Abstract
Objective
To investigate transcriptional alterations in human semen samples associated with COVID-19 infection.
Design
Retrospective observational cohort study.
Setting
City hospital.
Patient(s)
Ten patients who had recovered from mild COVID-19 infection. Eight of these patients had different sperm abnormalities that were diagnosed before infection. The control group consisted of 5 healthy donors without known abnormalities and no history of COVID-19 infection.
Intervention(s)
We used RNA sequencing to determine gene expression profiles in all studied biosamples. Original standard bioinformatic instruments were used to analyze activation of intracellular molecular pathways.
Main Outcome Measure(s)
Routine semen analysis, gene expression levels, and molecular pathway activation levels in semen samples.
Result(s)
We found statistically significant inhibition of genes associated with energy production pathways in the mitochondria, including genes involved in the electron transfer chain and genes involved in toll-like receptor signaling. All protein-coding genes encoded by the mitochondrial genome were significantly down-regulated in semen samples collected from patients after recovery from COVID-19.
Conclusion(s)
Our results may provide a molecular basis for the previously observed phenomenon of decreased sperm motility associated with COVID-19 infection. Moreover, the data will be beneficial for the optimization of preconception care for men who have recently recovered from COVID-19 infection.
Key Words: COVID-19, gene expression analysis, molecular pathways, RNA sequencing, sperm
Discuss: You can discuss this article with its authors and other readers at https://www.fertstertdialog.com/posts/xfss-d-21-00043
According to the World Health Organization, more than 100 million married couples face the problem of infertility, with 40% of cases due to male infertility. The main causes of male infertility are genital infections, varicocele, and idiopathic oligo-, astheno-, and teratozoospermia. Infectious diseases account for 15% of cases of male fertility (1). Thus, investigating the potential impact of novel coronavirus COVID-19 infection on male reproductive health is a particularly relevant topic.
According to Holtmann et al. (2), mild COVID-19 infection is not associated with impaired function of the testes and epididymis, whereas patients with moderate COVID-19 infection have changes in semen parameters: decreased sperm concentration, decreased total number of sperm in the ejaculate, and decreased sperm motility. Ma et al. (3) found the following abnormal semen parameters in patients who had recovered from COVID-19 infection: increased rate of DNA fragmentation and decreased sperm motility. Several other studies have described cases of decreased libido and loss of morning erection in patients with COVID-19 infection. Ruan et al. (4) found decreased sperm concentration and motility in patients with COVID-19 infection from analysis of semen parameters from 55 patients and 145 controls.
Hajizadeh Maleki and Tartibian (5) investigated semen samples of 84 patients who had recovered from COVID-19 infection and 105 controls. The semen of patients was analyzed every 10 days for 2 months. The investigators found statistically significant decreases in ejaculate volume, sperm motility, and sperm concentration and changes in sperm morphology in the COVID-19 patients. It is interesting to note that analysis of the semen at 30, 40, 50, and 60 days after COVID-19 infection revealed a progressive increase in sperm motility. The study also found that ejaculate volume, sperm morphology, and sperm concentration in the ejaculate did not change during the entire observation period (5). Analysis of a wide range of molecular abnormalities in semen of patients with COVID-19 infection can reveal the potential mechanisms of the influence of SARS-CoV-2 on male fertility and can be used to optimize the treatment plan for these patients.
In this study, we profiled gene expression in semen samples obtained from 10 patients after mild COVID-19 infection. All the patients had different sperm abnormalities that were diagnosed before infection. We were also able to profile the semen specimens from 5 patients obtained before infection, thus giving 5 matched sample pairs before and after COVID-19 infection. As the control group, we included 5 healthy donors with no known abnormalities or previous history of COVID-19 infection. We used RNA sequencing to establish gene expression profiles in these 20 biosamples. Gene ontology analysis of differentially regulated genes and in-depth study of activation of molecular pathways revealed statistically significant inhibition of genes associated with toll-like receptor (TLR) pathways and with energy production pathways in the mitochondria, including genes involved in the electron transfer chain. In addition, all protein-coding genes encoded by the mitochondrial genome were significantly down-regulated in postinfection samples. This finding may provide a molecular basis for the previously observed phenomenon of decreased sperm motility after COVID-19 infection.
Materials and methods
Ethical Statement
The Committee of Biomedical Ethics of the World-Class Medical Center, Moscow (Institutional Review Board) approved all procedures and study methods. Informed written consent was obtained from all patients included in the study. The study was conducted in accordance with the Declaration of Helsinki, 1975.
Study Population and Design
The participants were divided into 3 groups according to the time of sample collection. Group 1 consisted of 5 patients who had recovered from mild COVID-19 infection and provided semen samples before infection and after proven recovery (10 samples in total). The semen samples of these patients were stored before COVID-19 infection as they were preparing for in vitro fertilization (IVF). Four patients in this group had different sperm abnormalities before infection. Inclusion of these patients in the study allowed us to determine whether they needed additional treatment before IVF.
Group 2 consisted of 5 patients who had recovered from mild COVID-19 infection and provided semen samples after proven recovery. Four patients in this group also had different sperm abnormalities before infection. Inclusion of these patients in the study allowed us to determine whether they needed additional treatment before IVF. Group 3 consisted of 5 healthy controls.
The inclusion criteria were sexually active men of reproductive age (18 to 65 years) with proven recovery from mild COVID-19 infection by 2 consecutive negative nasopharyngeal swabs for SARS-CoV-2 RNA. The severity of COVID-19 infection was classified according to the World Health Organization guideline, which defines mild COVID-19 infection as infection without evidence of viral pneumonia or hypoxia (6). The patients complained of fever (38.5°C for 3 days) and fatigue. No treatment was administered except acetaminophen 500 mg. The exclusion criteria were ejaculatory disorders, a history of prostate surgery, treatment with antihypertensive medications, and inability to express informed consent. The diagnosis of COVID-19 infection was confirmed by the presence of SARS-CoV-2 RNA in a nasopharyngeal swab detected by molecular testing (reverse transcription polymerase chain reaction) according to medical records.
