Abstract
Background
Colposcopy is widely used to detect cervical cancer, but developing countries lack the experienced colposcopists necessary for accurate diagnosis. Artificial intelligence (AI) is being widely used in computer-aided diagnosis (CAD) systems. In this study, we developed and validated a CAD model based on deep learning to classify cervical lesions on colposcopy images.
Methods
Patient data, including clinical information, colposcopy images, and pathological results, were collected from Qilu Hospital. The study included 15,276 images from 7,530 patients. We performed two tasks in this study: normal cervix (NC) vs. low grade squamous intraepithelial lesion or worse (LSIL+) and high-grade squamous intraepithelial lesion (HSIL)− vs. HSIL+. The residual neural network (ResNet) probability was calculated for each patient to reflect the probability of lesions through a ResNet model. Next, a combination model was constructed by incorporating the ResNet probability and clinical features. We divided the dataset into a training set, validation set, and testing set at a ratio of 7:1:2. Finally, we randomly selected 300 patients from the testing set and compared the results with the diagnosis of a senior colposcopist and a junior colposcopist.
Results
The model that combines ResNet and clinical features performs better than ResNet alone. In the classification of NC and LSIL+, the area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were 0.953, 0.886, 0.932, 0.846, 0.838, and 0.936, respectively. In the classification of HSIL− and HSIL+, the AUC, accuracy, sensitivity, specificity, PPV, and NPV were 0.900, 0.807, 0.823, 0.800, 0.618, and 0.920, respectively. In the two classification tasks, the diagnostic performance of the model was determined to be comparable to that of the senior colposcopist and exhibited a stronger diagnostic performance than the junior colposcopist.
Conclusions
The CAD system for cervical lesion diagnosis based on deep learning performs well in the classification of cervical lesions and can provide an objective diagnostic basis for colposcopists.
Keywords: Computer-aided diagnosis (CAD), cervical lesion, colposcopy, ResNet
Introduction
Cervical cancer is the fourth female malignant tumor in incidence and mortality (1). In 2018, there were an estimated 570,000 new cases and 311,000 deaths worldwide (1). Human papillomavirus (HPV) infection is the main cause of cervical cancer (2). There are large differences in the incidence and mortality of cervical cancer worldwide. The incidence in low- and middle-income countries is 7–10 times higher than that in developed countries, partly due to the existence of large-scale screening and prevention services in various regions (1,3). Vaccination against HPV and secondary prevention, using screening tests to facilitate early detection and treatment of cervical intraepithelial neoplasia are effective strategies for cervical cancer prevention (4). According to the severity of cervical precancerous lesions, it can be divided into three types of cervical intraepithelial neoplasms (CIN I, CINII and CINIII). With the deepening of the understanding of HPV infection and cervical cancer, the three-level classification system could be replaced with a two-level classification system, applying the term CINI to low-grade squamous intraepithelial lesions (LSIL) and the term CINII, III to high-grade squamous intraepithelial lesions (HSIL) (5). To date, addressing barriers to HPV vaccination and cervical screening remains a major challenge for most countries, particularly low- and middle-income countries (LMICs) (6). The combination of cytology and HPV testing has become an option for screening programs (7). However, cervical cancer screening is still a problem with low diagnostic sensitivity and specificity, especially in LMICs. Some people are undertreated, and some people are overtreated (3,7). Colposcopy is used to evaluate abnormal or uncertain cervical cancer screening tests. Colposcopy helps to identify precancerous lesions and cervical cancer that can be treated (8). Some underdeveloped areas support screening of the cervix by visual inspection after application of acetic acid to highlight precancerous or cancerous abnormalities (5).Although colposcopy plays an important role in preventing cervical cancer, its accuracy and repeatability are limited. The accuracy problem is mainly caused by the inconsistency between the visible changes of cervical epithelium and the severity of precancerous lesions (9). The diagnostic performance of colposcopy largely depends on the subjective experience of the operator, which requires the operator to be able to identify and deal with changes in the acetic acid white epithelium according to standards (10). In LMICs, the lack of experienced colposcopists and the heavy workload of colposcopists exacerbate the inaccuracy of colposcopy diagnosis (10).
With the advancement of artificial intelligence (AI) technology, computer aided diagnosis (CAD) has become one of the leading research topics of medical imaging in recent decades. CAD has shown great potential in diagnosing malignant tumors such as breast cancer, colorectal cancer, and gastrointestinal tumors (11-14). In the meantime, there is an urgent need for computer-aided detection and classification of images acquired with colposcopy to reduce the burden of colposcopists. Some impressive results have been achieved. Early work usually used feature extraction methods to extract discriminative features from colposcopy images and then machine learning methods to classify the images (15-18). In recent years, deep learning methods, especially deep convolutional neural networks (CNNs), have shown greater advantages over traditional machine learning methods and have achieved remarkable results in the development of various types of CAD systems (19). Miyagi et al. (20) built a CNN with 11 layers by using 310 images. This CNN showed a high accuracy of 82.3%, sensitivity of 80%, and specificity of 88.2% for the classification of LSIL and HSIL+. Zhang et al. (21) proposed a CAD method for automatic classification of HSIL or higher-level lesions in colposcopic images based on transfer learning and pretrained densely connected CNN. This method achieved an accuracy of 73.08% over 600 test images. Cho et al. (22) applied deep learning methods to automatically classify cervical neoplasms on colposcopic photographs based on pretrained CNN and achieved an AUC of 0.781 for the system. Li et al. (23) proposed a deep learning-based CAD system for LSIL+ identification. They collected colposcopic images captured at different times during an acetic acid test and proposed a novel graph convolutional network to fuse the features extracted from these time-lapsed colposcopic images. They achieved a classification accuracy of 78.33%, which was comparable to that of experienced colposcopists.
