Abstract
Background
Significant racial disparities in pancreatic cancer incidence and mortality rates exist, with the highest rates in African Americans compared to Non-Hispanic Whites and Hispanic/Latinx populations. Computer-derived quantitative imaging or “radiomic” features may serve as non-invasive surrogates for underlying biological factors and heterogeneity that characterize pancreatic tumors from African Americans, yet studies are lacking in this area. The objective of this pilot study was to determine if the radiomic tumor profile extracted from pretreatment computed tomography (CT) images differs between African Americans, Non-Hispanic Whites, and Hispanic/Latinx with pancreatic cancer.
Methods
We evaluated a retrospective cohort of 71 pancreatic cancer cases (23 African American, 33 Non-Hispanic White, and 15 Hispanic/Latinx) who underwent pretreatment CT imaging at Moffitt Cancer Center and Research Institute. Whole lesion semi-automated segmentation was performed on each slice of the lesion on all pretreatment venous phase CT exams using Healthmyne Software (Healthmyne, Madison, WI, USA) to generate a volume of interest. To reduce feature dimensionality, 135 highly relevant non-texture and texture features were extracted from each segmented lesion and analyzed for each volume of interest.
Results
Thirty features were identified and significantly associated with race/ethnicity based on Kruskal-Wallis test. Ten of the radiomic features were highly associated with race/ethnicity independent of tumor grade, including sphericity, volumetric mean Hounsfield units (HU), minimum HU, coefficient of variation HU, four gray level texture features, and two wavelet texture features. A radiomic signature summarized by the first principal component partially differentiated African American from non-African American tumors (area underneath the curve = 0.80). Poorer survival among African Americans compared to Non-African Americans was observed for tumors with lower volumetric mean CT [HR: 3.90 (95% CI:1.19–12.78), p=0.024], lower GLCM Avg Column Mean [HR:4.75 (95% CI: 1.44,15.37), p=0.010], and higher GLCM Cluster Tendency [HR:3.36 (95% CI: 1.06–10.68), p=0.040], and associations persisted in volumetric mean CT and GLCM Avg Column after adjustment for key clinicopathologic factors.
Conclusions
This pilot study identified several textural radiomics features associated with poor overall survival among African Americans with PDAC, independent of other prognostic factors such as grade. Our findings suggest that CT radiomic features may serve as surrogates for underlying biological factors and add value in predicting clinical outcomes when integrated with other parameters in ongoing and future studies of cancer health disparities.
Keywords: radiomics, cancer disparities, pancreatic cancer, quantitative imaging, blacks
Introduction
Pancreatic cancer is the deadliest malignancy in the United States, with a 5-year relative survival rate of only 10% (1). Due to the lack of effective strategies for prevention, early detection, and treatment, pancreatic cancer is projected to become the second leading cancer killer by 2030 (2). Coinciding with the rise in pancreatic cancer diagnoses and deaths is a notable health disparity, with African Americans/Blacks having significantly higher pancreatic cancer incidence and mortality rates than Non-Hispanic Whites and Hispanic/Latinx (2–12). Biological reasons for these disparities are underexplored and often rely on biomarkers from tissue biopsies, which may not be representative of the entire tumor and its microenvironment. Easily accessible minimally invasive methods that can reflect tumor heterogeneity and correlate with clinical outcomes are urgently needed to advance personalized care for the racially and ethnically diverse population of patients diagnosed with pancreatic cancer each year.
Computed tomography (CT) images are routinely obtained as part of the diagnostic work-up for pancreatic cancer and can be repurposed to support quantitative imaging analyses (13). Radiomics refers to high-throughput extraction and analysis of quantitative features from standard-of-care medical images, many of which are “invisible to the human eye,” to generate mineable data (14). Whereas standard “semantic” radiologic features are typically subjectively and qualitatively measured, computer algorithm-generated radiomic features such as tumor signal intensity, texture, shape, and volume have many advantages (15–21): they represent quantitative, objective measures; reflect tumor heterogeneity and subregional habitats; and are reproducible, stable, and strongly linked to clinical outcomes and underlying molecular data. Radiomic evaluations of pancreas CT scans have been conducted by our team (22–24) and others (15, 25–38), but to date none of these studies have focused on evaluating radiomic features present in pancreatic tumors from AA compared to other ethnic populations. Furthermore, we are unaware of published investigations that specifically compare racial and ethnic differences in radiomic features of different types of non-pancreas tumors. The objective of this study was to compare pretreatment CT radiomic features from a racially and ethnically diverse cohort of cases with pancreatic ductal adenocarcinoma (PDAC), the main histologic subtype of exocrine pancreatic cancers. The implications of this body of work could be far-reaching if radiomic features suggestive of a poor prognosis are identified in the pretreatment setting, in turn influencing clinical decision-making so that more aggressive treatments could be administered earlier to reduce disparities in historically underserved groups.
Methods
Study Population
This retrospective cohort was derived from a radiological records database search of individuals with available pretreatment multiphase CT scans and a corresponding histologic diagnosis of PDAC. Cases were diagnosed and treated for PDAC at Moffitt Cancer Center and Research Institute (Tampa, Florida) between 1/2008 and 8/2018. Subjects were excluded if postcontrast venous phase CT imaging was not available or if pathology reports were not available. Race and ethnicity and other covariates were based on self-report. The final analytic dataset included CT images from 71 unique patients ( Table 1 ). Ethics approval and written consent to participate were reviewed and approved by Advarra IRB (MCC# 19431; IRB #:Pro00024543).
Table 1.
Select demographic and clinical characteristics of the pancreatic ductal adenocarcinoma CT radiomic study cohort (N=71).
