Abstract
Background
Differentiating malignant lung tumors from benign pulmonary nodules is a great challenge. While the analysis of bronchoalveolar lavage fluid (BALF) is used for diagnosing infections and interstitial lung diseases, there is limited evidence to support its use for lung cancer diagnosis. This study aimed to interrogate the potential of using BALF cell-free DNA (cfDNA) to discriminate malignant lesions from benign nodules.
Methods
Fifty-three patients with solid pulmonary nodules (≤2 cm) were prospectively enrolled, including 21 confirmed with benign disease and 32 with malignant tumors. Mutations were profiled for 30 tumor tissues and 40 BALFs. Paired BALFs and plasma from 48 patients underwent DNA methylation profiling. A methylome-based classification model was developed for BALF and plasma separately.
Results
Among the 30 patients with paired tissues and BALFs, 96.7% and 70% had alterations detected from their tissues (79 alterations) and BALFs (53 alterations), respectively. Using tissues as references, BALFs revealed 14 new alterations and missed 41. BALF mutation displayed a sensitivity of 71%, specificity of 77.8%, and accuracy of 72.5% in detecting lung cancer. BALF methylation achieved an accuracy of 81.3%, with both sensitivity and specificity being 81%. Plasma methylation showed a 66.7% sensitivity, 71.4% specificity, and 68.8% accuracy. BALF methylation also demonstrated 82.4% sensitivity in stage I patients. Parallel bronchoscopy, lavage cytology, and bronchial brushing demonstrated an inferior sensitivity of 23%, 3.1%, and 9.7%, respectively, compared with BALF methylation and mutation (P<0.0001).
Conclusions
BALF cfDNA can serve as a liquid biopsy media for both mutation and methylation profiling, demonstrating better sensitivities in distinguishing small malignant tumors from benign nodules than conventional methods.
Keywords
Lung cancer diagnosis; pulmonary nodule; bronchoalveolar lavage fluid (BALF); methylation; genomic mutation
Introduction
As most lung cancer patients are diagnosed at an advanced stage when they present with symptoms, the option of curative treatment is often missed (1). Therefore, an effective screening method for early detection has been highly sought. As low-dose CT (LDCT) screening demonstrates a sensitivity of 93.7% for high-risk people defined as 55–75 years old with a smoking history of over 30 pack-years and has been proved to reduce mortality from lung cancer by 20% (2,3), it has been recommended for high-risk individuals (4,5). When suspicious nodules are identified, subsequent medical procedures such as bronchoscopy, transthoracic needle biopsy and surgical excision, or a long-term follow-up with repeated CT examination are required depending on a number of factors, including but not limited to lesion size and location, the ability to biopsy, risks associated with surgery, as well as the patient’s preferences (6). However, LDCT introduces excessive false positives of up to 96% (3), resulting in unnecessary medical care and causing great anxiety, as even for patients with surgical interventions, 20–30% are diagnosed with benign nodules (3,7,8). Therefore, it is necessary to develop screening technologies with greater sensitivity and specificity (9).
Plasma peripheral circulating free DNA (cfDNA) mainly derives from apoptotic or necrotic cells and is often present in a minimal amount in healthy people. Its amount can increase in cancer patients and is associated with tumor burden. cfDNA often exists in double-stranded DNA fragment with the size ranging from 18 to 10,000 bp (10). cfDNA derived from tumor cells, known as cell-free circulating tumor DNA (ctDNA) carries tumor-associated genetic and epigenetic alterations, making it a potential liquid biopsy material for tumor diagnosis, monitory and therapeutic evaluations (11). Plasma ctDNA has demonstrated its clinical utility in advanced lung cancer as a tissue biopsy surrogate for the noninvasive evaluation of tumor-associated alterations, therefore offering prognostic and predictive information (12). ctDNA-based plasma testing assays have also been extensively investigated for screening and early detection of lung cancer, including quantification of ctDNA, detection of genomic abnormities, and cancerous methylation signatures (13-16). However, due to the limited amount of ctDNA present in early-stage patients, the sensitivities of such tests are limited. Investigators have also evaluated other source of liquid biopsy materials and biomarkers such as sputum, microRNA, and gene expression, but the diagnostic performance of these assays either remain unsatisfactory or unreliable due to the lack of large validation studies (16-19), and none have been recommended for clinical utility.