Inspired by the above works, we proposed a deep learning-based CAD system to classify colposcopic images. We collected data from more than 7,000 patients, which is more than most existing literature. In addition, we collected detailed clinical features and combined these features with deep learning to build models. To verify the performance of our proposed CAD system, we compared the CAD with the diagnosis results of different levels of colposcopists.
We present the following article in accordance with the TRIPOD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-885).
Methods
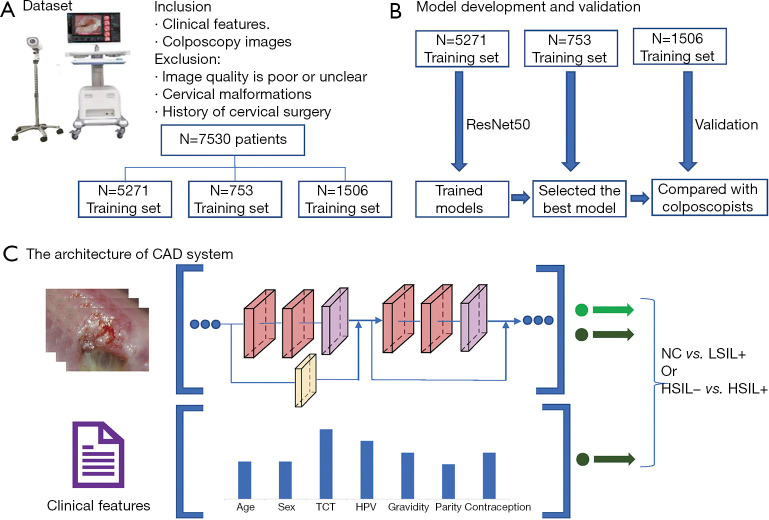
In this section, we introduce the main methods involved in the proposed CAD system. Figure 1 shows a flowchart of the study. Using certain inclusion and exclusion criteria, we collected a dataset containing 7,530 patients. Two models were constructed to distinguish NC vs. LSIL+ (LSIL, HSIL, and cancer), and HSIL− (Normal and LSIL) vs. HSIL+ (HSIL and cancer), respectively.
Figure 1.
Flowchart of the study. (A) Dataset; (B) model development and validation; (C) the architecture of CAD system. CAD, computer aided diagnosis.
Patients
This retrospective study collected information and colposcopy images of patients who underwent colposcopies at Qilu Hospital of Shandong University from May 2018 to August 2020. All participants had clinical information and image information. The retrospective study was performed according to the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethical Committee of Qilu Hospital in Jinan, Shandong Province, China (No. 2019095) and individual consent for this retrospective analysis was waived. All images in the study were taken with a Leisegang 3ML LED colposcopy camera (Leisegang, Germany) with a resolution of 480×320 pixels and stored in JPEG format. Patients who met the following criteria were excluded: (I) poor or unclear image quality of the colposcopy, (II) lack of cytology and HPV results, (III) presence of cervical malformations (such as double cervix), (IV) presence of cervical polyps or cervical benign neoplasms (such as uterine fibroids, inflammatory fibrous hyperplasia), (V) difficult cervical exposure, (VI) cervical surgery, (VII) visual LSIL or HSIL but no pathological results. Except for patients whose colposcopists believe that the colposcopy is normal and do not need to take a biopsy, the remaining patients have pathology reports. The dataset contains 7,530 patients (ages from 15 to 85 years old), which can be classified as normal (n=3,966), LSIL (n=1,411), HSIL (n=1,966) and cancer (n=187). A total of 15,276 colposcopy images from 7,530 patients were included in this study, including normal/benign (n=7,433), LSIL (n=2,916), HSIL (n=4,458), and cancer (n=469) images. We randomly divided the dataset into a training set, validation set, and testing set at a ratio of 7:1:2. Table 1 shows the detailed dataset used in this study.
Table 1. The detailed dataset used in this study.
| Category | Training set | Validation set | Testing set | |||||
|---|---|---|---|---|---|---|---|---|
| Patients | Images | Patients | Images | Patients | Images | |||
| NC | 2,760 | 5,144 | 394 | 744 | 812 | 1,545 | ||
| LSIL | 998 | 2,058 | 143 | 290 | 270 | 568 | ||
| HSIL | 1,378 | 3,115 | 204 | 463 | 384 | 880 | ||
| Cancer | 135 | 333 | 12 | 34 | 40 | 102 | ||
| Total | 5,271 | 10,650 | 753 | 1,531 | 1,506 | 3,095 | ||
NC, normal cervix; LSIL, low-grade squamous intraepithelial lesions; HSIL, high-grade squamous intraepithelial lesions.
