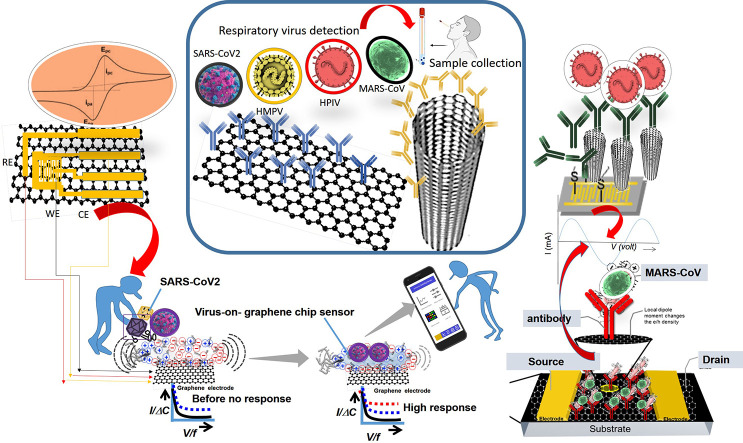
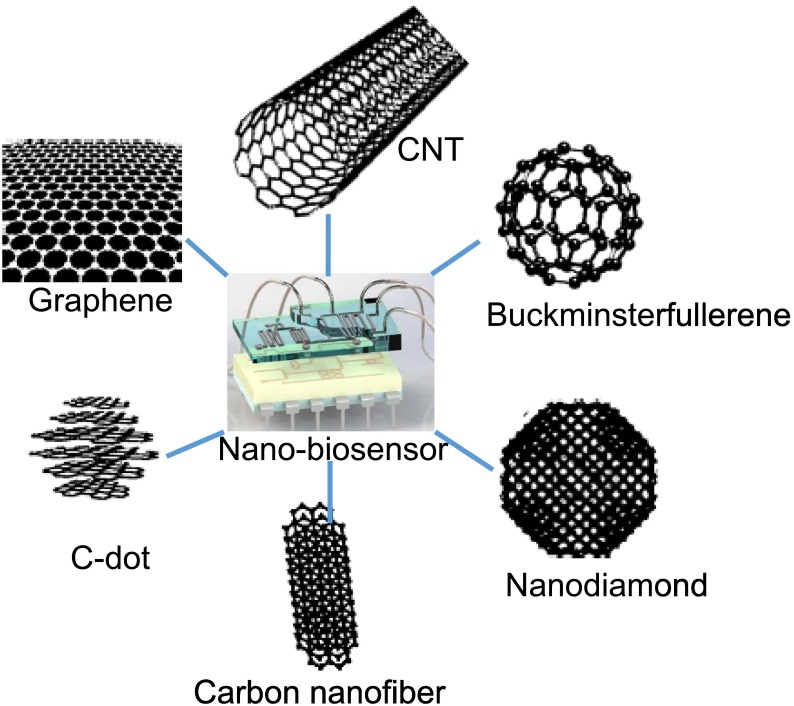
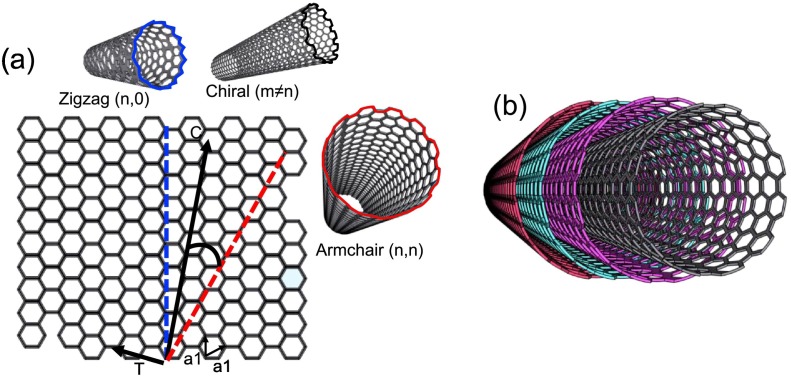
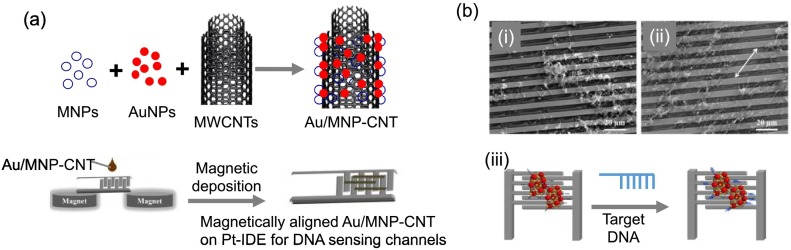
Abstract
Recent COVID-19 pandemic has claimed millions of lives due to lack of a rapid diagnostic tool. Global scientific community is now making joint efforts on developing rapid and accurate diagnostic tools for early detection of viral infections to preventing future outbreaks. Conventional diagnostic methods for virus detection are expensive and time consuming. There is an immediate requirement for a sensitive, reliable, rapid and easy-to-use Point-of-Care (PoC) diagnostic technology. Electrochemical biosensors have the potential to fulfill these requirements, but they are less sensitive for sensing viruses/viral infections. However, sensitivity and performance of these electrochemical platforms can be improved by integrating carbon nanostructure, such as graphene and carbon nanotubes (CNTs). These nanostructures offer excellent electrical property, biocompatibility, chemical stability, mechanical strength and, large surface area that are most desired in developing PoC diagnostic tools for detecting viral infections with speed, sensitivity, and cost-effectiveness. This review summarizes recent advancements made toward integrating graphene/CNTs nanostructures and their surface modifications useful for developing new generation of electrochemical nanobiosensors for detecting viral infections. The review also provides prospects and considerations for extending the graphene/CNTs based electrochemical transducers into portable and wearable PoC tools that can be useful in preventing future outbreaks and pandemics.
Keywords: Electrochemical nanobiosensor, Graphene, Carbon nanotubes, Respiratory viruses, Diagnosis
Graphical abstract
1. Introduction
The latest World Health Organization (WHO) report revealed over hundreds of million positive cases with millions of deaths occurred worldwide due to lower respiratory tract infections (LRTI). The ongoing virus infection of new severe acute respiratory syndrome (SARS) coronavirus-2 (CoV-2) cases reported above 177 million and claimed ~3.84 million deaths as of this review submission. Other common among the respiratory viruses are Influenza A and B viruses (FluA and FluB), Human adenovirus (HAdV), and Respiratory syncytial viruses (RSV) tend to exhibit more frequent mutations and potentially cause future pandemics that may surface with the signs and symptoms similar or more severe to that of COVID-19. Infants, children, elderly, and those with pre-existing chronic disease conditions or with compromised immune system are more vulnerable to LRTI. These new emerging viral variants may cause future morbidity and mortality and pose serious threat to public health and the global economy. Rapid and early diagnosis of viral LRTI is the only option to prevent future outbreak and spread of viruses.
Conventional diagnostic approaches for detecting viral infections are limited to only detecting viral nucleic acids using PCR or RT-qPCR, which limits in its accessibility, assay speed and cost. Therefore, it is imperative to explore the existing diagnostic approaches, tools and techniques to improving or re-purposing for early monitoring and disease prevention. In the last decade, nanotechnology has opened several avenues to improving current diagnostic assays and developing point-of-care devices by interfacing with new nanomaterials to improve sensitivity and detection speed. Nanomaterials can attach to living cells, viruses, proteins or other molecules allowing their early detection in a sample, and they simultaneously exhibit unique physico-chemical or electrical properties that make them suitable to address the needs of current challenges in rapid diagnosis of infections derived from viruses.
In this review, we explored the uses and application of functional nanomaterials in biosensing with special emphasis to carbon-based nanostructures, including lightweight two-dimensional (2D) graphene and one-dimensional carbon nanotubes (CNTs) that are electrically conductive, chemically stable with large surface-to-volume ratios. These nanomaterials in conjunction with specific bio−/chemical-receptors provide them with improved properties in developing future electrochemical nanobiosensors for PoC detection ability with cost-effectiveness, sensitivity, and fast detection of various respiratory viruses. Further, enhanced sensitivity and specificity for virus detection can be achieved through graphene and CNTs' surface modification via suitable functional groups and/or combination with other materials in hybrid nanostructures. This review covers all the above elements highlighting; (a) key advancement made in functional surface modification of graphene and CNTs toward the development of electrochemical nanobiosensors that can be potentially applied for respiratory virus electrochemical detection platforms, (b) current understanding on classical detection methods for respiratory viruses and fabrication processes of graphene, and (c) CNTs interfaced electrochemical biosensors for sensitive detection. Finally, the advantages and future challenges of graphene and CNTs based electrochemical sensor devices.
2. Respiratory viruses and epidemiology
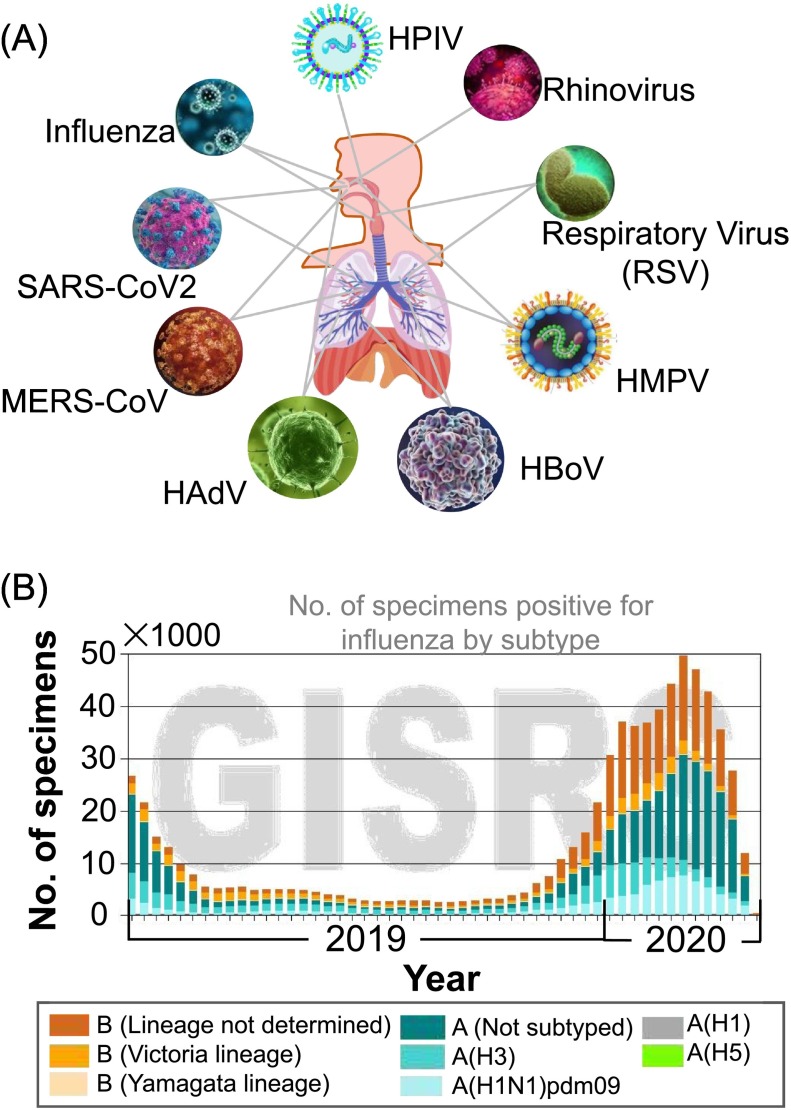
Biosensing of any viral infections requires a thorough understanding of viruses, their mode of transmission or port of entry, and associated diseases caused in humans or other living beings. There are a variety of different portals through which viruses gain entry into the body, such as respiratory tract, gastrointestinal and genital tracts, subcutaneous (skin), placenta and eyes [1]. Most common viral transmission and infection occurs via the respiratory tract in humans, which is the main portal entry focused in this review. Respiratory viruses are small microorganisms and leading causative agents of diseases in humans with severe impact on morbidity and mortality worldwide. There have been diverse pathogenic respiratory viruses taking part in both lower and upper Respiratory Tract Infections (LRTI, URTI) that include RSV, influenza viruses, Human Adenovirus (HAdV), Human Rhinovirus (HRV), Human Metapneumovirus (HMPV), Human Parainfluenza Virus (HPIV), Human Bocavirus (HBoV), Severe Acute Respiratory Syndrome (SARS) Coronavirus-1 (SARS-CoV-1), Middle East Respiratory Syndrome Coronavirus (MERS-CoV), and SARS-CoV-2 (Table 1 ) [2,3].
Table 1.
A summary of distinct respiratory viral features, such as size, genome, morphology, disease symptoms and related outbreaks in the past decades.
| Virus | Size | Genome | Morphology | Disease Symptoms | Outbreak Examples | Ref. |
|---|---|---|---|---|---|---|
| Respiratory syncytial (RSV) | 120–300 nm | -ssRNA | Enveloped, spherical or filamentous, F, G and SH proteins protruding from the envelope |
Cold like symptoms, wheezing, bronchiolitis |
Most of outbreaks reported in NICUs [6,7] | [2,16] |
| Influenza | 80–120 nm | Segmented -ssRNA | Enveloped, Elliptical, (H) and (N) proteins protruding from the envelope | Mild fever, myalgia, sore throat, cough, and headache severe: pneumonia, ARDS |
In 1918, “Spanish Flu” pandemic. In 2009, “Swine Flu” pandemic |
[8,17,18] |
| Human adenovirus (HAdV) |
90 nm | dsDNA | Non-enveloped Icosahedral |
Mild fever, sore throat, cough, chest pain, headache, or severe pneumonia or bronchitis | In 2007, United States [2] | [2] |
| Human rhinovirus (HRV) | 30 nm | +ssRNA | Nonenveloped, Icosahedral | Sore throat, cough, sneezing, congestion, fatigue, body aches and loss of appetite | In 2009, Canada, in long-term care facilities [19] | [2] |
| Human parainfluenza virus (HPIV) | 150-200 nm | -ssRNA | Enveloped, Elliptical, HN, F glycoproteins protruding from the surface |
Mild common cold with fever, croup, bronchiolitis, pneumonia. |
In 2018, residential care facility for the disable (HPIV-3) Croatia [20] | [2] |
| Human meta-pneumovirus (HMPV) | 150–200 nm | -ssRNA | Enveloped, Elliptical,F, G, SH glycoproteins protruding from the envelope |
Fever, cough, tachypnoea, dyspnoea, hypoxia, rhinitis and sore throat |
In 2006, Canada in long term care facility [21] In 2005, Taiwan [22] |
[2] |
| Human bocavirus (HBoV) |
18–26 nm | -ssDNA or + ssDNA | Non-enveloped, icosahedral |
Cough, fever, rhinorrhoea, asthma exacerbation, bronchiolitis, acute wheezing and pneumonia. |
In 2014, Japan [23] | [2] |
| SARS-CoV-1 | 100-160 nm | +ssRNA | Enveloped, spherical, or pleiomorphic | Mild flu-like, fever, cough, sore throat, lethargy, severe pneumonia, ARDS also vomiting, diarrhea |
SARS-CoV-1 Outbreak between 2002 and 2004 (China) [11] | [2] |
| MERS CoV | 120 nm | +ssRNA | Enveloped, spherical, or pleomorphic | Fever, diarrhea, and abdominal pain, severe pneumonia | MERS-CoV Outbreak in in middle east (Saudi Arabia) 2012 [10] | [2] |
| SARS-CoV-2(Covid19) | 120–160 nm | +ssRNA | Enveloped Helical “crown like” appearance “Spike” proteins (S) protruding from the envelope |
+Loss of smell or taste asymptomatic cases |
In 2019- present SARS-CoV-2 pandemic (China and worldwide) | [2,3] |
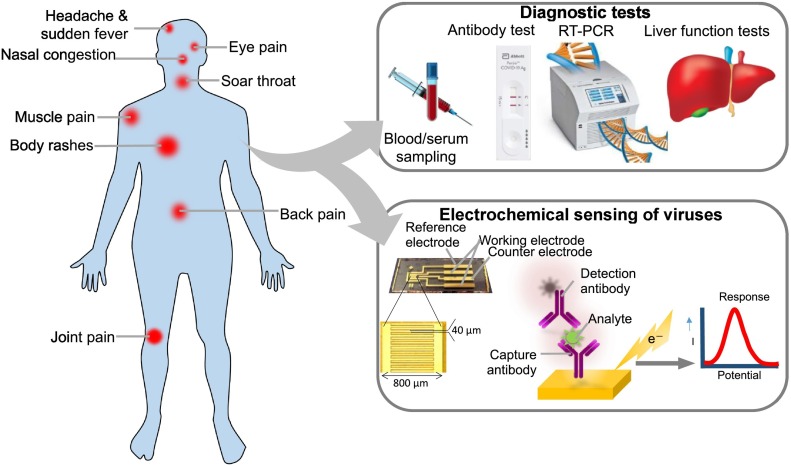
Respiratory viruses mostly cause URTIs, such as common cold, sinusitis, pharyngitis, epiglottitis and laryngotracheitis. However, they may progress and lead to LRTIs, such as pneumonia or bronchitis [2]. Viral transmission occurs through several routes, for e.g., aerosol (long range) or airborne (short range, caused by sneezing, coughing etc.) respiratory droplets, contact directly or indirectly [2]. URTIs caused by viruses presents general flu-like symptoms including cough, runny nose, sneezing, fever, chest pain, difficulty breathing (especially with epiglottitis). Symptoms of LRTIs include cough, fever, chest pain, sputum production, tachypnoea and more. Pneumonia may cause other symptoms such as headache, myalgia, nausea, vomiting and diarrhea. The primary cause of bronchiolitis in infants related outbreak is associated with RSV according to reports from neonatal intensive care units (NICUs) [4]. RSV causes annually, ~33.8-million new cases of acute LRTIs in children worldwide with <5 years of age with hospital admissions of 3.2 million and 59,600 in-hospital children's deaths in 2015 [5,6]. It has been reported that about 60–70% of children aged just a year and all of the children aged ~2 years have been infected by RSV [6,7]. Geographically, it is observed that the RSV travels south to north showing within March–June period in the countries located in southern hemisphere region and September–December in the northern hemisphere region [6]. RSV wave occurs during the rainy season in humid countries. A summary of distinct respiratory viral features such as size, genome, morphology, disease symptoms and related outbreaks in the past decades is presented in Table 1 (Fig.1 ).
Fig. 1.
(A) Schematic images showing different portal entry of viruses through nose, throat and lungs. Respiratory viruses are commonly spread through inhaling the virus particles containing droplets (B) Influenza subtype profile in global influenza types in circulation in the order high to low number of incidence occurred with influenza A and B followed by subtypes, H1N1, H3 and B-Yamagata that are derived from influenza A [9].
In 20th century, three influenza pandemic outbreaks occurred in 1918, 1957 and 1968 termed Spanish, Asian, and Hong Kong influenza, respectively based on their presumed sites of origins [8]. They represent three distinct antigenic influenza A virus subtypes, such as H1N1, H2N2, and H3N2, respectively. The recent global influenza surveillance and response system (GIRS) WHO recorded during 2019–2020 showed that influenza A(H1N1) strain subtype found to be more frequent type of viral infections caused that reached to a highest peak in 2020 (Fig. 1B) [9]. Most viral pneumonia cases are reported to be caused by influenza viruses and in some cases, secondary bacterial infections after influenza may lead to serious outcomes [8].
Other large family of viruses that constitute over 50 distinct serotypes are double stranded DNA based adenoviruses [2]. These virions are non-enveloped that bear size of ~90 nm with icosahedral symmetry. Most adenoviruses infections occur in children aged less than 5-year or adults with compromised immune system. In 2007, HAdV viruses caused severe outbreaks, while in 2014, initially new HAdV-14 variant emerged with military staff (US) and then spread in the populations and led to pneumonia and acute respiratory distress syndrome (ARDS) [2]. Human Rhinoviruses (HRVs) are ~30 nm in diameter are other non-enveloped viruses made of single stranded +ve sense RNA (Table 1). These HRVs are identified with over 150 different strains that causes a higher frequency of colds affecting mostly children approx. 12 infections per year. Among other RNA viruses, single stranded –ve sense and non-segmented RNA viruses called Human parainfluenza virus (HPIV). These are larger viruses that measure 150– 200 nm in diameter and pleomorphic enveloped virions that infects young children and causes mild to severe pneumonia and bronchitis worldwide. Major risk factors for HPIV infections are associated with malnutrition, vitamin deficiency, and respiratory irritants. There are four classes of HPIVs (1–4), while the HPIV-4 is again having subtypes a and b that infects about 5 million young children every year in the United States. Similar ~200 nm diameter size of another –ve sense, ssRNA enveloped virus that has been for over 50 years in the human population is called Human Metapneumovirus (HMPV). Globally, it causes respiratory tract infections about 20% among children.
Among DNA based viruses, Human Bocavirus (HBoV) is a small 18– 26 nm ssDNA virus is found to be active and causes infections especially during the winter and spring months (Table 1). According to an epidemiological report, HBoV is generally found to co-exist about 50% cases with other respiratory viruses [2]. HBoV mostly found to be associated with pneumonia in young children.
The human corona virus was discovered in the beginning of 1960 and it has been mostly circulating in the human population for a longtime [10,11]. The word “corona” was derived for these viruses because of its crown-like morphology viewed under a scanning under electron microscope. Genetic studies classified these viruses into alpha-, beta-, gamma-, and delta-CoV. There are five different human coronaviruses namely, alpha- (including 229E and NL63) and beta- (OC43, HKU1, and SARS-CoV) coronaviruses [10,11]. Interest in this group of viruses has drawn attention after 2003 global pneumonia outbreak (SARS-CoV-1), which resulted in ~800 deaths and affected people in ~30 countries. The second outbreak took place in 2012 caused by MERS-CoV virus and MERS-CoV with new flu-like respiratory illness that was reported in over 21 countries [10,11]. Further, a new virus was identified that caused recent corona virus disease 19 (COVID-19) that transmitted across global boundaries and caused respiratory tract infections. According to WHO, as of June 2021, there have been over 178 million COVID-19 confirmed cases and over 3.8 million deaths worldwide [12]. The fatality ratio was reported to be higher in male to female (2.4:1) [13]. The studies suggested that SARS-CoV-2 infections occur in all age groups. However, old age people (70 year) are at higher risk and vulnerable due to potential pre-existing health conditions. The median time from symptoms to death and incubation time is reported to be 14 and 5.2 days, respectively. The individuals with chronic pre-existing condition, especially cardiovascular disease, diabetes and hypertension and other compromised immune systems are major associated risk factors for COVID-19 [3]. SARS-CoV-2 particles are approximately 120–160 nm in diameter decorated with ~20 nm petal-shaped spike protein extending on viral peripheral surface and this virus belong to sub-genus Sarbecovirus and genus Betacoronavirus whose morphology varies from oval to spherical or pleomorphic [14]. A summary of distinct respiratory virus features such as size, genome, morphology, disease symptoms and related outbreaks in the past decades are presented in Table 1.
