Abstract
Since the start of the outbreak of coronavirus disease 2019 in Wuhan, China, there have been more than 150 million confirmed cases of the disease reported to the World Health Organization. The beta variant (B.1.351 lineage), the mutation lineages of SARS-CoV-2, had increase transmissibility and resistance to neutralizing antibodies due to multiple mutations in the spike protein. N501Y, K417N and E484K, in the receptor binding domain (RBD) region may induce a conformational change of the spike protein and subsequently increase the infectivity of the beta variant. The L452R mutation in the epsilon variant (the B.1.427/B.1.429 variants) also reduced neutralizing activity of monoclonal antibodies. In this study, we discovered that 300 μg/mL GB-2, from Tian Shang Sheng Mu of Chiayi Puzi Peitian Temple, can inhibit the binding between ACE2 and wild-type (Wuhan type) RBD spike protein. GB-2 can inhibit the binding between ACE2 and RBD with K417N-E484K-N501Y mutation in a dose-dependent manner. GB-2 inhibited the binding between ACE2 and the RBD with a single mutation (K417N or N501Y or L452R) except the E484K mutation. In the compositions of GB-2, glycyrrhiza uralensis Fisch. ex DC., theaflavin and (+)-catechin cannot inhibit the binding between ACE2 and wild-type RBD spike protein. Theaflavin 3-gallate can inhibit the binding between ACE2 and wild-type RBD spike protein. Our results suggest that GB-2 could be a potential candidate for the prophylaxis of some SARS-CoV-2 variants infection in the further clinical study because of its inhibition of binding between ACE2 and RBD with K417N-E484K-N501Y mutations or L452R mutation.
Abbreviations: ACE2, angiotensin converting enzyme 2; SARS-CoV-2, The severe acute respiratory syndrome coronavirus 2; COVID-19, coronavirus disease 2019; GU, glycyrrhiza uralensis Fisch. ex DC; T3G, Theaflavin 3-gallate; RBD, receptor binding domain
Chemical compounds studied in this article: Theaflavin (Pubchem CID: 135403798), Theaflavin 3-gallate (Pubchem CID: 136825044), (+)-Catechin (Pubchem CID: 9064)
Keywords: SARS-CoV-2, Spike protein, GB-2, Beta variant, Epsilon variant
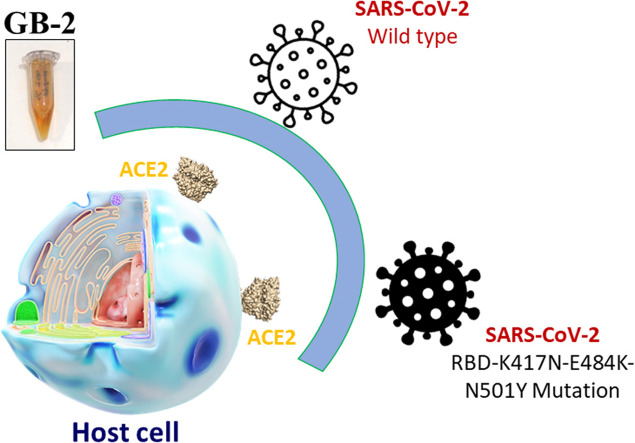
Graphical Abstract
1. Introduction
Since the start of the outbreak of coronavirus disease 2019 in Wuhan, China, there have been more than 150 million confirmed cases of the disease, including more than 3 million deaths worldwide, reported to the World Health Organization. SARS‐CoV‐2 is the seventh human coronavirus and the first human coronavirus with global pandemic disease [1]. In severe COVID-19, many patients suffered from severe pneumonia and acute respiratory distress syndrome. Moreover, older patients who had co-morbidities such as diabetes mellitus, cardiovascular disease, and dementia, such as Alzheimer's-like dementia, have higher rates of deaths in COVID-19 [2], [3].
In coronaviruses, spike protein on the surface envelope of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for promoting viral entry into host cells [4]. The spike protein is composed of two functional subunits: the S1 subunit that binds to host cell receptors, and the S2 subunit that mediates the fusion of the viral and host cellular membranes. The distal part of the S1 subunit contains the receptor binding domain (RBD) that directly binds to the peptidase domain of angiotensin-converting enzyme 2 (ACE2) [5], [6]. Moreover, heparan sulfate may facilitates SARS-CoV-2 spike protein binding to ACE2 [7]. RBD of SARS-CoV-2 spike protein binds to the three sub-domains through amino acids 22–42, 79–84, and 330–393 of ACE2 of the human host cells to initiate entry [8]. Moreover, the direct area of contact between RBD and ACE2 includes several regions. Gln498, Thr500, and Asn501 residues of the amino terminus (N) of RBD form several hydrogen bonds with the Tyr41, Gln42, Lys353, and Arg357 residues of ACE2. The Lys417 and Tyr453 residues of RBD interact with the Asp30 and His34 residues of ACE2, respectively [5]. Moreover, the S subunit of SARS-CoV-2 contains a cleavage site for furin and other proteases and accelerates viral entry [9].
Until now, most current mono-antibodies, vaccines and protease inhibitors are developing by targeting the structure of SARS-CoV-2 [10], [11]. However, SARS-CoV-2 belongs to the family of RNA viruses, which have higher mutation rates than DNA viruses [12], [13]. Among the mutation lineages of SARS-CoV-2, the beta variant (B.1.351 lineage), which was first detected in South Africa, has demonstrated higher transmissibility and resistance to neutralizing antibodies due to multiple mutations in the spike protein [14], [15]. The SARS-CoV-2 beta variant contains nine mutations in the spike protein: D614G, Δ242–Δ244, and R246I in the N-terminal domain; three mutations (K417N, E484K, and N501Y) in the RBD; and one mutation (A701V) near the furin cleavage site [14]. Among them, N501Y, K417N, and E484K in the receptor binding motif of the RBD region may induce a conformational change in the spike protein and subsequently increase the infectivity of beta variant [15]. Moreover, beta variant is resistant to neutralization by most monoclonal antibodies against the N-terminal domain and multiple individual monoclonal antibodies against the receptor-binding motif of the RBD due to E484K mutation [14]. Beta variant is also refractory to neutralization by sera from vaccinated persons [14]. In addition, the SARS-CoV-2 epsilon variant (the B.1.427/B.1.429 variant of concern) was first reported for in California and has been detected in more than 30 countries till now [16], [17]. Moreover, L452R mutation in spike protein of epsilon variant could increase infectivity in vitro and loss neutralizing activity of monoantibodies [17], [18], [19]. These results suggest that the protective effects of current SARS-CoV-2 vaccines and monoclonal antibody therapies are threatened by these variants.
In our previous studies, we discovered that GB-2, a prophylaxis formula against SARS-CoV-2 and from Tian Shang Sheng Mu of Chiayi Puzi Peitian Temple in Taiwan, inhibited ACE2 mRNA expression and ACE2 and TMPRSS2 protein expression in HepG2 and 293 T cells and decreased the ACE2 expression level of lung and kidney tissue in the animal model [20]. However, the effect of GB-2 on the interaction between ACE2 and wild-type and mutation-type spike proteins of SARS-CoV-2 remains unclear. In this study, we investigated the effects of GB-2 and the index compounds of Camellia sinensis var. assamica extract on the interaction between ACE2 and wild-type and mutation-type spike proteins of SARS-CoV-2.