Bronchoalveolar lavage (BAL) is a technique for sampling the cellular and non-cellular components of epithelial lining fluid from the alveolar and bronchial airspaces with both non-bronchoscopic and bronchoscopic types. Analysis of the returned fluid is used for the diagnosis of infections and interstitial lung diseases by evaluating microbiological and/or cellular components (20,21). Due to its minimal invasiveness and vicinity to tumor cells, BAL fluid (BALF) may represent an alternative source of more sensitive lung cancer biomarkers. However, BALF cytology alone displays a low sensitivity, ranging from 29–69% (22-25). Limited studies have explored the role of BALF for identifying lung cancer beyond cytology purposes (26-28). Carstensen et al. for the first time demonstrated the feasibility of isolating cell-free DNA (cfDNA) qualified for the detection of tumor genomic alterations from BALF supernatants of lung cancer patients (29). The present study aims to interrogate the potential of using BALF cfDNA to discriminate malignant lesions from benign solid nodules at both genetic and epigenetic levels.
We present the following article in accordance with the STARD reporting checklist (available at https://dx.doi.org/10.21037/atm-21-2579).
Methods
Patient information and study design
Consecutive patients were prospectively recruited from the Department of Respiratory Medicine of the First Affiliated Hospital of Soochow University according to the following criteria: (I) presence of 8 mm–2 cm solid pulmonary nodules detected by CT scan; (II) bronchoscopy was recommended by the physician. The exclusion criteria included: (I) intolerance to bronchoscopy; (II) failed to provide written informed consent; (III) recovered BALF <20 mL. Patients were followed until a diagnosis was established, and a diagnosis of lung cancer was confirmed by a histopathological test using a surgical biopsy, bronchoscopy, or a transthoracic needle. Ultimately, a total of 53 patients with solid pulmonary nodules (1–2 cm in diameter) were enrolled, including 49 patients with solitary pulmonary nodules at initial diagnosis and four patients with pulmonary nodules after curative surgery or complete response to anti-tumor treatment (Table 1). The study was approved by the institutional review board of the First Affiliated Hospital of Soochow University (No. 2020188) and all patients provided written informed consent, in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1. Clinical characteristics of patients (N=53).
| Characteristics | Value |
|---|---|
| Age, years | |
| Median [range] | 57 [32–84] |
| Sex, n (%) | |
| Male | 29 (54.7) |
| Female | 24 (45.3) |
| Smoking history, n (%) | |
| Current (1–20 packs/day) | 5 (9.4) |
| Current (>20 packs/day) | 3 (5.7) |
| Former | 2 (3.8) |
| Never | 42 (79.2) |
| Unknown | 1 (1.9) |
| Nodule diameter, cm | |
| Median (range) | 1.3 (1.0–2.0) |
| Clinical diagnosis, n (%) | |
| Benign | 21 (39.6) |
| Stage I adenocarcinoma | 20 (37.7) |
| Stage II–IIIA adenocarcinoma | 5 (9.4) |
| Stage IIIB–IV adenocarcinoma | 7 (13.2) |
| Treatment history, n (%) | |
| Treatment naïve | 49 (92.5) |
| Relapse after surgery/anti-tumor therapy | 4 (7.5) |
| Other complemental assay, n (%) | |
| Bronchoscopy biopsy | 38 (71.7) |
| Bronchial brushing cytology | 51 (96.2) |
| Lavage cytology | 53 (100.0) |
DNA extraction
We used a QIAamp Circulating Nucleic Acid kit and QIAamp DNA FFPE tissue kit to extract cfDNA and genomic DNA from plasma/BALF and tissue samples, respectively, according to the manufacturer’s standard protocol (Qiagen, Hilden, Germany). DNA was quantified using the Qubit dsDNA assay (Life Technologies, Carlsbad, CA, USA).
DNA sequencing and genomic profiling
Capture-based targeted sequencing for somatic mutation profiling was carried out using a panel including 168 lung cancer-related genes (Burning Rock Biotech, Guangzhou, China). The next-generation library was prepared as described previously (30) and subsequently sequenced on a NextSeq 500 (Illumina, Inc., San Diego, CA, USA) with a median depth of 2,104X for genomic DNA and a median depth of 47,304X for cfDNA. We used BWA aligner 0.7 (31) to align the FASTQ sequencing data to the human genome (hg19). Genome Analysis ToolKit (GATK) 3.2 (32), Picard (http://picard.sourceforge.net/) and VarScan (33) were used to perform local alignment optimization, mark duplication, and variant calling. Gene translocations were analyzed using FACTERA (34) and the copy number variation (CNV) was identified using an in-house algorithm based on sequencing depth (35).