Despite of major advancement in science and medicine, the potential for spread of viral diseases is ever increasing and these outbreaks risks escalate into epidemics or pandemics. Delayed detection of infectious diseases presents risks in controlling the spread of disease or pandemics. There are many preventable non-medical reasons, of which are urbanization, global travel, climate change and human-animal contact that are the likely causes for future pandemics to occur. However, most importantly, medical intervention, such as by early disease diagnosis, treatment and prevention offers effective controlling of disease, allow rapid isolation of diseased individuals, reporting and surveillance of episodes for infection control measures [15]. Current practices in contagious disease diagnosis approaches are inadequate and therefore fail in very first step of early disease diagnosis and prevention, and therefore required re-evaluation of current diagnostic practices to develop new diagnostic tools. In this review, we first began assessing the most common traditional diagnostic methods employed for viral disease diagnosis. Later, explored on possible means by which these approaches can potentially be further improved with respect to sensitivity, speed of detection, and cost-effectiveness with special emphasis to interfacing carbon-based nanostructured materials.
3. Conventional/laboratory-based methods for diagnosis viral diseases
Currently, there are various methods employed for viral diagnosis (Fig. 2 ). All those existing methods can be classified into five categories, such as virus detection via viral isolation from cell cultures, detecting viral-antigens and their viral nucleic acids, electron microscopy or detection of antibodies (serology) [24].
Fig. 2.
Schematic illustration of conventional clinical laboratory based diagnostic methods for viral diagnosis.
3.1. Virus isolation from cell cultures
Viruses are cultured in disaggregated cells derived from tissue samples and isolated following chemical, mechanical and enzymatic treatments as gold standard approach for isolating virus. It is used to compare new methods and the only method that provides a supply of isolated viruses for further study through other methods [24]. There are many different cell lines used for virus isolation, for example, primary monkey kidney cell line, which is used for respiratory viruses and human epithelial cell line such as HEp-2 used for RSV isolation. Proliferation of viruses in cell culture can be monitored by cytopathic effect (CPE), which involves the visualization of morphological changes (such as swelling, shrinking etc.) in the cell culture. This method allows detection of multiple viruses however requires trained laboratory staff and it is difficult to accurately interpret CPE. The time required for cell cultures varies from long incubation periods of 1–4 weeks for classical methods to 1–2 days for rapid culture (e.g., shell vial assay) methods.
3.2. Direct detection via viral antigens
In our recent review, [25] we have comprehensively described different approaches to detecting viral or bacterial infections using host-biomarker proteins or antibodies that are not covered in this review. However, some of the approaches described for host biomarker proteins are also applicable to detection of viral antigens. Antigen detection methods, such as immunofluorescence (IF), immunochromatography, immunohistochemistry and enzyme-immunoassays (EIA or ELISA), provide direct evidence of viral infection in clinical samples. In immunofluorescence assays, viruses are detected by the interaction of fluorochrome tagged antibodies binding to target antigens, which then absorbs and emits light at a specific wavelength that is detected optically. IF is widely used for detection of respiratory virus infections, especially with parainfluenza viruses and adenoviruses; rapid diagnosis (less than 2 h) is possible [24]. Immunohistochemistry works on a similar principle as that of IF; here antibodies are tagged instead of fluorochrome with colored complex formed by an enzyme and substrate reaction. Generally, a horseradish peroxidase enzyme is used as a secondary probe for this method. One advantage of immunohistochemistry over IF is that immunohistochemistry stained slides can be kept for longer time than IF stained ones, making it possible to carry out several observations and a disadvantage is that immunohistochemistry method requires more time than IF, usually more than 24 h. One property of both IF and immunochromatography (IC) methods is that viral antigens are detected in-situ [24].
EIAs, or ELISAs can be used to detect viral antigens or antibodies and are very commonly used methods. There are different types of ELISA tests with various properties, detection and amplification systems, and sensitivities [25]. For example, with direct ELISA method, the antigen is detected through enzyme-labelled antibody directly; therefore, it is faster than indirect ELISA, although it is also less sensitive due to no signal amplification step. Indirect ELISA tests require an unlabeled primary antibody and a secondary, labelled antibody, which increases the sensitivity of the test through signal amplification, but it also leads to less diagnostic specificity. IC assays are widely used for virus detection, such as influenza A, B and RSV. The technique is based on the lateral movement of antigen or antigen-antibody complexes through the test strip by capillary action. Unlike ELISA, there are no reagent additions or washing steps required for IC assays. They require moderate expertise, equipment, and takes relatively short time (~30 min) to see the results, and some are approved as “point-of-care (PoC)” tests. However, IC is less sensitive compared to other antigen-based methods [26]. These isolation/separation methods are less sensitive as compared to viral culture or DNA/RNA (nucleic acids)-based polymerase chain reaction (PCR). Another downside is that some viruses, specifically Influenza A, have antigenic variations from year to year making the antigenic test kits unreliable or limited when a novel subtype of virus emerges.
3.3. Detection by electron microscopy
Electron microscopy (EM) is a rapid method used for virus diagnosis and it is the only method that can be used for direct visualization of viruses. EM is also important in detection of novel and non-culturable viruses, such as SARS-CoV, which was first identified in 2003 [24,26,27]. One advantage of EM over serologic and molecular methods is that there is no need for organism-specific reagents for viral detection. Thin sectioning and negative staining are two sample preparation methods that are used for examining viruses. Thin section EM takes around 3 days, whereas negative stain EM takes around 1 h. However, EM has the limitations of low sensitivity compared to other methods and requires expensive equipment and high expertise for operation of electron microscopy equipment [24,26].
3.4. Viral nucleic acids detection
Another method to viral antigen or antibody detection is by RNA/DNA from viruses that can be isolated from infected-cells and analyzed by classical methods such as Southern Blotting (DNA), Northern Blotting (RNA), dot blot, in situ hybridization or nucleic acid amplification methods (e.g. PCR, RT-PCR) [24,27]. Classical hybridization techniques are sensitive however, they are not very suitable for routine testing. Nucleic acid detection is especially useful for the cases where the viruses cannot be cultured easily or with latent viral infections. It is also more rapid and safer than viral culture methods. Development of PCR testing has evolved as a most sensitive molecular technique for detecting virus. Conventional PCR tests take 5–9 h and are more suitable for multiplex testing (detection of multiple viruses at once). However, it is prone to cross contamination because PCR tubes must be opened before the detection step, and toxic substances like ethidium bromide are used for amplicon (PCR product) detection. Real-time PCR has a shorter assay time of 1–5 h, less prone to cross contamination as it combines the amplification and detection steps in one-step and thus the test tube is not required to be open for detection. It is more sensitive than conventional PCR [24,26,27]. For RNA virus detection, reverse transcriptase PCR (RT-PCR) is applied, where reverse transcriptase enzyme is used to convert RNA to DNA first. An advantage of PCR technique is that it can be quickly adapted for novel viruses through a specifically designed primer/probes. Most commonly, RT-PCR method is utilized to demonstrate the detection of SARS-CoV-2 SARS virus [27].
3.5. Detection of antibodies in blood (serological assays)
Serological tests are widely used for detecting immunoglobulins (Ig), such as IgM and IgG antibodies. There are several methods used to detect viral antibodies described in previous section and most common methods include EIA-ELISA, immunoblotting (Western Blotting), serum neutralization assay, indirect immunofluorescence, hemagglutination-inhibition. Serological tests can detect both recent and past infections, providing information of the cause or origin of the disease (etiology). Time required for serologic tests depends on the type of method and it can range from 30 min to 24 h [26]. Antibody based ELISA tests are commonly used for viral detection, as they are rapid and relatively cheaper alternatives to other methods. Hemagglutination inhibition test is based on inhibition of hemagglutinin molecules by antiviral antibodies and therefore, highly useful for influenza virus detection as it shows enhanced sensitivity [28]. There are limitations to serological assays such as in some infection cases, antibody response is not strong enough to detect viral antigens or with immunocompromised patients sample the assay response was found to be weaker and cross reactivity between similar viruses was also reported [24,26].
So far, various methods have been reported for detecting causative viral infections by means of probing viral antigens, their nucleic acids, host antibodies or by physically examining the whole virus itself. However, the main problem in detecting viral causative agents at the early onset of infection is extremely challenging because of their low titer values. Another challenge is viruses such as SARS-2 are likely to mutate and with signs and symptoms that severely affects elderly people, infants, pregnant women, children and individuals with chronic pre-existing disease conditions or compromised immune system. Until now, only nucleic acid-based detection such as PCR/RT-PCR are the reliable methods used for accurately detecting viral infections at the early onset of disease. These nucleic acids-based methods require prior knowledge of viral genome sequence or run parallel tests for a range of known pathogenic viral nucleic acid probes for diagnosis of viral infections. There is always a need and demand to develop new, simple, rapid and accurate PoC detection device for pathogenic viruses, or by adding new functionality to existing sensing regimes. Elaboration of biosensor with nanomaterials-assisted biosensing can potentially address the future PoC diagnostic devices that can function remotely and in resource-limited settings without relying on centralized clinical laboratories, including those described in former sections of this review. Development of biosensor incorporating carbon nanomaterials (NMs) such as graphene and CNTs provides an exceptional advantage with their biological compatibility as well as, superior chemical properties. Graphene and CNTs nanostructures long been used for bio−/chemical sensing of a variety of targets, such as biomolecules, chemicals, and gases. There are several transduction methods for designing of carbon NMs integrated biosensors and a majority of these rely on electrical and fluorescence transduction schemes. Electrical (electrochemical) biosensing is another potentially most suitable type of transduction scheme that can be employed for infectious viral disease diagnosis. Integrating carbon nanomaterials provides unique physico-chemical properties and surface chemical modifications on electrochemical biosensors, making carbon nanomaterials most suitable nano-candidates for biosensing viral pathogens with greater speed, and sensitivity.
4. Electrochemical biosensors
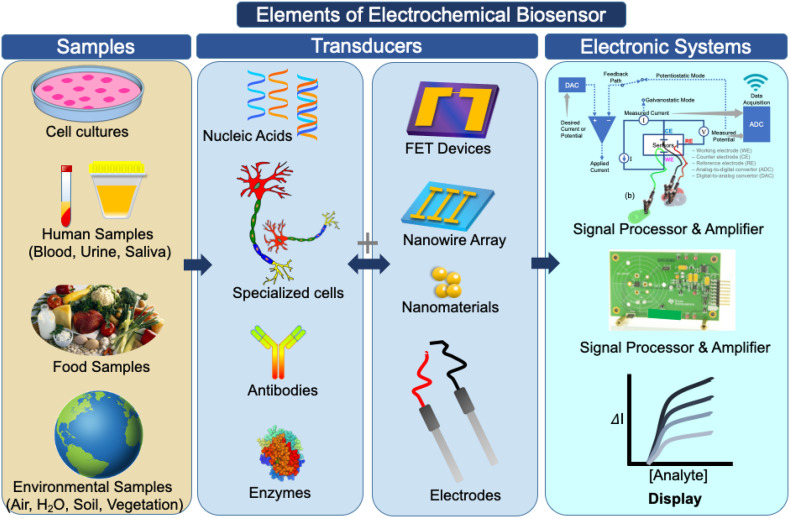
Biosensors play an essential role in disease diagnosis and health care monitoring applications and there has been a continuous effort made toward the improvement of health care technologies. A biosensor is an analytical device that detect and quantify biomolecular interaction that takes place between an immobilized probe with target/analyte molecules on a transducer platform (Fig. 3 ). Biosensors are powerful analytical tools in point-of-care diagnostics that facilitates personalized medicine therapeutics and cost-effective medical treatments. Biosensor devices are also widely used for detecting nucleic acids, various proteins, or for monitoring target (antigen) – probe (antibody) interactions. A biosensor is constructed by interfacing its transducer surface with a bio-receptor element, mainly antibodies, receptor, DNA/RNA that translate biochemical signal into a quantifiable electronic/electronic, electrochemical, optical, acoustic, or magnetic signal.
Fig. 3.
Scheme showing biosensor design components employed for sensing of target analyte samples, with special emphasis to utilizing an electrochemical biosensing platform that transforms biochemical information into current or voltages signals on an electrochemical transducer surface.
Electrochemical biosensors so far are the most widely used and highly preferred type of sensing platforms. As per the IUPAC definition, electrochemical biosensor is a self-contained integrated device that is capable of providing specific quantitative or semi-quantitative analytical information [29]. In other words, an electrochemical biosensor, which transforms a biochemical reaction into a measurable analytical signal, which is usually in terms of changes in current or voltages, whereby an interfaced bio-electrochemical element serves as transduction element (Fig. 6b). Electrochemical biosensing devices are designed by using different recognition elements, such as living whole-cells, micro-organisms, nucleic acids, antibodies, or enzymes [30]. Enzyme based electrochemical biosensing detection techniques are most widely used techniques, whereas immunosensors were developed by using antibodies, fragments of antibodies or antigens to monitor molecular binding in bio-electrochemical reactions [31]. The electrochemical biosensing research development in fact began after Clark first published papers on the development of oxygen electrode and modified oxygen electrode system in 1955–1956 and 1962, respectively [[32], [33], [34]]. These works sparked the development of different electrochemical transduction signal modes such as polarographic, potentiometric and conductometric based on enzyme reaction based biosensing platforms. These developments lead to the evolution of a first electrochemical biosensing technology, now commercially marketed as glucose biosensor device first produced by Clark's group Yellow Spring Instrument Company in 1975 [33]. The performance of electrochemical biosensor and analyte detection ability is heavily dependent on electrode material, electrode design, dimension, geometry and surface because electrochemical reaction signal are generally measured in close proximity of electrode surface area. Electrochemical sensor system development conventionally requires a working electrode (redox electrode), a counter or auxiliary and a reference electrode. Ag/AgCl is commonly used as reference electrode in the electrochemical sensor because it maintains a known and steady potential. Typically, in the electrochemical sensor, working electrode is used as transducing element and a counter electrode serves to establish a contact to the electrolyte solution in order to apply a current on the working electrode. In the electrochemical biosensor development, conductive and chemically stable materials such as, carbon based nanostructures (graphene, GO, CNTs, and graphite), gold, platinum and silicon-based materials were generally used to fabricate working and counter electrodes. The electronic transducing working electrode surface is chemically modified with nanostructures and referred to as chemically modified electrode (CME). The CME surface modified/immobilized with nanostructure provides excellent electronic, ionic or semiconducting properties with a biochemical-film on transducer surface [29] as shown in scheme Fig. 4 .
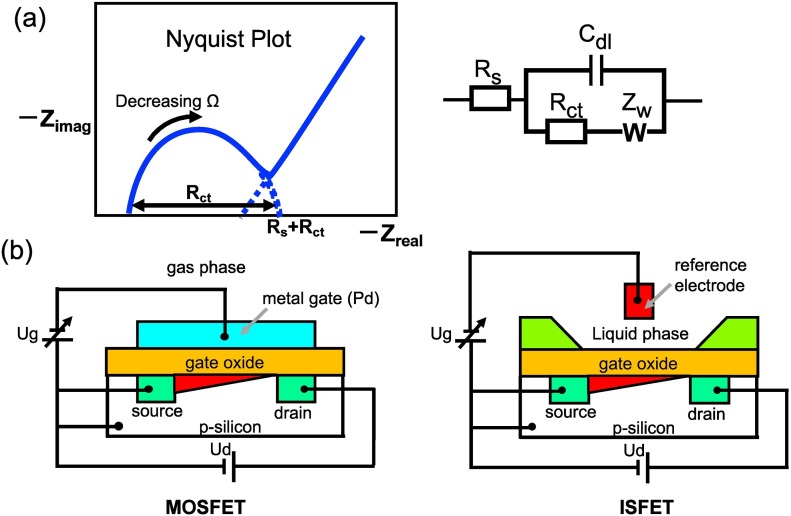
Fig. 6.
Schematic illustration of (a) EIS Nyquist plot (Zimag/Zreal) and Randles circuit consist of W, which is Warburg element that corresponds to diffusion processes. (b) Structures of MOSFET and an ISFET, where Ugand, Ud are gate voltage the source-drain voltage, respectively.
Fig. 4.
Scheme showing the typical components of a electrochemical biosensor including probe, target and interfacing of nanostructure electrode for biosensing of target molecules [29].
Electrochemical biosensors are popularly used for detecting a variety of biomolecules because they can be easily integrated with multiple modules such as; (i) miniaturized lab-on-a-chip, (i) low-cost microelectronic circuits, (iii) interfacing with electronic read-out, and (iv) a signal processing unit. Electrochemical biosensors being sensitive, easy to miniaturize, require small analyte volumes, excellent limit of analyte detection and display on-site results are most desired in medical diagnostics and in many other research areas including food safety and environmental monitoring.
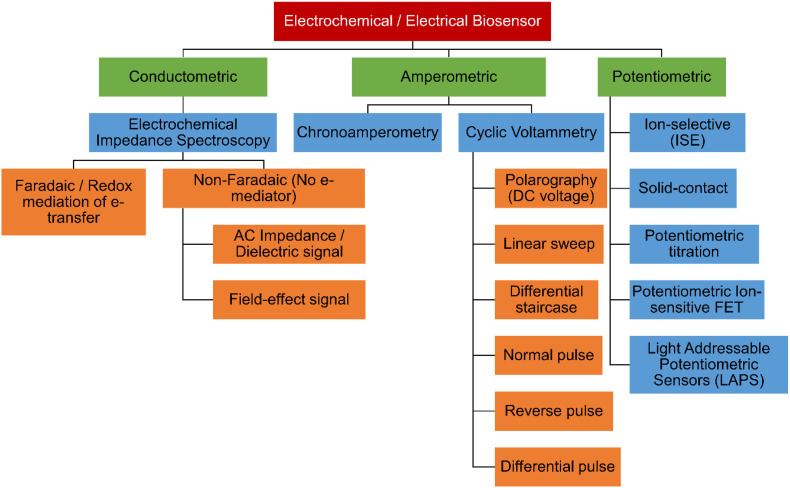
Electrochemical biosensor can be classified based on type of transducer modes used for signal measurements. These includes conductometry and surface charge, amperometry and potentiometry transduction platforms (Fig. 5 ) [29,30]. General principle of electrochemical biosensing (bio-electrochemistry) is based on electrochemical reaction taking place on or at the proximity of electrode and or between the electrodes that lead to; (i) a measurable current signal (amperometric), (ii) cumulate charge or potential(potentiometric), or (iii) changes in the conductivity of the medium (conductometric) [30].
Fig. 5.
Classification of electrochemical biosensors based on type of transducer and signal modes.
4.1. Conductometric biosensor
In the conductometric electrochemical biosensor, analyte induced signal is measured through electrolyte solution on transducing electrode that conduct an electric current between electrodes. These conductometric electrochemical biosensors with electrochemical transducer formats have been conventionally employed for sensing various biomolecules, disease biomarkers, proteins or antibodies. Most of designed conductometry-sensors are based on electrochemical impedance spectroscopy (EIS) or conductometric measurement technique. In the following section, basic principle and mechanism of EIS are discussed.
4.2. Electrochemical impedance spectroscopy basic principle and mechanism
EIS method measures the multiple microscopic electrochemical reaction processes between electrode and electrolyte system with an applied electrical stimulus. The transfer of electrons takes place due electrical perturbation, and it initiates oxidation and reduction reactions between electrode to electrolyte or vice-versa. The EIS sensor signal transduction is originating as a result of charges flow-rate and electrons as a function of electrode resistance, electrolyte as well as electrochemical reaction rates at the interface of sensor electrode [35,36]. In principle, AC current or potential is measured as function of frequency by applying AC current (galvanostatic) or potential (potentiostatic) to the electrochemical sensor. The electrochemical impedance parameter Z(ω,t) is a ratio of applied potential E(ω,t) or DC constant potential and current I(ω,t), which is described by following Eqs. (1a), (1b);
| (1a) |
| (1b) |
where, ωis an angular frequency (ω = 2πf, f is AC frquency) and t is time. The magnitude of impedance is |Z|, while the real and imaginary impedance are represented by Z′ and Zʺ respectively. Here, j represents an imaginary number √-1, and φ shows the phase angle between V and I.
A Nyquist plot generally represents the impedance data, where real impedance (Z') profile is plotted as a function of imaginary impedance (Z"). Another representation is shown by Bode plot where |Z| and/or φ is represented as a function of applied AC frequency. These data representation allow defining the electrochemical sensor signal transduction mechanism and characteristics such as interfacial and conductive properties of electrode and electrolyte. In EIS, representation of complex impedance data values is generally molded using an equivalent electrical circuit to analyze sensor performance parameters such as inductive, capacitive and resistive components. EIS method is generally classified in to two EIS sensor categories: (a) Faradaic electrochemical sensor and (b) non-Faradaic electrochemical sensor (Fig. 5). Faradaic sensor refers to electrical charge transfer across a sensor electrode and electrolyte in the presence redox species, whereas non-Faradaic sensor system does not require redox mediator to initiate the charge transfer between the electrodes. Conductometric sensors design can function in both Faradaic and non-faradaic types of EIS detection regimes.