2. Methods and materials
2.1. Cell culture and treatment
293 T cells (human embryonic kidney cell line) were obtained from the Bioresource Collection and Research Center, Taiwan. 293 T cells were cultured in Dulbecco's Modified Eagle's medium (DMEM, Invitrogen Corp., Carlsbad, Cat. Number: 11965–048), supplemented with 10% FBS at 37 ℃ and 5% CO2. The GB-2 formula was designed from Tian Shang Sheng Mu of Chiayi Puzi Peitian Temple. The compositions of GB-2 included dry root of glycyrrhiza uralensis Fisch. ex DC. (10 g, brought form Chang Gung Memorial Hospital, Taiwan) and Camellia sinensis var. assamica (black tea, 25 g, brought form Chang Gung Memorial Hospital and Xiamen Qingheyu Biological Technology Co., Ltd). The formula was soaked in 2000 mL water and boiling hot water for 25 min in thermal flasks, respectively. The sample was filtered with filter paper to remove particulate matter. The water extracts were evaporated through reducing pressure to obtain viscous masses (GB-2: 6 g). These samples were stored at − 80 °C. For all experiments, final concentrations of the tested compound were prepared by diluting the stock with water. Theaflavin 3-gallate and theaflavin were purchased from ChromaDex Corp. (Irvine, CA, USA, Product ID: ASB-00020253 and ASB-00020252). (+)-Catechin was purchased from Sigma-Aldrich Co., Ltd. (CAS number: 154–23–4). pCEP4-myc-ACE2 plasmid was a gift from Erik Procko (Addgene plasmid # 141185; http://n2t.net/addgene:141185;RRID:Addgene_141185). pcDNA3-SARS-CoV-2-S-RBD-sfGFP plasmid was a gift from Erik Procko (Addgene plasmid # 141184; http://n2t.net/addgene:141184; RRID:Addgene_141184). Before treatment, 293 T cells were cultured to 60–70% confluence. Then, cultured medium was replaced with fresh medium containing indicated compounds in water at the indicated concentrations. 293 T cells treated with water were used as controls. 293 T cells without treatment were used as blank control.
2.2. XTT assay
293 T cell lines were cultured at a density of 1 × 103 per well of 96-well plates, in DMEM with 10% FBS. Once attached, the cultured medium was replaced with fresh medium with 10% FBS. 293 T cell were then treated with indicated drugs for indicated hours; and absorbance were measured using the XTT assay kit (Roche, catalog number: 11465015001) depending on the manufacturer’s instructions. The XTT formazan complex was quantitatively measured at 492 nm using an ELISA reader (Bio-Rad Laboratories, Inc.).
2.3. ACE2/SARS-CoV-2 spike inhibitor screening assay kit
The ACE2/SARS-CoV-2 Spike Inhibitor Screening Assay were performed as described previously [21] using the ACE2: SARS-CoV-2 Spike Inhibitor Screening Assay Kit (BPS Bioscience Cat. number #79936) according to the manufacturer’s instructions. Briefly, ACE2 solution was used to coat a 96-well nickel-coated plate and washing the plate.Then, the plate was incubated with a Blocking Buffer. Next, the indicated compounds were added and incubated for 1 h at room temperature with slow shaking. After the incubation, SARS-CoV-2 Spike (RBD)-Fc was added to each well, except to the blank and incubated the reaction with slow shaking. After incubation with a Blocking Buffer, an Anti-mouse-Fc-HRP and incubate and an HRP substrate was added to the plate to produce chemiluminescence, which then can be measured using microplate reader.
2.4. Flow cytometry analysis of ACE2-spike protein binding
The flow cytometry analysis of ACE2-S Binding were performed as described previously [22]. Briefly, 293 T cells were transfected with pCEP4-MYC-ACE2 or pcDNA3-SARS-CoV-2-S-RBD-sfGFP plasmids (500 ng DNA per mL of culture at 2 ×106 / mL) using lipofectamine 2000 (ThermoFisher, Cat. Number: 11668–019). The the medium with indicated RBD-sfGFP were collected from 293 T cells which transfected with pcDNA3-SARS-CoV-2-S-RBD-sfGFP plasmid after 48 h. After pretreated by indicated drugs for 24 h, 293 T cells which transfected with pCEP4-MYC-ACE2 plasmid were washed with ice-cold PBS-BSA, and incubated for 30 min on ice with medium containing RBD-sfGFP and the anti-MYC Alexa 647 (clone 9B11, Cell Signaling Technology, Cat. Number: 2233 S). Then, cells were washed twice with PBS-BSA and analyzed by the flow cytometer BD FACSCanto (Becton Dickinson).
2.5. Site-directed mutagenesis
The QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies Inc., Santa Clara, CA, Cat. Number: 210519) was used to introduce amino acid substitutions (N501Y, E484K, E417N or L452R) in pcDNA3-SARS-CoV-2-S-RBD-sfGFP plasmid by following the manufacturer’s instructions.
2.6. High-performance liquid chromatography (HPLC)
For theaflavin, HPLC analysis was performed on the LC-10Avp system (Shimadzu) equipped with a Luna ® C18(2)100 A,LC column (5-μm particle size, 150-mm length × 2-mm internal diameter; Luna, Cat. Number: 00 F-425-B0); 0.1% trifluoroacetic acid was used as the mobile phase, the flow rate was 0.4 mL/min, the detection wavelength was 205 nm, and the column temperature was 60 °C. For theaflavine-3-gallate, HPLC analysis was performed on the LC-10Avp system (Shimadzu) equipped with a SUPELCO Discovery® C18 column (5-μm particle size, 150-mm length × 4.6-mm internal diameter; Supelco, Cat. Number: 504955); 0.1% phosphoric acid was used as the mobile phase, the flow rate was 0.6 mL/min, the detection wavelength was 280 nm, and the column temperature was 25 °C.
2.7. Statistical analyses
All values were the means ± standard error of mean (SEM) of the replicate samples (n = 3–6, depending on the experiment). These experiments were repeated by a minimum of three times. Differences between two groups were assessed using the unpaired two-tailed Student’s t-test or by ANOVA if more than two groups were analyzed. For testing the significance of pairwise group comparisons, the Tukey test was used as a post-hoc test in ANOVA. For all comparisons, P values of < 0.05 were considered statistically significant. SPSS version 13.0 (SPSS Inc., Chicago, IL, USA) was used for all calculations.