| AA (n = 23) | H/L (n = 15) | NHW (n = 33) | Overall | P-value | |
|---|---|---|---|---|---|
| Gender, N (%) | 0.993 | ||||
| Female | 12 (52.2%) | 8 (53.3%) | 17 (51.5%) | 37 (52.1%) | |
| Male | 11 (47.8%) | 7 (46.7%) | 16 (48.5%) | 34 (47.9%) | |
| Age at diagnosis, mean (SD) | 64.9 (10.2) | 61.8 (12.7) | 64.9 (10.1) | 64.2 (10.6) | 0.611 |
| Vital status, N (%) | 0.560 | ||||
| Alive | 4 (17.4%) | 5 (33.3%) | 9 (27.3%) | 18 (25.4%) | |
| Dead | 19 (82.6%) | 10 (66.7%) | 24 (72.7%) | 53 (74.6%) | |
| Smoking status, N (%) | 0.971 | ||||
| Ever | 11 (47.8%) | 9 (60.0%) | 17 (51.5%) | 37 (52.1%) | |
| Missing | 1 (4.35%) | 0 (0.00%) | 1 (3.03%) | 2 (2.82%) | |
| Never | 11 (47.8%) | 6 (40.0%) | 15 (45.5%) | 32 (45.1%) | |
| Marital status, N (%) | 0.516 | ||||
| Divorced | 1 (4.35%) | 3 (20.0%) | 1 (3.03%) | 5 (7.04%) | |
| Married | 17 (73.9%) | 10 (66.7%) | 24 (72.7%) | 51 (71.8%) | |
| Separated | 2 (8.70%) | 1 (6.67%) | 1 (3.03%) | 4 (5.63%) | |
| Single | 1 (4.35%) | 0 (0.00%) | 2 (6.06%) | 3 (4.23%) | |
| Unknown | 0 (0.00%) | 1 (6.67%) | 1 (3.03%) | 2 (2.82%) | |
| Widowed | 2 (8.70%) | 0 (0.00%) | 4 (12.1%) | 6 (8.45%) | |
| Primary site, N (%) | 0.265 | ||||
| C241 Ampulla of vater | 0 (0.00%) | 0 (0.00%) | 2 (6.06%) | 2 (2.82%) | |
| C250 Pancreas Head | 16 (69.6%) | 12 (80.0%) | 25 (75.8%) | 53 (74.6%) | |
| C251 Pancreas Body | 3 (13.0%) | 1 (6.67%) | 1 (3.03%) | 5 (7.04%) | |
| C252 Pancreas Tail | 0 (0.00%) | 2 (13.3%) | 4 (12.1%) | 6 (8.45%) | |
| C257 Pancreas Other Specified | 1 (4.35%) | 0 (0.00%) | 0 (0.00%) | 1 (1.41%) | |
| C258 Pancreas Overlapping | 1 (4.35%) | 0 (0.00%) | 1 (3.03%) | 2 (2.82%) | |
| C259 Pancreas, Not otherwise specified | 2 (8.70%) | 0 (0.00%) | 0 (0.00%) | 2 (2.82%) | |
| SEER Derived Stage, N (%) | 0.257 | ||||
| Localized | 2 (9.09%) | 2 (18.2%) | 6 (18.2%) | 10 (15.2%) | |
| Regional, by direct extension only | 4 (18.2%) | 1 (9.09%) | 5 (15.2%) | 10 (15.2%) | |
| Regional, to lymph nodes only | 4 (18.2%) | 0 (0.00%) | 0 (0.00%) | 4 (6.06%) | |
| Regional, direct extension and lymph nodes | 10 (45.5%) | 6 (54.5%) | 20 (60.6%) | 36 (54.5%) | |
| Distant | 2 (9.09%) | 2 (18.2%) | 2 (6.06%) | 6 (9.09%) | |
| Tumor grade, N (%) | 0.091 | ||||
| Well differentiated | 1 (4.6%) | 0 (0.0%) | 2 (6.01%) | 3 (4.35%) | |
| Moderately differentiated | 8 (36.4%) | 11 (78.6%) | 20 (60.6%) | 39 (56.5%) | |
| Poorly differentiated | 6 (27.3%) | 3 (21.4%) | 8 (24.2%) | 17 (24.6%) | |
| Not determined or Not available | 7 (31.8%) | 0 (0.0%) | 3 (9.1%) | 10 (14.5%) | |
| Clinical tumor size (cm), median (1st ~ 3rd quantile) | 3.20 [2.65;4.68] | 2.90 [2.40;9.05] | 3.65 [2.42;16.0] | 3.20 [2.50;9.60] | 0.796 |
| Pathological tumor size (cm), median (1st ~ 3rd quantile) | 3.0 [2.3;4.7] | 3.2 [3.0;4.9] | 3.0 [2.5;4.9] | 3.0 [2.5;5.2] | 0.602 |
| Regional nodes examined, median (1st ~ 3rd quantile) | 16.0 [0.0;27.2] | 26.0 [21.0;36.5] | 17.0 [13.0;22.0] | 18.5 [13.0;28.0] | 0.005 |
| Regional nodes positive, median (1st ~ 3rd quantile) | 1.0 [0.5;2.5] | 1.0 [0.0;5.5] | 1.0 [0.0;2.0] | 1.0 [0.0;3.0] | 0.840 |
| Survival time (months) median (1st ~ 3rd quantile) | 15.0 [9.0;22.5] | 24.0 [16.0;27.0] | 31.0 [15.0;43.0] | 22.0 [13.0;36.0] | 0.028 |
AA, African Americans; H/L, Hispanic/Latinx; NHW, Non-Hispanic White; CT, computed tomography; SD, standard deviation; SEER, surveillance, epidemiology, end results program
Some numbers and percentages may not add up to the total due to missing data.
Statistically significant differences are noted in bold font.
CT Scanner Types, Acquisitions, and Procedures
CT exams were performed on different scanners as represented in Table 2 , with most scans being performed on a Siemens Sensation 16 (n=31, 43.6%) (Siemens Healthcare, Erlangen, Germany). The post contrast venous phase series was used in this study due to the homogenous availability of this series within our cohort and the superior ability to visualize and segment tumors. The venous phase was generally acquired following weight-based Iopamidol 76% (Bracco Diagnostics Inc., Monroe Township, NJ, USA) dosing to achieve venous phase approximately 60 s post injection. Contrast dosing generally ranged from 75 ml for patients below 55 kg, to 150 ml for patients above 110 kg with gradient increases every 5 kg. Field of view (FOV) ranged from 299 to 500 mm × 299–500 mm based on patient size. The matrix was 512 × 512 for each exam. Slice thickness was 3.0 ± 0.3 mm. Mean venous phase voxel volumes were 1.61, 1.71, and 1.65 mm3, for AA, H/L, and NHW, respectively ( Table 2 ). At our institution, arterial phase bolus triggering is achieved via placement of the contrast tracking region of interest (ROI) over the abdominal aortic lumen at the level of the celiac trunk, with image acquisition triggered at a measured Hounsfield Unit density of 120, and venous phase ensues after a 30 s delay to achieve a 60 s venous phase.
Table 2.
Scanner type and voxel volumes measured for the study cohort.
| AA (n = 23) | H/L (n = 15) | NHW (n = 33) | P value | |
|---|---|---|---|---|
| Scanner model | 0.512 | |||
| Brilliance 64 | 1 (4.55%) | 0 (0.00%) | 0 (0.00%) | |
| Lightspeed pro 32 | 1 (4.55%) | 1 (6.67%) | 3 (9.09%) | |
| Lightspeed ultra | 0 (0.00%) | 1 (6.67%) | 1 (3.03%) | |
| Lightspeed VCT | 1 (4.55%) | 0 (0.00%) | 1 (3.03%) | |
| Sensation 16 | 13 (59.1%) | 5 (33.3%) | 13 (39.4%) | |
| Sensation 40 | 3 (13.6%) | 3 (20.0%) | 9 (27.3%) | |
| Sensation 64 | 3 (13.6%) | 3 (20.0%) | 6 (18.2%) | |
| Somatom definition AS | 0 (0.00%) | 2 (13.3%) | 0 (0.00%) | |
| Voxel volume (mm3), mean (range) | 1.6 [1.4;1.8] | 1.7 [1.6;2.0] | 1.7 [1.4;2.1] | 0.303 |
AA, African Americans; H/L, Hispanic/Latinx; NHW, Non-Hispanic White.
CT Segmentation and Radiomic Feature Extraction/Reduction
Archived non-contrast and contrast-enhanced CT images were acquired from Moffitt’s GE Centricity Picture Archiving and Communication System (PACS). Our experienced board-certified abdominal oncologic radiologists (JC and DJ) were blinded to patient characteristics and outcomes. For each case, the standardized imaging reporting template for PDAC staging (39) was completed to collect information on “semantic” qualitative-based radiologic features related to morphology, arterial and venous enhancement, and evaluation of extra-pancreatic structures. Whole lesion semi-automated segmentation was performed on each slice of the lesion on all pretreatment (within 3 months prior to treatment) venous phase CT exams using Healthmyne Software (Healthmyne, Madison, WI, USA). The venous phase was chosen in part because this phase was most consistent across all exams. To reduce feature dimensionality, 135 highly relevant non-texture (which measure tumor size, shape, and location) and texture features (which measure properties such as smoothness, coarseness, and regularity) were extracted from each segmented lesion and analyzed for the venous contrast phase. Additionally, CT specifications including scanner type, slice thickness, pixel size were recorded given the known variability that can occur with different scanners and settings (40, 41).
