas13a to detect single nucleotide differences was significantly improved by introducing additional mismatches in the crRNAs (Gootenberg et al., 2017). Therefore, we designed crRNAs with additional nucleotide mismatches from the -3 to +3 positions centered around the A23403G site (Fig. 1C) and obtained crRNAs which can distinguish G614 from D614 (Fig. 1D). When coupled with a synthetic mismatch primer, whose 3′ end is located at the D614G SNP (A23403G) site, the D614G mutation was selectively amplified. Hence, the fluorescence ratio of G614/D164 template detected by the dsmCRISPR system was increased by 73.53% (Fig. 3C). Therefore, dsmCRISPR can efficiently increase the detection specificity of D614G variant by 23.12-fold while isolated study reported to increase by 13-fold (Meng et al., 2021). Furthermore, when combined with RT-PCR, the dsmCRISPR system could detect as low as 6 copies of synthetic RNA, whereas isolated studies reported the detection limitation of 10 copies (Mayuramart et al., 2021; Meng et al., 2021).
The rapid spread of SARS-CoV-2 virus globally is reported to link with the occurrence of highly transmissive variants including D614G, E484Q, L452R, E484K etc (Verghese et al., 2021). Our dsmCRISPR assay utilized dual synthetic crRNA to enable sensitive, specific, and robust detection of D614G, the most dominant SARS-CoV-2 variant. Furthermore, this cas12-based dsmCRISPR detection could also be utilized to discriminate any other SARS-CoV-2 variants by introducing synthetic mismatch crRNA and synthetic mismatch primer. If there is no PAM sequence in the targeting sequence, PAM sequence could be introduced by using primers.
Previously reported cas12as-based detection of SARS-CoV-2 (Chen et al., 2020; Ding et al., 2020; Huang et al., 2020; Wang et al., 2021) have successfully detected SARS-CoV-2 by targeting the conservative sequence in N gene or E gene of SARS-CoV-2, yet the introduced methods do not discriminate against variants of the virus. Although Meng and colleagues recently reported a cas12a-based detection of D614G (Meng et al., 2021), our dsmCRISPR method is more effective as it could be extended to discriminate other SARS-CoV-2 variants with high specificity and sensitivity. Despite the potential of dsmCRISPR method, there are several limitations in this study. Firstly, internal control was not included in the study, however, studies have reported the accuracy of utilizing crRNA targeting E gene or N gene as internal control to confirm the infection of SARS-CoV-2 (Chen et al., 2020; Ding et al., 2020; Huang et al., 2020; Wang et al., 2021). Besides, the validation of the dsmCRISPR system on clinical sample was not performed due to the lack of eligibility for clinical sample collection.
In summary, dsmCRISPR system is an efficient and effective assay to detect the SARS-CoV-2 variant with D614G mutation. The assay is of great potential as it could be extended to detect any variants of SARS-CoV-2 by combining with RPA and RT-LAMP (Chen et al., 2020; Wang et al., 2021). Additionally, the results of this assay can be easily and conveniently visualized under UV light or a portable instrument, making it suitable for POC diagnostics.
Authors’ statement
X.H., F.Z. and W.M. conceived and planned the experiments; X.H., F.Z., K.Z. and W.L. performed the experiments and analyzed the data; X.H. and W.M. wrote the manuscript.
Declaration of Competing Interest
Kun Zhu was employed by Beijing Origingene-tech biotechnology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
This work was funded by the Science and Technology Development Fund, Macau SAR, China (File no. 0039/2020/A, 0036/2020/A1 and 0013/2019/A1).
References
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shi Y., Chen Y., Yang Z., Wu H., Zhou Z., Li J., Ping J., He L., Shen H. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: a promising method in the point-of-care detection. Biosens. Bioelectron. 2020;169 doi: 10.1016/j.bios.2020.112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Q., 2017. An application of a Cas protein, and a method and kit for detecting a target nucleic acid molecule. CN107488710A.
- Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Escarmís C., Sevilla N., Moya A., Elena S.F., Quer J., Novella I.S., Holland J.J. Basic concepts in RNA virus evolution. FASEB J. 1996;10(8):859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S., Kellogg D., McKinney N., Spasic D., Goda L., Levenson C., Sninsky J. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18(4):999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. e1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Cheng Q., Wang J., Li X., Zhang Z., Gao S., Cao R., Zhao G., Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4(1):1–4. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37(7):730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Ma P., Meng Q., Sun B., Zhao B., Dang L., Zhong M., Liu S., Xu H., Mei H., Liu J. MeCas12a, a highly sensitive and specific system for COVID-19 detection. Adv. Sci. 2020;7(20) doi: 10.1002/advs.202001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayuramart O., Nimsamer P., Rattanaburi S., Chantaravisoot N., Khongnomnan K., Chansaenroj J., Puenpa J., Suntronwong N., Vichaiwattana P., Poovorawan Y. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp. Biol. Med. 2021;246(4):400–405. doi: 10.1177/1535370220963793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Wang X., Wang Y., Dang L., Liu X., Ma X., Chi T., Wang X., Zhao Q., Yang G., Liu M., Huang X., Ma P. Detection of the SARS-CoV-2 D614G mutation using engineered Cas12a guide RNA. Biotechnol. J. 2021;16(6) doi: 10.1002/biot.202100040. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020:1–6. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese M., Jiang B., Iwai N., Mar M., Sahoo M.K., Yamamoto F., Mfuh K.O., Miller J., Wang H., Zehnder J., Pinsky B.A. Identification of a SARS-CoV-2 variant with L452R and E484Q neutralization resistance mutations. J. Clin. Microbiol. 2021 doi: 10.1128/JCM.00741-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N., Patel D., Pandya A. Emerging diagnostic tools for detection of COVID-19 and perspective. Biomed. Microdevices. 2020;22(4):1–18. doi: 10.1007/s10544-020-00534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2020. WHO director-general's remarks at the media briefing on 2019-nCoV on 11 February 2020. 2020. https://www. who. int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020.
- WHO, 2021. WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/.
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]