3. Results
3.1. Effect of GB-2 on cell variability in the 293T cell line
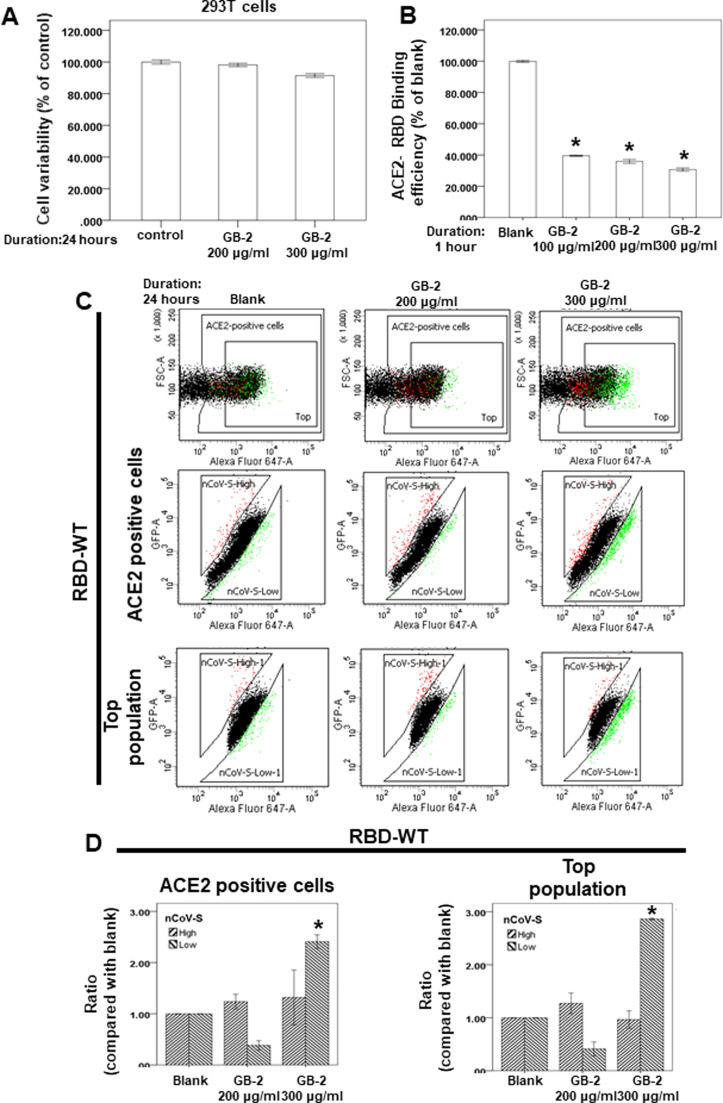
293T cells was used as a model of SARS-CoV-2 entry in the previous studies [22], [23]. To investigate the effect of GB-2 on the interaction between ACE2 and the spike protein of SARS-CoV-2, we used 293T cells as the model in our study. First, we used a 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)− 2 H-tetrazolium-5-carboxanilide assay to investigate the cytotoxicity of GB-2–293 T cells. Our results showed that GB-2 treatment with concentrations 200–300 μg/mL had not inhibited the cell variability of the 293 T cells after the indicated number of hours ( Fig. 1A). These data suggested that 200–300 μg/mL GB-2 did not have significant cytotoxicity against the cell variability of the 293 T cell line.
Fig. 1.
Effect of GB-2 on cell variability of 293 T cells and interaction between ACE2 and SARS-CoV-2 Spike. (A) 293 T cells were measured by XTT assay after indicated hours of culturing in the presence of GB-2. (B) The indicated compounds were tested to evaluate their ability to inhibit the binding of spike protein to immobilized ACE2 by the ACE2/SARS-CoV-2 spike inhibitor screening assay. (C) Flow cytometry analysis of ACE2-Spike protein binding. 293 T cells with pCEP4-MYC-ACE2 plasmid were incubated with RBD (wild type)-sfGFP-containing medium and co-stained with anti-MYC Alexa 647 to detect surface ACE2 by flow cytometry. During analysis, the top population were chosen from the ACE2-positive population. Then, two subsets of the ACE2-positive population were collected: the top population (nCoV-S-High sort, red gate) and the bottom population (nCoV-S-Low sort, green gate) based on the fluorescence of bound RBD-sfGFP relative to ACE2 surface expression. (D) Quantitative results of nCoV-S-High sort and nCoV-S-low sort, which were presented as ratio compared with blank, in the top population or ACE2-positive population. All the results are representative of at least three independent experiments. (Error bars=mean±S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.05).
3.2. Effect of GB-2 on the interaction between ACE2 and wild-type RBD of spike protein
To investigate the effect of GB-2 on the interaction between ACE2 and the spike protein of wild-type SARS-CoV-2, we used a spike/ACE2 inhibitor screening assay kit to determine the impact on the binding of Spike protein to the ACE2 receptor by GB-2 [10]. As illustrated in Fig. 1B, we found that incubating the spike RBD with GB-2 resulted in blocking of the binding of the wild-type RBD the spike protein to the ACE2 receptor in a concentration-dependent manner over the concentration range 100–300 μg/mL (Fig. 1B).
To confirm the previous results, we used dual-color flow cytometric analysis to investigate ACE2–spike protein binding in 293 T cells [22]. 293 T cells expressing MYC-tagged ACE2 were then incubated with medium containing the wild-type RBD of SARS-CoV-2 fused to superfolder green fluorescent protein [24]. After treatment of the cells with the indicated drugs, populations of cells that expressed MYC-tagged ACE2 at the cell surface with high or low binding to RBD were collected through fluorescence-activated cell sorting. After treatment of the cells with 300 μg/mL GB-2, the number of 293 T cells with low binding to RBD was significantly increased in both ACE2-positive cells and the top population (Fig. 1C, D). These results suggested that 300 μg/mL GB-2 inhibited the binding between ACE2 and the wild-type RBD of the spike protein.
3.3. Effect of GB-2 on the interaction between ACE2 and RBD of the spike protein with triple mutation (K417N-E484K-N501Y)
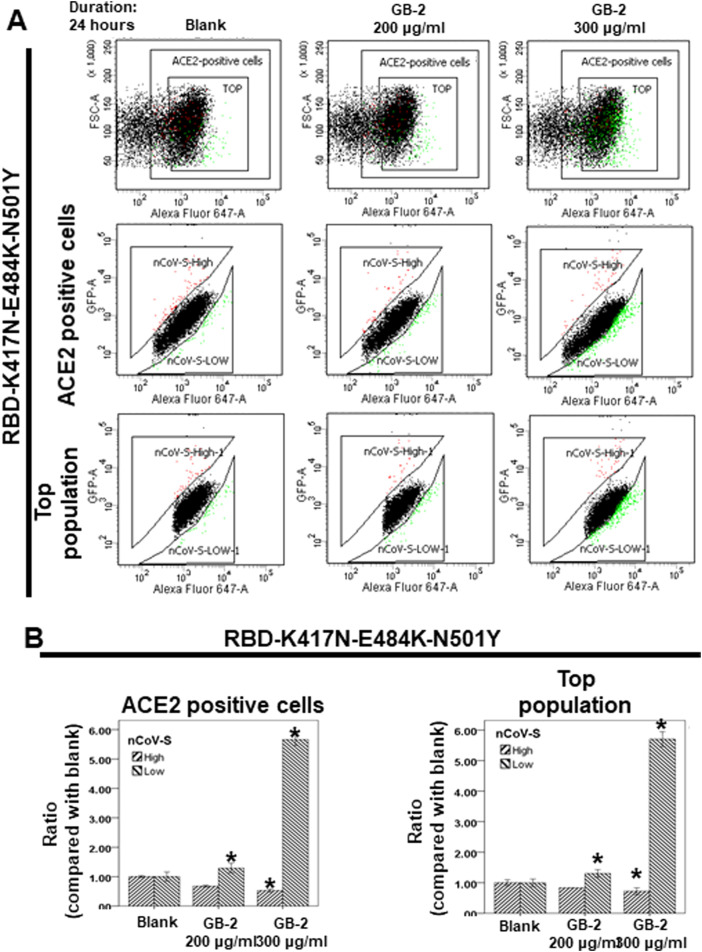
Previous study reported that triple mutations (N501Y, K417N, and E484K) in the RBD of beta variant may increase the infectivity and resistance to neutralization of monoclonal antibodies [15]. Next, through dual-color flow cytometric analysis, we investigated the effect of GB-2 on the binding between ACE2 and the RBD of the spike protein with the triple mutation (K417N-E484K-N501Y). After treatment of the cells with the indicated concentration of GB-2, the number of 293 T cells with low binding to RBD with a triple mutation (K417N-E484K-N501Y) was significantly increased in both ACE2-positive cells and the top population in a dose-dependent manner ( Fig. 2A, B). The number of 293 T cells with high binding to the RBD with a triple mutation (K417N-E484K-N501Y) was also decreased in ACE2-positive cells and the top population in a dose-dependent manner (Fig. 2A, B). These results suggested that 200–300 μg/mL GB-2 inhibited the binding between ACE2 and RBD with a triple mutation (K417N-E484K-N501Y).