4.2.1. Faradaic electrochemical sensor
In Faradaic electrochemical sensor, electrochemical transducer measures the impedance (resistance and reactance) signal through charge transfer capability of an analyte between the electrode and redox electrolyte solution, or redox medium between the electrodes or reference nodes over a wide range of applied alternating current frequency (Hz to MHz). In Faradaic EIS technique, an electroactive species (redox reagent) reduces and oxidizes at the electrochemical sensor electrode surface to induce the electrical signal (current, impedance). Developing Faradaic electrochemical sensor or Faradaic impedimetric sensor requires a redox reagent/probe and a constant applied DC in order to generate signal from electrochemical reactions. Therefore, difference between the applied potential and cell-equilibrium potential is generated (called an over-potential) through an applied constant or variable potential over the electrode cell/sensor. The over-potential generation results in promoting electron transfer in electrochemical redox reaction known as Faradaic process [37]. Faradaic sensor signal response is defined by Faraday's law (Eq. (2)), where the Faradaic current is directly proportional to; (i) number of electrons transferred in electrochemical redox reaction (n), (ii) Faraday constant(F), (iii) surface area of electrode (A) and, (iv) analyte/electroactive species flux (j) at the interfacial boundary.
| (2) |
The Faradaic sensors exhibits greater sensitivity than the non-Faradaic sensors because it measures current generated from the redox reactions. However, detection and biosensing of certain specific biomolecules using the Faradaic sensor can be difficult due to potential interactions of the target molecule with electrolyte. The Faradaic sensor signal response modeled through appropriate equivalent circuit with signal parameters such as impedance, which is a complex number, and often, real part is associated with resistive behavior (Z real), whereas the imaginary part (Z imag) is associated with capacitive process. Other parameters include (i) R s, resistance of electrolyte solution (ii) C dl , double layer capacitance and is also accompanied with a constant phase element (CPE), (iii) R ct, charge transfer resistance and (iv) Z w is Warburg impedance.
The R ct generates through potential due to oxidative and reductive process at the sensor electrode surface including the stearic effect or repulsion of charges species on conductive electrode surface. Warburg impedance (Z w) originates due to diffusion of electroactive charges through electrolyte solution to surface of sensor electrodes. Here, Nyquist plot provides the charge transfer resistance values (R ct), in which the R ct is the diameter of semicircle and Warburg impedance (Z w), which can be eliminated through an inclined 45° straight line at low AC signal frequencies (Fig. 6a) [35,36]. In the Faradaic electrochemical sensor, oxidation and reduction reactions are performed with a mediating redox chemical present in the medium. The most commonly employed redox mediator electrolyte is Fe(CN)6 3−/4- in the literature. A few Faradaic electrochemical sensors designs also utilized conductive polymer structure such as polyaniline, ploypirrole and polythiophene that facilitate the redox process through conducting charges along their chain structure due to doping mechanism.
4.2.2. Non-faradaic electrochemical sensor
Non-Faradaic based electrochemical sensor does not require redox intermediary chemicals/agents and it measures the change in dielectric parameters (capacitance, impedance, permittivity, current) in between the electrode and the medium (electrolyte) as well as at the sensor electrode interface. Non-Faradaic EIS method has exceptional advantage owing to its feasibility to miniaturization of sensor design, real-time monitoring and most importantly, it does not require additional electrodes and accessories. The impedance signal transduction in non-Faradaic type of sensor originates from change in dielectric properties between the electrodes which is ascribed from the formation of double-layer capacitance and its charging and discharging process. Here, sensing principle of non-Faradaic type of EIS sensors is therefore relied on changes in charge distribution or conductivity or dielectric properties as a result of formation of analyte–target complex on the surfaces of conductive electrodes [38]. The impedance signal directly depends on different parameters that mainly include double layer capacitance formation, nature of charge carries and their concentration in between the electrodes, the electrode design and geometry, as well as the type of conductive electrode material employed. The equivalent circuit for such sensor system consists of a combination of capacitors and resistors. The transducing sensor signal response can be described by an equivalent circuit that is comprised of elements such as capacitors and resistors. Here, the |Z| and φ components are adequate transducer parameters. As per the Helmholtz model, the double layer capacitance (C) is given by the following Eq. (3).
| (3) |
C depends on the relative permittivity (ε r) of the solution, permittivity of free space (ε 0), electrode surface area (A) and Helmholtz layer. For providing larger sensor surface, non-Faradaic conductor electrodes can also be made capacitive sensor that consist of interdigitated fingers pattern. Here, capacitance signal changes due to dielectric properties change in between the electrodes. This change in capacitance is correlated to the binding of analyte/target molecules and the concentration of captured target by chem−/bio-receptors tethered on sensor electrodes.
Recently, immunoassays utilizing capacitive array sensors have been employed for detection of multi-analytes through both Faradaic and non-Faradaic electrochemical regimes. These arrays of sensors offers complex multi-analytes or multiplexed detection option with simple analytical procedure, short analysis time, require low sample volume, high test efficiency, and less expensive as compared with that of single-analyte assays. Several immunoassays have been successfully designed that utilize capacitive label-free biosensors platforms without/with involving a redox mediator (non-Faradaic or Faradaic, respectively) [[39], [40], [41]]. However, there are major challenges in the development of capacitive label-free biosensors mainly because of their poor reproducibility which is associated with non-uniformity in fabricated electrode designs that provides a large standard deviation and non-specific signal.
Further, other types of electrochemical sensor systems include field-effect transistors (FETs), which utilizes source-drain electrode terminals to fabricating ion-sensitive FETs (ISFETs) and metal-oxide FETs (MOSFETs) transistors. These FET sensors are designed to measure the current signal generated as a potentiometric effect at a gate electrode (Fig. 6b). In surface charge-based electrochemical transduction system, ISFETs are used to detect the ion concentrations [29,30]. Here FET gate electrode surface is modified with an ion-selective membrane (ISM) to measure the analyte signal. If these sensors are used for biological sensing purposes, they are called for instance, enzyme-FETs (ENFETS) or immunological FETs (IMFETs) for integrating FETs with enzyme or antibody, respectively. To understand ISE-based biosensors, ENFETs and IMFETs can be understood by further reading on biologically sensitive FETs reported by Schöning et al. [42]. Interested readers can find more information on FET sensors in a recent review by Sadighbayan et al. [43].
4.3. Amperometric sensors
Amperometric electrochemical sensing usually requires a three electrodes system made of a reference (Ag/AgCl), working electrode (Au, carbon), and a counter or auxiliary electrode (Pt). Here, the current signal is continuously measured by an amperometric transducer following redox reaction of an electroactive species (redox/electron mediator) in a bio−/chemical reaction taking place on the electrochemical transducer surface. In other words, the concentration of bulk electroactive species in the electrochemical sensor system is directly proportional to the generated current from oxidation and reduction of charge species produced/consumed at the bio-electrode-electrolyte interface [29,30]. This electrochemical biosensing method rely on use of redox reagents to mediate oxidation and reduction of the analyte at the working sensor electrode because not all biomolecules (protein, DNA, microorganisms) have ability to serve as redox mediator themselves. These sensors are known as amperometric sensors because the transducing signal is measured as changes in current signal at a certain value of applied potential. If the transducing current signal measurement is performed by applying different values of potential, such method is known as voltammetry sensing method, which is reviewed in the following section. The concentration of analyte/target molecule is directly proportional to the value of peak current with respect to selected potential range. According to literature reports, the amperometric devices exhibit superior sensitivity compared with the potentiometric devices [29,30,44].
4.3.1. Cyclic voltammetry (CV)
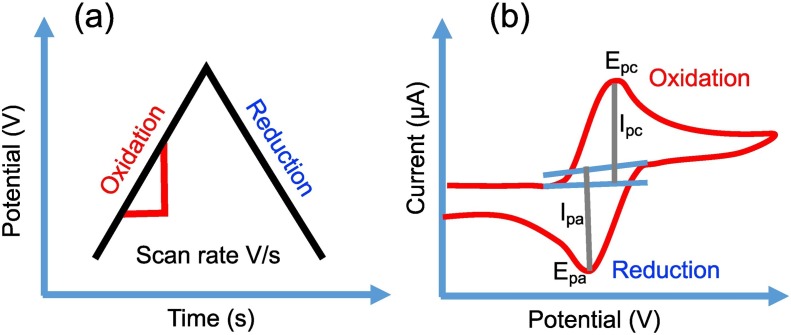
In the voltammetry electrochemical/bio sensing method, the analytical measurement of analyte concentrations is obtained by measuring transducing current against varying applied potential. Therefore, voltammetry is classified as an amperometric electrochemical technique. Further, voltammetry can be categorized based on the condition of applied potential that includes linear seep, differential staircase, pulse voltammetry (normal, reverse and differential pulse), and polarography (DC Voltage) voltammetry [29,30,44]. Cyclic voltammetry is a most commonly utilized amperometry technique that provide important chemical/biosensing parameters such as kinetics of rate of electrochemical reaction and redox potential. Typically, CV is operated by sweeping the voltage between two values (V 1 and V 2) at a fixed electrochemical reaction rate (voltage/s). In the electrochemical reaction sweep, initial voltage (V 1) is swept to final voltage (V 2) and then the voltage sweep is returned to V 1 as shown in Fig. 7 (a-b). The electrochemical scan rate is defined by a ratio (V 2 -V 1 )/(t 2 -t 1) to generate an electrochemical signal in a given time of interval. In CV, potential (voltage) and current (I) parameters are measured between the working-reference and working-counter electrodes, respectively. A voltammogram is generated by plotting the measured current signal as a function of applied voltage. The current signal increases when the potential voltage (V 1) increases toward the oxidation potential of the target analyte molecule. After the potential voltage V2 reaches to reduction potential, the current begins to decrease which is displayed as a peak shaped curve at which the analyte concentration diminishes on the surface of electrode (Fig. 7b). In a reverse voltage sweep, the voltage scan turns back to voltage (V 1) through reduction reaction as the current decreases, which is seen as a second, reduction peak with opposite polarity in comparison with forward scan until finally voltage scan reaches to V 1. The information related to the reversibility of electrochemical reactions can be obtained in the reverse voltage scan sweep at a specific scant rate. The nature of voltammogram shape provides information about analyte concentration on the working electrode. Performance of voltammogram depends on several factors such as scan rate, electrode material, adsorption, and concentration of analyte.
Fig. 7.
(a) An illustration of scan rate activity with potential over time and (b) cyclic voltammetry curve for detailed oxidation and reduction reaction illustrated with potential over current.
4.3.2. Chrono-amperometry
Chrono-amperometry is based on amperometric principle in which a square wave potential is applied at working electrode to measure the steady-current signal [29,30,44]. The change in sensor electrode current signal response originates due to change in thickness of diffusion layer on the working electrode surface. The chrono-amperometry technique is introduced by Nernst and its principle is based on the formation of a static thin film which is in contact with electrode surface. This arrangement allows controlled diffusion process through which transfer of analyte occur from electrolyte solution to the electrode. In this case, the transfer of analyte concentration (c 0) in the bulk-solution while balancing a gradient concentration in the electrochemical reaction between electrolyte solution and electrode surface. In the chrono-amperometry, the current response can be defined by Cottrell equation, where linear electrochemical diffusion process on planar electrode is monitored by measuring the current as a function of time. Cottrell equation exhibits that the current I is dependent on Faraday's constant (F), the number of electrons transferred per molecule (n), electrode area (A), the concentration of analyte (c 0), diffusion coefficient (D), time (t) were all expressed in the following Eq. (4).
| (4) |
Chrono-amperometry electrochemical technique does not relay on reactants labelling and it is a sensitive and supportive technique utilized for time dependent CV measurements.
4.4. Potentiometric sensors
In potentiometric sensors, the transducers measure potential due to the accumulation of charges on the working electrode where bio−/chem-receptors interact with analytes. The potential measurement is performed between the reference and working electrodes when negligible current flows through them. Typically, potentiometric devices transduce the ion activity signal in the electrochemical reactions [29,30]. The detection of analyte is usually determined by Nernst equation, which shows the correlation of potential with concentration of analyte and known as direct potentiometry method. In this technique, cell potential (E cell) or electromotive force (EMF) is measured at working electrode at zero current and define by Eq. (5)
| (5) |
where, parameters in Eq. (5) represents absolute temperature (°K) (T), universal gas constant (R), ratio of ion concentration (Q) between electrode, number of charges (n) and Faraday constant (F).
Potentiometric transducers can be designed for different transducing platforms that includes ion-selective electrode with polymer or membrane, wire electrodes based solid electrodes, potentiometric titration, ISEFTs and light addressable potentiometric sensors (LAPS). Most of the potentiometric electrochemical sensors utilize ion-selective electrodes (ISE) to achieve lower analyte detection limit. Potentiometric sensor offers advantages of measuring small concentrations in small sample volumes, minimized or no chemical cross talk during measurement of a sample. In the potentiometric titration method biochemical reaction point is measured in which equal quantities of opposing solutions, such as 0.1 M HCl and 0.1 M NaOH gain equilibrium state. Here, end point is determined by measuring changes in electrode potential at steady or negligible current which is brought through variation in solution concentration. ISFETs are another potentiometric sensor formats to measure changes in pH values, selective ion-concentrations and the enzyme involved biocatalytical reaction kinetics. Further, LAPS is another example of novel optical/electrochemical hybrid potentiometric technique. LAPS combines a focused light source scanning that allows determining the potential distribution at the sample and substrate interface. Potential distribution along the interface of the sample and substrate surfaces. Bratov et al. provided detailed information about potentiometric sensor arrays, their design, and development of different approaches for further reading [45].
In recent years, the conventional electrochemical transducers provided opportunity to integrate them with carbon-based nanomaterials to enhance the sensitivity, surface-to-volume ratio, charge-carrier density that constitute new generation of powerful electrochemical sensing platforms that find application in sensitive detection of analytes, such as SARS-CoV-2 which otherwise, not possible by conventional electrochemical sensors. The major issues in the development of electrochemical nanobiosensors include designing of transducer biosensing interface, high selectivity and sensitivity of biosensor, response time, compatibility with complex biological samples, confined operating environmental conditions, multiplexing biosensor with multi-analyte detection capability within one biosensor device. Recently, nanomaterial interfaced electrical biosensors with new design and strategies have been attracted much attention to apply them for respiratory virus detection. Advancement in structure and functional properties of nanomaterials, especially carbon-based nanostructures with the nanofabrication approaches include electron beam lithography, photolithography, nano-printing lithography played a crucial role in designing and specification of new generation of electrochemical biosensors [46,47]. Nanomaterials, specifically carbon based nanomaterials such as graphene, CNTs and its derivatives offer significant advantages in electronic biosensing of infectious viruses and other biomolecules that include label free, real-time, high sensitivity, interfacing with wafer-scale semiconductor chip fabrication, increase spatial resolution of sensor signal and ease of scale-up production.
The following sections describe on the advantages of 2D graphene and CNT nanostructures and their unique physicochemical properties, various approaches employed for their surface modification/chemistry and linker molecule characteristics that facilitate in the development of electrochemical nanobiosensors. Further, advantages of sensors interfaced with these carbon-based nanostructures and their unique physicochemical properties in designing new nanobiosensors with special emphasis to detecting viral or other disease causative agents are presented.
5. Carbon nanomaterials: properties and surface modification
Carbon materials are applicable in every aspect of our daily life because they are abundant and lightweight materials that can be used for a variety of applications [48]. These carbon-based nanomaterials can be classified based on their dimensionalities (D) as zero-D (0D) such as carbon dots, one-D (1D) as in CNTs, and two-D (2D) in graphene nanostructures (Fig. 8 ). These nanomaterials bear wider operating temperature, sensitivity and wider dynamic transducing signal range even in harsh environmental conditions. The emerging applications of CNTs and graphene in sensor and energy storage fields has led to their large-scale industrial productions. Both graphene and CNTs are important polymorphic forms of carbon and possess excellent electronic (electron transfer), mechanical properties and excellent biocompatibility. These useful features of graphene and CNTs make them excellent carbon-based materials for the fabrication of sensor electrode.
Fig. 8.
Schematic illustration of certain allotropes of carbon nanomaterials for nanobiosensor development.
Interfacing graphene and CNTs nanostructures in electrochemical sensors enhances sensitivities by several orders of magnitude, enabling fabrication of an entirely new generation of biosensor devices. Graphene and CNTs nanostructures carry extraordinary structural, electrical, physical, and optical properties and following section highlight their unique characteristics that make them superior nanomaterials for the development of electrochemical biosensing transducing platforms. The following sections also describe individual nanostructure properties, their surface functionality and important case studies successfully employed for biological sensing.
5.1. Graphene nanostructure properties
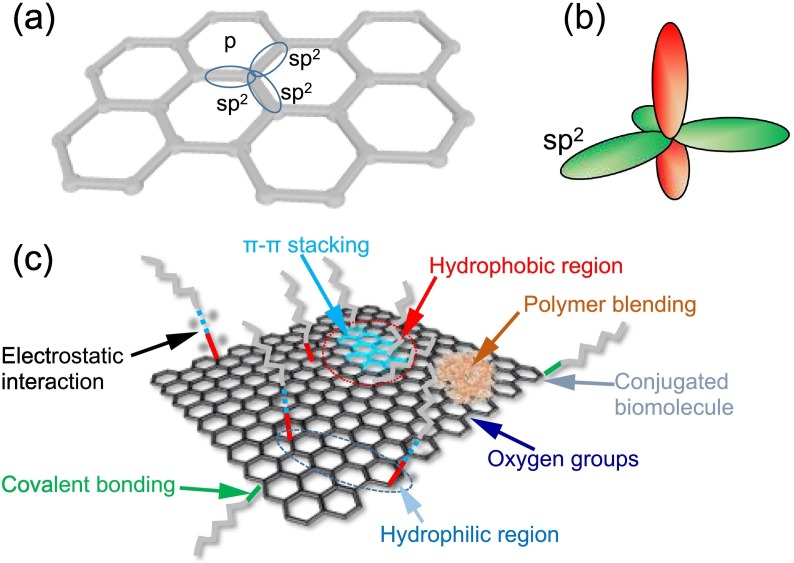
Graphene is identified as a new class of 2D material with an atom-thick honeycomb sheet made of carbon atoms. Novoselov's group (University of Manchester) received a Nobel Prize (physics) (2010) for their outstanding work on synthesis of graphene and isolation, identification, and characterization. Graphene being a 2D material has unique properties most desired in the development of electrical sensor and other future applications. The diameter of a carbon atom is 0.33 nm, which implies that there are approximately 3 million graphene layers in a one-mm of graphite material [49]. Graphene is one atom thin transparent conductor and suitable nanostructure especially for sensor technologies today because, it has high electron mobility that is four-times larger than Group III-V semiconductors and ~ 200-times higher than Si, much higher electrical conductivity than copper, and has a large 2630 m2/g surface area [50]. In graphene, thin layer sp2 carbon atoms are connected covalently forming a honeycomb lattice structure, which makes graphene zero-gap or semimetal semiconductor arising from valance and conductive bands touch Dirac points in its electronic structure [50]. This allows graphene to be a very attractive nanomaterial for sensitive biosensors and high-speed transistors platforms.
The unique electronic feature of graphene makes the incident light to constantly transmit through its nanostructure in the range of visible-IR. The optical opacity of graphene is about 2.3% and its transmittance is proportional to the number of graphene layers [50]. Graphene has the ability to quench photoluminescence, which enables its usefulness in designing optical sensor devices. In the graphene structure, carbon atoms are connected with strong σ-bonds of sp2 hybridized orbitals, which gives rise to a 2D- hexagonal networks of carbon (Fig. 9a-b). Graphene exhibits outstanding electronic and photonic properties due to the presence of π-orbitals orthogonal in the hexagonal plane [50,51].
Fig. 9.
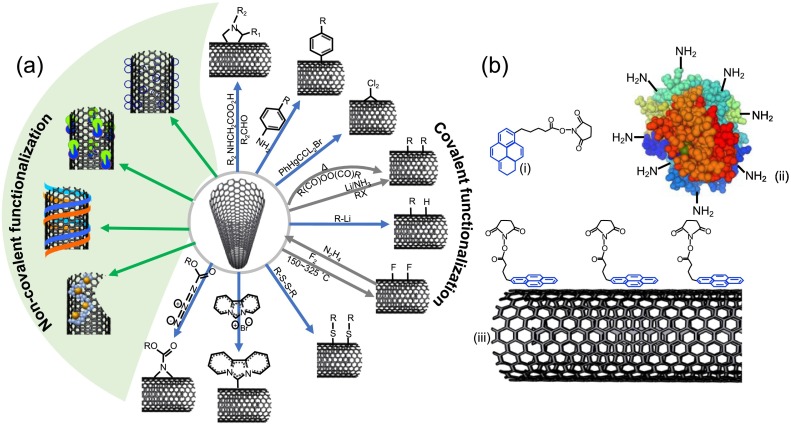
Schematic illustration of graphene structure with; (a) sp2 hybrid carbon-atoms, (b) π and σ bonds formation and (c) Schematic illustration of different chemical surface modifications on graphene by non-covalent and covalent functionalization [51].
The superior thermal transport properties of graphene is attributed to its out of plane quadratic dispersion of phonon in its 2D crystal structure. Graphene's thermal conductivity is one order higher compared to Cu and measured to be 5000 W mK−1. Similarly, graphene found to be applicable for heat management in electronics due to its excellent heat conductivity. Interfacial thermal resistance can be minimized by strongly coupling graphene and polymer. Graphene is used to fabricate highly conductive transparent films because of its unique carrier mobility and electrical conductivity [52]. Further, graphene structure composed of sp2 carbon hybrids, which establishes σ bonds with the neighboring atoms and the length each carbon bond is 0.142 nm. This σ bond property in graphene influences on the strength or Young's modulus (1 TPa with a thickness of ~0.33 nm) that makes mechanical strength of graphene ~100-times greater than for steel. Graphene is also stretchable flexible conductor [50], graphene can also exhibit magnetic properties such as paramagnetic, and exhibits magnetic switching (ferromagnetic or antiferromagnetic) phenomena [[52], [53], [54]]. Recent studies show externally induced magnetic property of graphene through its modification by coupling with magnetic nanoparticles either by in-situ chemical reduction of metal precursors such as iron, cobalt, or nickel-salt precursors, or coupling graphene with magnetic nanoparticles synthesized [52]. These hybrid features enable tuning the graphene surfaces to adsorb guest molecules and segregation that is most desired properties for fabricating high sensitivity graphene-based sensors.
Graphene has been utilized as electrochemical nanomaterial for the fabrication of sensors such as gas, chemical, and bio- sensors because of its superior physico-chemical and biological properties. Graphene can be used to design electrical sensor as a silicon analog because of its advantages of flexibility, miniaturization, and cost-effectiveness. However, the major challenges to fabricating sensor devices using graphene are poor chemical functionality and lack of energy band gap in its electronic energy structure. Therefore, development of further approaches/methods are required to engineer electronic and surface functionality of graphene structure. The following sections describe various approaches employed for surface modification of graphene, their surface chemistry and linker molecule characteristics and ways to utilize them as superior nanomaterials for fabricating electrochemical biosensing transducing platforms.