Bisulfite sequencing and data analysis
We used a capture-based bisulfite sequencing panel to sequence cfDNA extracted from plasma and BALF as described previously (36). The brELSATM method (Burning Rock Biotech, Guangzhou, China) was used to prepare the bisulfite sequencing (BS-seq) library. Briefly, sodium bisulfite treatment first converted unmethylated C was into U. The converted single-strand DNA was subsequently ligated to an adapter and amplified with an uracil-tolerating DNA polymerase. The amplified whole-genome BS libraries were subjected to target enrichment using methylation profiling RNA probes covering 80,672 CpG sites and spanning 1.05 megabase of the human genome (Burning Rock Biotech, Guangzhou, China). The target libraries were subsequently quantified by real-time PCR (Kapa Biosciences Wilmington, MA, USA) and sequenced on NovaSeq 6000 (Illumina, San Diego, CA, USA) with an average 2,000× sequencing depth. Custom adaptor sequences and low-quality bases were removed using Trimmomatic (v.0.32). BWA-meth (v.0.2.2) (37) aligned paired-end reads to C to T- and G to A-transformed hg19 genome. After alignment, samblaster (v.0.1.20) (38) marked duplicate reads, and sambamba (v.0.4.7) (39) removed low mapping quality (MAPQ <20) or improper pairing reads from further downstream analyses. To avoid double-counting of methylation calls, paired reads were merged by clipping overlapping reads.
Methylome-based classification model
MethylDackel (v.0.2.1) was used to quantify the methylation status of CpG sites. Each site was covered with reads that contributed either the methylated signal noted as M or the unmethylated signal noted as U, and the methylation level of each CpG site was calculated as M/(M+U), where both forward and reversed strands were comprised. Lung-specific markers were achieved from an internal database, where 31 pairs of lung cancer tissues and normal tissues were sequenced, and the methylation level of each CpG site was collected. Using software “limma (v2.0)”, each group of CpG sites close in position which showed either continuously higher or continuously lower methylation level in cancer tissues compared to normal tissues were gathered into one differentially methylated region (DMR) and the cutoff was set as adjusted FDR <0.05. As a DMR can contain only one CpG site, the minimal length of DMR was set as 10 bp. Ultimately, 3,420 regions including 42,176 CpG sites were chosen as candidate features to develop classification models of malignant/benign lesions. The average methylation level of all CpG sites in each DMR was applied to represent the signal of each feature in the model. The differential signal was visually confirmed by a supervised clustering heatmap (metric for clustering: “euclidean” distance) using R packages of “FactoMineR” and “heatmap. plus” respectively, and the heatmap represents the DNA methylation level with red denoting hyper-methylation and blue denoting hypo-methylation.
The DMR signal features of the plasma/BALF samples were collected from both malignant and benign lung tumor patients and Scikit-learn (v.0.20.4) was used to perform a machine learning algorithm and build the classification model to distinguish malignant and benign samples (40). As five-fold cross-validation was repeated five times, all data in the held-out folds were accessed independently of training. The classification model was built for plasma and BALF separately. The plasma model was trained with the feature vectors of plasma from training subjects with cancer (labeled with 1) and the feature vectors of plasma from training subjects without cancer (labeled with 0). The model thereby generated a function that computes the cancer prediction using learned weights for each DMR feature. The function obtained from the current fold of training subjects was further applied to provide the cancer prediction (cancer as 1 and non-cancer as 0) for the current fold of testing subjects according to their feature vectors. Similarly, the BALF model was trained with the training group of BALF feature vectors and applied to predict testing groups of BALF subjects.
Statistical analysis
Receiver operating characteristic (ROC) curves were plotted for both BALF and plasma methylation signatures to determine the optimal cut-offs. For BALF mutation, we defined patients who had mutations detected from BALF cfDNA as positive. The performance of assays was evaluated by using the estimates of sensitivity and specificity and differences in the groups were calculated and presented using Fisher’s exact test. Statistical analysis was performed with R software, version 3.01 (R Project for Statistical Computing).
Results
Patient characteristics
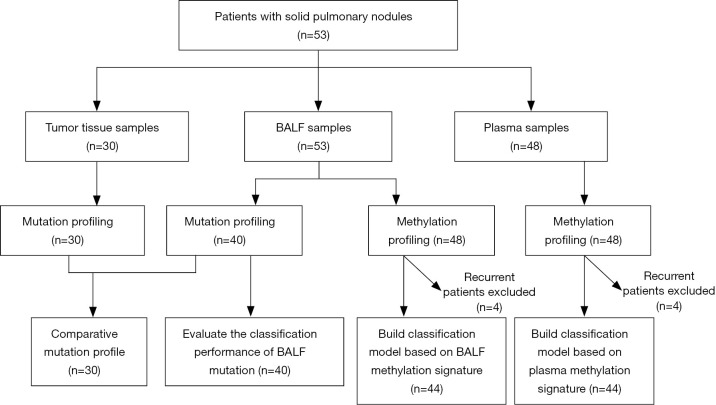
BALF and plasma samples were collected from 53 and 48 patients, respectively. Mutation profiling was performed on 30 tumor tissues and 40 BALF samples with 30 paired samples for comparison and BALF and plasma samples from 48 patients were subjected to DNA methylation profiling. A methylome-based classification model was developed for BALF and plasma separately based on a training set of 44 patients after excluding the four patients with recurrent diseases. The discriminating performance of BALF mutation was assessed in the 40 patients with BALF mutation profiling available. This study design is shown in Figure 1. Of the 53 patients enrolled, 54.7% were male and 45.3% were female (Table 1). The cohort had a median age of 57 years and a median nodule size of 1.3 cm in diameter, and the majority (79.2%) had no smoking history. Solitary pulmonary nodules were identified in 49 (92.5%) patients at initial diagnosis and in another four (7.5%) after curative surgery or complete response to anti-tumor treatment. Ultimately, 21 (39.6%) patients were diagnosed with benign diseases, and 32 (60.4%) were confirmed with malignant tumors, with 25 at early stages and seven at advanced stages (IIIB–IV). Of the 25 patients with early-stage diseases, 20 were at stage I.