Collection of Biosamples
The participants received clear instructions about collecting their semen samples into a sterile container following at least 2 days of sexual abstinence (but no more than 5 to 7 days). All patients included in the study provided semen samples in the hospital. Semen samples of patients in the COVID-19 groups were collected within less than 75 days after proven recovery from infection.
Information regarding serum levels of testosterone, follicle-stimulating hormone (FSH), and luteinizing hormone (LH) was obtained from the medical records of the patients. Routine laboratory analyses were performed with commercially available kits. The laboratory’s reference ranges were 8.33–30.19 nmol/L for testosterone, 0.95–11.95 IU/L for FSH, and 0.57–12.07 IU/L for LH.
RNA Isolation and Sequencing
RNA was extracted from semen samples with the QIAGEN RNeasy Plus Universal Mini Kit (Qiagen, Germantown, MD) following the manufacturer’s protocol. RNA 6000 Nano (Agilent, Santa Clara, CA) or Qubit RNA Assay kits (Life Technologies, Bleiswijk, Netherlands) were used to measure RNA concentration. The RNA integrity number was measured with an Agilent 2100 Bio-Analyzer. For depletion of ribosomal RNA and library construction, the KAPA RNA Hyper with rRNA erase kit (KAPA, Cape Town, South Africa) (HMR only) was used. Different adaptors were used for multiplexing samples in one sequencing run. Library concentrations and quality were measured with the Qubit ds DNA HS Assay kit (Life Technologies, Eugene, OR) and Agilent TapeStation (Agilent, Santa Clara, CA). RNA sequencing was done with Illumina NextSeq 550 equipment for single-end sequencing, 75-bp read length, for approximately 30 million raw reads per sample. Demultiplexing was performed with the Illumina Bcl2fastq2 v 2.17 program.
Bioinformatic Analysis
RNA sequencing FASTQ files were processed with STAR aligner in “GeneCounts” mode with the Ensembl human transcriptome annotation (Build version GRCh38 and transcript annotation GRCh38.89) (7). Ensembl gene IDs were converted to HUGO Gene Nomenclature Committee (HGNC) gene symbols with the use of the Complete HGNC data set (https://www.genenames.org/; database version from July 13, 2017). Expression levels were established for 36,596 annotated genes with the corresponding HGNC identifiers. The minimum number of uniquely mapped reads was 5.66 million for the studied biosamples, with a mean value of 21.66 million. Differential gene expression analysis was performed with DESeq2 software with the following thresholds: false discovery rate-adjusted P<.05 and |log2(fold change)|>1 (8). Volcano plots were built with the use of “EnhancedVolcano” R package (9). Gene ontology (GO) analysis was performed with the R “enrichgo” package (10). Pathway activation levels and corresponding visualization were performed with Oncobox software and pathway collection (11, 12). We checked all biosamples for the presence of SARS-CoV-2–specific reads with fastv utility (13).
Results
In this study, we compared gene expression profiles in semen samples of patients with different diagnosed sperm abnormalities before COVID-19 infection and after proven recovery. All patients in groups 1 and 2 suffered from mild COVID-19 infection (Supplemental Table 1, available online). Other patient clinical characteristics, including hormone profiles, are presented in Supplemental Table 1. Biosamples taken both before and after infection were available for 5 patients (10 samples in total). For another 5 patients, only biosamples collected after recovery from COVID-19 infection were available for further analysis.
In addition, we included 5 control archival semen samples obtained from healthy donors who were not affected by COVID-19 before sample collection. These samples were used as the reference cohort for the molecular analyses.
We performed RNA sequencing (RNAseq) of all mentioned semen samples. For one patient (ID P4), both pre- and postinfection biosamples were available in triplicate, and we performed RNAseq for all biosamples. Altogether we obtained 24 RNAseq profiles; the mapping statistics are shown in Supplemental Table 2. We used the fastv program to check our transcript profiles for the presence of SARS-CoV-2–specific reads and found no such calls in any samples tested (13).
The replicates of P4 biosamples formed distinct clusters both on the dendrogram (Supplemental Fig. 1A, available online) and on principal component analysis plots (Supplemental Fig. 1B). Samples obtained before infection and healthy donor samples also tended to cluster together, whereas postinfection samples showed distinct clustering patterns and a higher degree of heterogeneity (Supplemental Fig. 1).
Paired Analysis
First, we performed differential gene expression analysis between replicates of patient P4. This resulted in 5,411 and 3,846 significantly down- and up-regulated genes, respectively (Supplemental Table 3 and Supplemental Fig. 2). Gene ontology analysis of up-regulated genes revealed biological processes associated with cation transmembrane transporter activity, ion channel activity, extracellular matrix, and others (Supplemental Fig. 3). Gene ontology analysis of down-regulated genes identified processes associated with nucleoside binding and cellular energy metabolism (Supplemental Fig. 4). Quantitative molecular pathway activation assay revealed Janus kinase and signal transducer and activator of transcription pathway Janus kinase degradation, and peptide chain elongation as the most significantly up- and down-regulated pathways, respectively, in P4 postinfection samples normalized on samples before infection (Supplemental Fig. 5).