Statistical Analysis
Data analysis was performed to evaluate racial/ethnic differences in (a) study population characteristics, (b) CT procedures and standard NCCN imaging criteria, and (c) radiomic features. The Kruskal-Wallis test was used for continuous variables, and Chi-squared test or Fisher’s exact test was used for categorical variables to compare the difference among racial/ethnic groups. Significant race/ethnicity-associated radiomic features were determined using a false discovery rate (42) at a threshold of 20%. Spearman correlation analysis was applied to evaluate the correlation between radiomic features. High correlated features were filtered out based on the absolute correlation coefficient above 0.9. Statistically significant radiomic features were summarized by principal component analyses (PCA) to derive a race/ethnicity-associated radiomic signature score as we described previously (43). Receiver operating characteristic (ROC) curve analysis was used to evaluate the prediction efficacy for race/ethnicity using the derived radiomic signature score. Cox proportional hazard regression was performed to evaluate the association between overall survival and each radiomic feature, including interaction terms between median-dichotomized radiomic features and race/ethnicity group (AA versus non-AA). Hazards ratios (HR) and 95% confidence intervals (CI) were estimated. Overall survival (OS) was calculated from the date of diagnosis to the date of death or last follow-up using the Kaplan–Meier method. Survival time was censored if patients were lost to follow up or after 4 years. Cox regression analysis was used to identify radiomic features independently prognostic for OS after adjustment for the following clinicopathological variables: age at diagnosis, gender, tumor size, tumor grade, and stage of disease. Statistical tests were two-sided and significant at alpha = 0.05. All statistical analyses were performed using the R 3.6.0 software (https://www.R-project.org).
Results
Study Population Characteristics
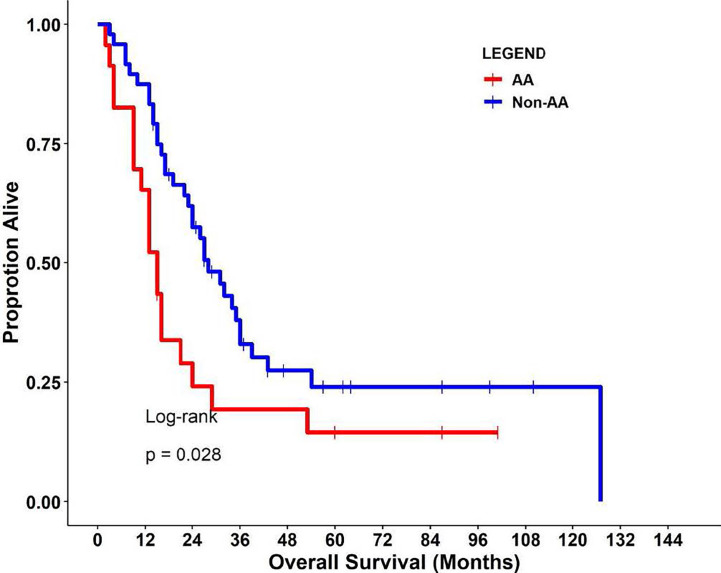
This retrospective cohort included 71 individuals diagnosed and treated for PDAC at Moffitt Cancer Center and Research Institute (Tampa, Florida) frequency-matched on age-group (+/− 5 years) and gender. Select characteristics of the study population are shown in Table 1 . There were 23 AA, 15 H/L, and 33 NHW represented, with a slightly higher percentage of females (52%, n=37). The average age at diagnosis was 64.2 years (standard deviation=10.6), and most patients had regional or distant disease. H/L cases had significantly higher numbers of regional nodes examined than AA and NHW (p=0.005), though node positivity was similar between groups (p=0.84). Finally, AA had a significantly shorter average survival time (15 months) compared to Non-AA populations (p=0.028) ( Figure 1 ).
Figure 1.
Kaplan-Meier curves (log-rank test) of overall survival in the study cohort.
CT Procedures and Standard NCCN Imaging Criteria
No significant differences were observed between racial/ethnic groups in the scanner types used (p=0.512) or in the venous phase voxel volumes (p=0.303) ( Table 2 ). Evaluation of standard imaging reporting criteria revealed three parameters that appeared to differ significantly between the three racial/ethnic groups. CT images from the AA group were found to have greater tumor involvement of the superior mesenteric vessels, as measured by degree of superior mesenteric artery (SMA) solid soft tissue contact (p=0.002), extension to the first SMA branch (p=0.036), and superior mesenteric vein (SMV) vessel narrowing and/or contour irregularity (0.033), when compared to NHW ( Table 3 ).
Table 3.
PDAC radiologic reporting template parameters for the study cohort.
| Parameter | AA (n = 20) | H/L (n = 14) | NHW (n = 33) | P value |
|---|---|---|---|---|
| Appearance (vs. parenchyma), N (%) | 0.15 | |||
| Hypodense | 18 (90.0%) | 11 (78.6%) | 22 (66.7%) | |
| Isodense | 2 (10.0%) | 3 (21.4%) | 11 (33.3%) | |
| Size (cm) [range] | 2.6 [2.0;4.3] | 2.6 [2.1;3.0] | 2.5[1.8;3.1] | 0.734 |
| Location, N (%) | 0.385 | |||
| body/tail | 5 (25.0%) | 2 (14.3%) | 4 (12.1%) | |
| head/neck | 14 (70.0%) | 12 (85.7%) | 29 (87.9%) | |
| uncinate | 1 (5.0%) | 0 (0.0%) | 0 (0.0%) | |
| Pancreatic duct narrowing/abrupt cutoff N (%) | 16 (80.0%) | 8 (57.1%) | 26 (78.8%) | 0.244 |
| Biliary tree abrupt cutoff N (%) | 0.729 | |||
| absent | 6 (30.0%) | 5 (35.7%) | 9 (27.3%) | |
| present | 8 (40.0%) | 3 (21.4%) | 9 (27.3%) | |
| stent | 6 (30.0%) | 6 (42.9%) | 15 (45.5%) | |
| Arterial evaluation | ||||
| Superior mesenteric artery (SMA) N (%) | ||||
| Solid soft tissue contact | 7 (35.0%) | 0 (0.0%) | 1 (3.0%) | 0.002 |
| Hazy attenuation/stranding contact | 4 (20.0%) | 3 (21.4%) | 4 (12.1%) | 0.232 |
| Focal vessel narrowing or contour irregularity | 1 (5.0%) | 0 | 0 | 0.309 |
| Extension to first SMA branch | 6 (30.0%) | 2 (14.3%) | 2 (6.1%) | 0.036 |
| Celiac axis N (%) | ||||
| Solid soft tissue contact | 2 (10.0%) | 0 | 0 | 0.092 |
| Hazy attenuation/stranding contact | 2 (10.0%) | 1 (7.1%) | 1 (3.0%) | 0.591 |
| Focal vessel narrowing or contour irregularity | 0 | 0 | 0 | . |
| Common Hepatic Artery (CHA) N (%) | ||||
| Solid soft tissue contact | 3 (15.0%) | 1 (7.1%) | 1 (3.0%) | 0.246 |
| Hazy attenuation/stranding contact | 3 (15.0%) | 2 (14.3%) | 2 (6.1%) | 0.49 |
| Focal vessel narrowing or contour irregularity | 1 (5.0%) | 0 | 0 | 0.309 |
| Extension to celiac axis | 1 (5.0%) | 0 | 0 | 0.309 |
| Extension to bifurcation of hepatic arteries | 1 (5.0%) | 0 | 0 | 0.309 |
| Arterial variant N (%) | ||||
| Present | 3 (15.0%) | 3 (21.4%) | 3 (9.1%) | 0.515 |
| Venous evaluation | ||||
| Main portal vein (MPV) N (%) | ||||
| Solid soft tissue contact | 7 (35.0%) | 2 (14.3%) | 9 (27.3%) | 0.398 |
| Hazy attenuation/stranding contact | 7 (35.0%) | 2 (14.3%) | 9 (27.3%) | 0.398 |
| Focal vessel narrowing or contour irregularity | 8 (40.0%) | 1 (7.1%) | 4 (12.1%) | 0.274 |
| Superior mesenteric vein (SMV) N (%) | ||||
| Solid soft tissue contact | 12 (60.0%) | 3 (21.4%) | 10 (30.3%) | 0.055 |
| Hazy attenuation/stranding contact | 4 (20.0%) | 4 (28.6%) | 8 (24.2%) | 0.846 |
| Focal vessel narrowing or contour irregularity | 8 (40.0%) | 1 (7.1%) | 5 (15.1%) | 0.033 |
| Extrapancreatic evaluation N (%) | ||||
| Liver lesions | 3 (15.0%) | 2 (14.3%) | 3 (9.1%) | 0.779 |
| Peritoneal or omental nodules | 1 (5.0%) | 1 (7.1%) | 1 (3.0%) | 0.818 |
| Ascites | 1 (5.0%) | 0 | 0 | 0.309 |
| Suspicious lymph nodes | 8 (40.0%) | 3 (21.4%) | 8 (24.2%) | 0.385 |
| Venous collaterals | 5 (25.0%) | 3 (21.4%) | 4 (12.1%) | 0.465 |
Possible program errors were observed when contouring inferior margin of mass for one H/L case having a tumor with a cystic component.