Preprocessing
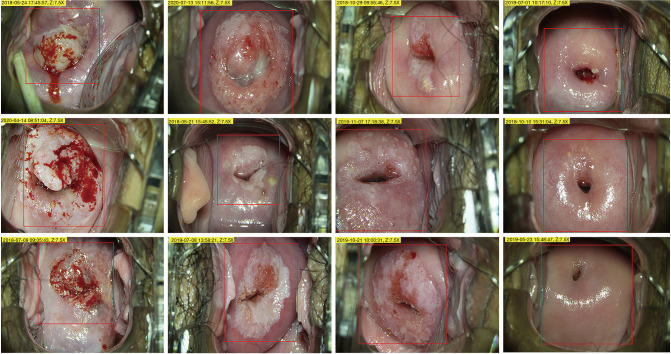
Since the original colposcopy image has high resolution and contains a large amount of irrelevant content, sending it directly to the computer will cause too much calculation and reduce the model's performance. Therefore, we asked a junior colposcopist to outline the region of interest (ROI), which is the suspicious lesion area. Figure 2 shows the ROI contours of different grades. Second, data augmentation methods are used to increase the volume of training data, including flip/mirror input images horizontally and vertically with a probability of 0.5, rotation by −10 to +10 degrees, and shear by −4 to +4 degrees. Data augmentation can generate more training samples to enhance the robustness of the model and reduce overfitting. Finally, we resized all images to 224×224 pixels to fit the input size of the CNN.
Figure 2.
ROI contours of different grades outlined by colposcopist. Red contours denote the ROIs. From left to right: cancer, HSIL, LSIL, and NC. ROI, region of interest; HSIL, high-grade squamous intraepithelial lesions; LSIL, low-grade squamous intraepithelial lesions; NC, normal cervix.
CNN architecture
We selected ResNet (24) as the main structure of our CAD system. Compared to plane CNN architectures, ResNet can effectively avoid the problems of vanishing gradients and exploding gradients by introducing residual blocks. Specifically, skip connections are added between some layers, which is the core idea of residual blocks. With skip connections, the information of the convolutional layer output and the input is fused; thus, the gradient can flow through this shortcut path to solve the vanishing gradient problem. In addition, these connections can act as identity functions to ensure that the higher layer to performs at least as well as the lower layer and not worse. We applied ResNet50 to construct our CAD system, which contains 50 convolutional layers.
ResNet models were performed on the workstation of an Ubuntu20.4 64-bit operating system with 256 GB memory and an NVIDIA GeForce Titan RTX GPU. The input size was 224×224. We used a batch size of 100 and trained 300 epochs. The optimizer used was stochastic gradient descent (SGD) with an initial learning rate of 0.1 and a momentum of 0.9. After every 50 epochs of training, the learning rate was reduced by 50%. After each training epoch, we calculated the performance of the model on the validation set. Finally, we choose the model with the highest accuracy on the validation set as the final model to evaluate the model in the test set.
Interpretability of deep learning model
Among various deep learning based applications related to medical imaging, it is essential to investigate the interpretability of the deep learning model. One of the most prominent methods to provide an interpretable view of a deep learning model is gradient weighted class Activation mapping (Grad-CAM) (25). Grad-CAM can effectively visualize the region of interest, which can produce a coarse heat map that highlights the contribution to the classification task.
Development and validation of combination model
Multivariable analysis was used to combine the ResNet50 probability with clinical features through a multivariable logistic regression model. The clinical features involved age, cytology, HPV, gravidity, parity, and contraception. The output probability of ResNet50 was combined with these clinical features, and then a logistic regression model was constructed to output the final probability.
Comparison with colposcopists
To verify whether our CAD system can improve the level of colposcopists who do not have extensive experience, we randomly selected 300 samples from the testing set and asked a junior colposcopist to make a diagnosis. These diagnoses of the CAD system, the junior colposcopist, and the senior colposcopist were compared.
Performance metrics and statistical analysis
We used the measures of accuracy, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) to evaluate the binary predictors. We also plotted the receiver operating characteristic (ROC) curve to assess the performance of the CAD system over a range of possible cutpoints. Then, the AUCs were evaluated. In addition, the diagnostic performance was demonstrated using confusion matrix, which records the correct and incorrect predictions on each class in the form of a matrix.
Statistical analysis was performed using R language version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria) or MedCalc Statistical Software version 19.4.1 (MedCalc Software Ltd, Ostend, Belgium). Continuous variables were compared using unpaired t-test or Wilcoxon rank-sum test, while categorical variables were compared using Chi-squared test, as appropriate. The Delong test was used to compare different AUCs. A P value <0.05 was considered statistically significant.
Results
Patient demographics
In the classification of NC vs. LSIL+, there were 2,760 NCsamples and 2,511 LSIL+ samples in the training set. There were 812 NC samples and 694 LSIL+ samples in the test set. The clinical information of the patient is displayed in Table 2. In the classification of HSIL− vs. HSIL+, there were 3,758 HSIL− samples and 1,513 HSIL+ samples in the training set. There were 1,082 HSIL− samples and 424 HSIL+ samples in the test set. The clinical information of the patient is displayed in Table 3. There was no statistically significant difference between the training set and testing set. Since only cytology and HPV have significant differences in both classifications, we selected these two features as the final clinical features for modeling.