5.2. Graphene surface modification
Understanding the process/methods of graphene synthesis is important to appropriately modify the graphene nanostructure surface. There are different approaches to synthesize graphene and the most simplest method being first reported by Novoselov and Geim in 2004 is by exfoliation of graphene layers from graphite using a scotch-tape [55]. Among other methods, chemical vapor deposition (CVD) method that utilizes carbon precursors, such as methane (CH4) or ethanol (C2H6O) for their thermal decomposition on a transition metal foil as a catalyst such as Cu, Ni or Fe at high temperatures (~1000 °C) [56]. Then, transfer polymer method is used to separate or peel off the synthesized graphene from the surface of catalyst metal foil, where polymethylmethacrylate (PMMA) or polydimethylsiloxane (PDMS) served as suitable polymers for graphene transfer process [50]. Further, considerable research effort has been made for the preparation of graphene oxide (GO) [57]. GO differs from graphene because of the presence of additional oxygen atoms bound with the carbon networks making GO hydrophilic in nature and easy to prepare its dispersion in water, unlike graphene which is hydrophobic in nature. GO can be chemically reversed to reduced graphene oxide (rGO) by reducing by means of different chemical or thermal reduction processes in high yields. However, the major challenge is to control the structural defects by reduction process that limit the electrical performance of rGO. Despite, it is most preferred approach for surface modification of graphene nanomaterial and synthesis in large- scale of graphene and its nanocomposites.
Unlike GO, it is difficult to handle graphene's hydrophobicity or preparation of its dispersions in water or polar media, which is essential for biosensor surface modifications, and thus making it a challenging task for fabrication of graphene biosensor. The reactive chemical functional groups on pure graphene are essential for the covalent or non-covalent coupling of specific molecules, however pure graphene does not possess any additional chemical functionally on its surface. Therefore, different approaches have been introduced for surface modification of graphene with tunable surface chemistry. This includes covalent and non-covalent functionalization of graphene nanostructures as shown in schematic Fig. 9c. Additionally, surface properties of graphene nanostructures are chemically modified by various approaches, such as doping graphene with other elements, changing the number of graphene layers, inducing defects, and forming different sizes, such as in the form of nanoplatelets, nanoflakes and nanoribbons. The chemically modified graphene forms a strong interface for the biological systems to be tethered without geometric restrictions or compromising the integrity of the attached biomolecule/chemical, which is most desired feature for electrochemical biosensing of analytes, especially for example, respiratory viruses or the disease diagnosis.
There have been numerous studies reporting surface modifications of GO and graphene based on covalent and non-covalent interactions. In covalent modifications, the sp2 structure of graphene honeycomb lattices forms covalent adducts with reactive species and induce defects that causes loss of the electronic properties [58]. The degree of functionalization in graphene lattices can be identified by the ratio of sp2 and sp3 hybridized carbon atoms, which can be observed in its Raman spectra. Noncovalent immobilization of graphene/GO is highly preferred because of reactive surface, dispersibility, biocompatibility, coupling capacity to capture the desired analyte for sensing properties. The most common strategies for non-covalent immobilization/functionalization utilize Van der Waals, π-π interactions, hydrogen bonding and ionic interactions facilitates non-covalently modifying graphene and GO [59]. The graphene surface can be non-covalently functionalized without altering the graphene's electronic and structural properties, and it is easy to surface-modify simultaneously to introduce new desired surface chemical groups.
Graphene and GO surfaces can be non-covalently modified with desired organic molecules or polymers via Van der Waals or π-π interactions. These interactions can induce high hydrophobic character of graphene/GO. The most common interactions between graphene/GO with organic molecules are π–π interactions that varies from small to most expanded π network. Presence of uniform oxygen functional groups on the surface of and edges of GO promotes the hydrogen bonding and ionic interactions. Georgakilas et al. [59] has provided a comprehensive review on non-covalent interactions in graphene and GO in relation to the Van der Waals, π-π interactions, hydrogen bonding and ionic interactions.
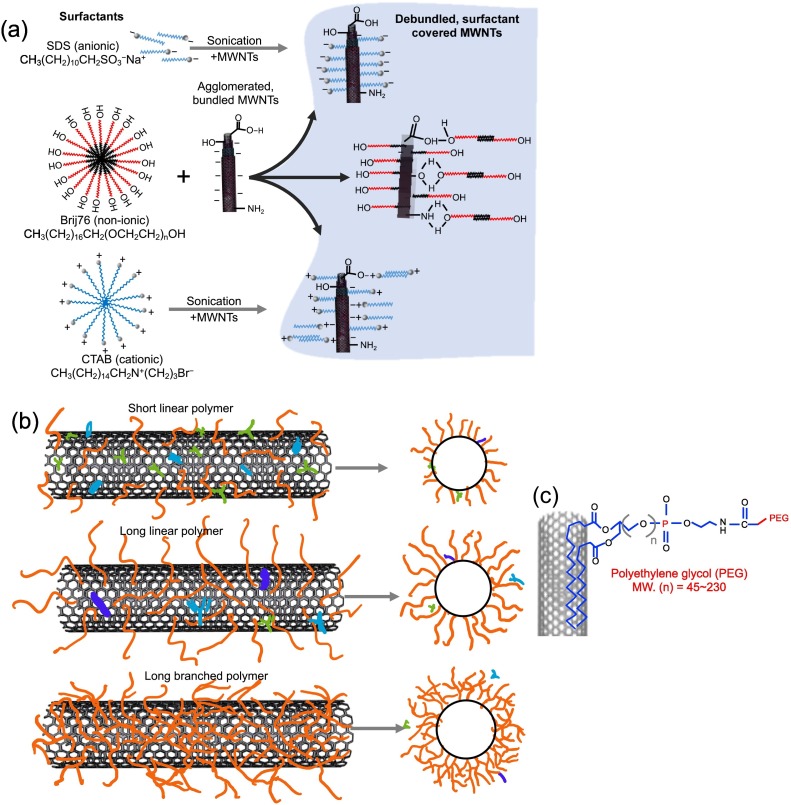
For non-covalent functionalization, mostly aromatic derivatives such as pyrene and perylenediimide derivatives are utilized as electron acceptor molecules and or electron donor on graphene/rGO sheets through π–π interactions that leads to induced repulsive forces between the layers and provide highly dispersible nanostructures in aqueous phase with remarkable charge transfer capability. The most suitable and common aromatic compounds used for functionalization of graphene/rGo through noncovalent π–π interactions is listed in Table 2 . These include porphyrin derivatives [[60], [61], [62]], pyrene [[63], [64], [65], [66], [67], [68]], pyridine and fluorinated benzene derivatives [69], ionic liquids [[70], [71], [72], [73], [74]] and polymer ionic liquid [75], quinolone [76], anthracene [77], coronene derivative [78] and triphenylene [79]. These aromatic molecules have ability of donating or withdrawing electron, stacking capability (planarity) and electrostatic repulsion with graphitic materials (graphite/graphene/rGO) which can result in the stabilized and highly dispersed exfoliated graphene sheets (EGS) from graphite/GO/rGO/graphene materials. Further attempts have been made for enhancing dispersion stability of hydrophobic G or rGO in water by utilizing amphiphilic organic molecules (surfactants) and macromolecules such as sodium dodecyl benzenesulfonate, [80,81] zwitterionic (quaternary ammonium salts) [82,83] and non-ionic [84], polar functional groups [85], sodium cholate [86], cellulose [87,88] (Table 2). The hydrophobic interaction graphene/rGO surfaces with hydrophilic/ionic/polar/non-ionic functional groups of surfactants and macromolecules allow exfoliate/charge transfer and stability of surface modified graphitic nanomaterials.
Table 2.
The list of most suitable aromatic/surfactants/amphiphilic compounds for functionalization of graphene/rGo through noncovalent π–π interactions reported in the literature.
| Surface functional molecule for non-covalent functionalization | Carbon nanomaterial | Interaction | Reference |
|---|---|---|---|
| Pyridinium-functionalized porphyrin | Graphene | Size and planarity with graphene by π stacking | [60] |
| Protoporphyrins of iron (FePP) and zinc (ZnPP) | Graphene | Large bandgap (0.45 eV) yielded bv FePP π–π stacking in graphene, while ZnPP physi-sorbed (0.23 eV). | [61] |
| Porphyrin derivatives with sulfonate (negatively charged TPP-SO3Na) and ammonium groups (positively charges TPP-ammonium) | rGO | Non-covalent functionalization induced repulsive forces between the negative charges. | [62] |
| 3.4.9.10-perylenetetracarboxylic diimidebisbenzenesulfonicacid (PDI) -acceptor and Pyrene-1–sulfonic acid (Pys)-donor | rGO | Electron donor or acceptor π-π interactions for remarkable charge-transfer ability | [65] |
| Different polar molecules: naphthalene, 1-napthylamine, and 1-naphthol | GO/FeO.Fe2O3 and multi-walled carbon nanotubes (MWCNTs)/ FeO.Fe2O3 | Polar nature of molecules with the adsorption ability | [89] |
| Four pyrene units and a laterally-grafted oligo ether dendron | Graphite exfoliation | Tetrapyrene aromatic rings interacted with basal plane of graphene, hydrophilicity due to oligo ether chains | [63] |
| Sulfonated pyrene derivatives:PS1(1-pyrenesulfonic acid sodium), PS2 (6,8-dihdroxy-1,3-pyrenedisulfonic acid disodium), PS3 (8-hydroxypyrene-1,3,6-trisulfonic acid trisodium) and PS4 (pyrene-1,3,6,8-tetrasulfonic acid tetrasodium) | Graphite exfoliation | PS2 is the most asymmetric functionalization with highest dipole moment for graphite exfoliation | [64] |
| PS1: 1-pyrenesulfonic acid sodium salt | In-situ exfoliation of graphite, graphene nanosheets (GNS) | GNS surface decorated with functional molecules via with π–π stacking force | [66] |
| 1-pyrenecarboxylic acid (PCA) | Graphite exfoliation into single-, few-, and multilayered graphene flakes | PCA induces separation of graphitic layers with the help of its carboxylic groups and prevent π-stacking reform reversing. PCA also exhibits a hydrophobic pyrene group which mimics graphene with π-π interaction | [67] |
| Lamination of PCA-functionalized graphene onto flexible-transparentpolydimethylsiloxane (PDMS) | Multifunctional hybrid structure for electronic device | [90] | |
| Pyrene butanoic acid succidymidyl ester (PBSA) | Few layers of CVD graphene | Interactions (π − π) between PBSA and graphene | [91] |
| Pyrenebutanoic acid-succinimidyl ester | Noncovalent functionalization of Epitaxial graphene | Graphene non-covalently binds with pyrene and succinimide ester reactive group | [92] |
| 1,3,6,8-pyrenetetrasulfonic acid (Py-SO3) tetrasodium hydrate,1-pyrenemethylamine (Py-NH2) hydrochloride | Exfoliation of graphite | Strong anchoring of planar aromatic structures of pyrene moles on hydrophobic graphene surface via π–π interactions yielding a stable graphene/pyrene hybrid | [68] |
| Sulfonated aluminum phthalocyanine (aromatic planar component) | GO, rGO, single wall carbon nanotubes (SWNTs) | π–π interactions of aromatic planar component | [93] |
| Pynidinium tribromide (aromatic with hydrophilic groups) | highly oriented pyrolytic graphite (HOPG) | Hydrophilic or lipophilic chains electrostatic repulsions | [69] |
Imidazolium ionic liquids (IL) with phyneyle:
|
Graphene | IL-4 exhibited most effective graphene stabilizer via π–π stacking with its two phenyl groups was | [70] |
| 1-allyl-methylimidazolium chloride ionic liquid | rGO | Prevented aggregation between graphene nanosheets with cation–π stacking/π–π interaction and electrostatic repulsion | [71] |
Hydrophilic ILs:
|
rGO | Dispersion and stabilization are the driving forces caused by the cation-π and/or π–π interactions | [72] |
Imidazolium salt (functional amphiphilic ionic liquid):
|
GO | Interaction with imidazolium moieties and graphitic structure occur by π-cation stacking electrostatic interactions and post-reduction | [73] |
| Two vinyl-benzyl groups containing imidazolium ionic liquids (Imi-ILs). | GO | Exchange of ions between -vely charged GO and + ve imidazolium of Imi-ILs. Imi-ILs attachment via non-covalent π-stacking on graphene | [74] |
Polymer IL-
|
Exfoliation of graphite into graphene | Interaction of GO sheet edges with -COO of imidazolium cations in PIL via electrostatic attractions and also cation−π and/or π − π interactions | [75] |
| Quinoline | Non oxidized graphene flakes (NOGFs) from exfoliation of graphite | Binding of benzoic portion of quinoline to basal plane of NOGFs via strong via π–π interaction. | [76] |
| 9-anthracene carboxylic acid (ACA) | Surface modification of rGO for enhanced electrochemical properties | Stacking (π-π interaction) of the benzene ring in ACA anion on surface of rGO, while -COO− assisted their dispersion and water-solubility promoted by hydrogen bonding. | [77] |
Derivative of coronene (Anionic):
|
Stabilization and functionalization of graphene sheet | π-π stacking and non-covalent charge transfer between CS and graphene sheet | [78] |
Stabilizer C10:
|
Expanded graphite into few-layer graphene (FLG) dispersion | C10, an amphiphilic aromatic molecule with core triphenylene six-acid groups. Strong affinity interaction between π-electron rich aromatic core and graphene via π–π interaction. | [79] |
| Sodium dodecyl benzenesulfonate (SDBS) | Exfoliation of graphite to graphene | Anionic SDBS surfactant induce uniform dispersion in water preventing π-π stacking | [81] |
Ionic surfactants:
|
Exfoliation, stable dispersion, surface modification GO, rGO | SDBS modified rGO showed excellent dispersibility and electrical conductivity | [80] |
| Exfoliation and surface modification of GO | SO3− groups provided electrostatic repulsion, Sulfonated graphene forms a stable suspension followed by replacement of SO3− with a cationic nonylphenyl-PEG-quaternary ammonium salt (NPEQ) to form a stable suspension. | [82] | |
Ionic surfactants:
Zwitterionic:
|
rGO stabilization | Interaction of rGO with hydrophobic tail (alkyl vs. aromatic); or polar heads (sulfonic vs. carboxylic groups) of different surfactants. PSS and Tween 80 modified rGO exhibited significant increase in capacitance | [85] |
| Sodium cholate | Exfoliation of graphene | Graphene interaction with carboxylic polar head groups | [86] |
| Polyvinylpyrrolidone (non-ionic macromolecule) | Exfoliation of graphite | hydrophilic polymer confers exfoliation and colloidal stability graphene in water | [84] |
| Anionic polysaccharide: Carboxymethyl cellulose (CMC) | Porous rGO | Active functional groups of CMC such as carbonyl and hydroxyl groups which show a strong affinity toward metal ions as well as stable dispersion | [87] |
Another most frequently employed approach for surface modification of graphene is based on covalent chemistry to immobilize specific functional molecules, which is most essential feature in biological/chemical sensor applications. Covalent surface modification of graphene induces interaction with sp2 carbon-atoms, re-hybridization occurs from sp2 to sp3 carbon-atoms hybridization that facilitate graphene edges be easily functionalized covalently. Graphene's surface modification via covalent functionalization enhances graphene's properties, such as tuning electrical conductivity, controlled surface chemical functionality, opening its band gap, stability and dispersibility. A summary of various chemical linkers/molecules required for covalent functionalization of graphene/GO/derivatives including the chemical functionality characteristics is listed in Table 3 . A detailed mechanism of covalent binding of graphene/GO/derivatives is previously reported that are notable, [[94], [95], [96]] and a comprehensive review provides various routes for covalent functionalization of graphene on solid substrates [97]. Generally, surface functionalization approaches for graphene/GO/derivatives were classified in five groups that include; (a) functionalization of organic molecules, such as free radicals, (b) oxidation, (c) cycloadditions, (d) hydrogenations and (e) halogenations. The most suitable common chemical compounds for functionalization of graphene/rGO via covalent approach are summarized in Table 3.
Table 3.
The list of most suitable covalent coupling linkers for functionalization of graphene/rGo through covalent interactions reported in the literature.
| Surface functional molecule for covalent functionalization | Carbon nanomaterial functionalization feature | Interaction | Reference |
|---|---|---|---|
| Free radical addition | |||
| Aryl diazonium salt | GO reduction in presence of a surfactant (SDBS) with hydrazine followed by treatment with aryl diazonium salts | Aryl radical formation from aryl diazonium ion by elimination of nitrogen and the radical aryl moiety interact/bind to graphene sp2 surface which donates an electron | [108] |
| 4-nitrophenyl diazonium (NPD) tetrafluoroborate | Aryl groups grafting to epitaxial graphene via reduction of 4-nitrophenyl diazonium (NPD) tetrafluoroborate | Transfer of electron to the diazonium salt from the graphene layer and its substrate | [109] |
| 4-nitrobenzene diazonium tetrafluoroborate | Graphene nanoribbons (GNRs) and chemical functionalization with diazonium salts by oxidative unzipping of carbon nanotubes | Chemical functionalization of 4-nitrophenyl on GNR through in-situ electrolytic reduction to alter the electrical properties | [110] |
| 4-nitrobenzene diazonium tetrafluoroborate | A layer formed on epitaxial graphene with Sic substrate by spontaneous reduction of-nitrophenyl diazobium (4-NPD) tetrafluoroborate | Band gap was observed in covalent attachment sites on modified graphene | [111] |
| Electrografting of aryl diazonium salts | Electrochemical reduction of diazonium salt with a supportive electrolyte medium on carbon electrode | Electrode surface modified with phenyl radical through producing the electron transfer and facilitated by the separation of dinitrogen yielding phenyl radical on carbon substrate | [112] |
| (4-nitrophenyl) diazonium tetrafluoroborate (NDTB) | Electrochemical controlled functionalization of active aromatic radicals CVD grown graphene on SiO2 | Selectively setting of electrochemical potential bias allow the ratio control and tuning electrical properties of graphene | [113] |
| bis(4-trifluoromethylphenyl) iodonium tetrafluoroborate [(CF3Ph)2I+BF4−] | Anchoring of CF3C6H4-moieties by electrochemical reduction of[(CF3Ph)2I+BF4−] on epitaxial graphene-SiC substrate | covalent bonding of trifluoromethylphenylene (CF3Ph) on graphene functionalization-induced defect density through rehybridization of sp2 to sp3 state and useful for transparent electrode and sensing application | [114] |
| Electrochemical oxidation of α-naphthylacetate for generatingradical addition of α-naphthylmethyl groups (Naph-CH2-) | Grafting α-naphthylmethyl radicals on epitaxial graphene /SiC by electrochemical oxidation of α-naphthylacetate | Preparation of electrochemically erasable organic dielectric film and reversible bandgap engineering in graphene | [115] |
| In situ generation of aryl diazonium salts by treatment of 4-methoxyaniline with isopentylnitrite | Surface/chemical multi-walled carbon nanotubes (MWCNT) | Controlled molar ratio of reactants can tune the chemical modification of MWCNTs | [116] |
| 4-nitrophenyl diazonium tetrafluoroborate | Chemical modification of monolayered graphene supported on bare SiO2, self-assembled monolayer of octadecyltrichlorosilane (OTS) on SiO2, Al2O3 (sapphire), reactive imprint lithography on hexagonal boron nitride (hBN) | Covalent functionalization of graphene on SiO2 and Al2O3 with aryl diazonium salts was more reactive compared with hBN. | [117] |
| Aryl functionalization on epitaxial graphene on SiC | Aryl-functionalized graphene network reorganize the π-bonds of graphene forming C − C additional bonds at the sp3 basal plane centers thereby introducing a band gap | [118] | |
| Covalently bonded phenyl substituent on single and multi-layers graphene on Si wafers | Chemical reactivity of graphene edges is greater as compared with single sheet of bulk graphene due to differences in electron transfer rates | [119] | |
| Selective functionalization graphene with high local curvature on aryl radical-SiO2 NPs with silicon substrate | Enhanced chemical reactivity in high local curvature of graphene facilitated by NPs – because of mechanical deformation and increases the strain energy | [120] | |
| 4-nitrobenzendiazonium (4-NBD) 4-bromobenzenediazonium (4-BBD) 4-methoxybenzendiazonium (4-MBD) |
Fast functionalization reactions of diazonium salts with chemical reactive CVD graphene on polydimethylsiloxane (PDMS) stretching via applying external strain | Electronic structure alteration in graphene by strain induced distortion of lattice attributing to high reactivity rate to facilitate functionalization. | [121] |
| Benzoyl peroxide | Functionalization of benzoyl peroxide (a precursor of phenyl radical) on mechanically exfoliated single and double layer graphene on SiO2 under laser illumination | Reaction is initiated by an electron transfer from photo-excited graphene on field-effect-transistor (GFET) to benzoyl peroxide that physisorbs and photo-excited. The photo-excited species is then decomposed into a phenyl radical that readily reacts with graphene's sp2 carbon atoms enabling introduction of sp3 defect centers | [122] |
| In-situ diazonium addition reaction | Covalent polystyrene chains grafting on graphene nanoplatelets | Covalent grafting on graphene with hydroxylated aryl groups through addition reaction of diazonium that polymerizes styrene through atomic transfer radical polymerization (ATRP) process | [123] |
Photopolymerization:
|
Styrene based UV-induced polymerization on graphene; yielding layers of homogeneous polystyrene (PS) brush on epitaxial graphene-SiC substrate and CVD grown graphene on Cu | Self-organized PS brushes covalently bound to graphene with exceptional electrical transport properties and useful for ultra-capacitors and graphene-based biosensors | [124] |
Photografting and photopolymerization (SIPGP) of monomers:
|
Photopolymerization of CVD grown graphene transistor on PMMA that carry pH sensitivity with DMAEMA, whereas tBMA incorporates -COOH groups for immobilization of enzyme | Polymer brush functionalization scaffold produced on graphene transistor for biosensor development | [125] |
| Cycloaddition reactions | |||
|
Single layer and few-layers of graphene | Diels−Alder (DA) reaction with graphene | [99] |
| 1,3-dipolar cyclo-addition of azomethine ylide. | Graphene by graphite in pyridine followed by reaction of N-methyl-glycine and 3,4-dihydroxybenzaldehyde | OH-functionalized graphene with better dispersibility | [126] |
| Benzyne species generation when of carbon reacts with o-trimethylsilylphenyl triflate (TMST) and cesium fluoride (CsF) | Benzyne functionalization on epitaxial graphene and generation of graphite by fluoride-induced elimination with TMS and triflate precursors | Benzene moieties on graphene generates fluorescence signal and useful for optoelectronic applications | [127] |
| 2-(trimethylsilyl)aryl triate | Aryl-modified graphene sheets via aryne cycloaddition through a four-membered ring formation that attached to the graphene surface | Aryn ring attachment covalently to graphene surface and useful for tuning electric and magnetic properties of graphene | [128] |
Azide compounds, e.g.,
|
Different functional moieties (−OH, −COOH, −NH2, and − Br) and polymers (PEG, PSS) | GO is used as a graphene precursor and the reduction of GO to graphene with simultaneous. The electrically conductive graphene showedexcellent dispersibility | [129] |
| Azidotrimethylsilane | Chemical modification of graphene (epitaxial) with azidotrimethylsilane (ATS) | Covalent bond formation between thermally generated nitrene radicals and epitaxial graphene | [130] |
| Hydrogenation | |||
| Cold hydrogen plasma | Graphane ‑hydrogenated graphene after exposing with the cold hydrogen plasma | Altered electronic properties graphene via attachment of sp2 carbons hydrogen atoms that changes their hybridization state to sp3 | [100] |
| Lithium/ammonia solution | Hydrogen coverage on single-layer CVD-grown graphene on SiO2/S by Birch reduction reaction Liquid-based method | Hydrogenation reduced the electronic conductivity and transformed graphene into an insulator and the reduction reaction is reversible. | [101] |
| Microwave plasma enhanced chemical vapor deposition | Introducing hydrogen in few layers CVD graphene on SiO2/Si | Turning graphene as ferromagnetic semiconductor material by hydrogen functionalization | [131] |
| Epitaxial graphene by heating an Ir(111) crystal | Patterned adsorption of atomic hydrogen on graphene, which is grown on Ir(111) substrate | Bandgap opening of graphene through adsorption of H atom. | [132] |
| Dissociation of hydrogen silsesquioxane (HSQ) induced by electrons | Chemisorbed hydrogenation of single or bilayered graphene on Si/SiO2 and photothermal activation | Localized generation of reactive species can engineer charge transport properties and electronic structure of graphene | [133] |
| Halogenation | |||
| SF6 and XeF2 Plasma | Plasma SF6 treatment to epitaxial graphene on SiC and plasma XeF2 fluorination on CVD graphene/SiO2 | Intermediate formation between the semi-ionic and covalent bonding on fluorinated graphene (F—C bond character) | [102] |
| Chlorine plasma treatments | Plasma-based chlorination of graphene flakes exfoliated and CVD-grown large-area graphene | Chlorinated graphene field-effect transistors exhibits a hole doping effect and electrical conductivity increase | [134] |
| Gas-phase photochlorination | Covalent linking of Cl radicals to the basal C-atoms of graphene flakes on Si/SiO2 substrate | C-Cl bonds on graphene and chemical modification by photochemical chlorination and chlorinated graphene field-effect transistors demonstrated band gap opening | [135] |
| Photochlorination and alkylation reactions | Cholorinating epitaxial graphene on SiC using Cl reaction under UV light followed by methylation using CH3MgBr | Chemical bonding of Cl and CH3 on the graphene basal-plane | [103] |
| Oxidation | |||
| Plasma treatment O2/Ar | Functionalization of epitaxial graphene surface on SiC by incorporating oxygen as ether, alcohol, and carboxyl groups | Graphene's covalent modification by O2/Ar changing its electronic properties | [104] |
| Covalent attachment of organic molecules: | |||
| Octadecylamine (ODA) | Covalently functionalize GO nanosheets with long alkyl chains | Enhanced lipophilicity and better dispersion of GO on polypropylene through -NH2 bonds formed between carboxylic groups of GO and octadecylamine (ODA) | [136] |
| p-phenylene diamine (PPD) | Conductive graphene synthesis using PPD as reducing agent on ITO substrate | Graphene surfaces absorbed with OPPD via π–π stacking + protonation to form –N+ during the reaction | [137] |
| 1-(3-aminopropyl)-3-methylimidazolium bromide | Covalent chemical attachment of 1-(3-aminopropyl)-imidazolium bromide GO nanoplatelet | Reaction between amine groups of ionic liquid and epoxy groups of GO resulting in imidazolium-modified GO that exhibited excellent dispersibility | [138] |
| 1-(3-aminopropyl)imidazole | Coupling by covalent interaction between the acyl chloride of activated GO and 1-(3-aminopropyl)imidazole followed by treating with 1-bromobutane and positive ion exchange with either protoporphyrin IX disodium salt or NaPF6 | Positively charged imidazolium functionalized graphene modulated wettability and photoactive anionic porphyrin for optoelectronic application | [139] |
| 3-aminopropyl triethoxysilane (APTS) | GO surface functionalized with APTS through the epoxy groups | APTS –GO exhibited improved mechanical properties | [140] |
| phenyl isocyanate | Mixing solution-phase of polystyrene and exfoliated phenyl isocyanate-treated GO sheets followed by chemical reduction | GO covalently attached with phenyl isocyanate through an ester bond with the epoxy group that enhanced the dispensability in polystyrene polymer and electrical properties by reduction | [140] |
| Grafting with polymer | |||
| Butyl amine | Plasma induced fluorination of a few layer of graphene on ITO and chemically functionalized with butylamine | PEDOT:PSS-graphene based transparent conductive electrode | [141] |
|
Graphene sheet on ITO were treated with plasma and covalently attached fluorine. The fluorinated graphene sheets were then exposed to a polymerization initiator such as butylamine |
Amino-functionalized graphene can be used to integrate with thermosetting polymers |
[142] | |
| Gamma-aminopropyltriethoxysilane (APTES) | APTES-graphene oxide was grafted with polyethylene (MA-g-PE) using maleic anhydride | Graphene as reinforcement for non-polar polymers (PE/polystyrene) | [143] |
Addition of free radicals trigger the formation of covalent bonds with aromatic structure of graphene via highly reactive organic species [98]. In this approach, the functionalization of graphene surfaces occurs through reduction of diazonium salts that generate aryl radicals. Here, diazonium salts reduction process is triggered by one of the following treatments, such as thermal, electrochemical, or by photochemical treatments. Aryl functionalized graphene surface can modify the surface and tune electronic properties (e.g., opening band gap, tuning the conductivity, and deformation) and free surface functional moieties to bind other biomolecules for sensing application. Chemical reactivity and modification of graphene with free organic radical groups depends on number of graphene layers, substrate type, flexibility of substrate, doping/decoration of nanoparticles and polymeric brushes, reaction conditions such as electrochemical oxidation/reduction potential, and photo polymerization conditions. These parameters enables to reengineer electronic properties of graphene essentially via rehybridization of sp2 to sp3 states that can be used as transparent electrodes or supercapacitors in chemical/bio sensor devices.