Fig. 2.
Effect of GB-2 on interaction between ACE2 and SARS-CoV-2 Spike (K417N-E484K-N501Y). (A) Flow cytometry analysis of ACE2-Spike protein binding. 293 T cells with pCEP4-MYC-ACE2 plasmid were incubated with RBD (K417N-E484K-N501Y)-sfGFP-containing medium and co-stained with fluorescent anti-MYC Alexa 647 to detect surface ACE2 by flow cytometry. During analysis, the top population were chosen from the ACE2-positive population. Then, two subsets of the ACE2-positive population were collected: the top population (nCoV-S-High sort, red gate) and the bottom population (nCoV-S-Low sort, green gate) based on the fluorescence of bound RBD(K417N-E484K-N501Y)-sfGFP relative to ACE2 surface expression. (B) Quantitative results of nCoV-S-High sort and nCoV-S-low sort, which were presented as ratio compared with blank, in the top population or ACE2-positive population. All the results are representative of at least three independent experiments. (Error bars=mean±S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.05).
3.4. Effect of GB-2 on the interaction between ACE2 and the RBD of the spike protein with a single mutation (K417N, E484K, N501Y, L452R)
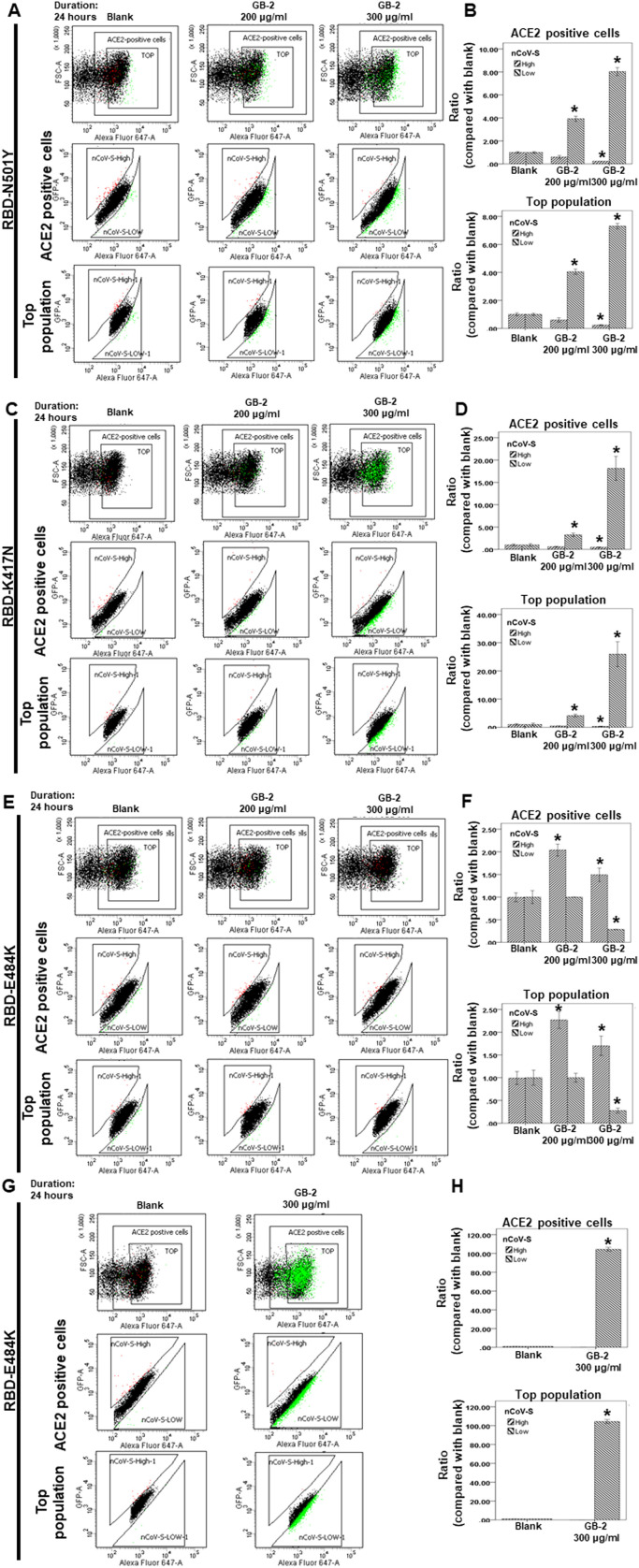
N501Y in the RBD was discovered in both alpha variant (B.1.1.7 lineage) and beta variant. N501 is the key residue in the RBD of the spike protein of SARS-CoV-2 that is involved in critical contact with some critical residues at the interface of ACE2 [5], [25]. The mutation of N501Y was discovered to increase the ACE2 binding and infectivity of SARS-CoV-2 [25], [26], [27]. Moreover, the K417N and E484K mutations dramatically enhanced cell–cell fusion and may be more transmissible than the N501Y mutation–containing SARS-CoV-2 variant [15].
In our previous results, GB-2 could inhibit the binding between ACE2 and the RBD of the spike protein with a triple mutation (K417N-E484K-N501Y). Next, using dual-color flow cytometric analysis, we investigated the effect of GB-2 on the interaction between ACE2 and the different single mutations of the RBD. After treatment of the cells with the indicated concentration of GB-2, the number of 293 T cells with low binding to the RBD with a single mutation (N501Y or K417N) was significantly increased in both ACE2-positive cells and the top population in a dose-dependent manner ( Fig. 3A–D). The number of 293 T cells with high binding to the RBD with a single mutation (N501Y or K417N) was also decreased in both ACE2-positive cells and the top population in a dose-dependent manner (Fig. 3A–D). However, the number of 293 T cells with high binding to the RBD with a single mutation (E484K) was slightly increased in both ACE2-positive cells and the top population (Fig. 3E, F). These results suggested that 200–300 μg/mL GB-2 inhibited the binding between ACE2 and the RBD with a single mutation (K417N or N501Y) except the E484K mutation.