Table 2. Patients and clinical feature of NC vs. LSIL+.
| Clinical features | Training set | Testing set | |||||
|---|---|---|---|---|---|---|---|
| NC (n=2,760) | LSIL+ (n=2,511) | P | NC (n=812) | LSIL+ (n=694) | P | ||
| Age | *** | 0.0008 | |||||
| Mean | 41.2 | 39.5 | 41.2 | 39.5 | |||
| Range | 16–75 | 15–79 | 18–77 | 18–79 | |||
| Cytology | *** | *** | |||||
| Negative | 2,411 | 1,375 | 697 | 383 | |||
| Positive | 349 | 1,136 | 115 | 311 | |||
| HPV | *** | *** | |||||
| Negative | 321 | 60 | 91 | 13 | |||
| Positive | 2,439 | 2,451 | 721 | 681 | |||
| Gravidity | 0.008 | 0.350 | |||||
| ≤2 | 1,304 | 1,094 | 373 | 302 | |||
| >2 | 1,456 | 1,417 | 439 | 392 | |||
| Parity | 0.799 | 0.818 | |||||
| ≤2 | 2,626 | 2,385 | 770 | 656 | |||
| >2 | 134 | 126 | 42 | 38 | |||
| Contraception | 0.539 | 0.228 | |||||
| Condom | 906 | 845 | 259 | 242 | |||
| No condom | 1,854 | 1,666 | 553 | 452 | |||
***, P<0.001. NC, normal cervix; LSIL, low-grade squamous intraepithelial lesions; HPV, human papillomavirus.
Table 3. Patients and clinical feature of HSIL− vs. HSIL+.
| Clinical features | Training set | Testing set | |||||
|---|---|---|---|---|---|---|---|
| HSIL− (n=3,758) | HSIL+ (n=1,513) | P | HSIL− (n=1,082) | HSIL+ (n=424) | P | ||
| Age | 0.014 | 0.331 | |||||
| Mean | 40.6 | 39.9 | 40.2 | 40.8 | |||
| Range | 16–75 | 15–79 | 18–77 | 21–79 | |||
| Cytology | *** | *** | |||||
| Negative | 3,079 | 707 | 878 | 202 | |||
| Positive | 679 | 806 | 204 | 222 | |||
| HPV | *** | *** | |||||
| Negative | 357 | 24 | 101 | 3 | |||
| Positive | 3,401 | 1,489 | 981 | 421 | |||
| Gravidity | 0.001 | 0.038 | |||||
| ≤2 | 1,764 | 634 | 503 | 172 | |||
| >2 | 1,994 | 879 | 579 | 252 | |||
| Parity | 0.362 | 0.162 | |||||
| ≤2 | 3,579 | 1,432 | 1,030 | 396 | |||
| >2 | 179 | 81 | 52 | 28 | |||
| Contraception | 0.383 | 0.504 | |||||
| Condom | 1,262 | 489 | 366 | 135 | |||
| No condom | 2,496 | 1,024 | 716 | 289 | |||
***, P<0.001. HSIL, high-grade squamous intraepithelial lesions; HPV, human papillomavirus.
Prediction of patients between NC and LSIL+
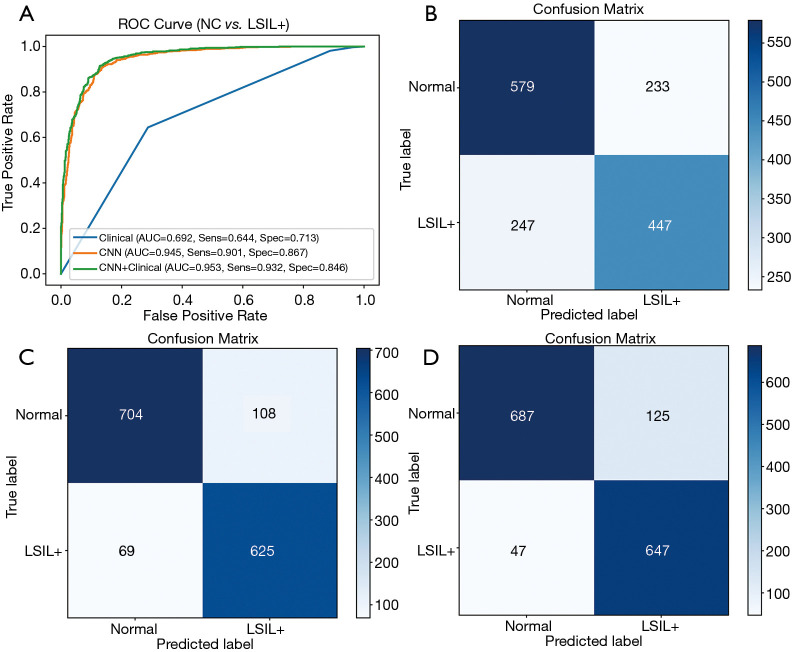
In the classification of NC vs. LSIL+, the evaluation metrics of the testing set are listed in Table 4. The AUC, accuracy, sensitivity, specificity, PPV and NPV were 0.692, 0.681, 0.644, 0.713, 0.657, and 0.710, respectively, for the model based on clinical features only. When modeled with ResNet50, the AUC, accuracy, sensitivity, specificity, PPV and NPV were 0.945, 0.882, 0.901, 0.867, 0.853 and 0.910, respectively. The AUC of ResNet50 was significantly higher than the AUC of the clinical based model (P<0.001). When using clinical factors and ResNet50 for joint modeling, the AUC, accuracy, sensitivity, specificity, PPV and NPV were 0.953, 0.886, 0.932, 0.846, 0.838, and 0.936, respectively. Compared with ResNet50, the joint model had a higher AUC value (P<0.001). The ROCs and confusion matrixes are shown in Figure 3. The confusion matrix is a 2-dimensional array comparing predicted category labels to the true label. The columns represent the predicted category labels, and the rows represent the true labels, which can provide an insight into the errors being made by the classifier. For the joint model, the true positive (TP) is 647, which is higher than the 625 of the ResNet model and the 447 of the clinical model, but the true negative (TN) is slightly lower than that of the ResNet.