Another method for surface modification of graphene is to attach functional group of cyclohexane derivatives via the Diels–Alder reaction. A substituted alkene and conjugated diene chemically react, which is also called the dienophile that forms a substituted cyclohexene derivative. Addition of dienophiles lead to transforming sp2 carbons of graphene to sp3 and enhances dispersibility with alter graphene's electronic properties [99]. Further, hydrogenation as well as halogenation processes on graphene have been applied to covalently modify the graphene's optical, electrical and magnetic properties. Hydrogenation of graphene is generally carried out through exposing plasma and by using liquid-based hydrogenation methods [100,101]. In halogenation treatment of graphene, most commonly Fl and Cl atoms are functionalized by methods such as decomposition of chemical precursors, gaseous plasma treatment and photochemical reduction methods [102,103]. Oxidation of graphene can be easily triggered using oxygen plasma treatment that alters the electronic properties of graphene [104]. Table 3 summarizes various chemical crosslinking routes and coupling linker agents in free radical addition, cycloaddition reactions, hydrogenation, halogenation, oxidation and grafting with polymer for covalent surface modification of graphene/rGO/GO.
Introducing surface modification can improve graphene's functionality, which can be exploited mainly in biosensing applications. Direct interaction of molecules or analytes with pure graphene without surface modification poorly transduce a significant response or signal. Therefore, electrochemical functionalization of graphene can offer improvement in the sensitivity of graphene's surface toward capturing of molecules or specific analytes. Electrochemical surface modification of graphene/rGO can be obtained by different approaches that include electrodeposition, electrophoretic deposition (EPD), cathodic reduction of GOs, electrochemical polymerization, electrospinning and electrochemical doping [105]. For example, electrochemical synthesis of graphene nanocomposite, such as decoration of ferromagnetic metal particles in array on graphene surface may yield phenomena of tuned quantum interference during the process of electrical transport via carbon network [106,107]. Electrochemical synthesis and surface modification routes could be cost effective to synthesize large amount of graphene nanomaterials, however their chemical modification mechanism such as oxidation/reduction are still under investigation.
5.3. Carbon nanotubes and their properties
Carbon nanotubes (CNTs) were first introduced as the helical microtubules of graphitic carbon by Sumio Iijima in 1991 [144]. They are needle-like hollow cylindrical nanotube structures made of 2 to 50 layers of graphitic carbon with sp2 arrangements with size range 1 to 100 nm in diameter. CNTs are one-dimensional structures because they have a high length/diameter ratio and structurally classified based on the numbers of graphene sheet planes, namely single walled CNTs (SWCNTs) and multi walled CNTs (MWCNTs) (Fig. 10a-b). SWCNTs made of a single rolled graphene sheet and the MWCNTs can be depicted as several concentric and coaxial cylindrical graphene sheet planes instead of one plane sheet [145]. The structure of unrolled honeycomb SWCNT lattice is shown with its chiral vector, translational vector, the unit vectors and and chiral angle θ (Fig. 10a). The chiral vector, which is also called circumferential vector, always perpendicular to the translational vector. The structure of SWCNT can be defined on the basis of tube axis orientation on hexagonal carbon lattice through its chiral vector defined as for chiral indices (m,n) as shown in Fig. 10a. The structure of SWCNT is shown with one carbon atom to another arrangement in cylinders on an unrolled graphene plane, in which SWCNTs structures are shown with zigzag (m = 0), armchair (m = n) and chiral (m ≠ n) configurations.
Fig. 10.
Schematic diagram showing structure of (a) SWCNTs and (b)MWCNs [145,146].
SWCNTs and MWCNTs both have greater tensile strength, Young's modulus, and a bending constant, which make them one of the strongest and most flexible structures existed. CNTs exhibits high mechanical strength, surface to volume ratio, optical, electrical, stable chemical and thermal properties. Especially the superior electrical properties of CNTs make them very useful for electrochemical sensing, since they can exhibit completely conductive or semi-conductive features because of the arrangements of the carbon atoms and curved sheets of the graphene within the CNTs [145]. CNTs are highly preferred materials in biosensors because they promote high sensitivity, exhibit low noise and a broad absorption spectrum, simple to operate that is most desirable in real-time monitoring. In addition, CNTs can provide a large electroactive area that can help sensing of many molecules at the same time. The unique properties of CNTs make them great conducting nanoscale electrode materials suitable for biosensor transducing mechanisms, for instance, when directly interfaced with redox enzymes in electrochemical enzyme biosensors [47,147].
5.4. Surface modification of CNTs by covalent and non-covalent forces
Like GO and graphene, surface modification of CNTs can be performed by non-covalent and covalent modifications. Covalent modifications of CNTs are carried out through chemical reactions that may include halogenation, cycloaddition, oxidation and electrochemical reactions. Detailed modification approaches with CNTs have been highlighted in many previous reports [[148], [149], [150]]. These surface modification reactions can alter the shape, length, structure of the CNTs, or introduce entirely new chemical groups on the CNTs, for instance, covalent attachment of carboxy, amine, and sulfhydryl functional groups. CNTs often carry free carboxyl groups at their edges that are formed because of defects and these defects may be found in the interior of nanotube structures. These carboxyl groups are the most common cross-linking sites for covalent coupling with amino groups of desired chemical−/biological molecules, which is mediated by a carbodiimide cross-linker. The most popular water-soluble cross-linker used for such reactions is a carbodiimide, N-ethyl-N-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC). In carbodiimide cross-linker EDC based coupling chemistry, carboxy group in carboxylated-CNT reacts with EDC to form a transient o-acylisourea intermediate ester, which is readily replaced by formation of a peptide bond via reaction between carboxy group of CNTs and primary amine of a desired chemical/biomolecule to be conjugated as illustrated in Fig. 11 [148]. Carbodiimide compound is popularly used in covalent coupling of carboxylated CNTs with biomolecules.
Fig. 11.
Covalent chemistry of carboxylated CNTs based on carbodiimide linker EDC and NHS covalent coupling of biomolecule [148].
The most suitable and common cross linking agents other than EDC and NHS used for surface functionalization of CNTs through covalent chemistry for biosensing in the literature are ethylenediamine (EDA), poly(allylamine) (PAH), polypyrrole-NHS hydroxysuccinimido(pyrrol-1-yl)undecanoate, and glutaraldehyde [148], [[150], [151], [152], [153], [154], [155]]. Surface modification by covalent coupling of carboxylated CNTs with amine functional groups were commonly used for biomolecules include enzymes, proteins or antibodies. CNTs functionalized with amine groups were also crosslinked using homo-bifunctional cross-linkers such as glutaraldehyde or active ester, or epoxy compounds depending upon the compatibility of functional groups. Designing of multimodal CNTs by covalent multi-functionalization thoroughly reviewed by Dinesh et al. [156]. By these covalent reactions, CNTs can be made more biocompatible and hydrophilic (aqueously soluble) that find application in biomedical fields. However, covalent modifications can distort the CNT structure and its properties. In order to avoid this, non-covalent modifications can be adapted such as by interacting the benzene ring structured CNTs with aromatic compounds, polymers and/or affinity molecules, such as surfactants through π-π or electrostatic interactions, or CH-π interactions [157].
In Table 4 , the most suitable common aromatic molecules, surfactants and polymers used for surface functionalization of CNTs through non-covalent chemistry in the literature were compiled and shown in scheme (Fig. 12, Fig. 13 ). Polymers are considered efficient dispersing agents due their long chain structure which allow Van der walls force interaction and wrapping on CNTs [158].Further, biological and chemical properties of CNTs can be manipulated including toxicity [159].
Table 4.
The list of most suitable aromatic/surfactants/amphiphilic compounds/polymers for functionalization of CNTs through noncovalent π-π and electrostatic interactions reported in the literature.
| Surface functional molecule for noncovalent functionalization | Carbon nanomaterial | Interaction | Reference |
|---|---|---|---|
| Porphyrin | SWCNTs | Porphyrin functionalized onto SWCNT by π-π interactions | [162] |
| Hydroxyferriproto- porphyrin (hematin) | SWCNTs | π-π interactions | [163] |
| PBSE | MW and SW CNTs | π-π interactions | [158,[164], [165], [166], [167], [168], [169]] |
| Adamantane-pyrene, biotin-pyrene and nitrilotrilotriacetic acid (NTA) pyrene | SWCNTs | π-π interactions | [170] |
| [bis(2-anthraquinonyl)-aminomethyl]pyrene and 1-(2-Anthraquinonylamino-methyl)pyrene | MWCNTs | π-π interactions | [169] |
| Ferrocene | SWCNTs | π-π interactions | [[171], [172], [173]] |
| Anthracene | SWCNTs | π-π interactions | [174] |
| Thionine | MWCNTs | π-π interactions | [175,176] |
| Naphthalen-1-methyl-phosphonic acid | SWCNTs | π-π interactions | [177] |
| Polymers | |||
| Polyethyleneimine | MWCNTs | electrostatic | [178] |
| Polydiallyl dimethyl ammonium chloride (PDDA) | SWCNTs | electrostatic | [179] |
| Polysaccharides - amylase (with only -OH groups), sodium alginate (with -OH, -COOH groups) and chitosan (with -OH, −-NH2 groups) | SWCNTs | electrostatic | [180] |
| Synthetic glycolipids- pyrene–polyethylene–lactose, glycolipid | SWCNTs | electrostatic | [168,181] |
| Synthetic glycopolymers, glycodendrimers | SWCNTs | electrostatic | [181,182] |
| Porphyrin-based glycoconjugates | SWCNTs | electrostatic | [183] |
| Polyfluorene (PFO) and their derivatives | [182] | ||
| Polypyrrole (PPy) and polyaniline (PANI) |
[182,184] | ||
| Polymers with pyrene block | SWCNTs | electrostatic | [182] |
Fig. 12.
Schematic representation of; (a)surface modification of CNTs by covalent and non-covalent functionalization, (b) Systematic stacking with aromatic compound 1-pyrenebutanoic acid succinimidyl ester (PBSE) interaction with SWCNT sidewall surface (π-stacking) [158],
Fig. 13.
Schematic representation of; (a) possible interactions between CNTs and different surfactants (CTAB and Brij76) upon sonication [160], (b and c) Interactions between CNTs and different polymers such as short and long linear and branched polymers [161].
In this review, we explored on a range of electrochemical sensor platforms that utilized graphene and CNTs nanomaterials with special emphasis to detecting respiratory viruses. The nanobiosensors and electrochemical platforms developed for the most prominent respiratory viruses that integrate graphene and CNTs nanostructures are summarized in Table 5, Table 6 .
Table 5.
Recent studies based on graphene interfaced electrochemical biosensors for detection and quantification of RSV particles.
| Target Analyte | Technique | Limit of Detection (LOD) | Derivative of Graphene | Additional Features | Citation |
|---|---|---|---|---|---|
| NP of SARS-CoV-2, CRP, IgG, IgM of S1 | Graphene electrochemical | Range of NP in serum = 0.1–0.8 μg/mL and in saliva 0.5–2.0 ng/mL of COVID-19 patients. Range of S1-IgG in serum = 20–40 μg/mL and in saliva 0.2–0.5 μg/mL of COVID-19 patients. Range of S1-IgM in serum =20–50 μg/mL and in saliva 0.6–5.0 μg/mL of COVID-19 patients Range of CRP = 10–20 μg/mL in serum and 0.1–0.5 μg/mL in saliva of COVID-19 patients. |
Graphene sensor electrode on flexible PI substrate | Multiplex detection with remote assessment of COVID-19 biomarkers | [186] |
| SARS-Cov-2 | Electrochemical biosensor | 200 copies/mL RNA ofSARS-CoV-2 | Reduced graphene oxide composite (Au@SCX8-TB-RGO) | It requires 10 μL of SARS-CoV-2 (2 copies) for an assay | [185] |
| SARS-CoV-2 | Field-Effect Transistor-Based Biosensor | SARS-CoV-2 -1.6 × 101 pfu/mL and 1 fg/mL of SARS-CoV-2 spike antigen | Graphene | Without any cross-reactivity with MERS-CoV antigen | [187] |
| Influenza A (H9N2) | Graphene-Au hybrid nanocomposite modified biosensor based on neuraminidase activity | 10−8 U/mL of N | Graphene-Au | More sensitive than ELISA assay | [189] |
| Influenza A (H1N1) and (H5N1) |
Dual immunosensor based on methylene blue-electro adsorbed graphene oxide | 9.4 pM for H1N1 8.3 pM for H5N1 |
Graphene Oxide | Designed sensor has <1 min response time, reproducibility | [191] |
| Influenza A (H1N1) |
Reduced graphene oxide-based immuno-electrochemical sensor integrated with a microfluidic platform | 0.5 PFU/mL | Reduced Graphene Oxide | Sensor selectivity was proved using MS2 virus. | [192] |
| Avian influenza H7 |
Silver nanoparticles coated graphene electrochemical sensor | 1.6 pg/mL | AuNP-G and AgNP-G |
[193] | |
| Influenza A H1N1 |
Low-cost synthesis of reduced graphene oxide using biopolymer | 26–33 pfu/mL for phosphate saline and diluted saliva | rGO | No chemical oxidation-reduction step with toxic materials are used | [194] |
| Influenza A H5N1 | rGO transistor integrated with flow-through microfluidic chip for the detection of influenza gene | 5 pM | rGO | Detection time is 1 h | [195]. |
Table 6.
Recent studies based on CNTs based electrochemical biosensors for detection and quantification of RSV particles.
| Target Analyte | Technique | LOD | Speed | Additional Features | Citation |
|---|---|---|---|---|---|
| Influenza virus H1N1 | Stencil-printed carbon electrodes modified with SWNTs and chitosan | 113 PFU mL−1 | 30 min | disposable paper based immunosensor | [197] |
| influenza virus H1N1 DNA | Au/iron-oxide magnetic nanoparticle-decorated CNTs (Au/MNP-CNT) hybrid nanomaterial as a sensing channels | 8.4 pM | Au-F3O4-CNTs on IDES and magnetic alignment | [198] | |
| Avian Influenza (H5N1) |
SWCNTs or nitrogen-doped N-MWCNTs that were placed between IDE electrodes making the individual nanotubes remain in contact with electrodes | 2 pM-2 nM | 15 min | Easy-to-fabricate disposable small, flexible suitable PoC diagnostics applications | [199] |
| Influenza A virus | The CNTs field effect transistor (CNTFET) based DNA sensor | 1 pM | < 1 min | High sensitivity, accuracy, precision and integration for lab-on-a-chip development | [196] |
| Swine Influenza Virus (H1N1) |
Electric immunoassay based on electrical properties of SWCNTs underlying antibody-virus complexes | 180 TCID50/mL | ~1 h | low-cost, label-free, ultra-sensitive | [90] |
| AIV H7 | CNTs/MoSx aerogen modified GCE | 1.6 pg/mL | [200] |
6. Graphene and CNTs interfaced electrochemical sensing of respiratory viruses
6.1. Graphene interfaced electrochemical nanobiosensors for virus detection
6.1.1. Graphene interfaced electrochemical detection of SARS-CoV-2
Viruses or any living microbial pathogens can be detected by electrochemical transducing platforms using their specific biorecognition elements. There are several alternative routes for electrochemical detection of disease-causing agents. For example, (a) detecting specific gene fragments from disease causing agents in lysed bio-fluid using DNA/RNA probes, (b) directly detecting whole virus or microbial pathogen in biological fluids, (c) by indirect detection of relevant biochemical markers, such as proteins or metabolites, or (d) detecting the host antibodies against a virus or pathogen. Detecting genetic markers using electrochemical sensing platforms is not sensitive for viral detections due to their undetectable viral titers, especially at the early onset of the viral infections. In order to achieve early detectable levels, additional steps are required to be integrated with electrochemical detection, such as that similar to the PCR/RT-PCR reaction for amplifying viral DNA/RNA copies. In rare cases, such as in one study, where the amplification of viral RNA was not required for the detection of viral specific sequence, rather enriched the electrochemical redox mediator to generate detectable signal [185]. However, electrochemical detection systems are more suitable and sensitive for detecting biochemical markers and host antibodies. Electrochemical sensing was by far only limited to glucometers to measuring blood glucose levels. Their application began to expand for the detection of various other analytes, including whole viruses or other pathogenic living microbes, which is not yet fully matured. In recent years, several attempts have been made toward applying strategies similar to electrochemical glucometers for detection of viruses or viral infections. The following sections describe on a few recently reported notable advancements made toward electrochemical sensors for detecting of viruses or causative agents for viral infections that involve carbon-based nanomaterials.