We then performed differentially expressed gene (DEG) analysis for all 5 pairs of matched samples obtained before COVID-19 infection and after proven recovery from infection. The adjusted P value cutoff was set at .05, and the |log2 fold change| cutoff was set higher than 1. The analysis resulted in 57 statistically significantly down-regulated and, remarkably, no up-regulated genes after infection (Fig. 1 A). Of these 57 genes, 52 were identical to the down-regulated differential genes obtained previously in the analysis of P4 replicates (Fig. 1B). We further tested the significance of this intersection by repetitively intersecting random gene sets of the same sizes 1,000 times and registering the numbers of randomly intersected genes (Fig. 1C). The randomness analysis showed that the observed actual intersection could not be obtained by chance (P<.001).
Figure 1.
(A) Distribution of DEGs between P1, P2, P3, P4 (averaged), and P5 samples before COVID-19 and after proven recovery by log2 fold change and log10P value. The adjusted P value cutoff was set at .05, and the |log2 fold change| cutoff was set higher than 1. (B and C) Intersection of DEGs. (B) Venn diagram showing intersection of down-regulated DEGs from 2 analyses: matched samples from patients P1–P5 before COVID-19 and after proven recovery, and triplicates of P4 patient biosamples before and after COVID-19 infection. (C) Distribution of overlapped genes for randomly selected groups (1,000 random permutations). Red dashed line denotes actual number of intersected down-regulated DEGs. (D) Significantly enriched GO terms for the intersected set of down-regulated DEGs (n = 52). All terms passed the Benjamini-Hochberg adjusted P value threshold of .05. DEG = differentially expressed gene; FC = fold change; GO = gene ontology; NADH = nicotinamide adenine dinucleotide; NAD(P)H = nicotinamide adenine dinucleotide phosphate; RAGE = advanced glycosylation end-product specific receptor.
Analysis of Common DEGs
The following genes were statistically significantly down-regulated in both types of the DEG analysis: AIF1, APOC1, C1QA, C1QB, CCL2, CCR1, CSF3R, CXCL2, CXCL8, CXCR4, FCER1G, FGR, HCK, HLA-DRA, HMOX1, IFI44, IFI44L, IGSF6, IL1B, LAPTM5, LYZ, MNDA, MPEG1, MT-ATP6, MT-ATP8, MT-CO1, MT-CO2, MT-CO3, MT-CYB, MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND4L, MT-ND5, MT-ND6, MT1A, NDUFB8, PI3, POLR2K, RGS1, RPL39, S100A8, S100A9, SAMSN1, SDS, SOCS3, SPN, SRGN, TGFBI, TNFRSF1B, and TYROBP. The results of GO analysis for this gene set are shown in Figure 1D. The most significant GO terms were associated with reduced nicotinamide adenine dinucleotide dehydrogenase activity, followed by cytokine binding and fatty acid binding.
We then used Oncobox software to calculate pathway activation levels for 3,044 molecular pathways. Using the set of 52 intersected genes in the averaged post–COVID-19 infection sample normalized on the averaged pre–COVID-19 infection sample, a specific pathway activation pattern was obtained (Fig. 2 A). Overall, the extent of down-regulation was higher than the extent of up-regulation of the pathways interrogated. The most strongly up-regulated pathway was the interferon gamma (IFN-γ) signaling pathway (Fig. 2B), and the most strongly down-regulated pathways were 3 branches of the endogenous TLR signaling pathway: regulation of interleukin-1-beta production, regulation of cell proliferation, and regulation of apoptosis. The latter 3 subpathways contained identical gene sets, which explains the identity of their pathway activation levels (Fig. 2C). However, the next down-regulated pathway was for the respiratory electron transfer chain, which is in good agreement with the results of GO terms analysis (Fig. 1D).
Figure 2.
(A) Top 10 up-regulated and inhibited pathways sorted by statistically significantly different PAL values for comparison between samples obtained after and before COVID-19 infection. Pathways up-regulated after recovery from COVID-19 infection are shown on the top, pathways down-regulated after recovery on the bottom. The t test P value and PAL value for each pathway are shown on the right. (B) Activation scheme of the reactome interferon gamma signaling pathway. Color indicates the log2-transformed ratio of mean gene expression values for each pathway node in samples after recovery from COVID-19 infection normalized on expression levels in samples before infection. (C) Activation scheme of NCI endogenous TLR signaling pathway (regulation of interleukin-1-β production, regulation of cell proliferation, and regulation of apoptosis). Color indicates the log2-transformed ratio of mean gene expression values for each pathway node in samples after recovery from COVID-19 infection normalized on expression levels in samples before infection. ATP = adenosine triphosphate; FC = fold change; IFNG = interferon gamma; IFNGR = interferon gamma receptor; IL = interleukin; JAK = Janus kinase; KEGG = Kyoto Encyclopedia of Genes and Genomes; NCI = National Cancer Institute; PAL = pathway activation level; PKC = protein kinase C; SHP = Src homology region 2 domain-containing phosphatase-1; SOCS = suppressor of cytokine signaling; STAT = signal transducer and activator of transcription; TLR = toll-like receptor.