Figure 1.
Flowchart of the study design.
Mutations in tissue and BALF
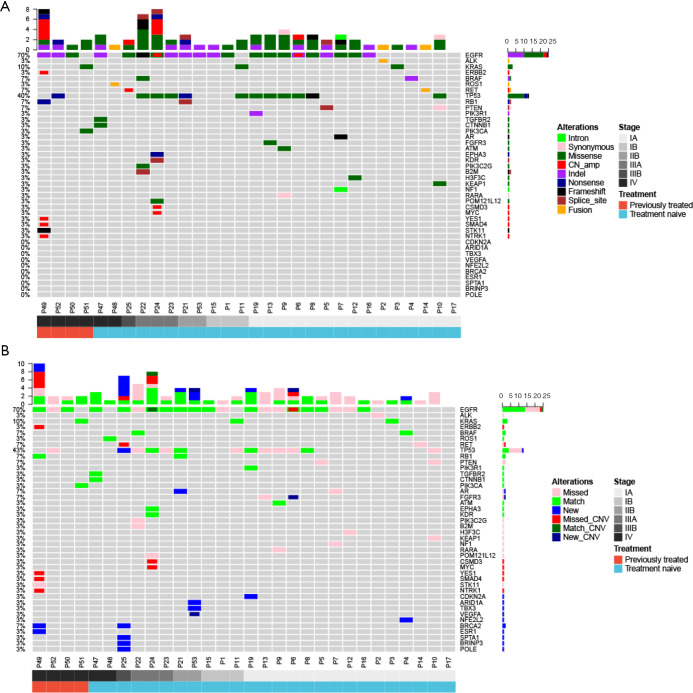
The genomic profile between tumor tissue and paired BALF cfDNA from 30 patients with a definitive lung cancer diagnosis were compared. We found that 96.7% (29/30) and 70% (21/30) patients had genomic alterations detected from their tissue and BALF samples, respectively, resulting in a concordance of 70%. A total of 79 and 53 alterations were identified from tissue and BALF samples, respectively. Using tissue samples as references, BALF samples revealed 14 new alterations (13 mutations and one CNV) and missed 41 alterations (33 mutations and eight CNVs). For EGFR mutation, BALF exhibited a detection rate of 66.7% (14/21). All three KRAS mutations and one ROS fusion identified in tissue samples were detected in BALF cfDNA samples, and one ERBB2 amplification, one ALK fusion and one RET fusion were missed in BALF (Figure 2A,B).
Figure 2.
Mutational landscapes of paired tumor tissue genomic DNA and BALF cfDNA (N=30). (A) Tumor tissue genomic DNA; (B) BALF cfDNA. BALF, bronchoalveolar lavage fluid; cfDNA, cell-free DNA.
Methylome-based classification models
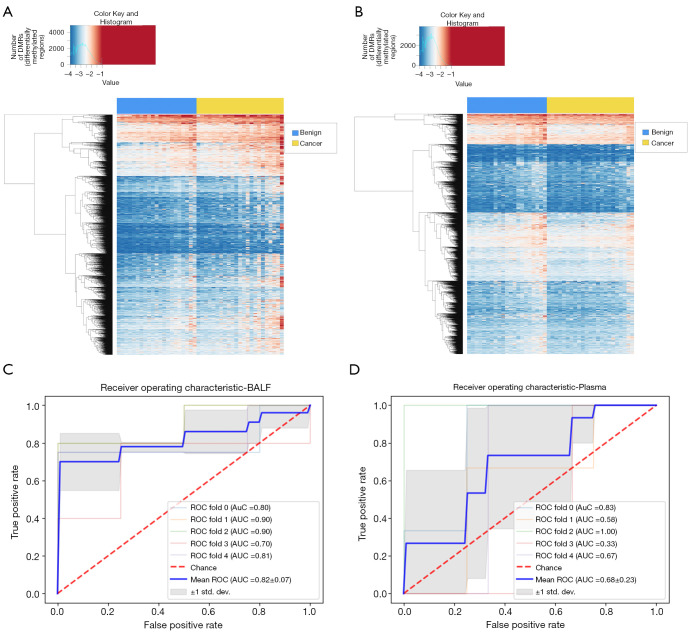
Targeted bisulfite sequencing was performed with BALF and plasma cfDNA from 48 patients to profile DNA methylation signatures (Figure 1A). The supervised clustering heatmap for BALF revealed that a higher frequency of cancer patients presented with a hypermethylated pattern in certain regions compared with benign patients (Figure 3A). On the other hand, the methylation signature derived from plasma cfDNA lacked significant differential patterns between malignant and benign patients (Figure 3B). To eliminate the impact of systemic treatment on the baseline methylation profile of patients, we developed methylome-based classification models for both BALF and plasma based on the 44 patients with solid nodules at initial diagnosis (21 benign and 23 malignant) using a machine learning algorithm and performed cross-validations. The ROC analyses showed an area under curve (AUC) of 0.82 (95% CI: 0.75–0.89) for BALF methylation signature compared with an AUC of 0.68 (95% CI: 0.45–0.91) for plasma signature (P=0.56) (Figure 3C,D).