A low cost graphene integrated portable electrochemical biosensor is reported by Torrente-Rodríguez et al. for rapid diagnosis and monitoring biochemical markers in serum and saliva samples for COVID-19 [186]. The electrochemical sensor electrodes were made of graphene engraved on a flexible polyimide (PI) polymeric substrate for multiplexed detection of viral infection biomarkers (antigens and antibodies). This study demonstrated quantitative detecting specific biomarkers of COVID-19, such as SARS-CoV-2 spike protein (S1), nucleocapsid protein of SARS, CRP, a protein biomarker for inflammation within physiologically relevant ranges in both blood and saliva and specific immunoglobulins (Igs) such as S1-IgM and S1-IgG, and (Fig. 14 ). This platform utilizes captured antigens and antibodies on graphene electrodes with high sensitivity with a multiplexing capability for sensing multiple SARS-Co-V2 makers, while the resulting response data is communicated wirelessly to a portable mobile device. This type of a miniaturized electrochemical platforms shows a great promise for the future PoC electrochemical and personalized health care devices.
Fig. 14.
Schematic illustration of (a-b) graphene engraved electrochemical sensor platform for multiplex sensing of SARS-CoV-2 markers such as proteins from SARS-CoV-2, IgM and IgG antibodies and CRP as an inflammatory biomarker in blood and saliva. (c-f) Image of a flexible and disposable laser engraved graphene electrochemical sensor that can be interfaced with a PCB for processing signal reading and a mobile user interface to receive results via wireless transmission [186].
Sensing viral RNA using electrochemical sensing is further improved toward ultrasensitivity in detection of viral-RNAs. For instance, in a recent study, ultrasensitive detection of RNA from SARS-CoV-2 was achieved by applying a modified electrochemical method that did not required amplification and reverse-transcription steps, which is a significant step forward in reducing the time and cost for diagnosing viral-infections that has a potential for PoC based SARS-CoV-2 detection. The designed bioassay detect target RNA by surface functionalization of graphene oxide with calixarene [185]. The super sandwich-type bioassay in question utilizes a combination of (a) Au@Fe3O4 nanoparticles conjugated with capture probes (CP), and (b) graphene functionalized with p-sulfocalix [8] arene (SCX8-RGO) that enriches electrochemical mediator toluidine blue (TB) for ultrasensitive RNA detection from SARS-CoV-2 (Fig. 15a-b). Here, the target RNA extracted from the virus is first captured by CP/Au@Fe3O4 which upon mixing with Au@CX8-RGO-TB previously conjugated with a label probe (LP) and an auxiliary probe (AP) gives rise to a complex of CP-target-LP that facilitated ultrasensitive electrochemical detection of virus (Fig. 15 a-b). Around 88 RNA extracts from 25 patients positively tested for virus and eight-recovery patients were tested with detectable ratios 85.5% and 46.2% that proved to be greater as compared with RT-qPCR (56.5 and 7.7%). Each assay using above method exhibited detection limit of SARS-CoV-2 approx. 200 copies/mL in clinical samples.
Fig. 15.
Scheme showing development steps of electrochemical biosensor for SARS-CoV-2 detection. (a) capture probes (CPs) composed of thiol modified Au@Fe3O4 NPs to form CP with Au@Fe3O4 followed by hexane-1-thiol (HT) addition to obtain premix A; rGO functionalized with the host-guest complexes (SCX8-TB) for the formation of Au@SCX8-TB-rGO, which was incubated with LP (label probe) and AP (auxiliary probe) bioconjugate to form long concatamers premix B.(b) electrochemical detection was carried out formation of CP-target-LP sandwich by incubating target viral RNA extraction-premix A with premix B [185].
FET type sensors have the advantage of greater sensitivity and rapid measurements with low concentrations of target analyte molecules. Recently, FET based electrochemical biosensors have been developed to detect the virus in human nasopharyngeal swap specimens [187]. Here, the FET based sensor surface was first coated with graphene and then functionalized with a spike (S) antibody coupled to PBASE probe linker. The honeycomb-like pyrene structure in PBASE-linker systematically stacks on FET-graphene surface that prevents from any interference in the electron transfer between source and gate electrodes that contribute to the ultra-sensitivity of this device (Fig. 16 ). The detection of the virus is proportional to the change in the current upon interaction between antibody and the respective antigen on graphene surface. The LOD of this FET biosensor for the spike protein was highly sensitive with 1 fg/mL in buffer (PBS) and 100 pg/mL in transport medium. This level of sensitivity is by far lower than that measured using a standard ELISA test. Further, LOD established using the cultured SARS-CoV-2 virus was found to be 16 plaque forming units (pfu)/mL in the growth medium and 242 copies/mL in clinical sample and it demonstrated the ability to distinguish between SARS-CoV-2 and MERS antigens [187].
Fig. 16.
FET-based immunosensor for Covid-19 detection in specimens collected from human nasopharyngeal swab. The schematic diagram shows graphene FET-biosensor for SARS-CoV-2 detection. Graphene served as an electron carriers on to which antibodies for spike (S) protein of is conjugated via a linker 1-pyrenebutyric acid N-hydroxysuccinimide ester that systematically stacks with its honeycomb like structure without any damage to the graphene nanostructure [187].
6.1.2. Graphene interfaced electrochemical detection of influenza A
Detection and measuring the change in the antigenic portions of H and N proteins is crucial for detecting influenza A virus due to their high mutagenic nature causing serious antigenic drift. Conventional electrochemical approaches utilizing antibodies may not be sufficient to accurately detect or measure changes in these antigenic drifts. Therefore, a different approach can be adapted, where neuraminidase activity (NA) was measured to detect the influenza virus A (H9N2) unlike those less-specific hemaglunin activity (HA) or conventional immunoassay-type reactions. There are 18 HA(1–18) and 10 NA(1−11) viral subtypes investigated. The antigenic properties of influenza virus is classified based on the HA and NA surface glycoprotein activities and both recognizes sialic acid and only the HA type being more commonly used for viral detection [188]. Recently, influenza A virus (H9N2 subtype) was detected based on measuring the changes in the H and N proteins antigenic portions due to mutagenic nature. This allow to measure the antigenic drift by NA using a graphene modified gold electrode by EIS. Here, the graphene modified gold electrode was previously coated with fetuin A glycoprotein followed by a neuraminidase glycoprotein and peanut agglutinin (PNA) lectin in a sequential order on the sensor electrode surface (Fig. 17A) [189,190]. Neuraminidase (N) activity from the influenza A virus cleaves the sialic acid ends of fetuin-A glycoproteins, producing galactose molecules which then reacts with PNA lectin. These reactions were monitored via resistance change as a function of varying viral concentrations. Here, influenza A virus detected with a LOD 10−8 U/mL NA by graphene interfaced EIS.
Fig. 17.
Scheme shows (A) functionalization of gold electrode with graphene coated with fetuin A glycoprotein followed by neuraminidase glycoprotein that reacts with PNA. Electrochemical detection of influenza A virus is monitored for the neuraminidase protein cleavage on sensor by from the virus [189]. (B) scheme of electrochemical immunosensor development for detection of H1N1 and H5N1 subtypes of influenza A [191]. It shows three electrodes based electrochemical sensor chip and step wise surface modification GO for the development of immunosensor platform. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Electrochemical immunosensor has been tested for detecting subtypes of influenza A virus including H5N1 and H1N1 viruses. The electrochemical reaction is mediated by methylene blue (MB) which is electro adsorbed with graphene oxide (GO) on electrode surface [191]. The positively charged MB is used as a redox probe like in many common electrochemical sensor applications while the viral subtype specific antibodies immobilized on electrodes previously coated with chitosan and protein-A (Fig. 17B). The influenza A virus subtypes H5N1 and H1N1 detection principle was based on the interactions between immobilized antibody and viral antigens resulting in a change in peak current intensity corresponding to the specific viral antigen. The sensor reported detection limits for H1N1 antigen and H5N1 were 9.4 pM, 8.3 pM, respectively and demonstrating that different viral subtypes can detected simultaneously [191].
Other electrochemical immunosensors utilizing a microfluidic platform with reduced GO (rGO) for influenza virus H1N1 detection (Fig. 18 ) [192]. The microfluidics assisted continuous monitoring of signals from the injected samples, while the viral particles were captured by antibodies on rGO-modified gold electrodes. The detection of electrochemical signal was measured by chronoamperometry technique, where different solutions containing varying H1N1 concentrations were injected to the microchannel, and the observation was that the chronoamperometric current increased linearly with increasing virus loads. Detection of H1N1 viruses with the developed sensor tested with 1– 104 pfu/mL and the LOD was found to be 0.5 pfu/mL. The above method demonstrated a promising approach with portable multi-analyte sensing capabilities using small volume samples [192].
Fig. 18.
Scheme representing microfluidic platform integrated with three electrode based electrochemical sensor chip integrated with rGO coated working electrode and its EDC/NHS coupling for antibody functionalization using for influenza virus H1N1 detection [192].
Huang et al. developed sandwich based electrochemical immunoassay to detect avian influenza H7 type virus [193]. In this study, gold electrodes coated with gold nanoparticle-graphene composite material (AuNPs-G) was used as a working electrochemical sensor electrode. The AuNPs were immobilized on graphene surface using chitosan as a linker for the formation of AuNPs-G nanocomposite (Fig. 19 a-c). The immunosensor was developed by immobilizing H7-monoclonal antibodies (MAb) on the (Au-G) electrode surface. On the other side, silver nanoparticle-graphene composite material was prepared, and H7-polyclonal antibodies (PAb) were functionalized to AgNP/graphene nanocomposite surfaces. For sandwich assay, first different concentrations of avian H7 influenza virus containing solutions were treated with Mab-AuNPs-G‑gold and then incubated with PAb-AgNPs-G bioconjugates (Fig. 19 a-c). By using this signal amplification model, LOD of 1.6 pg/mL was achieved for viral detection [193].
Fig. 19.
Scheme illustration of a sandwich immunosensor fabrication for avian influenza type H7 virus detection.(a) Preparation of AuNP-graphene functionalized with chitosan, (b) Preparation of PAb-AgNP-graphene bioconjugates and (c) sandwiching signal amplification by capturing of avian influenza H7 by secondary probe PAb-AgNP-graphene [193].
Joshi et al. developed indium tin oxide (ITO) modified with rGO electrodes were employed in the electrochemical biosensor [194]. Here, rGO was obtained from Shellac, which is a cheap (2 USD/Kg) polymeric material, thermally treated to synthesize rGO (TrGO) films with altered structural, chemical, and electrical properties (Fig. 20a). After the heat treatment, TrGO showed low sheet resistance with rich carbon and enhanced crystalline structure. The TrGO was used as transducing sensor material on the ITO/glass that was later immobilized with H1N1 antibodies with PBSE linkages for detecting influenza viruses (Fig. 20a-b). The LOD as per this study was 26 pfu/m in buffer and 33 pfu/mL in diluted saliva. Sensor signal sensitivity is influenced by the high adhesion of phenolic-OH moieties in TrGO that lead to covalent bonding at the interface. The influenza viral loads normally reported to be in the range of 103–105 TCID50/mL, therefore this sensor potentially applicable for clinical applications.
Fig. 20.
Schematic showing; (a) thermal treatment of Shellac biopolymer to fabricate TrGO films; and (b) immobilization ofH1N1 antibodies on TrGO-ITO electrochemical electrode surface for development of influenza virus sensor [194].
Graphene has been previously used in biosensor development, where functionalization of bio-probes was carried out via π-π stacking with linker-aromatic rings, such as pyrene butyric acid. For example, Chan et al. demonstrated rGO transistor integrated microfluidic flow-through chip for detecting Influenza virus [195]. In case of DNA based graphene-transistor biosensor, direct probe immobilization can be possible by π-stacking between graphene surface and nucleic acid. The stacked nucleic acid on graphene then hybridize DNA that induces affinity disassociation from the graphene surface that is triggered by the negative charge repulsion, and therefore decreases the sensor response. Using this phenomenon, researchers have demonstrated detection of H5N1 virus gene sequence using microfluidics integrated rGO transistor biosensor (Fig. 21a-c) [195]. For this, a capture-probe extended is made of two parts, where first part was designed for hybridizing the complementary sequence to the DNA target, while the second part extends and allow interaction of dsDNA and graphene through π-π interactions. This approach allowed retention of the target dsDNA-probe complex on surface of rGO in μfluidic environment (Fig. 21a-c). The interaction between target and probe gives rise to the signal on the rGO transistor due to the change in conductance across source-drain electrodes and sensor measures the relative conductance change. The results of this study demonstrated sensitive viral detection in real-time environment with stability. Here, integrated μfluidic device with rGO based transistor and with functional extended long capture probe achieved notably low levels of LOD = 5 pM LOD for influenza virus ~1 h interval in real-time [195].
Fig. 21.
Schematic shows (a) immobilization approach for long extended and short capture probe on rGO functionalized between source and drain electrodes rGO transistor chip, (b) Integration of microfluidic on rGO transistor chip using PDMS and (c) Matching short sequenced capture probes following target hybridization drained away from the surface of rGO that gives rise to change in the conductance, while rGO surface retained functional probes [195].
In summary, all the above electrochemical biosensors reported are promising approaches with specificity at a given time. However, their performance most likely to be hampered due to the constant viral mutations and antigenic drifts over time, which makes it extremely difficult for their application in the long run. For instance, seasonal changes bring different variants of the same viral subtype and that the antibodies generated for a particular subtype may fail to recognize the small variations in viral subtypes. Most recent development on graphene interfaced electrochemical biosensors for detection and quantification of RSV particle are summarized in Table 6:.
6.2. CNT interfaced electrochemical nanobiosensors
6.2.1. Influenza A detection
Recently, many electrochemical biosensors were fabricated by integration CNTs nanostructure for the detection of virus infection or viral load. For instance, a CNTs-based field effect transistor (CNTFET) was developed to detect DNA of influenza A viruses [196]. The viral DNA biosensor utilizes modified ion sensitive FET (ISFET) with single stranded DNA probes immobilized on to the CNT networks. Because of probe-target hybridization ion concentration changes and as result the voltage/current at the interface of CNTFET changed which is corresponding to the target viral DNA that was measured (Fig. 22 ). The developed CNT transistor biosensor exhibited LOD of 1 pM. This study showed that the CNTFET transuding signal depends on the probe concentration, time and temperature of the hybridization reaction and capability to integrate DNA on chip based biosensor platform.
Fig. 22.
Scheme shows the fabrication of CNTFET based DNA biosensor for. Typical fabrication procedure for FET with source and drain electrode and CNT network between the electrodes as a transducing element. CNTs network was immobilized with probe single DNA strands and the probe hybridization with the DNA of target virus induces change in the ion concentration at the interface of sensor that lead in turn the voltage of the CNTFET [196].
6.2.2. Influenza virus H1N1 detection
Devarakonda et al. reported fabrication of an immunosensor based on handmade paper [197] to electrochemically detect H1N1 influenza virus. The fabrication of immunosensor involved spraying of hydrophobic silica nanoparticles by a glass vaporizer to modify the surface of the paper to generate hydrophobicity (Fig. 23I). Working and counter electrodes were fabricated using conductive carbon-paste material, while the reference electrode was made of Ag/AgCl ink. Electrodes were deposited onto the hydrophilic part of the paper using a low-cost custom made self-adhesive polyester stencil. Carbon paste and chitosan was extruded through A 100 μm thin polyester stencil to print the electrodes that were later modified with single-walled CNTs that increased the sensitivity of the sensor because of their excellent electronic, mechanical properties of CNTs while chitosan provided excellent biocompatibility, adhesion, and high water permeability. Monoclonal antibodies specific to influenza viruses H1N1antibodies were immobilized by cross-linking on CNT –chitosan modified working electrode and the sensor repose was measured with differential pulse voltammetry against the virus. The fabricated paper based electrochemical sensor tested at different viral concentrations ranging from 1– 104 pfu/mL with LOD 113 pfu/mL.
Fig. 23.
Schematic diagram of; (a) fabricating a hydrophobic paper with the help of a glass vaporizer and a polyester stencil, and (b) Functionalization of the CNT–chitosan modified working electrode for H1N1 virus detection and three electrode paper-based electrochemical immunosensor embedded with a PDMS well carrying electrolyte [197]. (II) Schematic shows the developed CNTs based immunochips: (a) immunoassay showing thin film formed by self-assembled CNTs random network and capturing of virus on modified electrode surface, (b) an optical immunoassay CNT chip, (c) SEM-images showed metal electrode (Cr/Au) pattern, (d) A thin file of multilayered SWCNTs between the two electrodes and random network of SWNCTs in SEM, and (e) a microtiter plate in which immunochips were sorted for use in immunoassay [90].
Lee et al. fabricated self-assembled CNTs based electric immunoassay to detect swine influenza virus (SIV) H1N1 [90]. In this study, a thin film formed by self-assembled CNTs random network was used as a transducing material, which was immobilized with SIV-specific antibody for electrically detecting SIV H1N1 virus (Fig. 23 IIa). A thin film was constructed using polyelectrolytes poly(diallyldimethyammonium chloride) (PDDA) and poly(stylenesulfonate) (PSS) and assembled layer-by-layer to form polycation and polyanion, respectively. The transducing thin film was fabricated on substrate by first preparing a precursor layer made by self-assembly of bi-layers of (PDDA/PSS) that enhances charges followed by a second assembly of (PDDA/SWCNT) (Fig. 23 IIa). The immobilization of SIV antibodies were facilitated through modification of SWCNTs surface with poly-l-lysine. The immuno reaction was carried out in a microchannel integrated with SCNT biosensor structure, where formation of antibody–virus complex lead to conductance change in SWCNTs electrode (Fig. 23 IIa-d). CNTs networks-based biosensor enabled detection of SIV at a considerably low level (180 TCID 50/mL).
Lee et al. fabricated electrical biosensor using magnetic Au/Fe3O4 NPs-decorated CNTs (Au/MNP-CNT) hybrid nanomaterial arranged as a sensing channels and detected influenza H1N1 and norovirus DNA (Fig. 24 a-b) [198]. For this first, an external magnetic field is used to align nanohybrid material a platinum based interdigitated electrodes (IDEs) followed by immobilization of DNA probe through interactions of Au metal of hybrid and thiol groups of DNA probe (Fig. 24b). The hybridization of target and DNA probe on the surface of nanohybrid led to change in the electrical conductivity of the sensor, and sensor achieved LOD of 8.4 pM and 8.8 pM for influenza virus and norovirus, respectively.
Fig. 24.
(a) Schematic diagram showing synthesis of Au/MNP-CNTs hybrid nanomaterial and their alignment on the Pt-interdigitated electrodes as a sensing channel for DNA sensing. (b) SEM images showing Au/MNP-CNTs on the Pt-interdigitated electrodes; (í-ii) without and after magnetic alignment, and (iii) process showing DNA detection with hybrid materials as a sensing element [198].
6.2.3. Influenza virus H5N1 detection
Fu et al. fabricated semiconducting (sc) SWCNTs and nitrogen-doped MWCNTs (N-MWCNTs) integrated chemiresistor-type electrical sensors for viral DNA sequences of AIV subtype H5N1 detection [199]. The integrated nanotubes on the sensor electrodes were 5 μm in length because they influenced field effect electronic structure with charged DNA molecules on sensor (Fig. 25a-b). The N-MWCNTs (vertical with height 5 to 10 μm) were ink printed to interface on interdigitated electrode pattern on glass/quartz and flexible polyimide substrate (Fig. 25a). However, in case of sc-SWCNTs, the sensors were fabricated using CVD grown SWCNTs with short fragments of sc-SWCNTs (5–20 μm). Here, sensing mechanism of virus DNA biosensor device was based on change in sensor device resistance after DNA target molecules hybridized with their complementary DNA-probes. The change in resistance is postulated due to detachment of non-covalently functionalized double-stranded DNA helical structure probes on the sidewalls of CNTs after hybridizing with the complementary DNA. The complementary viral DNA target sequences of AIV H5N1 can be detected with dynamic range 2 pM to 2 nM using synthesized functionalized-sc-SWCNTs and N-MWCNT integrated nanotube sensors within 15 min.
Fig. 25.
(a) (i) SEM image of closely-packed CVD grown N-SWCNTs on silicon substrate, (ii) Vertically aligned CNTs by contact printing method, (iii) SEM image showing chemiresistor sensor device containing N-MWCNT on Kapton® substrate; the inset image shows interfacing of horizontal N-MWCNTs on interdigitated electrode, (iv) SEM image of sc-SWCNTs interfaced on patterned interdigitated electrode on quartz substrate, (v) Optical imaging of CNTs based fabricated sensor arrays with 400 sensors on quartz, glass and polyimide (left to right), (b) (i) Functionalization of CNT based chemiresistive biosensor (ii) Probe station during electrical characterization of CNT interfaced biosensor [199].
Further, in an another study, electrochemical immunosensor was developed based on AuNPs-CNTs/MoSx aerogel on GCE to detect AIV H7 virus [200]. Here, molybdenum sulfide was utilized as an efficient catalytic support for conductive MWCNTs to prepare aerogel as sensing material on working electrode. The modified electrochemical sensor electrode surface of CNTs/MoSx with the adsorption of H7-polyclonal antibodies specific to H7 and exhibited LOD for AIV H7 of 1.6 pg/mL.
7. Conclusions and future perspectives
In this review, notable examples of graphene and CNT interfaced electrochemical biosensors specification, design, and fabrication were discussed and summarized the detection of a variety of respiratory viruses. Both graphene and CNT interfaced electrochemical biosensing platforms are relatively rapid, apart from being sensitive, low-cost, reliable and feasible for easy-to-use PoC diagnostic health technology for detection of viruses. It is noticed that the main setback has been associated with inadequate surface bio-receptor functionality on graphene and CNTs electrodes that limits their performance in electrochemical biosensing applications. However, covalent and non-covalent modification strategies can provide sufficient active surface functionality for the fabrication of micro-nano electrodes and chips for electrochemical biosensing. Strategies that stand out for surface functionalization among graphene derivatives without structural and functional alteration is π-π stacking followed by functionalization through aromatic linkers, surfactants and covalent attachment of organic molecules. Functionalization of graphene/CNTs induce their dispersibility in aqueous medium with a well-defined stable conductive chemical nature to further re-engineer design for the development of tailored graphene/CNT-based biosensor platforms. The most suitable and popular approach for surface modification of CNTs however is through carbodiimide cross-linking chemistry for covalent surface attachment. Non-covalent interactions on CNTs such as by aromatic molecules, surfactants and polymers have been used for surface functionalization of CNTs but their stability remains a question.