Three AA cases also do not have these parameters generated and are not included in this table.
PDAC, pancreatic ductal adenocarcinoma; AA, African American; H/L, Hispanic/Latinx; NHW, non-Hispanic White; cm, centimeters.
Bold font indicates a P value < 0.05.
Radiomic Features
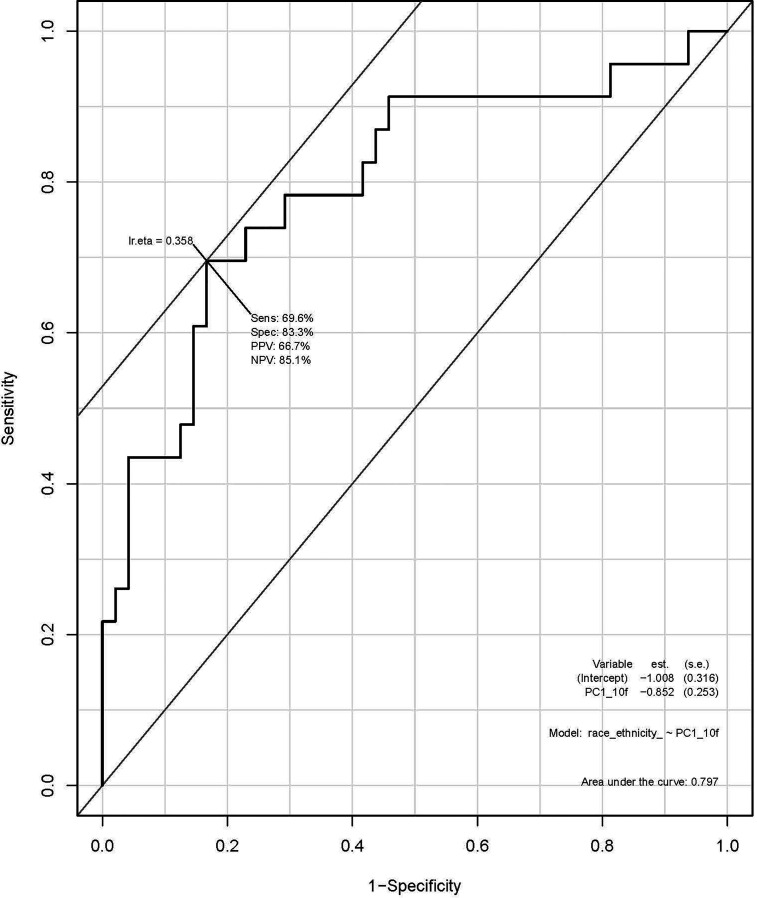
A total of 135 textural and non-textural radiomic features were evaluated for their association with race/ethnicity. Kruskal-Wallis test results indicated that 30 features were significantly associated with race/ethnicity (adjusted p<0.02; Table 4 ). Furthermore, 10 radiomic features were highly associated with race independent of tumor grade and included sphericity, volumetric mean Hounsfield units (HU), minimum HU, coefficient of variation HU, four gray level texture features, and two wavelet texture features ( Supplementary Table 1 ). A multivariable model using principal component analysis to represent the radiomic signature yielded an area underneath the curve (AUC)=0.80 in differentiating AA versus non-AA ( Figure 2 ).
Table 4.
Radiomic features evaluated in this study and their univariate association with race/ethnicity.