Mitochondrial Gene Expression
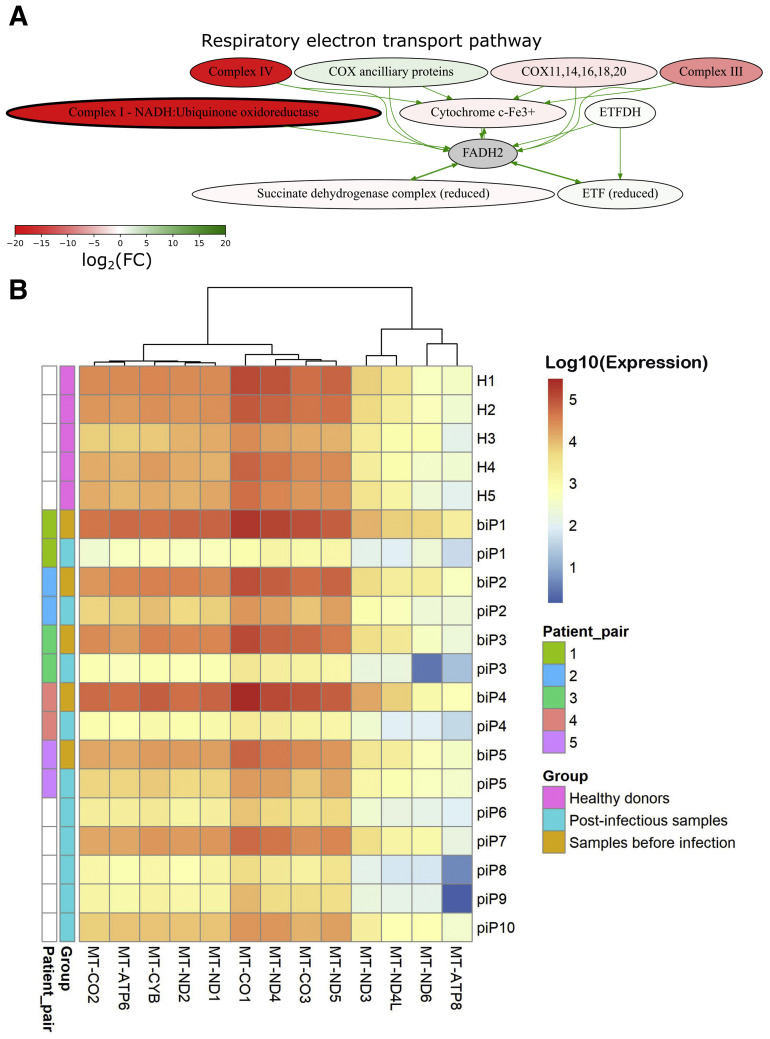
The second most strongly down-regulated pathway in semen samples collected after recovery from COVID-19 infection was the respiratory electron transport pathway. Detailed visualization of the pathway showed that the most down-regulated genes in postinfection samples were involved in the nodes Complex I and Complex IV (Fig. 3 A). These complexes contain 7 and 3 genes, respectively, which are encoded by the mitochondrial genome.
Figure 3.
(A) Activation scheme of respiratory electron transport pathway. Color indicates the log2-transformed ratio of mean gene expression values in samples after recovery from COVID-19 infection normalized on gene expression in samples before COVID-19 infection. (B) Heat map of mitochondrial genes that were differentially expressed in pairwise analysis of P1, P2, P3, P4 (averaged), and P5 samples. Color marker on the left indicates IDs of matched patient pairs and pre/post COVID-19 infection status of samples. COX = cytochrome c oxidase; ETF = electron transfer flavoprotein; ETFDH = electron transfer flavoprotein dehydrogenase; FADH = flavin adenine dinucleotide; FC = fold change; NADH = nicotinamide adenine dinucleotide.
We then analyzed mitochondrial genes and found that 13 of 13 (100%) protein-coding genes encoded in the mitochondrial genome were significantly down-regulated after infection. In all 5 paired samples, we observed decreased expression of the 13 mitochondrial genes compared with preinfection levels. However, both baseline and postinfection levels were highly heterogeneous between different patients (Fig. 3B).
We then compared the expression of all mitochondrial genes between postinfection samples vs. healthy donors and preinfection samples (Fig. 4 ). All group comparisons were significant (Fig. 4A–L), except for the borderline significance of MT-ND6 gene expression in postinfection samples in comparison with healthy donors (Fig. 4M). We also calculated the cumulative expression of 13 mitochondrial genes by summarizing the logarithms of normalized gene counts. We found that the cumulative expression was significantly lower in postinfection than in preinfection samples (P = 7.1∗10−5, t test) (Fig. 4N) and samples from healthy donors (P = .001, t test) (Fig. 4N).
Figure 4.
Expression of 13 protein-coding genes encoded in the mitochondrial genome and their cumulative expression (sum of log-transformed normalized gene counts) in samples from patients before COVID-19 infection, patients after recovery from COVID-19 infection, and healthy donors.
We further hypothesized that the expression level of mitochondrial genes was associated with the clinical characteristics of patients who had recovered from COVID-19 infection. We calculated fold changes in hormone levels before and after COVID-19 infection for 10 patients (Supplemental Table 1). The FSH level was increased in 5 patients and decreased in 5 patients after COVID-19 infection. However, none of the FSH levels before or after infection exceeded reference ranges. Interestingly, we observed a borderline significant negative correlation between FSH fold change and the cumulative expression of all mitochondrial genes (P = .067, Spearman test) (Supplemental Fig. 6). It is worth mentioning that no correlations were found between the cumulative expression of all mitochondrial genes and LH, testosterone, prolactin, or estradiol levels.
Finally, we investigated the expression levels of the genes associated with sperm motility according to previous studies. Previously, the expression of NRF2 (NFE2L2), ROPN1I , CABYR, FAM71D, RBMY1 (RBMY1A1), and sNHE (SLC9A1) was shown to be positively correlated with sperm motility, whereas expression of TP53 and HIF1A was negatively correlated with sperm motility (14, 15, 16, 17, 18, 19). CABYR and FAM71D were down-regulated in postinfection samples compared with levels before infection and in healthy donors (Supplemental Fig. 7A and B). There were no significant differences between the groups for other genes (Supplemental Fig. 7C to H).