Figure 3.
Visualization and ROC curves of methylation signatures in plasma and BALF cfDNA. (A) BALF supervised clustering heatmap; (B) plasma supervised clustering heatmap; (C) BALF ROC curve; (D) plasma ROC curve. ROC, receiver operating characteristic; BALF, bronchoalveolar lavage fluid; cfDNA, cell-free DNA.
Diagnostic performance of different assays
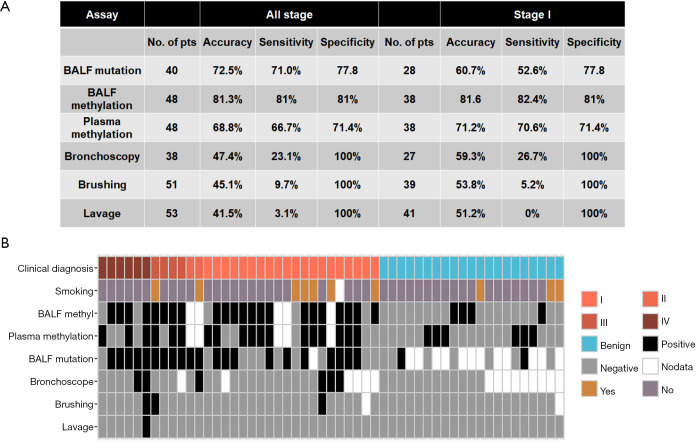
We next evaluated the performance of BALF ctDNA mutation profiling and methylation signatures in both BALF and plasma in differentiating malignant from benign pulmonary nodules. For BALF mutation, we defined patients who had mutations detected from BALF cfDNA as positive. A total of 40 patients including 31 with malignant nodules (six with stage IIIB or IV; 25 with stage IA–IIIA, and nine with confirmed benign nodules) were included in mutation profiling (Figure 1), resulting in a sensitivity of 71%, a specificity of 77.8% and an accuracy of 72.5% (Figure 4A). On the other hand, the model for BALF ctDNA-derived methylation profiling demonstrated both sensitivity and specificity of 81% and an accuracy of 81.3% in the 48 patients with methylation profiling, while the plasma-derived ctDNA methylation model showed a sensitivity of 66.7%, specificity of 71.4%, and accuracy of 68.8%. The difference in discriminating performance was not significant between any two of the three assays. Notably, in the 21 benign patients, BALF and plasma methylation profile misdiagnosed four and six patients with malignant disease (Figure 4B), respectively, and all false-positive cases had no smoking history. In addition, all three benign patients with a smoking history were classified as benign by both methylation profiles, suggesting that the methylation methodology used in this study has high specificity to discriminate cancer-specific signatures from tobacco use-related methylation signatures.
Figure 4.
Comparison of diagnostic performances of different assays. (A) The accuracies, sensitivities, and specificities of different assays in the whole cohort (all stage) and in the subset of stage I patients; (B) overview of the diagnostic outcomes of different assays in 53 patients.
We also compared ctDNA-based mutation and DNA methylation profiling with several conventional methods, including bronchoscope biopsy (N=38), bronchial brushing (N=51), and lavage cytology (N=53), and the sensitivity, specificity and accuracy of each method are summarized in Figure 4A. Collectively, conventional methods (bronchoscopy, brushing, and lavage) have very high specificity but very low sensitivity, especially brushing and lavage, which had a sensitivity of 9.7% and 3.1%, respectively. In comparison, ctDNA-based mutation and DNA methylation profiling offered a much higher sensitivity ranged from 66.7% to 81%. (0.001<P<0.0001). The diagnostic results of different assays for each patient are summarized in Figure 4B.