Fig. 3.
Effect of GB-2 on interaction between ACE2 and SARS-CoV-2 Spike (K417N, E484K, N501Y or L452R). (A, C, E, G) Flow cytometry analysis of ACE2-Spike protein binding. 293T cells with pCEP4-myc-ACE2 plasmid were incubated with RBD (N501Y(A), K417N(C), E484K(E) or L452R(G))-sfGFP-containing medium and co-stained with fluorescent anti-MYC Alexa 647 to detect surface ACE2 by flow cytometry. During analysis, the top population were chosen from the ACE2-positive population. Then, two subsets of the ACE2-positive population were collected: the top population (nCoV-S-High sort, red gate) and the bottom population (nCoV-S-Low sort, green gate) based on the fluorescence of bound RBD (N501Y(A), K417N(C), E484K(E) or L452R(G))-sfGFP relative to ACE2 surface expression. (B, D, F, G) Quantitative results of nCoV-S-High sort and nCoV-S-low sort, which were presented as ratio compared with blank, in the top population or ACE2-positive population. All the results are representative of at least three independent experiments. (Error bars=mean±S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.05).
The previous studies reported that L452R mutation could increase transmissibility, infectivity, and resist to antibody neutralization [17], [18], [19]. Because 300 μg/mL GB-2 has the best inhibitory effect on the binding between ACE2 and the RBD. Next, we also investigated the effect of 300 μg/mL GB-2 on the interaction between ACE2 and L452R mutation of the RBD through dual-color flow cytometric analysis. After treatment of the cells with 300 μg/mL GB-2, the number of 293 T cells with low binding to the RBD with L452R mutation was significantly increased in both ACE2-positive cells and the top population in a dose-dependent manner (Fig. 3G, H). The number of 293 T cells with high binding to the RBD with L452R mutation was also decreased in both ACE2-positive cells and the top population in a dose-dependent manner (Fig. 3G, H). These results suggested that 300 μg/mL GB-2 could inhibit the interaction between ACE2 and the RBD with L452R mutation.
3.5. Effect of Glycyrrhiza uralensis Fisch. ex DC., Theaflavin and (+)-catechin on the interaction between ACE2 and wild-type RBD of spike protein
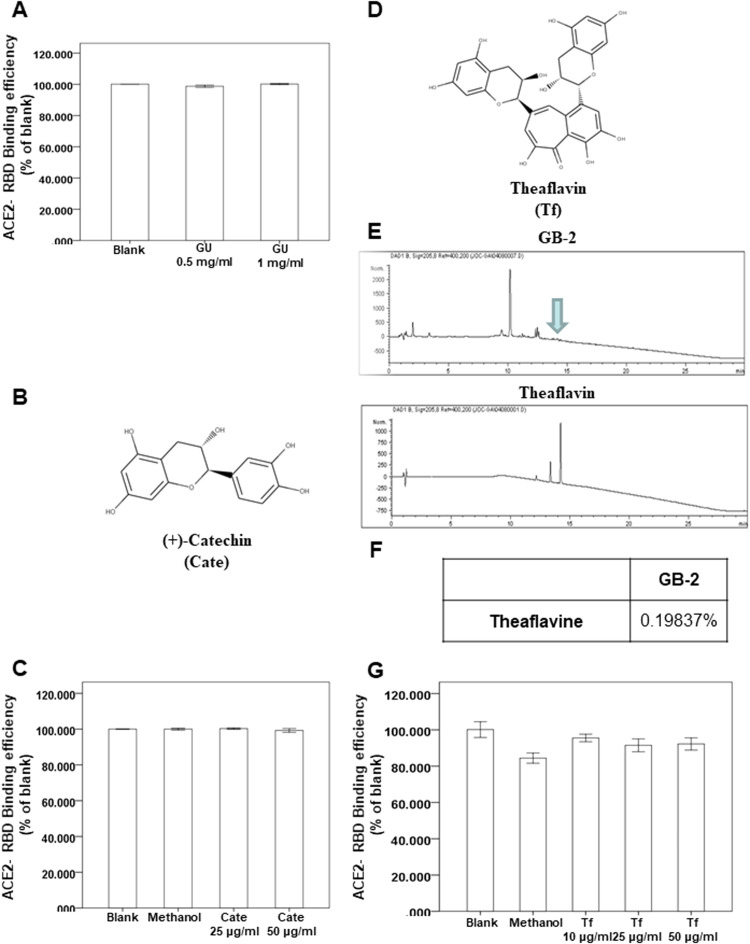
The dry roots of G. uralensis Fisch. ex DC. (GU) and C. sinensis var. assamica extract are the major components of GB-2. Our results indicate that 300 μg/mL GB-2 inhibited the binding between ACE2 and the wild-type RBD of the spike protein (Fig. 1B–D). However, the effect of GU on the interaction between ACE2 and the wild-type RBD of the spike protein is unclear. To investigate the effect of GU on the interaction between ACE2 and the spike protein of wild-type SARS-CoV-2, we used a spike/ACE2 inhibitor screening assay kit to discover the impact on the binding of spike protein to the ACE2 receptor [21]. Using the kit, we found that GU could not block the binding of the wild type of the RBD of the spike protein to the ACE2 receptor ( Fig. 4A).
Fig. 4.
Effect of glycyrrhiza uralensis Fisch. ex DC. extract (GU), theaflavin (Tf) and (+)-catechin (Cate) on interaction between ACE2 and SARS-CoV-2 Spike. (A, C, G) The indicated compounds were tested to evaluate their ability to inhibit the binding of spike protein to immobilized ACE2 by the ACE2/SARS-CoV-2 spike inhibitor screening assay. (B) The structure of (+)-catechin. (D) The structure of theaflavin. (E, F) HPLC chromatograms of theaflavin.
Next, we investigated the effect of the index compounds of C. sinensis var. assamica extract, (+)-catechin (Fig. 4B) on the binding between ACE2 and the wild type of the RBD of the spike protein. Using a spike/ACE2 inhibitor screening assay kit, we found that (+)-catechin could not block the binding of the wild type the RBD of the spike protein to the ACE2 receptor (Fig. 4C).
Several reports showed that theaflavin and the similar structures, index compounds extracted from C. sinensis var. assamica, could be the SARS-CoV-2 inhibtors through in vitro and in-silico methods [28], [29], [30], [31]. Because the effect of theaflavin on interaction between ACE2 and spike protein is still unclear, we investigated the binding between ACE2 and spike protein. First, the high-performance liquid chromatography (HPLC) results showed that the concentration of theaflavin in GB-2 was 0.19837% (wt/wt; Fig. 4E, F). Using the kit, we also found that theaflavin could not block the binding of the Wild type the RBD of the spike protein to the ACE2 receptor (Fig. 4G). These results suggested that both GU and theaflavin cannot inhibit the binding between ACE2 and the wild-type RBD of the spike protein.