Discussion
According to the literature, novel coronavirus SARS-CoV-2 enters cells through the interaction with angiotensin-converting enzyme receptor type 2 (ACE2) and type 2 transmembrane serine protease (TMPRSS2) (20). Analysis of ACE2 and TMPRSS2 expression patterns in male patients found that ACE2 was predominantly expressed in spermatogonia, Leydig cells, and Sertoli cells and TMPRSS2 was predominantly expressed in spermatogonia and spermatids. Wang and Xu (21) found that expression of the genes responsible for spermatogenesis was decreased in spermatogonia with high ACE2 expression.
Pan et al. (22) did not detect SARS-CoV-2 in the semen of patients with COVID-19 infection, possibly because of the nonsignificant expression of ACE2 and TMPRSS2 in these cells. Nevertheless, the investigators did not exclude the possibility of the presence of SARS-CoV-2 in the semen of patients in the acute phase of COVID-19 infection. In agreement with the study by Pan et al. (22), we did not detect SARS-CoV-2 RNA in our postinfection samples.
Testicular tissue can also be a target for SARS-CoV-2 because it coexpresses the receptors for ACE2 and TMPRSS2, which are necessary for invasion and further replication of the virus (21). Angiotensin-converting enzyme receptor type 2 expression was also detected in spermatogenic cells and somatic cells of the testes, which indicates a high susceptibility of the testes to damage by SARS-CoV-2 and impaired spermatogenesis, which in turn may have an impact on the sperm transcription profile.
Type 2 transmembrane serine protease is largely expressed in spermatogonia and spermatids. According to Huang et al. (23), coexpression of ACE2 and TMPRSS2 in spermatogonia and Leydig cells may be a risk factor for testicular degeneration and male infertility. Alternative receptor Basigin and protease cathepsin L, which can mediate the invasion of cells by SARS-CoV-2, have also been found in Leydig cells (23). Therefore, replication of SARS-CoV-2 in Leydig cells may potentially contribute to the disruption of testosterone production by the testes. However, only 5 of 10 patients had poorer sperm analysis after COVID-19 infection in our study.
Normally, gonadotropin-releasing hormone released from the hypothalamus stimulates production of FSH and LH by the anterior pituitary. Follicle-stimulating hormone then stimulates spermatogenesis in the Sertoli cells, whereas LH stimulates testosterone release by the Leydig cells. In turn, the rising level of testosterone inhibits release of gonadotropin-releasing hormone, FSH, and LH via a negative feedback loop. In cases of primary hypogonadism when the testosterone level is insufficient, gonadotropin-releasing hormone, FSH, and LH levels rise to compensate.
Luteinizing hormone and FSH increase significantly with age in men (24). In addition, previous results have shown statistically significant increases in mean FSH and LH levels in infertile men compared with fertile controls (25). In this study, we found mild elevations of FSH levels in 50% of patients who recovered from COVID-19 infection (5 of 10 patients), which were not correlated with testosterone levels. This elevation of FSH was associated with a decreased expression of mitochondrial genes in postinfection samples.
In agreement with this observation, Ma et al. (3) reported increased LH levels in male patients with COVID-19 infection, and Çayan et al. (26) reported decreased testosterone and elevated LH and FSH levels in male patients with COVID-19 infection and even showed that decreased testosterone levels were associated with the severity of COVID-19 infection. However, we did not find any mention of decreased gene expression in post–COVID-19–infection semen samples in the literature. We propose that this phenomenon may be associated with the decreased motility of sperm cells after COVID-19 infection.
Comparative analysis of signaling pathways showed that the most strongly up-regulated pathway in semen samples collected after recovery from COVID-19 infection when compared with semen samples of the same patients collected before infection was the IFN-γ signaling pathway. Like other viruses, SARS-CoV-2 induces the release of chemokines and cytokines from immune cells (27). Interferon gamma is a proinflammatory cytokine that can be elevated in semen samples during the inflammatory state and in patients with infertility (28, 29, 30). Recent studies revealed an elevated level of IFN-γ in the serum of patients with COVID-19 infection (31, 32). Furthermore, elevated levels of IFN-γ along with other cytokines can cause oxidative stress and increase membrane lipid peroxidation, which results in impaired sperm motility, capacitation, and the acrosome reaction (33, 34). Our results provide molecular evidence for elevated IFN-γ activity in semen samples collected after COVID-19 infection, which might be associated with decreased sperm motility in such patients.
The most strongly down-regulated pathways after recovery from COVID-19 infection are associated with regulation of interleukin-1-beta production, cell proliferation, and apoptosis (3 branches of the endogenous TLR signaling pathway). Toll-like receptors play a critical role in innate immunity, including protection of sperm cells from infection. Saedi et al. (35) reported expression of the genes for TLRs 1–10 in the testis, vas deferens, prostate, epididymis, and sperm cells (35). Nevertheless, more research needs to be done to investigate the impact of TLRs on sperm parameters.
In this study, we also found statistically significant inhibition of genes associated with energy production pathways in the mitochondria, including genes involved in the electron transfer chain, in semen samples from patients who had recovered from COVID-19 infection. Moreover, all protein-coding genes of the mitochondrial genome were significantly down-regulated in these samples.
The CABYR and FAM71D genes, which were previously shown to be correlated with sperm motility, showed decreased expression in postinfection samples in the current study. Moreover, the interleukin 10 pathway was up-regulated in samples obtained after COVID-19 infection. Kurkowska et al. (36) showed that elevated interleukin 10 was associated with reduced sperm motility in normal semen, which is in agreement with previous findings.