Furthermore, we analyzed the performance of these methods in patients with stage I diseases (Figure 4A) and found both BALF and plasma ctDNA methylation signatures demonstrated comparable sensitivities in stage I patients compared with that in all cancer patients (82.4% vs. 81.6% and 70.6% vs. 66.7%, respectively). Of note, BALF methylation profiling exhibited a trend of superior sensitivity (82.4% vs.52.6%, P=0.06) and accuracy (81.6% vs. 60.7%, P=0.06) than BALF mutation profile in the subset of stage I patients.
Discussion
Compared with other potential diagnostic biomarkers, the advantage of DNA methylation include its stability, its early occurrence during carcinogenesis, and that it provides a greater magnitude of markers since the number of CpG sites in the human genome is huge (9,15). Accordingly, DNA methylation has emerged as a promising biomarker for cancer detection and is being actively investigated in multiple cancers including lung cancer. Earlier studies often focused on examining the DNA methylation levels of pre-selected tumor-specific candidate genes in blood and have identified a number of potential biomarkers for lung cancer diagnosis, including RASSF1A, APC, and SHOCK2 (41-44). However, most studies enrolled rather late-stage patients and/or asymptomatic normal individuals, which tended to overestimate the sensitivity and specificity. With the rapid advancement of next-generation sequencing, high-throughput epigenomic studies provide a deeper characterization of methylation signatures in cancer along with the potential of discovering more robust markers. Diaz-Lagares et al. used two genome-wide DNA methylation datasets including early-stage lung cancers to identify a specific methylation signature including four genes: BCAT1, CDO1, TRIM58, and ZNF177, and validated its diagnostic value into independent cohorts with diverse sample types, including FFPE tissues, bronchial aspirates, and sputum from patients with lung cancer and cancer-free individuals (45). More recently, Liang et. al developed a blood-based diagnostic assay for lung cancer early detection by high throughput DNA bisulfite sequencing which demonstrated a sensitivity of 79.5% and a specificity of 85.2% for differentiating lung cancers from benign pulmonary nodules (15).
In the present study, we conducted a similar high throughput methylation sequencing study and developed parallel classification models for both plasma and BALF ctDNA samples. We also investigated the feasibility of using BALF ctDNA mutation profiling for classification. Our results revealed that the methylation and mutation profiling of BALF ctDNA demonstrated comparable sensitivities (81% vs. 71%, P=0.38) and specificities (81% vs. 77.8%, P=0.84). Interestingly, BALF methylation profiling exhibited a trend of superior sensitivity (82.4% vs. 52.6%, P=0.06) compared to BALF mutation on detecting early-stage cancer patients. On the other hand, the performance of the BALF methylation signature to differentiate malignant from benign lesions was comparable with that of the plasma methylation signature (sensitivity: 81% vs. 66.7%, P=0.24) and specificity: 81% vs. 71.4%, P=0.29) (Figure 4A), although the absolute numerical values for BALF were higher. Therefore, future studies in larger cohorts are required to further investigate whether BALF could serve as a superior liquid biopsy material over plasma for lung cancer early detection.
While in patients detected with chest radiographic abnormalities suspected to be lung cancer, bronchoscopy is a relatively safe approach for diagnostic tissue sampling and has been widely used (46), in approximately 30% of cases, it is non-diagnostic (47). Accordingly, cytology-based sampling techniques such as lavage and bronchial brushings are normally included with histopathological confirmation by biopsy to increases the diagnostic yield of bronchoscopy (26,48). In this study, the majority of our cohort also underwent bronchoscopy biopsy and cytology-based assays in parallel for comparing the performance of our newly developed assays with these clinically used techniques. Bronchoscopy biopsy and cytology analyses of lavage and bronchial brushing all demonstrated a specificity of 100% in our cohort. However, sensitivities were extremely low with 23% for bronchoscopy and 3–9% for cytology-based assays (Figure 4A), which were significantly inferior to that of BALF methylation (P<0.0001) and BALF mutation profiling (P<0.001), which suggests a large portion of malignant lesions would have been missed by these screenings. Our results clearly reveal the advantage of ctDNA-based profiling over cytology analysis of BALF for differentiating early-stage lung cancers from benign lung disease.
Compared to the invasive tissue-based test, BALF cfDNA provides a semi-invasive alternative for detecting genetic and epigenetic alterations in lung cancer. It also has the advantage of overcoming the intra-tumor heterogeneity to a certain extent. However, the ctDNA released in BALF may be limited especially in early stage-patients, which compromise its sensitivity in detecting early-stage diseases.
As a proof-of-principle investigation, our study has the major limitation of lacking an independent validation cohort, especially for the methylome-based classification model. Further studies recruiting larger cohorts are warranted to verify and improve the performance of the model we developed.