Table 4. The classification results of NC vs. LSIL+.
| Model | AUC | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| Clinical only | 0.692 (0.668–0.715) | 0.681 (0.657–0.705) | 0.644 (0.607–0.680) | 0.713 (0.681–0.744) | 0.657 (0.620–0.693) | 0.710 (0.668–0.732) |
| CNN only | 0.945 (0.933–0.956) | 0.882 (0.865–0.898) | 0.901 (0.876–0.922) | 0.867 (0.841–0.890) | 0.853 (0.825–0.878) | 0.910 (0.888–0.930) |
| CNN + clinical | 0.953 (0.941–0.963) | 0.886 (0.869–0.901) | 0.932 (0.911–0.950) | 0.846 (0.819–0.870) | 0.838 (0.810–0.863) | 0.936 (0.916– 0.953) |
Data were presented with 95% CIs. NC, normal cervix; LSIL, low-grade squamous intraepithelial lesions; AUC, receiver operating characteristic curve; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval; CNN, convolutional neural network.
Figure 3.
Classification results of NC vs. LSIL+. (A) ROC curves; (B) confusion matrix of the model based on clinical features; (C) confusion matrix of the model based on ResNet50; (D) confusion matrix of the model based on the combination of ResNet50 and clinical features. NC, normal cervix; LSIL, low-grade squamous intraepithelial lesions; ROC, receiver operating characteristic; ResNet, residual neural network.
Prediction of patients between HSIL− and HSIL+
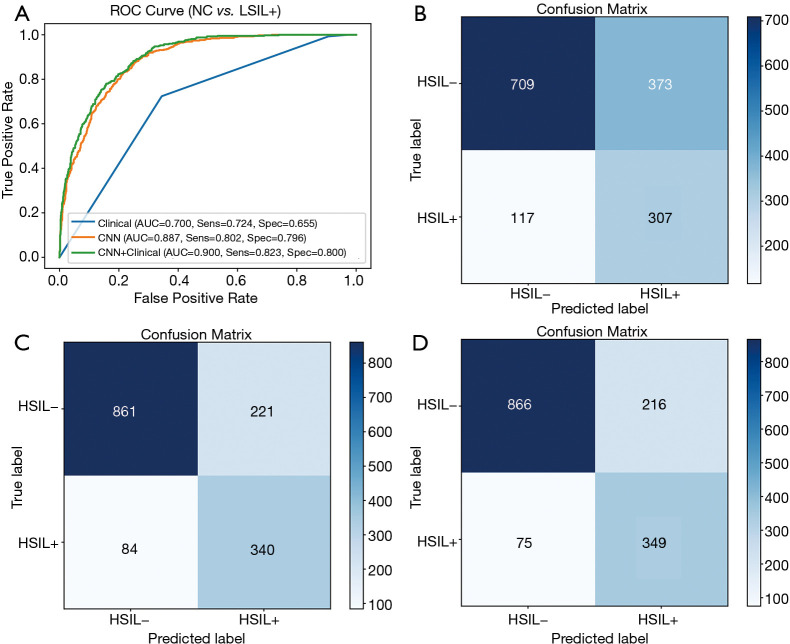
In the classification of HSIL− vs. HSIL+, the evaluation metrics of the testing set are listed in Table 5. In the testing set, the AUC, accuracy, sensitivity, specificity, PPV and NPV were 0.700, 0.675, 0.724, 0.655, 0.451, and 0.858, respectively, of the model based on clinical features only. When modeled with ResNet50, the AUC, accuracy, sensitivity, specificity, PPV and NPV were 0.887, 0.797, 0.802, 0.796, 0.606 and 0.911, respectively. The AUC of ResNet50 was significantly higher than the AUC of the clinical based model (P<0.001). When using clinical factors and ResNet50 for joint modeling, the AUC, accuracy, sensitivity, specificity, NPV and PPV were 0.900, 0.807, 0.823, 0.800, 0.618, and 0.920, respectively. Compared with ResNet50, the joint model had better performance and a higher AUC value (P<0.001). The ROCs and confusion matrixes are shown in Figure 4. For the joint model, the TP is 349 and the TN is 866, which are both higher than that of the clinical and ResNet models.
Table 5. The classification results of HSIL− vs. HSIL+.
| Model | AUC | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| Clinical only | 0.700 (0.676–0.723) | 0.675 (0.650–0.698) | 0.724 (0.679–0.766) | 0.655 (0.626–0.684) | 0.451 (0.414–0.490) | 0.858 (0.833–0.881) |
| CNN only | 0.887 (0.870–0.903) | 0.797 (0.776–0.818) | 0.802 (0.761–0.839) | 0.796 (0.770–0.819) | 0.606 (0.564–0.647) | 0.911 (0.891–0.928) |
| CNN + clinical | 0.900 (0.884–0.915) | 0.807 (0.786–0.826) | 0.823 (0.783–0.858) | 0.800 (0.775–0.823) | 0.618 (0.576–0.658) | 0.920 (0.901–0.937) |
Data were presented with 95% CIs. HSIL, high-grade squamous intraepithelial lesions; AUC, receiver operating characteristic curve; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval; CNN, convolutional neural network.
Figure 4.
Classification results of HSIL− vs. HSIL+. (A) ROC curves; (B) confusion matrix of the model based on clinical features; (C) confusion matrix of the model based on ResNet50; (D) confusion matrix of the model based on the combination of ResNet50 and clinical features. HSIL, high-grade squamous intraepithelial lesions; ROC, receiver operating characteristic; ResNet, residual neural network.