| AA N = 23 | H/L N = 15 | NHW N = 33 | P overall | |
|---|---|---|---|---|
| anterior_posterior_length_mm mean[95%CI] | 25.0 [18.0;28.5] | 27.0 [17.5;30.0] | 25.0 [19.0;28.0] | 0.853 |
| asphericity | 0.22 [0.17;0.33] | 0.22 [0.15;0.26] | 0.17 [0.14;0.23] | 0.090 |
| coefficient_of_variation | 0.34 [0.29;0.44] | 0.38 [0.28;0.66] | 0.49 [0.41;0.69] | 0.002 |
| cranial_caudal_length_mm | 27.0 [19.0;32.0] | 27.0 [21.0;30.5] | 24.0 [19.0;35.0] | 0.903 |
| elongation | 0.87 [0.71;0.93] | 0.79 [0.68;0.87] | 0.78 [0.68;0.88] | 0.413 |
| energy_intensity2 | 5.27 [2.81;10.32]x109 | 4.30 [2.55;10.39]x109 | 4.34 [2.51;12.06]x109 | 0.964 |
| energy_of_ct_number_hu2 | 3.03 [1.41;6.67]x107 | 3.50 [1.49;6.98]x107 | 2.56 [0.83;5.51]x107 | 0.458 |
| entropy_hu | 6.70 [6.55;6.90] | 6.70 [6.45;7.00] | 6.90 [6.70;7.00] | 0.042 |
| flatness | 0.58 [0.51;0.72] | 0.62 [0.55;0.69] | 0.65 [0.57;0.73] | 0.626 |
| glcm_avg_angular_second_moment | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | . |
| glcm_avg_column_mean | 82.2 [68.0;93.9] | 77.1 [46.1;90.7] | 65.0 [42.7;81.7] | 0.021 |
| glcm_avg_column_standard_deviation | 30.6 [28.1;40.5] | 29.9 [24.7;37.6] | 36.7 [31.2;43.9] | 0.037 |
| glcm_avg_column_var | 943 [798;1760] | 907 [611;1435] | 1370 [973;2113] | 0.062 |
| glcm_avg_contrast | 1207 [888;1637] | 1087 [761;1529] | 1580 [1051;2130] | 0.055 |
| glcm_avg_correlation | 0.38 [0.28;0.42] | 0.35 [0.28;0.40] | 0.40 [0.34;0.43] | 0.325 |
| glcm_avg_dissimilarity | 25.4 [22.5;28.8] | 25.3 [21.1;28.8] | 28.4 [25.2;33.0] | 0.031 |
| glcm_avg_energy | 0.02 [0.02;0.03] | 0.02 [0.01;0.03] | 0.02 [0.02;0.02] | 0.971 |
| glcm_avg_entropy | 11.5 [10.7;12.1] | 11.5 [10.6;12.1] | 11.5 [10.9;12.3] | 0.966 |
| glcm_avg_homogeneity | 0.04 [0.04;0.05] | 0.04 [0.04;0.05] | 0.04 [0.03;0.04] | 0.067 |
| glcm_avg_row_mean | 83.9 [67.2;93.8] | 76.7 [40.1;90.5] | 66.5 [41.0;79.5] | 0.027 |
| glcm_avg_row_standard_deviation | 27.2 [24.5;30.9] | 25.6 [22.5;31.8] | 32.2 [26.4;35.2] | 0.014 |
| glcm_avg_row_var | 741 [602;952] | 656 [507;1010] | 1039 [695;1237] | 0.014 |
| glcm_cluster_prominence†* | 136 [59.5;274] | 85.8 [47.0;185] | 187 [93.9;368] | 0.035 |
| glcm_cluster_shade†* | -1.70 [-6.45;-0.05] | -1.40 [-2.55;2.10] | -1.40 [-4.50;6.30] | 0.721 |
| glcm_cluster_tendency†* | 6.10 [4.20;7.40] | 4.90 [3.65;7.75] | 7.60 [5.20;8.70] | 0.029 |
| glcm_contrast†* | 1.70 [1.55;2.00] | 1.70 [1.30;2.15] | 2.10 [1.50;2.80] | 0.040 |
| glcm_correlation†* | 0.50 [0.45;0.60] | 0.50 [0.40;0.60] | 0.50 [0.50;0.60] | 0.374 |
| glcm_difference_average†* | 1.00 [0.90;1.05] | 1.00 [0.80;1.10] | 1.10 [0.90;1.20] | 0.096 |
| glcm_difference_entropy†* | 1.70 [1.70;1.85] | 1.70 [1.60;1.90] | 1.80 [1.70;2.00] | 0.111 |
| glcm_difference_variance†* | 0.70 [0.70;0.90] | 0.70 [0.60;0.90] | 1.00 [0.70;1.20] | 0.027 |
| glcm_dissimilarity†* | 1.00 [0.90;1.05] | 1.00 [0.80;1.10] | 1.10 [0.90;1.20] | 0.096 |
| glcm_first_measure_of_information_correlation†* | -0.10 [-0.15;-0.10] | -0.10 [-0.10;-0.10] | -0.10 [-0.10;-0.10] | 0.702 |
| glcm_inverse_difference†* | 0.60 [0.60;0.60] | 0.60 [0.60;0.65] | 0.60 [0.60;0.60] | 0.084 |
| glcm_inverse_difference_moment†* | 0.60 [0.55;0.60] | 0.60 [0.50;0.60] | 0.60 [0.50;0.60] | 0.077 |
| glcm_inverse_difference_moment_normalized†* | 1.00 [1.00;1.00] | 1.00 [1.00;1.00] | 1.00 [1.00;1.00] | . |
| glcm_inverse_difference_normalized†* | 0.90 [0.90;0.90] | 0.90 [0.90;0.90] | 0.90 [0.90;0.90] | 0.717 |
| glcm_inverse_variance†* | 0.50 [0.50;0.50] | 0.50 [0.50;0.50] | 0.50 [0.50;0.50] | 0.347 |
| glcm_joint_average†* | 6.90 [5.85;8.15] | 6.20 [5.90;7.70] | 7.30 [6.50;8.30] | 0.335 |
| glcm_joint_entropy†* | 4.70 [4.50;4.90] | 4.60 [4.30;4.95] | 5.00 [4.70;5.20] | 0.029 |
| glcm_joint_maximum†* | 0.10 [0.10;0.10] | 0.10 [0.10;0.10] | 0.10 [0.10;0.10] | 0.331 |
| glcm_joint_variance†* | 2.00 [1.55;2.25] | 1.60 [1.35;2.40] | 2.30 [1.80;2.90] | 0.027 |
| glcm_second_measure_of_information_correlation†* | 0.60 [0.50;0.70] | 0.60 [0.50;0.65] | 0.60 [0.50;0.70] | 0.470 |
| glcm_sum_average†* | 13.7 [11.8;16.3] | 12.4 [11.8;15.4] | 14.5 [12.9;16.5] | 0.347 |
| glcm_sum_entropy†* | 3.30 [3.10;3.40] | 3.20 [2.95;3.50] | 3.50 [3.20;3.60] | 0.059 |
| glcm_sum_variance†* | 5.20 [3.75;6.40] | 4.30 [3.30;7.00] | 6.40 [4.80;7.90] | 0.045 |
| gldzm_grey_level_nonuniformity | 41.6 [25.3;90.2] | 44.7 [23.1;83.9] | 42.3 [22.0;84.0] | 0.827 |
| gldzm_grey_level_nonuniformity_normalised | 0.10 [0.10;0.20] | 0.20 [0.10;0.20] | 0.10 [0.10;0.10] | 0.003 |
| gldzm_grey_level_variance | 5.60 [5.05;7.60] | 6.20 [4.65;8.00] | 7.60 [6.60;9.40] | 0.012 |
| gldzm_high_grey_level_zone_emphasis | 50.5 [42.0;79.0] | 48.8 [38.5;63.2] | 53.6 [44.3;78.8] | 0.442 |
| gldzm_large_distance_emphasis§ | 2.50 [1.65;3.65] | 2.70 [1.80;3.65] | 2.70 [1.80;6.10] | 0.709 |
| gldzm_large_distance_high_grey_level_emphasis§ | 107 [71.6;192] | 167 [80.7;216] | 129 [87.8;267] | 0.603 |
| gldzm_large_distance_low_grey_level_emphasis§ | 0.10 [0.10;0.15] | 0.10 [0.10;0.20] | 0.10 [0.10;0.20] | 0.584 |
| gldzm_low_grey_level_zone_emphasis | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.972 |
| gldzm_small_distance_emphasis§ | 0.80 [0.70;0.90] | 0.80 [0.70;0.90] | 0.80 [0.70;0.90] | 0.964 |