Conclusions
BALF can serve as liquid biopsy media for both mutation and DNA methylation profiling to distinguish small malignant tumors (≤2 cm in diameter) from benign pulmonary nodules, even in patients with early-stage disease. Both BALF mutation and methylation profiles demonstrated better sensitivities than conventional methods and have potential diagnostic value.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the Jiangsu Province Special Program of Medical Science (BE2016672) and Suzhou science and technology project (SLT201917).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the institutional review board of the First Affiliated Hospital of Soochow University (2020188) and all patients provided written informed consent, in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-2579
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-2579
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-2579). JX, LS, BL, HL, and BL are employees of Burning Rock Biotech. JJ received the funding from Jiangsu Province Special Program of Medical Science (BE2016672). The other authors have no conflicts of interest to declare.
(English Language Editor: B. Draper)
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Kakinuma R, Muramatsu Y, Asamura H, et al. Low-dose CT lung cancer screening in never-smokers and smokers: results of an eight-year observational study. Transl Lung Cancer Res 2020;9:10-22. 10.21037/tlcr.2020.01.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Lung Screening Trial Research Team , Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wender R, Fontham ET, Barrera E, Jr, et al. American Cancer Society lung cancer screening guidelines. CA Cancer J Clin 2013;63:107-17. 10.3322/caac.21172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo J, Wang C, Xu X, et al. DeepLN: an artificial intelligence-based automated system for lung cancer screening. Ann Transl Med 2020;8:1126. 10.21037/atm-20-4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould MK, Donington J, Lynch WR, et al. Evaluation of individuals with pulmonary nodules: when is it lung cancer? Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e93S-e120S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Klaveren RJ, Oudkerk M, Prokop M, et al. Management of lung nodules detected by volume CT scanning. N Engl J Med 2009;361:2221-9. 10.1056/NEJMoa0906085 [DOI] [PubMed] [Google Scholar]
- 8.Diederich S, Thomas M, Semik M, et al. Screening for early lung cancer with low-dose spiral computed tomography: results of annual follow-up examinations in asymptomatic smokers. Eur Radiol 2004;14:691-702. 10.1007/s00330-003-2200-5 [DOI] [PubMed] [Google Scholar]
- 9.Tseng TS, Gross T, Celestin MD, et al. Knowledge and attitudes towards low dose computed tomography lung cancer screening and smoking among African Americans—a mixed method study. Transl Cancer Res 2019;8:S431-42. 10.21037/tcr.2019.04.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 2010;38:6159-75. 10.1093/nar/gkq421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, Liang Y, Li S, et al. The interplay of circulating tumor DNA and chromatin modification, therapeutic resistance, and metastasis. Mol Cancer 2019;18:36. 10.1186/s12943-019-0989-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santarpia M, Liguori A, D'Aveni A, et al. Liquid biopsy for lung cancer early detection. J Thorac Dis 2018;10:S882-97. 10.21037/jtd.2018.03.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sozzi G, Roz L, Conte D, et al. Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study. Am J Respir Crit Care Med 2009;179:69-74. 10.1164/rccm.200807-1068OC [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Cuesta L, Perdomo S, Avogbe PH, et al. Identification of Circulating Tumor DNA for the Early Detection of Small-cell Lung Cancer. EBioMedicine 2016;10:117-23. 10.1016/j.ebiom.2016.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang W, Zhao Y, Huang W, et al. Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA). Theranostics 2019;9:2056-70. 10.7150/thno.28119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Huang X, Yin W, et al. Ultrasensitive DNA hypermethylation detection using plasma for early detection of NSCLC: a study in Chinese patients with very small nodules. Clin Epigenetics 2020;12:39. 10.1186/s13148-020-00828-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silvestri GA, Vachani A, Whitney D, et al. A Bronchial Genomic Classifier for the Diagnostic Evaluation of Lung Cancer. N Engl J Med 2015;373:243-51. 10.1056/NEJMoa1504601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sozzi G, Boeri M, Rossi M, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol 2014;32:768-73. 10.1200/JCO.2013.50.4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulbert A, Jusue-Torres I, Stark A, et al. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin Cancer Res 2017;23:1998-2005. 10.1158/1078-0432.CCR-16-1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radha S, Afroz T, Prasad S, et al. Diagnostic utility of bronchoalveolar lavage. J Cytol 2014;31:136-8. 10.4103/0970-9371.145636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Midulla F, Nenna R. Bronchoalveolar Lavage: Indications and Applications. Progress in Respiratory Research 2010;38:30-41. 10.1159/000314382 [DOI] [Google Scholar]