Interfacing carbon-based graphene and CNTs nanostructures with electrochemical sensors provides high biocompatibility, electrical conductivity, carrier mobility, large surface area, and better sensitivity. Graphene/CNT also allow flexibility and miniaturization of electrochemical transducers into wearable lab-on-a-chip (LoC) biosensors with multimodal capability for PoC based viral detection and continuous monitoring of vital health signs. In addition, graphene and CNTs interfaced LoC has potential for the development of hand-held device for quantitative analysis in PoC settings. This combines unique components such as; (a) integration of graphene transducers in multiple arrays with microfluidic channels to handle small volume of body fluids, (b) portability of such sensors are most desired in remote and resource-limited settings especially at homes during the times of pandemic, (c) integrating artificial intelligence (AI) components to PoC specially to handle the viral samples, and (d) integrating with a hand-held signal read-out circuit and data processing capability on smartphone. All the above features are essential for early-stage viral load detections and eliminate the time delay in diagnosis, and avoid the time required for sending samples to specialized laboratories.
Currently, electrochemical sensors are only limited to detecting blood glucose levels, such as in most popular and less-expensive commercial glucometers. With the advent of superior electronic/electric properties of carbon-based nanostructures especially, graphene and CNTs open new avenues to improving existing conventional electrochemical tools, such as glucometers. Extending glucometer-like functionality to measuring other biomarkers could not only bring down the healthcare cost, but also enable early monitoring of diseases, especially at times of COVID-19 pandemic. Electrochemical transducers interfaced with graphene/CNT serve as powerful tools for detection of a variety of disease biomarkers, including viral infections by means of nucleic acids (RNA/DNA) probes. Sensitive bioassays can then be coupled with electrochemical transducers for improved sensitivity and specific performance of electrochemical detection regimes. These electrochemical transducer platforms include; amperometric, potentiometric and field-effect transistors formats whose electrode surfaces are integrated with physico-chemically modified graphene and CNTs to serve as an electronic nose or electronic tongue components for diagnostics of respiratory viruses from samples (saliva, nasal swab, breath and aerosol). These electrochemical transducers were being made bio-specific by chemically tethering them with synthetic or natural bio-receptors. Sensing multiple viral components is timely and an ideal choice under current pandemic circumstances, such as by combining distinct bio-receptors. These include antibodies raised against the attenuated virus particle or with viral-coat proteins (e.g., spike proteins or nucleoproteins of MERS-CoV, SARS-CoV-2) apart from using their nucleic acids as targets. All the above receptors are now available commercially and that researchers are now engaged in improving the existing electrochemical transducing platforms for sensitively detecting viral RNA/DNA, coat proteins and/or nucleoproteins. Undetectable viral nucleic acids (RNA) form saliva required to be amplified into detectable copy numbers using a highly expensive and time consuming qRT-PCR test. For successful electrochemical detection of a viral loads or viral, it required improvement in its sensitivity because the viral load at the early onset of infection is too low to be electrochemically detected. Developing ultrasensitive hand-held electrochemical sensor PoC device for viral diagnosis like the glucometers could revolutionize the biosensors and diagnostics fields. The research community engaged in this direction, which is still limited, but growing with required convergence of far end disciplines. This review highlights the notable electrochemical sensor platforms that addressed detection of viral components through integrating with graphene and CNTs nanostructures and to improve sensitivity and performance.
Future directions of utilizing graphene derivatives and CNTs in biosensing are toward development of flexible graphene/CNTs ink printed electronics for faster, broader, and robust electrochemical biosensing devices. Other notable directions include doping metals with nanostructures or nanohybrids, and surface cross-linking chemistry with added functionality to electrochemical biosensing toward improved limit of detection, sensitivity, selectivity, reusability, cost-effectiveness and recyclability. Current challenges in electrochemical sensing is also associated with scaling up of device fabrication due to inadequate batch-to-batch reproducibility. However, it is not too far that these nano-enabled electrochemical sensors potentially be applicable for multiplexed detection of two or more bio-analytes for multiple disease diagnosis in a wearable sensor format. The aftermath of the recent and ongoing COVID-19 pandemic has given opportunity to the research community with necessary research funding for developing such tools. Nevertheless, it necessitates that researchers from different disciplines apart from biology and clinical sciences, mainly electronics and physics join in bridging the gap, bring these PoC tools to public health monitoring, and prevent future pandemics.
Data availability statements
The data that support the findings of this study are openly available in the reference section of manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to the PURE program of the Faculty of Engineering and Natural Sciences at Sabanci University.
References
- 1.Louten J. 2016. Essential Human Virology; pp. 71–92. [Google Scholar]
- 2.Weston S., Frieman M.B. In: Encyclopedia of Microbiology. Fourth edition. Schmidt T.M., editor. Academic Press; Oxford: 2019. Respiratory viruses; pp. 85–101. [Google Scholar]
- 3.Wang W.E., Tang J.M., Wei F.Q. J. Med. Virol. 2020;92:441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dizdar E.A., Aydemir C., Erdeve O., Sari F.N., Oguz S., Uras N., Dilmen U. J. Hosp. Infect. 2010;75:292–294. doi: 10.1016/j.jhin.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi T., McAllister D.A., O’Brien K.L., Simoes E.A., Madhi S.A., Gessner B.D., Polack F.P., Balsells E., Acacio S., Aguayo C. Lancet. 2017;390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obando-Pacheco P., Justicia-Grande A.J., Rivero-Calle I., Rodríguez-Tenreiro C., Sly P., Ramilo O., Mejías A., Baraldi E., Papadopoulos N.G., Nair H., Nunes M.C., Kragten-Tabatabaie L., Heikkinen T., Greenough A., Stein R.T., Manzoni P., Bont L., Martinón-Torres F. J. Infect. Dis. 2018;217:1356–1364. doi: 10.1093/infdis/jiy056. [DOI] [PubMed] [Google Scholar]
- 7.Stein R.T., Bont L.J., Zar H., Polack F.P., Park C., Claxton A., Borok G., Butylkova Y., Wegzyn C. Pediatr. Pulmonol. 2017;52:556–569. doi: 10.1002/ppul.23570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilbourne E.D. Emerg. Infect. Dis. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO Global Influenza Surveillance and Response System (GISRS) 2020. https://www.who.int/influenza/surveillance_monitoring/updates/latest_update_GIP_surveillance/en/ [DOI] [PMC free article] [PubMed]
- 10.Hajjar S.A., Memish Z.A., McIntosh K. Ann. Saudi Med. 2013;33:427–436. doi: 10.5144/0256-4947.2013.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoek L. Van Der. Antivir. Ther. 2007;12 [Google Scholar]
- 12.WHO 2020. https://covid19.who.int/
- 13.Jin J.-M., Bai P., He W., Wu F., Liu X.-F., Han D.-M., Liu S., Yang J.-K. Front. Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn J.S., McIntosh K. Pediatr. Infect. Dis. J. 2005;24:S223–S227. doi: 10.1097/01.inf.0000188166.17324.60. [DOI] [PubMed] [Google Scholar]
- 15.WHO . 2007. WHO Interim Guidelines. (WHO/CDS/EPR/2007.2006) [Google Scholar]
- 16.Schaffer K., Rosa A.M. La, Whimbey E. Infect. Dis. 2010:1598–1608. [Google Scholar]
- 17.WHO 2009. https://www.who.int/csr/disease/swineflu/en/
- 18.Vajda J., Weber D., Brekel D., Hundt B., Müller E. J. Chromatogr. A. 2016;1465:117–125. doi: 10.1016/j.chroma.2016.08.056. [DOI] [PubMed] [Google Scholar]
- 19.Longtin J., Marchand-Austin A., Winter A.L., Patel S., Eshaghi A., Jamieson F., Low D.E., Gubbay J.B. Emerg. Infect. Dis. 2010;16:1463–1465. doi: 10.3201/eid1609.100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Civljak R., Kosutic-Gulija T., Slovic A., Huljev E., Turcic N., Mestrovic T., Vranes J., Ljubin-Sternak S. Intervirology. 2019;62:174–181. doi: 10.1159/000503630. [DOI] [PubMed] [Google Scholar]
- 21.Boivin G., Serres G. De, Hamelin M.E., Côté S., Argouin M., Tremblay G., Maranda-Aubut R., Sauvageau C., Ouakki M., Boulianne N., Couture C. Clin. Infect. Dis. 2007;44:1152–1158. doi: 10.1086/513204. [DOI] [PubMed] [Google Scholar]
- 22.Tu C.C., Chen L.K., Lee Y.S., Ko C.F., Chen C.M., Yang H.H., Lee J.J. Scand. J. Infect. Dis. 2009;41:363–367. doi: 10.1080/00365540902849375. [DOI] [PubMed] [Google Scholar]
- 23.Obuchi M., Yagi S., Oguri A., Takizawa T., Kimura H., Sata T. Jpn. J. Infect. Dis. 2015;68:259–261. doi: 10.7883/yoken.JJID.2015.046. [DOI] [PubMed] [Google Scholar]
- 24.Burrell C.J., Howard C.R., Murphy F.A. 2017. Fenner and White's Medical Virology; pp. 135–154. [Google Scholar]
- 25.Qureshi A., Niazi J.H. Analyst. 2021;145:7825–7848. doi: 10.1039/d0an00896f. [DOI] [PubMed] [Google Scholar]
- 26.Peaper D.R., Landry M.L. In: Handbook of Clinical Neurology. Tselis A.C., Booss J., editors. Elsevier; 2014. Chapter 5 - laboratory diagnosis of viral infection; pp. 123–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., Veer B. van der, Brink S. van den, Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Eur. Surveill. 2020;25 [Google Scholar]
- 28.Meijer A., Bosman A., Kamp E.E.H.M. van de, Wilbrink B., Beest Holle M.D.R. van, Koopmans M. J. Virol. Methods. 2006;132:113–120. doi: 10.1016/j.jviromet.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Thévenot D.R., Toth K., Durst R.A., Wilson G.S. Biosens. Bioelectron. 2001;16:121–131. doi: 10.1016/s0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 30.Grieshaber D., MacKenzie R., Voros J., Reimhult E. Sensors-Basel. 2008;8:1400–1458. doi: 10.3390/s80314000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaubey A., Malhotra B.D. Biosens. Bioelectron. 2002;17:441–456. doi: 10.1016/s0956-5663(01)00313-x. [DOI] [PubMed] [Google Scholar]
- 32.Clark L. Trans. Am. Soc. Artif. Intern. Organs. 1956;2:41–48. [PubMed] [Google Scholar]
- 33.Wang J. Electroanalysis. 2001;13:983–988. [Google Scholar]
- 34.Clark L.C., Jr., Clark E.W. Int. Anesthesiol. Clin. 1987;25:1–29. doi: 10.1097/00004311-198702530-00004. [DOI] [PubMed] [Google Scholar]
- 35.Barsoukov E., Macdonald J.R. John Wiley & Sons; 2018. Impedance Spectroscopy: Theory, Experiment, and Applications. [Google Scholar]
- 36.Faria R., Heneine L.D., Matencio T., Messaddeq Y. Int. J. Bios. Bioelectron. 2019;5:29–31. [Google Scholar]
- 37.Hammond Jules L., Formisano N., Estrela P., Carrara S., Tkac J. Essays Biochem. 2016;60:69–80. doi: 10.1042/EBC20150008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christine B., Bjarni B., Gillis J. Electroanalysis. 2001;13:173–180. [Google Scholar]
- 39.Qureshi A., Niazi J.H., Kallempudi S., Gurbuz Y. Biosens. Bioelectron. 2010;25:2318–2323. doi: 10.1016/j.bios.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 40.Ceylan O., Mishra G.K., Yazici M., Qureshi A., Niazi J.H., Gurbuz Y. Vol. 12. 2018. IEEE Transactions on Biomedical Circuits and Systems; pp. 1440–1449. [DOI] [PubMed] [Google Scholar]
- 41.Jadon N., Jain R., Sharma S., Singh K. Talanta. 2016;161:894–916. doi: 10.1016/j.talanta.2016.08.084. [DOI] [PubMed] [Google Scholar]
- 42.Schöning M.J., Poghossian A. Analyst. 2002;127:1137–1151. doi: 10.1039/b204444g. [DOI] [PubMed] [Google Scholar]
- 43.Sadighbayan D., Hasanzadeh M., Ghafar-Zadeh E. Trends Analyt Chem. 2020;133:116067. doi: 10.1016/j.trac.2020.116067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beitollahi H., Khalilzadeh M.A., Tajik S., Safaei M., Zhang K., Jang H.W., Shokouhimehr M. ACS Omega. 2020;5:2049–2059. doi: 10.1021/acsomega.9b03788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bratov A., Abramova N., Ipatov A. Anal. Chim. Acta. 2010;678:149–159. doi: 10.1016/j.aca.2010.08.035. [DOI] [PubMed] [Google Scholar]
- 46.Wongkaew N., Simsek M., Griesche C., Baeumner A.J. Chem. Rev. 2019;119:120–194. doi: 10.1021/acs.chemrev.8b00172. [DOI] [PubMed] [Google Scholar]
- 47.Ray S.C., Jana N.R. Elsevier; 2017. Carbon Nanomaterials for Biological and Medical Applications. [Google Scholar]
- 48.Serrano-Aroca A., Takayama K., Tunon-Molina A., Seyran M., Hassan S.S., Choudhury P.P., Uversky V.N., Lundstrom K., Adadi P., Palu G., Aljabali A.A.A., Chauhan G., Kandimalla R., Tambuwala M.M., Lal A., El-Aziz T.M. Abd, Sherchan S., Barh D., Redwan E.M., Bazan N.G., Mishra Y.K., Uhal B.D., Brufsky A. ACS Nano. 2021;15:8069–8086. doi: 10.1021/acsnano.1c00629. [DOI] [PubMed] [Google Scholar]
- 49.CHAPTER 1 The World of Graphene,Nanoengineering: The Skills and Tools Making Technology Invisible. The Royal Society of Chemistry; 2020. pp. 1–60. [Google Scholar]
- 50.Radamson H.H. In: Springer Handbook of Electronic and Photonic Materials. Kasap S., Capper P., editors. Springer International Publishing; Cham: 2017. Graphene; pp. 1173–1183. [Google Scholar]
- 51.Kim S., Ihm J., Choi H.J., Son Y.-W. Phys. Rev. Lett. 2008;100 doi: 10.1103/PhysRevLett.100.176802. [DOI] [PubMed] [Google Scholar]
- 52.Wei W., Qu X. Small. 2012;8:2138–2151. doi: 10.1002/smll.201200104. [DOI] [PubMed] [Google Scholar]
- 53.Yazyev O.V., Helm L. Phys. Rev. B. 2007;75 [Google Scholar]
- 54.Matte H.S.S.R., Subrahmanyam K.S., Rao C.N.R. J. Phys. Chem. C. 2009;113:9982–9985. [Google Scholar]
- 55.Novoselov K.S., Geim A.K., Morozov S.V., Jiang D., Zhang Y., Dubonos S.V., Grigorieva I.V., Firsov A.A. Science. 2004;306:666–669. doi: 10.1126/science.1102896. [DOI] [PubMed] [Google Scholar]
- 56.Li X., Cai W., An J., Kim S., Nah J., Yang D., Piner R., Velamakanni A., Jung I., Tutuc E. Science. 2009;324:1312–1314. doi: 10.1126/science.1171245. [DOI] [PubMed] [Google Scholar]
- 57.Dimiev A.M., Tour J.M. ACS Nano. 2014;8:3060–3068. doi: 10.1021/nn500606a. [DOI] [PubMed] [Google Scholar]
- 58.Banhart F., Kotakoski J., Krasheninnikov A.V. ACS Nano. 2011;5:26–41. doi: 10.1021/nn102598m. [DOI] [PubMed] [Google Scholar]
- 59.Georgakilas V., Tiwari J.N., Kemp K.C., Perman J.A., Bourlinos A.B., Kim K.S., Zboril R. Chem. Rev. 2016;116:5464–5519. doi: 10.1021/acs.chemrev.5b00620. [DOI] [PubMed] [Google Scholar]
- 60.Xu Y., Zhao L., Bai H., Hong W., Li C., Shi G. J. Am. Chem. Soc. 2009;131:13490–13497. doi: 10.1021/ja905032g. [DOI] [PubMed] [Google Scholar]
- 61.A.A. Castellanos-Gomez, B.J. van Wees, (2013).