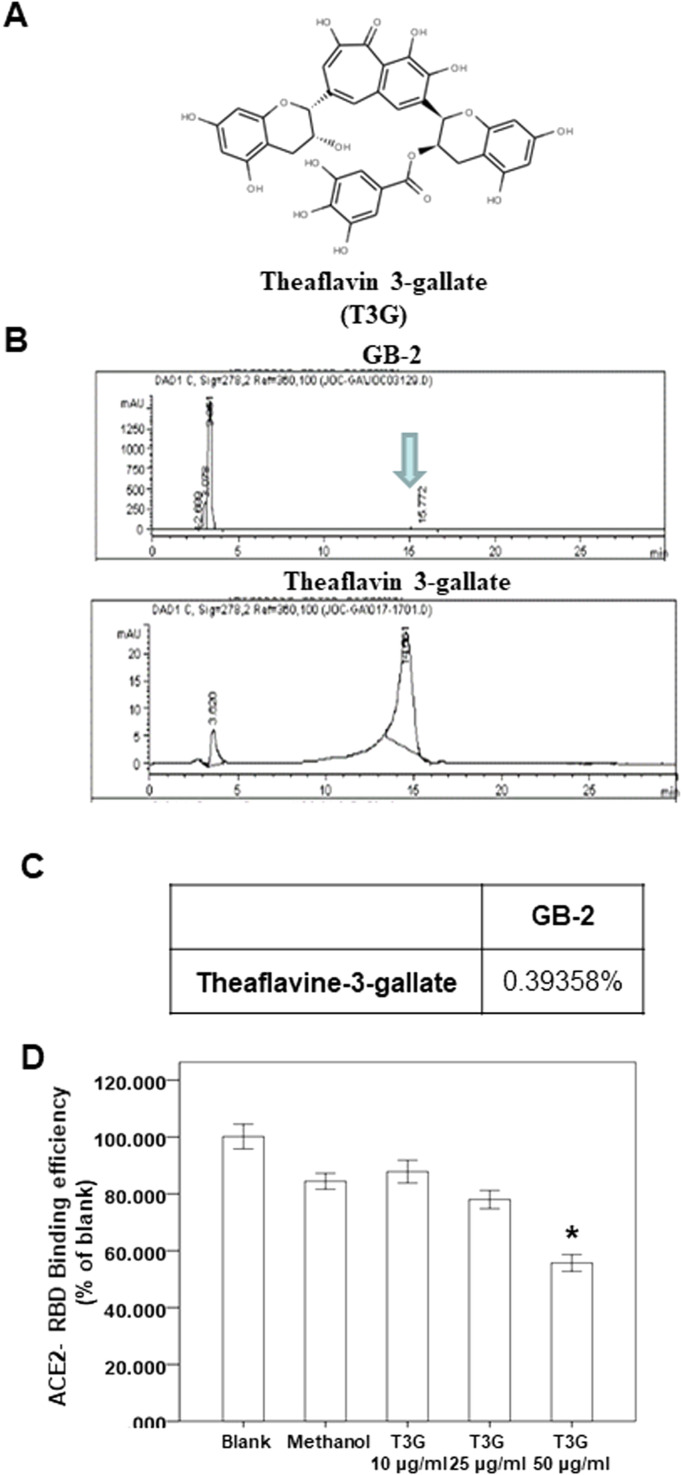
3.6. Effect of theaflavin 3-gallate on the interaction between ACE2 and wild-type RBD of spike protein
Using a spike/ACE2 inhibitor screening assay kit, we also investigated the effect of theaflavin 3-gallate ( Fig. 5A), another index compound of C. sinensis var. assamica extract, on the binding between ACE2 and the wild type the RBD of the spike protein. Our HPLC results showed that the concentration of theaflavin 3-gallate in GB-2 was 0.39358% (wt/wt; Fig. 5B, C). Using the kit, we found that only 50 μg/mL of theaflavin 3-gallate could partially block the binding of the wild type of the RBD of the spike protein to the ACE2 receptor (Fig. 5D). These results suggested that a higher concentration of theaflavin 3-gallate can partially inhibit the binding between ACE2 and the wild-type RBD of the spike protein.
Fig. 5.
Effect of theaflavin 3-gallate on interaction between ACE2 and SARS-CoV-2 Spike. (A) The structure of theaflavin 3-gallate. (B, C) HPLC chromatograms of theaflavin 3-gallate. (D) The indicated compounds were tested to evaluate their ability to inhibit the binding of spike protein to immobilized ACE2 by the ACE2/SARS-CoV-2 spike inhibitor screening assay.
4. Discussion
Recent studies have reported that SARS-CoV-2 alpha variant in the United Kingdom and beta variant in South Africa have significantly higher infectivity than other strains of SARS-CoV-2 [14], [32]. For beta variant, approximately 77% of mutations are located in the spike protein [32]. These mutations on spike proteins may affect the vaccine efficacy and treatments more than other variants. For epsilon variant, the two B.1.427 and B.1.429 lineages have the same spike protein mutations, inclunding S13I and W152C in the NTD and L452R in the RBD [17]. Moreover, L452R mutation of epsilon variant reduced neutralizing activity of several RBD-specific monoclonal antibodies [18]. Because these N501Y, K417N, E484K and L452R mutations in RBD play the important role in immune evasion, we focused on the effect of GB-2 on these mutations.
Both alpha and beta variants have the N501Y mutation. The N501Y mutation in the RBD domain of the spike protein increases both the binding affinity of the RBD to the ACE2 receptor and the viral transmission rate of both alpha and beta variants [5], [25], [32]. We discovered that GB-2 can block the binding between ACE2 and RBD with a single mutation (N501Y). The results suggested that GB-2 could decrease the viral transmission rate of both alpha and beta variants with the N501Y mutation. L452R mutation is the imporant mutatnt in RBD of epsilon variant. For L452R, previous studies showed that the L452R mutation may stabilize the binding between the spike protein and human ACE2 to increase infectivity [33], [34]. We found that GB-2 can block the binding between ACE2 and RBD with L452R mutation. The results suggested that GB-2 may decrease infectivity of epsilon variant with L452R mutation.
E484K can switch the charge on the flexible loop region of the RBD and induce the formation of new favorable contact. This is possibly why GB-2 cannot affect the binding between ACE2 and the RBD with a single mutation (E484K). In the other mutation, the K417 residue in the RBD of the spike protein of SARS-CoV-2 can interact with ACE2 and enhance the affinity of the virus for the ACE2 receptor. A previous study suggested that the K417N mutation minimally affects this binding between the RBD and the ACE2 receptor [26]. In our results, we also discovered that GB-2 can block the binding between ACE2 and the RBD with a single mutation (K417N). However, the combination of the E484K, K417N, and N501Y mutations could induce a higher degree of conformational alterations of the RBD when it was bound to the ACE2 receptor. GB-2 can inhibit the binding between ACE2 and RBD with the triple mutation (K417N-E484K-N501Y). The compounds in GB-2 may have binding affinity to conformational alterations of the triple mutation. In addition, beta variant has more mutations in the N-terminal domain region of the spike protein—including L18F, D80A, D215G, LAL 242–244 del, and R246I—within or near flexible variable loops without modifying the structure of the functional domains of the spike protein. Further study to investigate the effect of GB-2 on these mutations is necessary.