- 22.Binesh F, Pirdehghan A, Mirjalili MR, et al. Comparative assessment of the diagnostic value of transbronchial lung biopsy and bronchoalveolar lavage fluid cytology in lung cancer. Asian Pac J Cancer Prev 2015;16:201-4. 10.7314/APJCP.2015.16.1.201 [DOI] [PubMed] [Google Scholar]
- 23.Labbe C, Beaudoin S, Martel S, et al. Diagnostic yield of non-guided flexible bronchoscopy for peripheral pulmonary neoplasia. Thorac Cancer 2015;6:517-23. 10.1111/1759-7714.12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wongsurakiat P, Wongbunnate S, Dejsomritrutai W, et al. Diagnostic value of bronchoalveolar lavage and postbronchoscopic sputum cytology in peripheral lung cancer. Respirology 1998;3:131-7. 10.1111/j.1440-1843.1998.tb00111.x [DOI] [PubMed] [Google Scholar]
- 25.Bezel P, Tischler V, Robinson C, et al. Diagnostic Value of Bronchoalveolar Lavage for Diagnosis of Suspected Peripheral Lung Cancer. Clin Lung Cancer 2016;17:e151-6. 10.1016/j.cllc.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 26.Carvalho AS, Cuco CM, Lavareda C, et al. Bronchoalveolar Lavage Proteomics in Patients with Suspected Lung Cancer. Sci Rep 2017;7:42190. 10.1038/srep42190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uribarri M, Hormaeche I, Zalacain R, et al. A new biomarker panel in bronchoalveolar lavage for an improved lung cancer diagnosis. J Thorac Oncol 2014;9:1504-12. 10.1097/JTO.0000000000000282 [DOI] [PubMed] [Google Scholar]
- 28.Zhang C, Yu W, Wang L, et al. DNA Methylation Analysis of the SHOX2 and RASSF1A Panel in Bronchoalveolar Lavage Fluid for Lung Cancer Diagnosis. J Cancer 2017;8:3585-91. 10.7150/jca.21368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carstensen T, Schmidt B, Engel E, et al. Detection of cell-free DNA in bronchial lavage fluid supernatants of patients with lung cancer. Ann N Y Acad Sci 2004;1022:202-10. 10.1196/annals.1318.031 [DOI] [PubMed] [Google Scholar]
- 30.Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. 10.1016/j.jtho.2016.11.2235 [DOI] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568-76. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. 10.1093/bioinformatics/btu549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Ye F, Bao L, et al. Somatic alterations of TP53, ERBB2, PIK3CA and CCND1 are associated with chemosensitivity for breast cancers. Cancer Sci 2019;110:1389-400. 10.1111/cas.13976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang G, Chen K, Yang F, et al. Monitoring of circulating tumor DNA and its aberrant methylation in the surveillance of surgical lung Cancer patients: protocol for a prospective observational study. BMC Cancer 2019;19:579. 10.1186/s12885-019-5751-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedersen BS, Eyring K, De S, et al. Fast and accurate alignment of long bisulfite-seq reads. arXiv: 14011129v2 [q-bioGN]. 2014.
- 38.Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 2014;30:2503-5. 10.1093/bioinformatics/btu314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarasov A, Vilella AJ, Cuppen E, et al. Sambamba: fast processing of NGS alignment formats. Bioinformatics 2015;31:2032-4. 10.1093/bioinformatics/btv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825-30. [Google Scholar]
- 41.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. 10.1016/j.canlet.2010.12.011 [DOI] [PubMed] [Google Scholar]
- 42.Ponomaryova AA, Rykova EY, Cherdyntseva NV, et al. Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer 2013;81:397-403. 10.1016/j.lungcan.2013.05.016 [DOI] [PubMed] [Google Scholar]
- 43.Kneip C, Schmidt B, Seegebarth A, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol 2011;6:1632-8. 10.1097/JTO.0b013e318220ef9a [DOI] [PubMed] [Google Scholar]
- 44.Konecny M, Markus J, Waczulikova I, et al. The value of SHOX2 methylation test in peripheral blood samples used for the differential diagnosis of lung cancer and other lung disorders. Neoplasma 2016;63:246-53. 10.4149/210_150419N208 [DOI] [PubMed] [Google Scholar]
- 45.Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, et al. A Novel Epigenetic Signature for Early Diagnosis in Lung Cancer. Clin Cancer Res 2016;22:3361-71. 10.1158/1078-0432.CCR-15-2346 [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Pan X, Gu C, et al. The feasibility of navigation bronchoscopy-guided pulmonary microcoil localization of small pulmonary nodules prior to thoracoscopic surgery. Transl Lung Cancer Res 2020;9:2380-90. 10.21037/tlcr-20-1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Memoli JS, Nietert PJ, Silvestri GA. Meta-analysis of guided bronchoscopy for the evaluation of the pulmonary nodule. Chest 2012;142:385-93. 10.1378/chest.11-1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobler CC, Crawford AB. Bronchoscopic diagnosis of endoscopically visible lung malignancies: should cytological examinations be carried out routinely? Intern Med J 2009;39:806-11. 10.1111/j.1445-5994.2008.01882.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as