In 2 patients who had recovered from COVID-19 infection, semen analysis found completely normal results (normozoospermia), including normal sperm motility. Nevertheless, our results demonstrated statistically significantly decreased expression of mitochondrial genes in their semen samples. As in other cells, the main roles of the mitochondria in sperm cells include energy production, calcium uptake, and apoptosis initiation, while the generated energy is used for sperm motility, capacitation, and acrosome reaction (37, 38). Numerous studies have shown that the expression of mitochondrial genes is clearly correlated with important sperm parameters, including motility and sperm count (39). Moreover, mitochondrial dysfunction may be associated with oxidative stress, which causes reactive oxygen species–mediated sperm damage (40). Our findings indicate that even in the case of normal sperm parameters determined by routine semen analysis in patients who have recovered from COVID-19 infection, the expression of mitochondrial genes may be significantly decreased. This may be associated with sperm dysfunction and male fertility. Our results provide a molecular basis for the previously observed phenomenon of decreased sperm motility associated with COVID-19 infection. We believe that our findings will be beneficial for optimization of preconception care for men who have recently recovered from COVID-19 infection.
Limitations
At this point in time, to our knowledge, our study of gene expression in semen samples is the first and the largest conducted on patients with a history of COVID-19 infection. Nevertheless, further studies on larger groups of patients with and without a history of COVID-19 infection are needed to get more accurate results.
Patients who had recovered from mild COVID-19 infection and provided semen samples before infection and after proven recovery were included in this study. Eight patients had different sperm abnormalities before infection. Semen samples from these patients were stored before COVID-19 infection as they were preparing for IVF. Inclusion of these patients in the study allowed us to determine whether they needed additional treatment before IVF. Further studies on larger groups of patients with and without semen abnormalities before COVID-19 infection will provide more accurate results.
Semen samples from patients in the COVID-19 groups were collected within 75 days after proven recovery from infection, which represents the duration of spermatogenesis. Further studies including analysis of semen samples obtained at different times after recovery will provide additional data on the consistency of COVID-19–associated molecular changes in semen samples.
Footnotes
L.A. has nothing to disclose. V.E. has nothing to disclose. V.V. has nothing to disclose. A.S. has nothing to disclose. A.D. has nothing to disclose. D.D. has nothing to disclose. Y.A. has nothing to disclose. M.So. has nothing to disclose. M.Su. reports receiving a grant from World-Class Research Center “Digital Biodesign and Personalized Healthcare.” A.G. has nothing to disclose. A.B. reports receiving a grant from World-Class Research Center “Digital Biodesign and Personalized Healthcare.”
Supported by the Ministry of Science and Higher Education of the Russian Federation within the framework of state support for the creation and development of World-Class Research Center “Digital Biodesign and Personalized Healthcare” No. 075-15-2020-926 (to A.B. and M.Su.).
Data availability: The original sequencing data were deposited in the National Center for Biotechnology Information Sequencing Read Archive (https://www.ncbi.nlm.nih.gov/sra) with accession number PRJNA722736.
Supplementary data
Clustering and data quality assessment of experimental RNA sequencing profiles. (A) Dendrogram built using “complete” method and Euclidean distance in the space of log10-transformed normalized read counts for 19036 human protein-coding genes. (B) Principal Component Analysis of transcriptomic profiles. Samples obtained after COVID-19 are shown in red, before COVID-19 – in green, healthy controls are blue. P4 replicates are shown as triangles.
Supplemental Figure 2. Volcano plot showing distribution of observed differential genes by Log2fold change and log10p-value between Kr4 replicates before and after COVID-19 infection. Adjusted P-value was set 0.05, |Log2fold change| cut-off was set higher than 1.
Supplemental Figure 3. Significantly differential biological process terms for a set of up-regulated genes between P4 biosamples before and after COVID-19 infection. Terms are presented that passed Benjamini-Hochberg adjusted p-value threshold of 0.05.
Supplemental Figure 4. Significantly differential biological process terms for a set of down-regulated genes between P4 biosamples before and after COVID-19 infection. Terms are presented that passed Benjamini-Hochberg adjusted p-value threshold of 0.05.
Supplemental Figure 5. Top 10 upregulated and inhibited pathways sorted by PAL values for comparison of P4 samples before vs after COVID-19 infection. Pathways upregulated after COVID-19 infection are shown on the top, pathways downregulated after COVID-19 infection are shown on the bottom.
Supplemental Figure 6. Cumulative expression of mitochondrial genes in samples after COVID-19 infection vs FSH fold change (post infection / pre-infection FSH level). P-value = 0.067 according to Spearman test.
Supplemental Figure 7. Expression of 8 protein coding genes associated with sperm motility according to previous studies in samples from patients prior to COVID-19, after recovery from COVID-19, and from healthy donors.
Patients’ clinical characteristics.
Quality control and mapping statistics.
Differential gene expression and pathway analysis.
References
- 1.Brunner R.J., Demeter J.H., Sindhwani P. Review of guidelines for the evaluation and treatment of leukocytospermia in male infertility. World J Mens Health. 2019;37:128–137. doi: 10.5534/wjmh.180078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holtmann N., Edimiris P., Andree M., Doehmen C., Baston-Buest D., Adams O., et al. Assessment of SARS-CoV-2 in human semen—a cohort study. Fertil Steril. 2020;114:233–238. doi: 10.1016/j.fertnstert.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma L., Xie W., Li D., Shi L., Ye G., Mao Y., et al. Evaluation of sex-related hormones and semen characteristics in reproductive-aged male COVID-19 patients. J Med Virol. 2021;93:456–462. doi: 10.1002/jmv.26259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruan Y., Hu B., Liu Z., Liu K., Jiang H., Li H., et al. No detection of SARS-CoV-2 from urine, expressed prostatic secretions, and semen in 74 recovered COVID-19 male patients: a perspective and urogenital evaluation. Andrology. 2021;9:99–106. doi: 10.1111/andr.12939. [DOI] [PubMed] [Google Scholar]
- 5.Hajizadeh Maleki B., Tartibian B. COVID-19 and male reproductive function: a prospective, longitudinal cohort study. Reproduction. 2021;161:319–331. doi: 10.1530/REP-20-0382. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization. COVID-19 clinical management. Living guidance, 2021. WHO reference number: WHO/2019-nCoV/clinical/2021.1. Available at https://www.who.int/publications-detail-redirect/WHO-2019-nCoV-clinical-2021-1. Accessed January 25, 2021.