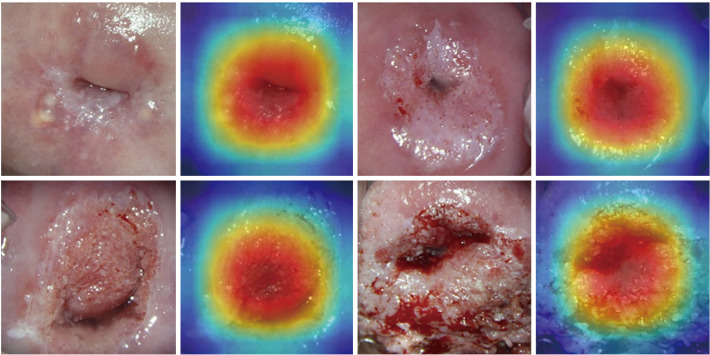
Representative Grad-CAM samples of different grades of colposcopy images are shown in Figure 5. Deep learning locates the most core lesion area, which proves the effectiveness of our proposed model.
Figure 5.
Grad-CAM maps. Each example shows the ROI and corresponding CAD map, and the red region represents a larger contribution for classification. Grad-CAM, gradient weighted class Activation mapping; ROI, region of interest; CAD, computer aided diagnosis.
Comparison with colposcopists
To evaluate the performance of our CAD, we randomly selected 300 samples from the testing set and asked a junior colposcopist and a senior colposcopist to make diagnoses. The related diagnosis results are listed in Table 6. For the diagnosis of NC and LSIL+, the performance of the CAD system is comparable to that of the senior colposcopist, and it can significantly improve the diagnostic level of the junior colposcopist. For the diagnosis of HSIL− and LSIL+, although the accuracy, specificity, and PPV of our CAD system are lower than those of the senior colposcopist, the sensitivity and NPV are much higher than those of the senior colposcopist, which we think is a great advantage in clinical applications. It is worth noting that the ROIs were outlined by a junior colposcopist. Therefore, with the help of our CAD system, the diagnostic performance of the junior colposcopist can be significantly improved.
Table 6. Comparison with colposcopists.
| Task | Model | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| NC vs. LSIL+ | CAD | 0.883 (0.841–0.917) | 0.954 (0.904–0.983) | 0.827 (0.762–0.881) | 0.812 (0.742–0.871) | 0.959 (0.912–0.985) |
| Senior | 0.857 (0.812–0.894) | 0.924 (0.865–0.963) | 0.803 (0.735–0.861) | 0.787 (0.714–0.849) | 0.931 (0.877–0.966) | |
| Junior | 0.750 (0.697–0.798) | 0.886 (0.819–0.935) | 0.642 (0.565–0.715) | 0.661 (0.586–0.730) | 0.878 (0.807–0.930) | |
| HSIL– vs. HSIL+ | CAD | 0.810 (0.761–0.853) | 0.828 (0.732–0.900) | 0.803 (0.743–0.854) | 0.632 (0.536–0.720) | 0.919 (0.870–0.954) |
| Senior | 0.833 (0.786–0.874) | 0.448 (0.341–0.559) | 0.991 (0.966–0.999) | 0.951 (0.835–0.994) | 0.814 (0.762–0.860) | |
| Junior | 0.757 (0.704–0.804) | 0.195 (0.118– 0.294) | 0.986 (0.959–0.997) | 0.850 (0.621–0.968) | 0.750 (0.695–0.799) |
Data were presented with 95% CIs. CAD, computer aided diagnosis; NC, normal cervix; LSIL, low-grade squamous intraepithelial lesions; HSIL, high-grade squamous intraepithelial lesions; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval.
Discussion
HPV screening, cytology, and colposcopy are the three strategies recommended by the WHO for cervical cancer screening (5). Although cytology has greatly reduced the incidence of cervical cancer, we must recognize the limitations of cytology in cervical cancer screening. The repeatability of cytology is poor, the sensitivity is between 43% and 96%, and the development of cytology requires a large number of cytopathologists (26). HPV has become a promising screening method due to its high negative predictive value and sensitivity. However, the false positive rate of HPV screening increases patients' subsequent unnecessary treatment (10,27). However, combined cytology and HPV screening is still the most recommended screening method at present. Obtaining a biopsy through colposcopy followed by histological analysis is the gold standard for detecting cervical disease. Although colposcopy is very successful in high-income countries, its application requires good organization, expensive equipment, and well-trained personnel, including colposcopists and pathologists (8).In view of the high incidence of cervical cancer and the importance of timely diagnosis and treatment, scholars are very interested in developing accurate and cost-effective screening and diagnosis methods. In our study, cytology and HPV are useful clinical factors. Therefore, we compared clinical features (cytology and HPV), ResNet, and the combination of clinical feature results with ResNet and finally evaluated their performance.
In our study, the accuracy of using clinical feature (cytology and HPV) data to identify LSIL+ and HSIL+ was low. This is mainly because the combined screening of cytology and HPV increases the false positive rate of diagnosis. Atypical squamous cells of unknown significance or higher (ASCUS+) or HPV positive referral colposcopy increased the workload of colposcopy, especially HPV positivity. A considerable proportion of HPV infections are transient infections. HPV combined with cytology screening reduces the false positive rate to a certain extent. However, in clinical applications, to reduce the missed diagnosis rate, the problem of a high false positive rate still exists. ResNet has shown good performance in our study. The combination of clinical feature results with ResNet performed better than ResNet alone. This result shows that the significance of colposcopy images in diagnosing cervical lesions is much greater than that of combined cytology and HPV screening. Our CAD model is more suitable for underdeveloped countries that cannot achieve combined cytology and HPV screening.