Black tea has the important medicinal values in many countries and areas in the world. The bioactive components form black tea are involved in the prevention and treatment of many diseases, including cardiovascular diseases, malignancy, digestive dysfunction, obesity, diabetes, chronic renal disease and renal fibrosis [35], [36], [37]. In a previous study, epigallocatechin-3-gallate, the major component of green tea, blocked the entry of SARS-CoV-2 with pseudotyped lentiviral vectors and inhibited viral infections in vitro by inhibiting the interaction between the SARS-CoV-2 spike protein and the ACE2 receptor [38]. Another report showed that (−)-gallocatechin gallate, a polyphenol in green tea, disrupted the liquid–liquid phase separation of the SARS-CoV-2 nucleocapsid protein and inhibited SARS-CoV-2 replication [39]. Moreover, another study showed that epigallocatechin-3-gallate and theaflavin inhibited the activity of the SARS-CoV-2 3CL protease [40]. These studies suggest that the tea extract and index compounds could play a major role in the treatment of SAR-CoV-2. However, the effects of the tea extract and index compounds on SARS-CoV-2 variants with mutations remain unclear. In this study, we discovered that GB-2 blocks the binding between ACE2 and RBD with a triple mutation (K417N-E484K-N501Y) and L452R mutation. The results suggest that GB-2 may play a useful role in the treatment of or prophylaxis against several variants. However, the further clinical trials of GB-2 and these index compounds should be investigated to confirm the clinical effort, even compare with the current vaccines.
In conclusion, our results suggest that GB-2 could be a candidate for prophylaxis against SARS-CoV-2 different variants infection because of its inhibition of the binding between ACE2 and the RBD with K417N-E484K-N501Y mutations and L452R mutation. However, the exact clinical effect remains unknown. Thus, further clinical research is necessary to confirm the protective effects of GB-2 against SARS-CoV-2 some variants.
CRediT authorship contribution statement
Ching-Yuan Wu: Conceptualization, Methodology, Software. Ming-Shao Tsai: Data curation, Writing- Original draft preparation. Yu-Shih Lin: Visualization, Investigation. Li-Hsin Shu: Visualization, Investigation. Yu-Ching Cheng: Visualization, Investigation. Hung-Te Liu: Visualization, Investigation. Cheng-Ming Hsu: Supervision, Revised the manuscript. Reming-Albert Yeh: Supervision, Revised the manuscript. Yao-Hsu Yang: Software, Analyzed the data, Validation. Geng-He Chang: Software, Analyzed the data, Validation. Rou-Chen Shen: Writing – review & editing. Yu-Huei Wu: Writing – review & editing. Yu-Heng Wu: Writing – review & editing. All authors reviewed and approved the final version.
Funding
Financial support was obtained from grant MOST 109-2320-B-182-021-MY3 from Ministry of Science and Technology (TW) and CORPG6K0211 from Chang Gung Memorial Hospital (Chiayi) to Dr. Ching Yuan Wu. The funding bodies allowed to the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgements
The authors acknowledge the Health Information and Epidemiology Laboratory at the Chiayi Chang Gung Memorial Hospital for the comments and assistance in data analysis. The authors also thank the research assists by Chang Gung Medical Foundation and Ministry of Science and Technology, Taiwan, R.O.C., especially Tse Hung Huang M.D. for his help (MOST 109–2327-B-182–002).
Availability of data and materials
All data generated or analyzed during this study are indicated in this article (with no patient data). The datasets generated during and /or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable (not contain any individual person’s data).
References
- 1.Seyran M., Pizzol D., Adadi P., El-Aziz T.M.A., Hassan S.S., Soares A., Kandimalla R., Lundstrom K., Tambuwala M., Aljabali A.A.A., Lal A., Azad G.K., Choudhury P.P., Uversky V.N., Sherchan S.P., Uhal B.D., Rezaei N., Brufsky A.M. Questions concerning the proximal origin of SARS-CoV-2. J. Med. Virol. 2021;93(3):1204–1206. doi: 10.1002/jmv.26478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vallamkondu J., John A., Wani W.Y., Ramadevi S.P., Jella K.K., Reddy P.H., Kandimalla R. SARS-CoV-2 pathophysiology and assessment of coronaviruses in CNS diseases with a focus on therapeutic targets. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalra R.S., Dhanjal J.K., Meena A.S., Kalel V.C., Dahiya S., Singh B., Dewanjee S., Kandimalla R. COVID-19, neuropathology, and aging: SARS-CoV-2 neurological infection, mechanism, and associated complications. Front Aging Neurosci. 2021;13 doi: 10.3389/fnagi.2021.662786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.J., Tortorici M.A., Wal A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan R., Zhang Y., Li Y., Xia L., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020 doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalra R.S., Kandimalla R. Engaging the spikes: heparan sulfate facilitates SARS-CoV-2 spike protein binding to ACE2 and potentiates viral infection. Signal Transduct. Target Ther. 2021;6(1):39. doi: 10.1038/s41392-021-00470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassan S.S., Ghosh S., Attrish D., Choudhury P.P., Aljabali A.A.A., Uhal B.D., Lundstrom K., Rezaei N., Uversky V.N., Seyran M., Pizzol D., Adadi P., Soares A., El-Aziz T.M.A., Kandimalla R., Tambuwala M.M., Azad G.K., Sherchan S.P., Baetas-da-Cruz W., Takayama K., Serrano-Aroca A., Chauhan G., Palu G., Brufsky A.M. Possible transmission flow of SARS-CoV-2 based on ACE2 features. Molecules. 2020;25(24) doi: 10.3390/molecules25245906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seyran M., Takayama K., Uversky V.N., Lundstrom K., Palu G., Sherchan S.P., Attrish D., Rezaei N., Aljabali A.A.A., Ghosh S., Pizzol D., Chauhan G., Adadi P., Abd El-Aziz T.M., Soares A.G., Kandimalla R., Tambuwala M., Hassan S.S., Azad G.K., Choudhury P.P., Baetas-da-Cruz W., Serrano-Aroca A., Brufsky A.M., Uhal B.D. The structural basis of accelerated host cell entry by SARS-CoV-2dagger. FEBS J. 2020 doi: 10.1111/febs.15651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandimalla R., John A., Abburi C., Vallamkondu J., Reddy P.H. Current status of multiple drug molecules, and vaccines: an update in SARS-CoV-2 therapeutics. Mol. Neurobiol. 2020;57(10):4106–4116. doi: 10.1007/s12035-020-02022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassan S.S., Attrish D., Ghosh S., Choudhury P.P., Uversky V.N., Aljabali A.A.A., Lundstrom K., Uhal B.D., Rezaei N., Seyran M., Pizzol D., Adadi P., Soares A., Abd El-Aziz T.M., Kandimalla R., Tambuwala M.M., Azad G.K., Sherchan S.P., Baetas-da-Cruz W., Lal A., Palu G., Takayama K., Serrano-Aroca A., Barh D., Brufsky A.M. Notable sequence homology of the ORF10 protein introspects the architecture of SARS-CoV-2. Int J. Biol. Macromol. 2021;181:801–809. doi: 10.1016/j.ijbiomac.2021.03.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294. doi: 10.1016/j.cell.2020.07.012. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., YIin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y.J., Jang U.S., Soh S.M., Lee J.Y., Lee H.R. The impact on infectivity and neutralization efficiency of SARS-CoV-2 lineage B.1.351 pseudovirus. Viruses. 2021;13(4) doi: 10.3390/v13040633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-Gonzalez A., Glasner D.R., Reyes K.R., Gliwa A.S., Reddy N.P., Sanchez San Martin C., Federman S., Cheng J., Balcerek J., Taylor J., Streithorst J.A., Miller S., Sreekumar B., Chen P.Y., Schulze-Gahmen U., Taha T.Y., Hayashi J.M., Simoneau C.R., Kumar G.R., McMahon S., Lidsky P.V., Xiao Y., Hemarajata P., Green N.M., Espinosa A., Kath C., Haw M., Bell J., Hacker J.K., Hanson C., Wadford D.A., Anaya C., Ferguson D., Frankino P.A., Shivram H., Lareau L.F., Wyman S.K., Ott M., Andino R., Chiu C.Y. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184(13):3426–3437. doi: 10.1016/j.cell.2021.04.025. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCallum M., Bassi J., De Marco A., Chen A., Walls A.C., Di Iulio J., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., Sprouse K.R., Agostini M., Pinto D., Culap K., Bianchi S., Jaconi S., Cameron E., Bowen J.E., Tilles S.W., Pizzuto M.S., Guastalla S.B., Bona G., Pellanda A.F., Garzoni C., Van Voorhis W.C., Rosen L.E., Snell G., Telenti A., Virgin H.W., Piccoli L., Corti D., Veesler D. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021 doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferreira I., Kemp S., Datir R., Saito A., Meng B., Rakshit P., Takaori-Kondo A., Kosugi Y., Uri K., Kimura I., Shirakawa K., Abdullahi A., Agarwal A., Ozono S., Tokunaga K., Sato K., Gupta R.K., CITIID-NIHR BioResource COVID-19 Collaboration, The Indian SARS-CoV-2 Genomics Consortium (INSACOG), Genotype to Phenotype Japan SARS-CoV-2 B.1.617 mutations L452 and E484Q are not synergistic for antibody evasion. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu C.Y., Lin Y.S., Yang Y.H., Shu L.H., Cheng Y.C., Liu H.T. GB-2 inhibits ACE2 and TMPRSS2 expression: in vivo and in vitro studies. Biomed. Pharmacother. 2020;132 doi: 10.1016/j.biopha.2020.110816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carino A., Moraca F., Fiorillo B., Marchiano S., Sepe V., Biagioli M., Finamore C., Bozza S., Francisci D., Distrutti E., Catalanotti B., Zampella A., Fiorucci S. Hijacking SARS-CoV-2/ACE2 receptor interaction by natural and semi-synthetic steroidal agents acting on functional pockets on the receptor binding domain. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.572885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369(6508):1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedelacq J.D., Cabantous S., Tran T., Terwilliger T.C., Waldo G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24(1):79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 25.Zhu X., Mannar D., Srivastava S.S., Berezuk A.M., Demers J.P., Saville J.W., Leopold K., Li W., Dimitrov D.S., Tuttle K.S., Zhou S., Chittori S., Subramaniam S. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021;19(4) doi: 10.1371/journal.pbio.3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310. doi: 10.1016/j.cell.2020.08.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X.F., Li J., Zhang N.N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D.Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.F., Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bello M., Hasan M.K. Elucidation of the inhibitory activity of plant-derived SARS-CoV inhibitors and their potential as SARS-CoV-2 inhibitors. J. Biomol. Struct. Dyn. 2021:1–13. doi: 10.1080/07391102.2021.1938234. [DOI] [PubMed] [Google Scholar]
- 29.Ohgitani E., Shin-Ya M., Ichitani M., Kobayashi M., Takihara T., Kawamoto M., Kinugasa H., Mazda O. Significant inactivation of SARS-CoV-2 in vitro by a green tea catechin, a catechin-derivative, and black tea galloylated theaflavins. Molecules. 2021;26(12) doi: 10.3390/molecules26123572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh R., Bhardwaj V.K., Sharma J., Purohit R., Kumar S. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J. Tradit. Complement. Med. 2021 doi: 10.1016/j.jtcme.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gogoi M., Borkotoky M., Borchetia S., Chowdhury P., Mahanta S., Barooah A.K. Black tea bioactives as inhibitors of multiple targets of SARS-CoV-2 (3CLpro, PLpro and RdRp): a virtual screening and molecular dynamic simulation study. J. Biomol. Struct. Dyn. 2021:1–24. doi: 10.1080/07391102.2021.1897679. [DOI] [PubMed] [Google Scholar]
- 32.Gomez C.E., Perdiguero B., Esteban M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines. 2021;9(3) doi: 10.3390/vaccines9030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Wang R., Wang M., Wei G.W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teng S., Sobitan A., Rhoades R., Liu D., Tang Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief. Bioinform. 2021;22(2):1239–1253. doi: 10.1093/bib/bbaa233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samanta S. Potential bioactive components and health promotional benefits of tea (Camellia sinensis) J. Am. Coll. Nutr. 2020:1–29. doi: 10.1080/07315724.2020.1827082. [DOI] [PubMed] [Google Scholar]
- 36.Kotze-Horstmann L.M., Sadie-Van Gijsen H. Modulation of glucose metabolism by leaf tea constituents: a systematic review of recent clinical and pre-clinical findings. J. Agric. Food Chem. 2020;68(10):2973–3005. doi: 10.1021/acs.jafc.9b07852. [DOI] [PubMed] [Google Scholar]
- 37.Kanlaya R., Thongboonkerd V. Molecular mechanisms of epigallocatechin-3-gallate for prevention of chronic kidney disease and renal fibrosis: preclinical evidence. Curr. Dev. Nutr. 2019;3(9):101. doi: 10.1093/cdn/nzz101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henss L., Auste A., Schurmann C., Schmidt C., von Rhein C., Muhlebach M.D., Schnierle B.S. The green tea catechin epigallocatechin gallate inhibits SARS-CoV-2 infection. J. Gen. Virol. 2021;102(4) doi: 10.1099/jgv.0.001574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao M., Yu Y., Sun L.M., Xing J.Q., Li T., Zhu Y., Wang M., Yu Y., Xue W., Xia T., Cai H., Han Q.Y., Yin X., Li W.H., Li A.L., Cui J., Yuan Z., Zhang R., Zhou T., Zhang X.M., Li T. GCG inhibits SARS-CoV-2 replication by disrupting the liquid phase condensation of its nucleocapsid protein. Nat. Commun. 2021;12(1):2114. doi: 10.1038/s41467-021-22297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jang M., Park Y.I., Cha Y.E., Park R., Namkoong S., Lee J.I., Park J. Tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-protease in vitro. Evid. Based Complement. Altern. Med. eCAM. 2020;2020 doi: 10.1155/2020/5630838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are indicated in this article (with no patient data). The datasets generated during and /or analyzed during the current study are available from the corresponding author on reasonable request.