- 7.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:617059. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blighe K., Rana S., Lewis M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. 2018. https://github.com/kevinblighe/EnhancedVolcano Available at:
- 10.Yu G. Gene ontology semantic similarity analysis using GOSemSim. Methods Mol Biol. 2020;2117:207–215. doi: 10.1007/978-1-0716-0301-7_11. [DOI] [PubMed] [Google Scholar]
- 11.Sorokin M., Borisov N., Kuzmin D., Gudkov A., Zolotovskaia M., Garazha A., et al. Algorithmic annotation of functional roles for components of 3,044 human molecular pathways. Front Genet. 2021;12 doi: 10.3389/fgene.2021.617059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borisov N., Sorokin M., Garazha A., Buzdin A. Quantitation of molecular pathway activation using RNA sequencing data. Methods Mol Biol. 2020;2063:189–206. doi: 10.1007/978-1-0716-0138-9_15. [DOI] [PubMed] [Google Scholar]
- 13.Chen S., He C., Li Y., Li Z., Melançon C.E. A computational toolset for rapid identification of SARS-CoV-2, other viruses and microorganisms from sequencing data. Brief Bioinform. 2021;22:924–935. doi: 10.1093/bib/bbaa231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K., Mai Z., Zhou Y., Gao X., Yu B. Low NRF2 mRNA expression in spermatozoa from men with low sperm motility. Tohoku J Exp Med. 2012;228:259–266. doi: 10.1620/tjem.228.259. [DOI] [PubMed] [Google Scholar]
- 15.Pelloni M., Paoli D., Majoli M., Pallotti F., Carlini T., Lenzi A., et al. Molecular study of human sperm RNA: ropporin and CABYR in asthenozoospermia. J Endocrinol Invest. 2018;41:781–787. doi: 10.1007/s40618-017-0804-x. [DOI] [PubMed] [Google Scholar]
- 16.Ma Q., Li Y., Luo M., Guo H., Lin S., Chen J., et al. The expression characteristics of FAM71D and its association with sperm motility. Hum Reprod. 2017;32:2178–2187. doi: 10.1093/humrep/dex290. [DOI] [PubMed] [Google Scholar]
- 17.Yan Y., Yang X., Liu Y., Shen Y., Tu W., Dong Q., et al. Copy number variation of functional RBMY1 is associated with sperm motility: an azoospermia factor-linked candidate for asthenozoospermia. Hum Reprod. 2017;32:1521–1531. doi: 10.1093/humrep/dex100. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z., Yang Y., Wu H., Zhang H., Zhang H., Mao J., et al. Sodium-hydrogen-exchanger expression in human sperm and its relationship with semen parameters. J Assist Reprod Genet. 2017;34:795–801. doi: 10.1007/s10815-017-0898-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghandehari-Alavijeh R., Zohrabi D., Tavalaee M., Nasr-Esfahani M.H. Association between expression of TNF-α, P53 and HIF1α with asthenozoospermia. Hum Fertil (Camb) 2019;22:145–151. doi: 10.1080/14647273.2018.1493750. [DOI] [PubMed] [Google Scholar]
- 20.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Xu X. scRNA-seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS-CoV-2 infection in spermatogonia, Leydig and Sertoli cells. Cells. 2020;9:1–9. doi: 10.3390/cells9040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan F., Xiao X., Guo J., Song Y., Li H., Patel D.P., et al. No evidence of severe acute respiratory syndrome-coronavirus 2 in semen of males recovering from coronavirus disease 2019. Fertil Steril. 2020;113:1135–1139. doi: 10.1016/j.fertnstert.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang C., Ji X., Zhou W., Huang Z., Peng X., Fan L., et al. Coronavirus: a possible cause of reduced male fertility. Andrology. 2021;9:80–87. doi: 10.1111/andr.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morley J.E., Kaiser F.E., Perry H.M., III, Patrick P., Morley P.M., Stauber P.M., et al. Longitudinal changes in testosterone, luteinizing hormone, and follicle-stimulating hormone in healthy older men. Metabolism. 1997;46:410–413. doi: 10.1016/s0026-0495(97)90057-3. [DOI] [PubMed] [Google Scholar]
- 25.Babu S.R., Sadhnani M.D., Swarna M., Padmavathi P., Reddy P.P. Evaluation of FSH, LH and testosterone levels in different subgroups of infertile males. Indian J Clin Biochem. 2004;19:45–49. doi: 10.1007/BF02872388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Çayan S., Uğuz M., Saylam B., Akbay E. Effect of serum total testosterone and its relationship with other laboratory parameters on the prognosis of coronavirus disease 2019 (COVID-19) in SARS-CoV-2 infected male patients: a cohort study. Aging Male. 2020;23:1493–1503. doi: 10.1080/13685538.2020.1807930. [DOI] [PubMed] [Google Scholar]
- 27.Shang J., Du L., Han N., Lv D., Wang J., Yang H., et al. Severe acute respiratory syndrome coronavirus 2 for physicians: molecular characteristics and host immunity (review) Mol Med Rep. 2021;23:1–11. doi: 10.3892/mmr.2021.11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikka S.C., Champion H.C., Bivalacqua T.J., Estrada L.S., Wang R., Rajasekaran M., et al. Role of genitourinary inflammation in infertility: synergistic effect of lipopolysaccharide and interferon-gamma on human spermatozoa. Int J Androl. 2001;24:136–141. doi: 10.1046/j.1365-2605.2001.00279.x. [DOI] [PubMed] [Google Scholar]