| gldzm_small_distance_high_grey_level_emphasis§ | 44.7 [37.1;50.3] | 41.1 [25.2;48.7] | 43.9 [36.6;65.0] | 0.515 |
| gldzm_small_distance_low_grey_level_emphasis§ | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.10] | 0.665 |
| gldzm_zone_distance_entropy | 3.80 [3.55;4.35] | 4.10 [3.60;4.55] | 4.20 [3.80;4.60] | 0.342 |
| gldzm_zone_distance_nonuniformity | 211 [98.3;270] | 159 [92.8;272] | 180 [120;242] | 0.970 |
| gldzm_zone_distance_nonuniformity_normalised | 0.60 [0.50;0.70] | 0.60 [0.50;0.70] | 0.60 [0.40;0.70] | 0.924 |
| gldzm_zone_distance_variance | 0.60 [0.20;1.05] | 0.60 [0.30;1.05] | 0.50 [0.30;1.50] | 0.867 |
| gldzm_zone_percentage§ | 0.10 [0.10;0.10] | 0.10 [0.00;0.10] | 0.10 [0.10;0.10] | 0.135 |
| glrlm_grey_level_nonuniformity‡ | 5986 [2823;10223] | 6001 [2708;10739] | 6170 [3615;13394] | 0.841 |
| glrlm_grey_level_variance‡ | 2.10 [1.75;2.70] | 1.90 [1.50;2.95] | 2.90 [2.00;3.40] | 0.012 |
| glrlm_high_grey_level_run_emphasis* | 46.6 [36.7;68.3] | 40.4 [37.1;60.9] | 53.9 [43.2;71.9] | 0.342 |
| glrlm_normalized_grey_level_nonuniformity‡ | 0.20 [0.20;0.20] | 0.20 [0.20;0.20] | 0.20 [0.20;0.20] | 0.567 |
| glszm_grey_level_nonuniformity‡ | 41.6 [25.3;90.2] | 44.7 [23.1;83.9] | 42.3 [22.0;84.0] | 0.827 |
| glszm_grey_level_variance‡ | 5.60 [5.05;7.60] | 6.20 [4.65;8.00] | 7.60 [6.60;9.40] | 0.012 |
| glszm_high_grey_level_zone_emphasis‡ | 50.5 [42.0;79.0] | 48.8 [38.5;63.2] | 53.6 [44.3;78.8] | 0.442 |
| glszm_large_zone_emphasis‡ | 13080 [4011;36509] | 12030 [3365;33727] | 7602 [2833;30157] | 0.742 |
| glszm_large_zone_high_grey_level_emphasis‡ | 3.89 [1.81;22.75]x105 | 4.01 [1.81;17.93]x105 | 3.88 [1.46;12.94]x105 | 0.884 |
| glszm_large_zone_low_grey_level_emphasis‡ | 344 [88.8;608] | 226 [120;932] | 174 [51.7;622] | 0.530 |
| glszm_low_grey_level_zone_emphasis‡ | 0.00 [0.00;0.10] | 0.00 [0.00;0.10] | 0.00 [0.00;0.10] | 0.814 |
| glszm_normalised_zone_size_nonuniformity‡ | 0.30 [0.30;0.35] | 0.30 [0.30;0.40] | 0.30 [0.30;0.40] | 0.677 |
| glszm_normalized_grey_level_nonuniformity‡ | 0.10 [0.10;0.20] | 0.20 [0.10;0.20] | 0.10 [0.10;0.10] | 0.003 |
| glszm_small_zone_emphasis‡ | 0.60 [0.50;0.60] | 0.60 [0.60;0.60] | 0.60 [0.60;0.60] | 0.023 |
| glszm_small_zone_high_grey_level_emphasis‡ | 29.8 [24.5;46.5] | 31.4 [24.7;37.2] | 33.1 [27.5;53.3] | 0.388 |
| glszm_small_zone_low_grey_level_emphasis‡ | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.445 |
| glszm_zone_percentage‡ | 0.10 [0.10;0.10] | 0.10 [0.00;0.10] | 0.10 [0.10;0.10] | 0.135 |
| glszm_zone_size_entropy‡ | 5.10 [4.80;5.50] | 4.80 [4.65;5.35] | 5.30 [5.00;5.50] | 0.085 |
| glszm_zone_size_nonuniformity‡ | 111 [46.7;169] | 129 [34.4;172] | 108 [63.4;190] | 0.981 |
| glszm_zone_size_variance‡ | 12621 [3911;35884] | 11642 [3255;33387] | 7458 [2756;29728] | 0.742 |
| hu_kurtosis | 3.58 [3.24;4.16] | 3.34 [3.13;4.08] | 3.49 [3.13;4.06] | 0.723 |
| hu_skewness | -0.20 [-0.35;0.02] | -0.05 [-0.26;0.13] | -0.10 [-0.30;0.28] | 0.486 |
| hu_uniformity | 66.2 [56.6;71.3] | 62.0 [34.1;72.3] | 50.6 [30.6;59.1] | 0.002 |
| hu_uniformity_acr | -33.30 [-65.15;-10.45] | -49.20 [-116.95;-1.35] | -85.70 [-117.80;-41.00] | 0.061 |
| maximum_ct_number_hu | 175 [160;210] | 190 [151;238] | 181 [159;231] | 0.999 |
| median_ct_number_hu | 83.0 [67.0;94.5] | 78.0 [39.0;90.5] | 67.5 [41.0;82.0] | 0.045 |
| mesh_compactness_1_mm | 20.9 [14.4;31.1] | 22.4 [16.5;33.4] | 23.9 [16.6;34.2] | 0.895 |
| mesh_compactness_2_mm | 0.55 [0.43;0.63] | 0.56 [0.50;0.67] | 0.62 [0.53;0.68] | 0.092 |
| mesh_sa_to_volume_ratio | 0.32 [0.26;0.42] | 0.31 [0.26;0.39] | 0.29 [0.24;0.38] | 0.804 |
| minimum_ct_number_hu | -30.00 [-57.50;-14.00] | -39.00 [-83.00;-15.00] | -58.00 [-74.00;-46.00] | 0.021 |
| ngldm_dependence_count_percentage‡ | 1.00 [1.00;1.00] | 1.00 [1.00;1.00] | 1.00 [1.00;1.00] | . |
| ngldm_dependence_energy‡ | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.034 |
| ngldm_dependence_entropy‡ | 5.80 [5.75;6.15] | 5.80 [5.70;6.10] | 6.00 [5.80;6.20] | 0.212 |
| ngldm_dependence_nonuniformity‡ | 380 [192;771] | 326 [167;675] | 331 [187;747] | 0.994 |
| ngldm_dependence_variance‡ | 11.4 [9.85;12.7] | 10.1 [9.75;14.6] | 10.4 [7.90;12.4] | 0.466 |
| ngldm_gl_nonuniformity‡ | 895 [485;2395] | 720 [406;2023] | 767 [386;1999] | 0.808 |
| ngldm_gl_variance‡ | 2.00 [1.55;2.45] | 1.70 [1.35;2.60] | 2.70 [1.80;3.20] | 0.013 |
| ngldm_high_dependence_emphasis‡ | 56.1 [45.9;67.8] | 58.8 [48.2;78.6] | 51.6 [37.8;62.7] | 0.361 |
| ngldm_high_dependence_high_gl_emphasis‡ | 2250 [1966;3719] | 2594 [1677;3588] | 2946 [1948;4005] | 0.964 |
| ngldm_high_dependence_low_gl_emphasis‡ | 1.10 [0.85;1.70] | 1.50 [0.80;2.40] | 1.00 [0.80;1.50] | 0.330 |
| ngldm_high_gl_dependence‡ | 49.9 [36.2;69.2] | 39.7 [37.0;62.2] | 54.4 [45.4;68.9] | 0.360 |
| ngldm_low_dependence_emphasis‡ | 0.10 [0.10;0.10] | 0.10 [0.10;0.10] | 0.10 [0.10;0.10] | 0.088 |
| ngldm_low_dependence_high_gl_emphasis‡ | 5.10 [3.45;5.75] | 4.40 [2.20;6.40] | 5.30 [3.80;8.20] | 0.186 |
| ngldm_low_dependence_low_gl_emphasis‡ | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | . |
| ngldm_low_gl_dependence‡ | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.879 |