- 62.Geng J., Jung H.-T. J. Phys. Chem. C. 2010;114:8227–8234. [Google Scholar]
- 63.Lee D.-W., Kim T., Lee M. Chem. Commun. 2011;47:8259–8261. doi: 10.1039/c1cc12868j. [DOI] [PubMed] [Google Scholar]
- 64.Schlierf A., Yang H., Gebremedhn E., Treossi E., Ortolani L., Chen L., Minoia A., Morandi V., Samorì P., Casiraghi C. Nanoscale. 2013;5:4205–4216. doi: 10.1039/c3nr00258f. [DOI] [PubMed] [Google Scholar]
- 65.Su Q., Pang S., Alijani V., Li C., Feng X., Müllen K. Adv. Mater. 2009;21:3191–3195. [Google Scholar]
- 66.Jang J.-H., Rangappa D., Kwon Y.-U., Honma I. J. Mater. Chem. 2011;21:3462–3466. [Google Scholar]
- 67.An X., Simmons T., Shah R., Wolfe C., Lewis K.M., Washington M., Nayak S.K., Talapatra S., Kar S. Nano Lett. 2010;10:4295–4301. doi: 10.1021/nl903557p. [DOI] [PubMed] [Google Scholar]
- 68.Zhang M., Parajuli R.R., Mastrogiovanni D., Dai B., Lo P., Cheung W., Brukh R., Chiu P.L., Zhou T., Liu Z., Garfunkel E., He H. Small. 2010;6:1100–1107. doi: 10.1002/smll.200901978. [DOI] [PubMed] [Google Scholar]
- 69.Chen I.-W.P., Huang C.-Y., Jhou S.-H. Saint, Zhang Y.-W. Sci. Rep. 2014;4:1–6. doi: 10.1038/srep03928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bari R., Tamas G., Irin F., Aquino A.J., Green M.J., Quitevis E.L. Colloids Surf. A Physicochem. Eng. Asp. 2014;463:63–69. [Google Scholar]
- 71.Ma W.-S., Wu L., Yang F., Wang S.-F. J. Mater. Sci. 2014;49:562–571. [Google Scholar]
- 72.Zhang B., Ning W., Zhang J., Qiao X., Zhang J., He J., Liu C.-Y. J. Mater. Chem. 2010;20:5401–5403. [Google Scholar]
- 73.Lonkar S.P., Bobenrieth A., Winter J. De, Gerbaux P., Raquez J.M., Dubois P. J. Mater. Chem. 2012;22:18124–18126. [Google Scholar]
- 74.Yang Y.-K., He C.-E., Peng R.-G., Baji A., Du X.-S., Huang Y.-L., Xie X.-L., Mai Y.-W. J. Mater. Chem. 2012;22:5666–5675. [Google Scholar]
- 75.Kim T., Lee H., Kim J., Suh K.S. ACS Nano. 2010;4:1612–1618. doi: 10.1021/nn901525e. [DOI] [PubMed] [Google Scholar]
- 76.Kim J., Song S.H., Im H.-G., Yoon G., Lee D., Choi C., Kim J., Bae B.-S., Kang K., Jeon S. Small. 2015;11:3124–3129. doi: 10.1002/smll.201403647. [DOI] [PubMed] [Google Scholar]
- 77.Khanra P., Uddin M.E., Kim N.H., Kuila T., Lee S.H., Lee J.H. RSC Adv. 2015;5:6443–6451. [Google Scholar]
- 78.Ghosh A., Rao K.V., George S.J., Rao C.N.R. Chem. Eur. J. 2010;16:2700–2704. doi: 10.1002/chem.200902828. [DOI] [PubMed] [Google Scholar]
- 79.Das S., Irin F., Ahmed H.S. Tanvir, Cortinas A.B., Wajid A.S., Parviz D., Jankowski A.F., Kato M., Green M.J. Polymer. 2012;53:2485–2494. [Google Scholar]
- 80.Uddin M.E., Kuila T., Nayak G.C., Kim N.H., Ku B.-C., Lee J.H. J. Alloys Compd. 2013;562:134–142. [Google Scholar]
- 81.Joo E.H., Kuila T., Kim N.H., Lee J.H., Kim S.A., Park E.G., Lee U.H. Adv. Mater. Res. Switz. 2013;747:246. [Google Scholar]
- 82.Li Q., Dong L., Sun F., Huang J., Xie H., Xiong C. Chem. Eur. J. 2012;18:7055–7059. doi: 10.1002/chem.201200596. [DOI] [PubMed] [Google Scholar]
- 83.Liang Y., Wu D., Feng X., Müllen K. Adv. Mater. 2009;21:1679–1683. [Google Scholar]
- 84.Bourlinos A.B., Georgakilas V., Zboril R., Steriotis T.A., Stubos A.K., Trapalis C. Solid State Commun. 2009;149:2172–2176. [Google Scholar]
- 85.Fernández-Merino M.J., Paredes J.I., Villar-Rodil S., Guardia L., Solís-Fernández P., Salinas-Torres D., Cazorla-Amorós D., Morallón E., Martínez-Alonso A., Tascón J.M.D. Carbon. 2012;50:3184–3194. [Google Scholar]
- 86.Lotya M., King P.J., Khan U., De S., Coleman J.N. ACS Nano. 2010;4:3155–3162. doi: 10.1021/nn1005304. [DOI] [PubMed] [Google Scholar]
- 87.Priya T., Dhanalakshmi N., Karthikeyan V., Thinakaran N. J. Electroanal. Chem. 2019;833:543–551. [Google Scholar]
- 88.Carrasco P.M., Montes S., García I., Borghei M., Jiang H., Odriozola I., Cabañero G., Ruiz V. Carbon. 2014;70:157–163. [Google Scholar]
- 89.Yang X., Li J., Wen T., Ren X., Huang Y., Wang X. Colloids Surf. A Physicochem. Eng. Asp. 2013;422:118–125. [Google Scholar]
- 90.Lee D., Chander Y., Goyal S.M., Cui T. Biosens. Bioelectron. 2011;26:3482–3487. doi: 10.1016/j.bios.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Y., Chen X.H., Zhong Y.L., Zhu F.R., Loh K.P. Appl. Phys. Lett. 2009;95 [Google Scholar]
- 92.Kodali V.K., Scrimgeour J., Kim S., Hankinson J.H., Carroll K.M., Heer W.A. De, Berger C., Curtis J.E. Langmuir. 2011;27:863–865. doi: 10.1021/la1033178. [DOI] [PubMed] [Google Scholar]
- 93.Zhang X.-F., Shao X. J. Photochem. Photobiol. A Chem. 2014;278:69–74. [Google Scholar]
- 94.Georgakilas V., Otyepka M., Bourlinos A.B., Chandra V., Kim N., Kemp K.C., Hobza P., Zboril R., Kim K.S. Chem. Rev. 2012;112:6156–6214. doi: 10.1021/cr3000412. [DOI] [PubMed] [Google Scholar]
- 95.Park J., Yan M. Acc. Chem. Res. 2013;46:181–189. doi: 10.1021/ar300172h. [DOI] [PubMed] [Google Scholar]
- 96.Vacchi I.A., Menard-Moyon C., Bianco A. Phys. Sci. Rev. 2017;2 [Google Scholar]
- 97.Criado A., Melchionna M., Marchesan S., Prato M. Angew. Chem. Int. Ed. 2015;54:10734–10750. doi: 10.1002/anie.201501473. [DOI] [PubMed] [Google Scholar]
- 98.Jiang D.-e., Sumpter B.G., Dai S. J. Phys. Chem. B. 2006;110:23628–23632. doi: 10.1021/jp065980+. [DOI] [PubMed] [Google Scholar]
- 99.Sarkar S., Bekyarova E., Niyogi S., Haddon R.C. J. Am. Chem. Soc. 2011;133:3324–3327. doi: 10.1021/ja200118b. [DOI] [PubMed] [Google Scholar]
- 100.Elias D.C., Nair R.R., Mohiuddin T., Morozov S., Blake P., Halsall M., Ferrari A.C., Boukhvalov D., Katsnelson M., Geim A. Science. 2009;323:610–613. doi: 10.1126/science.1167130. [DOI] [PubMed] [Google Scholar]
- 101.Whitener K.E., Lee W.K., Campbell P.M., Robinson J.T., Sheehan P.E. Carbon. 2014;72:348–353. [Google Scholar]
- 102.Zhou S., Sherpa S.D., Hess D.W., Bongiorno A. J. Phys. Chem. C. 2014;118:26402–26408. [Google Scholar]
- 103.Hossain M.Z., Razak M.B.A., Noritake H., Shiozawa Y., Yoshimoto S., Mukai K., Koitaya T., Yoshinobu J., Hosaka S. J. Phys. Chem. C. 2014;118:22096–22101. [Google Scholar]
- 104.Hernández S., Wheeler V., Osofsky M., Jernigan G., Nagareddy V., Nath A., Lock E., Nyakiti L., Myers-Ward R., Sridhara K. Surf. Coat. Technol. 2014;241:8–12. [Google Scholar]
- 105.Chakrabarti M.H., Low C.T.J., Brandon N.P., Yufit V., Hashim M.A., Irfan M.F., Akhtar J., Ruiz-Trejo E., Hussain M.A. Electrochim. Acta. 2013;107:425–440. [Google Scholar]
- 106.Sundaram R.S., Gómez-Navarro C., Balasubramanian K., Burghard M., Kern K. Adv. Mater. 2008;20:3050–3053. [Google Scholar]
- 107.Low C., Walsh F., Chakrabarti M., Hashim M., Hussain M. Carbon. 2013;54:1–21. [Google Scholar]
- 108.Lomeda J.R., Doyle C.D., Kosynkin D.V., Hwang W.-F., Tour J.M. J. Am. Chem. Soc. 2008;130:16201–16206. doi: 10.1021/ja806499w. [DOI] [PubMed] [Google Scholar]
- 109.Bekyarova E., Itkis M.E., Ramesh P., Berger C., Sprinkle M., Heer W.A. de, Haddon R.C. J. Am. Chem. Soc. 2009;131:1336–1337. doi: 10.1021/ja8057327. [DOI] [PubMed] [Google Scholar]
- 110.Sinitskii A., Dimiev A., Corley D.A., Fursina A.A., Kosynkin D.V., Tour J.M. ACS Nano. 2010;4:1949–1954. doi: 10.1021/nn901899j. [DOI] [PubMed] [Google Scholar]
- 111.Hossain M.Z., Walsh M.A., Hersam M.C. J. Am. Chem. Soc. 2010;132:15399–15403. doi: 10.1021/ja107085n. [DOI] [PubMed] [Google Scholar]
- 112.Pinson J., Podvorica F. Chem. Soc. Rev. 2005;34:429–439. doi: 10.1039/b406228k. [DOI] [PubMed] [Google Scholar]
- 113.Gan L., Zhang D., Guo X. Small. 2012;8:1326–1330. doi: 10.1002/smll.201102302. [DOI] [PubMed] [Google Scholar]
- 114.Chan C.K., Beechem T.E., Ohta T., Brumbach M.T., Wheeler D.R., Stevenson K.J. J. Phys. Chem. C. 2013;117:12038–12044. [Google Scholar]
- 115.Sarkar S., Bekyarova E., Haddon R.C. Angew. Chem. Int. Ed. 2012;51:4901–4904. doi: 10.1002/anie.201201320. [DOI] [PubMed] [Google Scholar]
- 116.Salice P., Fabris E., Sartorio C., Fenaroli D., Figà V., Casaletto M.P., Cataldo S., Pignataro B., Menna E. Carbon. 2014;74:73–82. [Google Scholar]
- 117.Wang Q.H., Jin Z., Kim K.K., Hilmer A.J., Paulus G.L., Shih C.-J., Ham M.-H., Sanchez-Yamagishi J.D., Watanabe K., Taniguchi T. Nat. Chem. 2012;4:724. doi: 10.1038/nchem.1421. [DOI] [PubMed] [Google Scholar]
- 118.Niyogi S., Bekyarova E., Itkis M.E., Zhang H., Shepperd K., Hicks J., Sprinkle M., Berger C., Lau C.N., deHeer W.A., Conrad E.H., Haddon R.C. Nano Lett. 2010;10:4061–4066. doi: 10.1021/nl1021128. [DOI] [PubMed] [Google Scholar]
- 119.Sharma R., Baik J.H., Perera C.J., Strano M.S. Nano Lett. 2010;10:398–405. doi: 10.1021/nl902741x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu Q., Wu Y., Hao Y., Geng J., Charlton M., Chen S., Ren Y., Ji H., Li H., Boukhvalov D.W. Chem. Commun. 2012;49:677–679. doi: 10.1039/c2cc36747e. [DOI] [PubMed] [Google Scholar]
- 121.Bissett M.A., Konabe S., Okada S., Tsuji M., Ago H. ACS Nano. 2013;7:10335–10343. doi: 10.1021/nn404746h. [DOI] [PubMed] [Google Scholar]
- 122.Liu H., Ryu S., Chen Z., Steigerwald M.L., Nuckolls C., Brus L.E. J. Am. Chem. Soc. 2009;131:17099–17101. doi: 10.1021/ja9043906. [DOI] [PubMed] [Google Scholar]
- 123.Fang M., Wang K., Lu H., Yang Y., Nutt S. J. Mater. Chem. 2009;19:7098–7105. [Google Scholar]
- 124.Steenackers M., Gigler A.M., Zhang N., Deubel F., Seifert M., Hess L.H., Lim C.H.Y.X., Loh K.P., Garrido J.A., Jordan R., Stutzmann M., Sharp I.D. J. Am. Chem. Soc. 2011;133:10490–10498. doi: 10.1021/ja201052q. [DOI] [PubMed] [Google Scholar]
- 125.Hess L.H., Lyuleeva A., Blaschke B.M., Sachsenhauser M., Seifert M., Garrido J.A., Deubel F. ACS Appl. Mater. Interfaces. 2014;6:9705–9710. doi: 10.1021/am502112x. [DOI] [PubMed] [Google Scholar]
- 126.Georgakilas V., Bourlinos A.B., Zboril R., Steriotis T.A., Dallas P., Stubos A.K., Trapalis C. Chem. Commun. 2010;46:1766–1768. doi: 10.1039/b922081j. [DOI] [PubMed] [Google Scholar]
- 127.Magedov I.V., Frolova L.V., Ovezmyradov M., Bethke D., Shaner E.A., Kalugin N.G. Carbon. 2013;54:192–200. doi: 10.1016/j.carbon.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhong X., Jin J., Li S., Niu Z., Hu W., Li R., Ma J. Chem. Commun. 2010;46:7340–7342. doi: 10.1039/c0cc02389b. [DOI] [PubMed] [Google Scholar]
- 129.He H., Gao C. Chem. Mater. 2010;22:5054–5064. [Google Scholar]
- 130.Choi J., Kim K.-j., Kim B., Lee H., Kim S. J. Phys. Chem. C. 2009;113:9433–9435. [Google Scholar]
- 131.Ray S.C., Soin N., Makgato T., Chuang C.H., Pong W.F., Roy S.S., Ghosh S.K., Strydom A.M., McLaughlin J.A. Sci. Rep. 2014;4:3862. doi: 10.1038/srep03862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Balog R., Jørgensen B., Nilsson L., Andersen M., Rienks E., Bianchi M., Fanetti M., Lægsgaard E., Baraldi A., Lizzit S., Sljivancanin Z., Besenbacher F., Hammer B., Pedersen T.G., Hofmann P., Hornekær L. Nat. Mater. 2010;9:315–319. doi: 10.1038/nmat2710. [DOI] [PubMed] [Google Scholar]
- 133.Ryu S., Han M.Y., Maultzsch J., Heinz T.F., Kim P., Steigerwald M.L., Brus L.E. Nano Lett. 2008;8:4597–4602. doi: 10.1021/nl802940s. [DOI] [PubMed] [Google Scholar]
- 134.Zhang X., Hsu A., Wang H., Song Y., Kong J., Dresselhaus M.S., Palacios T. ACS Nano. 2013;7:7262–7270. doi: 10.1021/nn4026756. [DOI] [PubMed] [Google Scholar]
- 135.Li B., Zhou L., Wu D., Peng H., Yan K., Zhou Y., Liu Z. ACS Nano. 2011;5:5957–5961. doi: 10.1021/nn201731t. [DOI] [PubMed] [Google Scholar]
- 136.Cao Y., Feng J., Wu P. Carbon. 2010;48:1683–1685. [Google Scholar]
- 137.Chen Y., Zhang X., Yu P., Ma Y. Chem. Commun. 2009:4527–4529. doi: 10.1039/b907723e. [DOI] [PubMed] [Google Scholar]
- 138.Yang H., Shan C., Li F., Han D., Zhang Q., Niu L. Chem. Commun. 2009:3880–3882. doi: 10.1039/b905085j. [DOI] [PubMed] [Google Scholar]
- 139.Karousis N., Economopoulos S.P., Sarantopoulou E., Tagmatarchis N. Carbon. 2010;48:854–860. [Google Scholar]
- 140.Yang H., Li F., Shan C., Han D., Zhang Q., Niu L., Ivaska A. J. Mater. Chem. 2009;19:4632–4638. [Google Scholar]
- 141.Valentini L., Cardinali M., Bon S. Bittolo, Bagnis D., Verdejo R., Lopez-Manchado M.A., Kenny J.M. J. Mater. Chem. 2010;20:995–1000. [Google Scholar]
- 142.Bon S.B., Valentini L., Verdejo R., Fierro J.L. Garcia, Peponi L., Lopez-Manchado M.A., Kenny J.M. Chem. Mater. 2009;21:3433–3438. [Google Scholar]
- 143.Lin Y., Jin J., Song M. J. Mater. Chem. 2011;21:3455–3461. [Google Scholar]
- 144.Iijima S. Nature. 1991;354:56–58. [Google Scholar]
- 145.R.E. Smalley, (2003).
- 146.Zhu H.W., Xu C.L., Wu D.H., Wei B.Q., Vajtai R., Ajayan P.M. Science. 2002;296:884–886. doi: 10.1126/science.1066996. [DOI] [PubMed] [Google Scholar]
- 147.Muhulet A., Miculescu F., Voicu S.I., Schutt F., Thakur V.K., Mishra Y.K. Mater. Today Energy. 2018;9:154–186. [Google Scholar]
- 148.Gao Y., Kyratzis I. Bioconjug. Chem. 2008;19:1945–1950. doi: 10.1021/bc800051c. [DOI] [PubMed] [Google Scholar]
- 149.Gaudino E.C., Tagliapietra S., Martina K., Barge A., Lolli M., Terreno E., Lembo D., Cravotto G. Org. Biomol. Chem. 2014;12:4708–4715. doi: 10.1039/c4ob00611a. [DOI] [PubMed] [Google Scholar]
- 150.Palomar Q., Gondran C., Marks R., Cosnier S., Holzinger M. Electrochim. Acta. 2018;274:84–90. [Google Scholar]
- 151.Singh C., Srivastava S., Ali M.A., Gupta T.K., Sumana G., Srivastava A., Mathur R.B., Malhotra B.D. Sensors Actuators B Chem. 2013;185:258–264. [Google Scholar]
- 152.Dias A.C.M.S., Gomes-Filho S.L.R., Silva M.M.S., Dutra R.F. Biosens. Bioelectron. 2013;44:216–221. doi: 10.1016/j.bios.2012.12.033. [DOI] [PubMed] [Google Scholar]
- 153.Silva M.M.S., Dias A.C.M.S., Silva B.V.M., Gomes-Filho S.L.R., Kubota L.T., Goulart M.O.F., Dutra R.F. J. Chem. Technol. Biotechnol. 2015;90:194–200. [Google Scholar]
- 154.Viswanathan S., Rani C., Anand A. Vijay, Ho J.-A.A. Biosens. Bioelectron. 2009;24:1984–1989. doi: 10.1016/j.bios.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 155.Periasamy A.P., Chang Y.-J., Chen S.-M. Bioelectrochemistry. 2011;80:114–120. doi: 10.1016/j.bioelechem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 156.Dinesh B., Bianco A., Menard-Moyon C. Nanoscale. 2016;8:18596–18611. doi: 10.1039/c6nr06728j. [DOI] [PubMed] [Google Scholar]
- 157.Kar T., Bettinger H.F., Scheiner S., Roy A.K. J. Phys. Chem. C. 2008;112:20070–20075. [Google Scholar]
- 158.Chen R.J., Zhang Y., Wang D., Dai H. J. Am. Chem. Soc. 2001;123:3838–3839. doi: 10.1021/ja010172b. [DOI] [PubMed] [Google Scholar]
- 159.Ray S.C., Jana N.R. 2017. Chapter 3 Application of Carbon-Based nanomaterials as Biosensor,Carbon Nanomaterials for Biological and Medical Applications; pp. 87–127. [Google Scholar]
- 160.Rausch J., Zhuang R.-C., Mäder E. Compos. A: Appl. Sci. Manuf. 2010;41:1038–1046. [Google Scholar]
- 161.Liu Z., Davis C., Cai W., He L., Chen X., Dai H. Proc. Natl. Acad. Sci. 2008;105:1410–1415. doi: 10.1073/pnas.0707654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Murakami H., Nomura T., Nakashima N. Chem. Phys. Lett. 2003;378:481–485. [Google Scholar]
- 163.Tu W., Lei J., Ju H. Chem Eur J. 2009;15:779–784. doi: 10.1002/chem.200801758. [DOI] [PubMed] [Google Scholar]
- 164.Wu P., Chen X., Hu N., Tam U.C., Blixt O., Zettl A., Bertozzi C.R. Angew. Chem. 2008;120:5100–5103. doi: 10.1002/anie.200705363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhou Y., Fang Y., Ramasamy R.P. Sensors (Basel) 2019;19:392. doi: 10.3390/s19020392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shim M., Kam N.W. Shi, Chen R.J., Li Y., Dai H. Nano Lett. 2002;2:285–288. [Google Scholar]
- 167.Kotagiri N., Kim J.-W. Int. J. Nanomedicine. 2014;9(Suppl. 1):85–105. doi: 10.2147/IJN.S51854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Assali M., Leal M.P., Fernández I., Baati R., Mioskowski C., Khiar N. Soft Matter. 2009;5:948–950. [Google Scholar]
- 169.Bourourou M., Elouarzaki K., Lalaoui N., Agnès C., Goff A. Le, Holzinger M., Maaref A., Cosnier S. Chem Eur J. 2013;19:9371–9375. doi: 10.1002/chem.201301043. [DOI] [PubMed] [Google Scholar]
- 170.Holzinger M., Baur J., Haddad R., Wang X., Cosnier S. Chem. Commun. 2011;47:2450–2452. doi: 10.1039/c0cc03928d. [DOI] [PubMed] [Google Scholar]
- 171.Yang X., Lu Y., Ma Y., Li Y., Du F., Chen Y. Chem. Phys. Lett. 2006;420:416–420. [Google Scholar]
- 172.Goff A. Le, Moggia F., Debou N., Jegou P., Artero V., Fontecave M., Jousselme B., Palacin S. J. Electroanal. Chem. 2010;641:57–63. [Google Scholar]
- 173.Huang X.-J., Im H.-S., Lee D.-H., Kim H.-S., Choi Y.-K. J. Phys. Chem. C. 2007;111:1200–1206. [Google Scholar]
- 174.Giroud F., Minteer S.D. Electrochem. Commun. 2013;34:157–160. [Google Scholar]
- 175.Li Q., Zhang J., Yan H., He M., Liu Z. Carbon. 2004;42:287–291. [Google Scholar]
- 176.Ghica M.E., Brett C.M. Talanta. 2014;130:198–206. doi: 10.1016/j.talanta.2014.06.068. [DOI] [PubMed] [Google Scholar]
- 177.Li P., Liu H., Ding Y., Wang Y., Chen Y., Zhou Y., Tang Y., Wei H., Cai C., Lu T. J. Mater. Chem. 2012;22:15370–15378. [Google Scholar]
- 178.Sanz V., Borowiak E., Lukanov P., Galibert A.M., Flahaut E., Coley H.M., Silva S.R.P., McFadden J. Carbon. 2011;49:1775–1781. [Google Scholar]
- 179.Liu G., Lin Y. Electrochem. Commun. 2006;8:251–256. [Google Scholar]
- 180.Zhang X., Meng L., Lu Q. ACS Nano. 2009;3:3200–3206. doi: 10.1021/nn9006362. [DOI] [PubMed] [Google Scholar]
- 181.Bandaru N.M., Voelcker N.H. J. Mater. Chem. 2012;22:8748–8758. [Google Scholar]
- 182.Fujigaya T., Nakashima N. Sci. Technol. Adv. Mater. 2015;16 doi: 10.1088/1468-6996/16/2/024802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Vedala H., Chen Y., Cecioni S., Imberty A., Vidal S., Star A. Nano Lett. 2011;11:170–175. doi: 10.1021/nl103286k. [DOI] [PubMed] [Google Scholar]
- 184.Oueiny C., Berlioz S., Perrin F.-X. Prog. Polym. Sci. 2014;39:707–748. [Google Scholar]
- 185.Zhao H., Liu F., Xie W., Zhou T.-C., OuYang J., Jin L., Li H., Zhao C.-Y., Zhang L., Wei J., Zhang Y.-P., Li C.-P. Sensors Actuators B Chem. 2021;327 doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14:12257–12258. doi: 10.1021/acsnano.0c06726. [DOI] [PubMed] [Google Scholar]
- 188.Jarocka U., Sawicka R., Gora-Sochacka A., Sirko A., Zagorski-Ostoja W., Radecki J., Radecka H. Biosens. Bioelectron. 2014;55:301–306. doi: 10.1016/j.bios.2013.12.030. [DOI] [PubMed] [Google Scholar]
- 189.Anik U., Tepeli Y., Sayhi M., Nsiri J., Diouani M.F. Analyst. 2018;143:150–156. doi: 10.1039/c7an01537b. [DOI] [PubMed] [Google Scholar]
- 190.Anik Ü., Tepeli Y., Diouani M.F. Anal. Chem. 2016;88:6151–6153. doi: 10.1021/acs.analchem.6b01720. [DOI] [PubMed] [Google Scholar]
- 191.Veerapandian M., Hunter R., Neethirajan S. Talanta. 2016;155:250–257. doi: 10.1016/j.talanta.2016.04.047. [DOI] [PubMed] [Google Scholar]
- 192.Singh R., Hong S., Jang J. Sci. Rep. 2017;7:42771. doi: 10.1038/srep42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Huang J., Xie Z., Xie Z., Luo S., Xie L., Huang L., Fan Q., Zhang Y., Wang S., Zeng T. Anal. Chim. Acta. 2016;913:121–127. doi: 10.1016/j.aca.2016.01.050. [DOI] [PubMed] [Google Scholar]
- 194.Joshi S.R., Sharma A., Kim G.-H., Jang J. Mater. Sci. Eng. C. 2020;108 doi: 10.1016/j.msec.2019.110465. [DOI] [PubMed] [Google Scholar]
- 195.Chan C., Shi J., Fan Y., Yang M. Sensors Actuators B Chem. 2017;251:927–933. [Google Scholar]
- 196.Tran T.L., Nguyen T.T., Tran T.T. Huyen, Chu V.T., Tran Q. Thinh, Mai A. Tuan. Phys. E. 2017;93:83–86. [Google Scholar]
- 197.Devarakonda S., Singh R., Bhardwaj J., Jang J. Sensors-Basel. 2017;17 doi: 10.3390/s17112597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lee J., Morita M., Takemura K., Park E.Y. Biosens. Bioelectron. 2018;102:425–431. doi: 10.1016/j.bios.2017.11.052. [DOI] [PubMed] [Google Scholar]
- 199.Fu Y., Romay V., Liu Y., Ibarlucea B., Baraban L., Khavrus V., Oswald S., Bachmatiuk A., Ibrahim I., Rümmeli M., Gemming T., Bezugly V., Cuniberti G. Sensors Actuators B Chem. 2017;249:691–699. [Google Scholar]
- 200.Tian J., Wang D., Zheng Y., Jing T. Int. J. Electrochem. Sci. 2017;12:2658–2668. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in the reference section of manuscript.