- 29.Seshadri S., Bates M., Vince G., Jones D.I. Cytokine expression in the seminal plasma and its effects on fertilisation rates in an IVF cycle. Andrologia. 2011;43:378–386. doi: 10.1111/j.1439-0272.2010.01042.x. [DOI] [PubMed] [Google Scholar]
- 30.Havrylyuk A., Chopyak V., Boyko Y., Kril I., Kurpisz M. Cytokines in the blood and semen of infertile patients. Cent Eur J Immunol. 2015;40:337–344. doi: 10.5114/ceji.2015.54596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong C.K., Lam C.W., Wu A.K., Ip W.K., Lee N.L., Chan I.H., et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Li J., Zhan Y., Wu L., Yu X., Zhang W., et al. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect Immun. 2004;72:4410–4415. doi: 10.1128/IAI.72.8.4410-4415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrasquel G., Camejo M.I., Michelangeli F., Ruiz M.C. IFN-gamma alters the human sperm membrane permeability to Ca(2+) Syst Biol Reprod Med. 2014;60:21–27. doi: 10.3109/19396368.2013.833658. [DOI] [PubMed] [Google Scholar]
- 34.Sanocka D., Jedrzejczak P., Szumała-Kaekol A., Fraczek M., Kurpisz M. Male genital tract inflammation: the role of selected interleukins in regulation of pro-oxidant and antioxidant enzymatic substances in seminal plasma. J Androl. 2003;24:448–455. doi: 10.1002/j.1939-4640.2003.tb02693.x. [DOI] [PubMed] [Google Scholar]
- 35.Saeidi S., Shapouri F., Amirchaghmaghi E., Hoseinifar H., Sabbaghian M., Sadighi Gilani M.A., et al. Sperm protection in the male reproductive tract by toll-like receptors. Andrologia. 2014;46:784–790. doi: 10.1111/and.12149. [DOI] [PubMed] [Google Scholar]
- 36.Kurkowska W., Bogacz A., Janiszewska M., Gabryś E., Tiszler M., Bellanti F., et al. Oxidative stress is associated with reduced sperm motility in normal semen. Am J Mens Health. 2020;14:1–8. doi: 10.1177/1557988320939731. 1557988320939731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boguenet M., Bouet P.E., Spiers A., Reynier P., May-Panloup P. Mitochondria: their role in spermatozoa and in male infertility. Hum Reprod Update. 2021;27:697–719. doi: 10.1093/humupd/dmab001. [DOI] [PubMed] [Google Scholar]
- 38.Piomboni P., Focarelli R., Stendardi A., Ferramosca A., Zara V. The role of mitochondria in energy production for human sperm motility. Int J Androl. 2012;35:109–124. doi: 10.1111/j.1365-2605.2011.01218.x. [DOI] [PubMed] [Google Scholar]
- 39.Amaral A., Lourenço B., Marques M., Ramalho-Santos J. Mitochondria functionality and sperm quality. Reproduction. 2013;146:R163–R174. doi: 10.1530/REP-13-0178. [DOI] [PubMed] [Google Scholar]
- 40.Losano J.D.A., Angrimani D.S.R., Ferreira Leite R., Simões da Silva B.D.C., Barnabe V.H., Nichi M. Spermatic mitochondria: role in oxidative homeostasis, sperm function and possible tools for their assessment. Zygote. 2018;26:251–260. doi: 10.1017/S0967199418000242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clustering and data quality assessment of experimental RNA sequencing profiles. (A) Dendrogram built using “complete” method and Euclidean distance in the space of log10-transformed normalized read counts for 19036 human protein-coding genes. (B) Principal Component Analysis of transcriptomic profiles. Samples obtained after COVID-19 are shown in red, before COVID-19 – in green, healthy controls are blue. P4 replicates are shown as triangles.
Supplemental Figure 2. Volcano plot showing distribution of observed differential genes by Log2fold change and log10p-value between Kr4 replicates before and after COVID-19 infection. Adjusted P-value was set 0.05, |Log2fold change| cut-off was set higher than 1.
Supplemental Figure 3. Significantly differential biological process terms for a set of up-regulated genes between P4 biosamples before and after COVID-19 infection. Terms are presented that passed Benjamini-Hochberg adjusted p-value threshold of 0.05.
Supplemental Figure 4. Significantly differential biological process terms for a set of down-regulated genes between P4 biosamples before and after COVID-19 infection. Terms are presented that passed Benjamini-Hochberg adjusted p-value threshold of 0.05.
Supplemental Figure 5. Top 10 upregulated and inhibited pathways sorted by PAL values for comparison of P4 samples before vs after COVID-19 infection. Pathways upregulated after COVID-19 infection are shown on the top, pathways downregulated after COVID-19 infection are shown on the bottom.
Supplemental Figure 6. Cumulative expression of mitochondrial genes in samples after COVID-19 infection vs FSH fold change (post infection / pre-infection FSH level). P-value = 0.067 according to Spearman test.
Supplemental Figure 7. Expression of 8 protein coding genes associated with sperm motility according to previous studies in samples from patients prior to COVID-19, after recovery from COVID-19, and from healthy donors.
Patients’ clinical characteristics.
Quality control and mapping statistics.
Differential gene expression and pathway analysis.