LSIL is considered to be a transient expression of HPV infection. Most of the LSIL will regress spontaneously (26).HSIL may develop into cervical cancer and requires further treatment (5). Based on this, we carried out two classification modes, NC vs. LSIL+ and HSIL− vs. HSIL+. We developed a CAD system based on deep learning for colposcopy images classification. Although this is not the first study on AI in the field of colposcopy, the performance of the diagnosis system we developed is better than previous studies. First, we asked a junior colposcopist to mark the cervical transformation area and the suspicious cervical lesion area, and used ResNet to diagnose the severity of the lesion. The accuracy, sensitivity, and specificity of ResNet are all approximately 0.8. Then, we added cytology and HPV to ResNet to build a joint model, and achieved better performance than ResNet alone.
Acetate whitening, vascular changes, lesion location, and lesion size are the most important features that distinguish LSIL from HSIL (8). However, these tiny features are not enough for the system to recognize. Therefore, as far as our system is concerned, it is still necessary to repeatedly correct and train to enhance the performance of the system. Nevertheless, the performance of ResNet in diagnosing LSIL+ and HSIL+ is still higher than that of colposcopists. We randomly selected 300 patients from the testing set and compared the results with the diagnosis of a senior colposcopist and a junior colposcopist. The diagnostic performance of the model is comparable to that of the senior colposcopist, and has a greater performance than the diagnostic level of the junior colposcopist. It is true that our proposed model can improve the diagnostic ability of colposcopists, enhance the judgment ability of colposcopists, and finally compensate for the lack of junior colposcopists.
Despite the encouraging performance of CAD in colposcopy imaging, there are still some challenges and obstacles that need to be resolved. For patients with cervical canal lesions and Type 3 transformation zone, the colposcopy doctor still needs to combine the patient's personal medical history to conduct a comprehensive assessment of the patient and make a final diagnosis. We analyzed patients ≥50 years of age, and the CAD system we developed had a lower sensitivity in both classifications (Tables S1 and S2). This is mainly because the group that age ≥50 years old has a higher proportion of type 3 transformation zone (62.7% vs. 19.1%), and is more likely to develop cervical canal lesions. Besides, CAD is applicable to the examination of a large range of people. In our study, after a junior colposcopist marked the cervix after acetic acid, the CAD model was applied to evaluate cervical lesions. The main purpose of this CAD is still to help colposcopists improve their diagnostic capabilities, not to replace them. The diagnosis result of CAD is regarded as a “second set of eyes” of human colposcopists, and human colposcopists are responsible for the final diagnosis result.
According to a recent report that made by the Australian Institute of Health and Welfare, the PPV of diagnose HSIL+ through colposcopies is 0.578 (28). The PPV of our CAD system is 0.618, which proves its potential for large-scale application. However, the relatively low PPV may have the problem of overtreatment. Positive patients diagnosed with CAD needed to be referred to colposcopists to finally determine whether a biopsy is needed. This is also the reason why CAD can only help colposcopists reduce work pressure and improve accuracy but cannot completely replace them. If we need to increase PPV to reduce overtreatment, we need higher quality colposcopy images and more advanced algorithms.
There are several limitations to this study. First, we only collected samples from a single medical center, which might introduce bias due to the lack of different types of colposcopy equipment. We will collect samples from different centers to enhance the diversity of samples. In addition, we will also establish a colposcope cloud platform based on our model to provide medical assistance to areas with resource shortages and narrow the diagnosis and treatment gap. Second, to obtain clear images, we excluded patients with cervical polyps, cervical benign neoplasms, cervical abnormalities, and cervical incomplete exposure, which will limit the application of the CAD model in actual clinical practice. Third, this is a retrospective study. We need to carry out prospective studies to validate the performance of our model. Fourth, the clinical features we included in the study were insufficient, and smoking history, age at first sex, and number of sexual partners should also be taken into consideration.
Conclusions
In this research, we developed a deep learning-based CAD system that combined colposcopy images and clinical features for colposcopy image classification. The proposed CAD system shows performance comparable to that of the senior colposcopist in colposcopy image classification. The CAD system can provide an objective diagnostic basis for colposcopists and has potential clinical application value. In the future, we will collect multicenter data and conduct more extensive research to apply this CAD model to clinical practice.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the Natural Science Foundation of China (U1806202), the Natural Science Foundation of Shandong Province of China (ZR2020ZD25), the Key Research Project of Shandong Province (2017CXGC1210) and Weifang Health Fund (2018-053).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The retrospective study was performed according to the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethical Committee of Qilu Hospital in Jinan, Shandong Province, China (No. 2019095) and individual consent for this retrospective analysis was waived.