| ngldm_normalized_dependence_nonuniformity‡ | 0.10 [0.10;0.10] | 0.10 [0.10;0.10] | 0.10 [0.10;0.10] | 0.034 |
| ngldm_normalized_gl_nonuniformity‡ | 0.20 [0.20;0.20] | 0.20 [0.20;0.30] | 0.20 [0.20;0.20] | 0.008 |
| rms_of_ct_number_hu | 89.6 [71.4;97.0] | 83.9 [49.3;94.8] | 74.3 [50.5;87.2] | 0.067 |
| sphericity | 0.82 [0.75;0.86] | 0.82 [0.79;0.87] | 0.85 [0.81;0.88] | 0.084 |
| standard_deviation_of_ct_number_hu | 27.2 [24.6;30.9] | 25.6 [22.7;31.8] | 32.3 [26.4;35.3] | 0.014 |
| surface_area_mm2 | 2077 [1241;3104] | 2215 [1464;3284] | 1771 [1408;3043] | 0.879 |
| transverse_length_mm | 26.0 [20.0;33.0] | 24.0 [22.5;31.5] | 25.0 [19.0;29.0] | 0.716 |
| volume_mm3 | 5522 [2863;11750] | 6748 [3892;13092] | 6075 [4018;12569] | 0.919 |
| volumetric_length_mm | 34.0 [25.0;39.0] | 35.0 [26.0;45.0] | 31.0 [28.0;38.0] | 0.810 |
| volumetric_mean_of_ct_number_hu | 83.9 [67.2;93.8] | 76.7 [40.1;90.5] | 66.5 [41.0;79.5] | 0.027 |
| wavelet_hhl_10th_percentile_hu | -4.80 [-5.80;-4.00] | -5.30 [-5.85;-4.70] | -5.2 0[-6.90;-4.30] | 0.310 |
| wavelet_hhl_90th_percentile_hu | 4.90 [4.10;5.85] | 5.30 [4.65;5.80] | 5.30 [4.30;7.00] | 0.454 |
| wavelet_hhl_coefficient_of_variation | 320 [-92.90;753] | 114 [-379.00;175] | -80.70 [-454.90;244] | 0.163 |
| wavelet_hhl_energy_hu2 | 5.90 [3.08;15.48]x104 | 1.04 [0.29;1.87]x105 | 9.86 [4.00;15.98]x104 | 0.707 |
| wavelet_hhl_entropy | 12.0 [11.2;13.3] | 11.7 [10.9;13.1] | 11.8 [11.1;13.2] | 0.980 |
| wavelet_hhl_excess_kurtosis | 0.10 [0.00;0.25] | 0.20 [0.10;0.40] | 0.20 [0.10;0.50] | 0.104 |
| wavelet_hhl_interquartile_range_hu | 5.10 [4.25;6.10] | 5.40 [4.90;5.95] | 5.60 [4.70;7.20] | 0.466 |
| wavelet_hhl_minimum_hu | -16.20 [-19.05;-12.35] | -19.50 [-23.70;-13.75] | -17.80 [-28.80;-14.40] | 0.163 |
| wavelet_hhl_maximum_hu | 16.6 [11.6;18.2] | 18.5 [15.3;22.7] | 18.3 [14.0;26.5] | 0.129 |
| wavelet_hhl_mean_deviation_hu | 3.00 [2.50;3.60] | 3.30 [2.90;3.55] | 3.30 [2.70;4.30] | 0.321 |
| wavelet_hhl_mean_hu | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.854 |
| wavelet_hhl_median_deviation_hu | 3.00 [2.50;3.60] | 3.30 [2.90;3.55] | 3.30 [2.70;4.30] | 0.326 |
| wavelet_hhl_median_hu | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.10] | 0.653 |
| wavelet_hhl_quartile_coefficient_of_dispersion | 45.7 [-105.15;289] | 34.2 [-81.30;63.6] | 63.6 [-66.20;179] | 0.315 |
| wavelet_hhl_range_hu | 32.4 [24.0;38.0] | 39.7 [28.8;50.5] | 35.9 [28.3;54.8] | 0.157 |
| wavelet_hhl_robust_mean_deviation_hu | 2.10 [1.75;2.50] | 2.20 [2.00;2.50] | 2.30 [1.90;3.00] | 0.442 |
| wavelet_hhl_rms_hu | 3.80 [3.20;4.60] | 4.30 [3.65;4.55] | 4.30 [3.40;5.50] | 0.254 |
| wavelet_hhl_skeweness | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.00 [0.00;0.00] | 0.419 |
| wavelet_hhl_variance_hu2 | 14.3 [10.2;20.9] | 18.3 [13.3;20.6] | 18.8 [11.6;30.2] | 0.218 |
Some P values are missing because they were unable to be estimated.
AA, African American; H/L, Hispanic/Latinx; NHW, non-Hispanic White; MM, millimeters; CT, computed tomography; HU, Hounsfield Units; GLCM, gray level cooccurrence matrix; AVG, average; VAR, variance; GLDZM, gray level distance zone matrix; American College of Radiology; SA, surface area; NGLDM, neighborhood gray-level different matrix; GL, gray level; RMS, root mean square; HHL, high-pass high-pass and low-pass filters.
†gray leveled image; *ibsi by slice with merging; ‡as volume with full merging; §with full merging.
Bold font indicates a P value < 0.05.
Figure 2.
Receiver operating characteristic (ROC) curve using principal component analysis to identify radiomic features predictive of race/ethnicity.
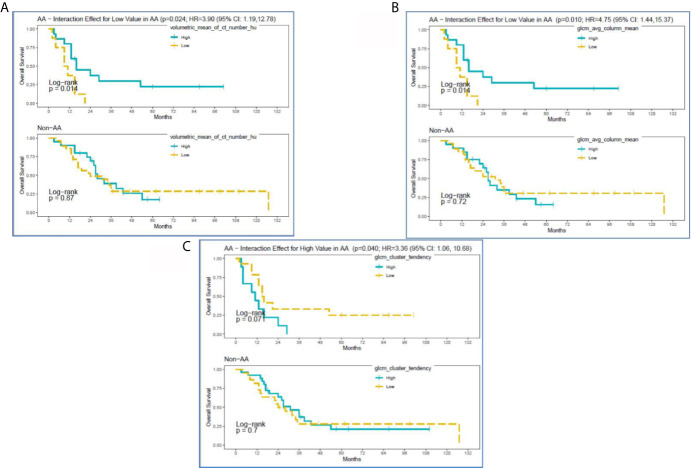
Survival analysis identified the following non-correlated radiomic features with a significantly different survival difference between AA and non-AA (interaction effect between radiomic features and race with p<0.05): Volumetric Mean CT (HU) (HR: 3.90 (95% CI:1.19–12.78), p=0.024), GLCM Avg Column Mean (HR:4.75 (95% CI: 1.44,15.37), p=0.010), and GLCM Cluster Tendency (HR:3.36 (95% CI: 1.06–10.68), p=0.040) ( Supplementary Table 2 ). Specifically, for Volumetric Mean CT and GLCM Avg Column Mean in tumors, low value of these radiomic features was associated with poorer survival among AA ( Figures 3A, B ). In contrast, survival curves overlapped between low and high groups of the radiomic features among non-AA. As a result, survival differences due to the radiomic features became differential between racial/ethnic groups (p=0.01–0.02). The GLCM Cluster Tendency ( Figure 3C ) had an opposite trend with high values associated with poorer survival among AA, but slightly improved survival among non-AA, leading to a significant differential survival difference between AA and non-AA (p=0.04). Furthermore, multivariate survival analysis indicated that Volumetric Mean CT (HU) and GLCM Avg Column Mean remain significantly associated with OS between AA and non-AA after adjustment for clinical-pathological features including age at diagnosis, gender, tumor size, tumor grade, and SEER-derived stage. Lower values of these radiomic features were associated with worse survival among AA ( Supplementary Table 3 ).