Footnotes
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-885
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-885
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-885). The authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2002;2:342-50. 10.1038/nrc798 [DOI] [PubMed] [Google Scholar]
- 3.Jeronimo J, Castle PE, Temin S, et al. Secondary Prevention of Cervical Cancer: American Society of Clinical Oncology Resource-Stratified Clinical Practice Guideline Summary. J Oncol Pract 2017;13:129-33. 10.1200/JOP.2016.017889 [DOI] [PubMed] [Google Scholar]
- 4.Canfell K. Towards the global elimination of cervical cancer. Papillomavirus Res 2019;8:100170. 10.1016/j.pvr.2019.100170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Guidelines Approved by the Guidelines Review Committee. WHO Guidelines for Screening and Treatment of Precancerous Lesions for Cervical Cancer Prevention. Geneva: World Health Organization 2013.; 2013. [PubMed] [Google Scholar]
- 6.Ogilvie G, Nakisige C, Huh WK, et al. Optimizing secondary prevention of cervical cancer: Recent advances and future challenges. Int J Gynaecol Obstet 2017;138:15-9. 10.1002/ijgo.12187 [DOI] [PubMed] [Google Scholar]
- 7.Saslow D, Solomon D, Lawson HW, et al. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. Am J Clin Pathol 2012;137:516-42. 10.1309/AJCPTGD94EVRSJCG [DOI] [PubMed] [Google Scholar]
- 8.Khan MJ, Werner CL, Darragh TM, et al. ASCCP Colposcopy Standards: Role of Colposcopy, Benefits, Potential Harms, and Terminology for Colposcopic Practice. J Low Genit Tract Dis 2017;21:223-9. 10.1097/LGT.0000000000000338 [DOI] [PubMed] [Google Scholar]
- 9.Reid R, Scalzi P. Genital warts and cervical cancer. VII. An improved colposcopic index for differentiating benign papillomaviral infections from high-grade cervical intraepithelial neoplasia. Am J Obstet Gynecol 1985;153:611-8. 10.1016/S0002-9378(85)80244-1 [DOI] [PubMed] [Google Scholar]
- 10.Xue P, Ng MTA, Qiao Y. The challenges of colposcopy for cervical cancer screening in LMICs and solutions by artificial intelligence. BMC Med 2020;18:169. 10.1186/s12916-020-01613-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng X, Yao Z, Huang Y, et al. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat Commun 2020;11:1236. 10.1038/s41467-020-15027-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou D, Tian F, Tian X, et al. Diagnostic evaluation of a deep learning model for optical diagnosis of colorectal cancer. Nat Commun 2020;11:2961. 10.1038/s41467-020-16777-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo H, Xu G, Li C, et al. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol 2019;20:1645-54. 10.1016/S1470-2045(19)30637-0 [DOI] [PubMed] [Google Scholar]
- 14.Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph 2007;31:198-211. 10.1016/j.compmedimag.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivasan Y, Corona E, Nutter B, et al. A Unified Model-Based Image Analysis Framework for Automated Detection of Precancerous Lesions in Digitized Uterine Cervix Images. IEEE J Sel Top Signal Process 2009;3:101-11. 10.1109/JSTSP.2008.2011102 [DOI] [Google Scholar]
- 16.Kim E, Huang X. A Data Driven Approach to Cervigram Image Analysis and Classification. In: Celebi ME, Schaefer G, editors. Color Medical Image Analysis. Dordrecht: Springer Netherlands, 2013:1-13. [Google Scholar]
- 17.Xu T, Kim E, Huang X, editors. Adjustable adaboost classifier and pyramid features for image-based cervical cancer diagnosis. 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI); 2015 16-19 April 2015. [Google Scholar]
- 18.Asiedu MN, Simhal A, Chaudhary U, et al. Development of Algorithms for Automated Detection of Cervical Pre-Cancers With a Low-Cost, Point-of-Care, Pocket Colposcope. IEEE Trans Biomed Eng 2019;66:2306-18. 10.1109/TBME.2018.2887208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan HP, Hadjiiski LM, Samala RK. Computer-aided diagnosis in the era of deep learning. Med Phys 2020;47:e218-27. 10.1002/mp.13764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyagi Y, Takehara K, Miyake T. Application of deep learning to the classification of uterine cervical squamous epithelial lesion from colposcopy images. Mol Clin Oncol 2019;11:583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T, Luo Y-m, Li P, et al. Cervical precancerous lesions classification using pre-trained densely connected convolutional networks with colposcopy images. Biomed Signal Process Control 2020;55:101566. 10.1016/j.bspc.2019.101566 [DOI] [Google Scholar]
- 22.Cho BJ, Choi YJ, Lee MJ, et al. Classification of cervical neoplasms on colposcopic photography using deep learning. Sci Rep 2020;10:13652. 10.1038/s41598-020-70490-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Chen J, Xue P, et al. Computer-Aided Cervical Cancer Diagnosis Using Time-Lapsed Colposcopic Images. IEEE Trans Med Imaging 2020;39:3403-15. 10.1109/TMI.2020.2994778 [DOI] [PubMed] [Google Scholar]
- 24.He K, Zhang X, Ren S, et al. editors. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 27-30 June 2016. [Google Scholar]
- 25.Selvaraju RR, Cogswell M, Das A, et al. editors. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. 2017 IEEE International Conference on Computer Vision (ICCV); 2017 22-29 Oct. 2017. [Google Scholar]
- 26.Fatahi Meybodi N, Karimi-Zarchi M, Allahqoli L, et al. Accuracy of the Triple Test Versus Colposcopy for the Diagnosis of Premalignant and Malignant Cervical Lesions. Asian Pac J Cancer Prev 2020;21:3501-7. 10.31557/APJCP.2020.21.12.3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoppenot C, Stampler K, Dunton C. Cervical cancer screening in high- and low-resource countries: implications and new developments. Obstet Gynecol Surv 2012;67:658-67. 10.1097/OGX.0b013e3182732375 [DOI] [PubMed] [Google Scholar]
- 28.Australian Institute of Health and Welfare. National Cervical Screening Program monitoring report 2020. Canberra: AIHW2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as