Figure 3.
Kaplan-Meier curves for significant interactions between radiomic features and overall survival among self-reported African American (AA) and Non-AA (Hispanic/Latinx, H/L; and Non-Hispanic White, NHW) groups according to (A) Volumetric Mean CT (HU), (B) GLCM Avg Column Mean, and (C) GLCM Cluster Tendency.
Figure 4 reveals pretreatment CT images for three PDAC patients matched on tumor grade, gender, and age-group; lower radiomic values were observed among tumors from AA in volumetric mean CT HU and two GLCM texture features, compared to non-AA. These observations suggest that although the pancreatic tumors may appear similar on CT images, they reflect significantly different radiomic values associated with race/ethnicity and are predictive of overall survival.
Figure 4.
Axial venous phase CT images are presented in PDAC patients matched for tumor grade, gender, and age-group. Image (A) from an AA patient and shows a poorly defined hypoenhancing tumor marked by the yellow arrows. Image (B) in a NHW shows a similar radiologic appearance of the tumor but with significantly different radiomic tumor values. Image (C) in a Hispanic patient also had radiomic values different from the AA case. Note that a common bile duct stent is present in each of these patients.
Discussion
We conducted the first investigation we are aware of to apply a radiomic approach to routine pretreatment CT scans from patients with PDAC to specifically explore associations with race/ethnicity and overall survival. Our analysis showed AA patients with low volumetric mean HU tumors had worse survival than similar tumors in non-AA. In PDAC, tumors with HU lower than surrounding pancreatic parenchyma have been correlated with worse outcomes (44). In our study, the low volumetric mean HU may be revealing a similar relationship to survival as the previously reported relative delta score, except that our measure is based in absolute HU as opposed to the delta score, which reflects relative differences in HU. Our analysis also demonstrated worse survival in AA patients having high coefficient of variation HU compared to similar tumors in non-AA, independent of key prognostic factors. The coefficient of variation HU is a reflection of tumor heterogeneity as it presents on CT based on voxel HU values, and it represents the standard deviation of the HU values within segmented tumors divided by the mean HU. Therefore, tumors with a wider range of different-appearing voxels within a tumor will have a larger coefficient of variation HU. In line with these findings, previous studies have shown that more heterogenous tumors are associated with high-grade dysplasia, resistance to anticancer therapies, and poorer prognoses (1, 45–48).
In this study, radiomics allowed us to preoperatively and non-invasively quantify the differences in appearance of pancreatic tumors across different racially and ethnically defined cohorts, even where the differences were not easy to visualize or describe qualitatively. We discovered multiple radiomic features that predict poor survival specifically in AA patients independent of other demographic and clinical factors. It is possible that these radiomic differences reflect inherent biological tumor differences specific to each ethnic group. Having potential poor prognostic biomarkers available in the pretreatment setting could influence clinical decisions and support earlier and more aggressive treatments that could reduce disparities for these underserved groups. Additionally, future studies correlating race/ethnicity-based radiomic features with tumor tissue-based biomarkers are needed to determine the capacity at which radiomics can be used in clinical decision-making workflows at the time of multidisciplinary tumor board.
We realize that the single-institutional retrospective design is prone to biases, but there is wealth in this exploratory investigation. Future prospective multicenter studies involving racially diverse cohorts of PDAC cases will be needed to continue to move PDAC disparities research forward. We plan to optimize and validate the most promising radiomic features and biomarkers in an independent cohort of AA PC cases using our multi-institutional Florida Pancreas Collaborative infrastructure (49). Furthermore, we plan to conduct a radiogenomic approach that integrates CT radiomic data with molecular biomarker data from pancreatic tumor tissue in order to uncover biological mechanisms to explain the disproportionate PDAC burden in AA.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Advarra IRB (MCC# 19431; IRB #: Pro00024543). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JP, DJ, and JC contributed to conception and design of the study. SV organized the database. JL and DT-C performed the statistical analysis. JP wrote the first draft of the manuscript, and JL, D-TC, DJ, and JC wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded in part through the George Edgecomb Society awarded to JP, JC, DJ, and D-TC. The research was also supported by the Quantitative Imaging Core and the Biostatistics and Bioinformatics Shared Resource at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.712950/full#supplementary-material
References
- 1. Burrell RA, McGranahan N, Bartek J, Swanton C. The Causes and Consequences of Genetic Heterogeneity in Cancer Evolution. Nature (2013) 501(7467):338–45. 10.1038/nature12625 [DOI] [PubMed] [Google Scholar]
- 2. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res (2014) 74(11):2913–21. 10.1158/0008-5472.Can-14-0155 [DOI] [PubMed] [Google Scholar]
- 3. Howlader N, Noone Am, Krapcho M, Miller D, Bishop K, Altekruse Sf, et al. Seer Cancer Statistics Review, 1975-2013. Bethesda, Md: National Cancer Institute. Based on November 2015 Seer Data Submission, Posted to the Seer Web Site, April (2016). [Google Scholar]
- 4. Abraham A, Al-Refaie WB, Parsons HM, Dudeja V, Vickers SM, Habermann EB. Disparities in Pancreas Cancer Care. Ann Surg Oncol (2013) 20(6):2078–87. 10.1245/s10434-012-2843-z [DOI] [PubMed] [Google Scholar]
- 5. Chang KJ, Parasher G, Christie C, Largent J, Anton-Culver H. Risk of Pancreatic Adenocarcinoma: Disparity Between African Americans and Other Race/Ethnic Groups. Cancer (2005) 103(2):349–57. 10.1002/cncr.20771 [DOI] [PubMed] [Google Scholar]
- 6. Riall TS, Townsend CM, Jr., Kuo YF, Freeman JL, Goodwin JS. Dissecting Racial Disparities in the Treatment of Patients With Locoregional Pancreatic Cancer: A 2-Step Process. Cancer (2010) 116(4):930–9. 10.1002/cncr.24836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singal V, Singal AK, Kuo YF. Racial Disparities in Treatment for Pancreatic Cancer and Impact on Survival: A Population-Based Analysis. J Cancer Res Clin Oncol (2012) 138(4):715–22. 10.1007/s00432-012-1156-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wray CJ, Castro-Echeverry E, Silberfein EJ, Ko TC, Kao LS. A Multi-Institutional Study of Pancreatic Cancer in Harris County, Texas: Race Predicts Treatment and Survival. Ann Surg Oncol (2012) 19(9):2776–81. 10.1245/s10434-012-2361-z [DOI] [PubMed] [Google Scholar]
- 9. Murphy MM, Simons JP, Hill JS, McDade TP, Chau Ng S, Whalen GF, et al. Pancreatic Resection: A Key Component to Reducing Racial Disparities in Pancreatic Adenocarcinoma. Cancer (2009) 115(17):3979–90. 10.1002/cncr.24433 [DOI] [PubMed] [Google Scholar]
- 10. Zell JA, Rhee JM, Ziogas A, Lipkin SM, Anton-Culver H. Race, Socioeconomic Status, Treatment, and Survival Time Among Pancreatic Cancer Cases in California. Cancer Epidemiol Biomarkers Prev (2007) 16(3):546–52. 10.1158/1055-9965.epi-06-0893 [DOI] [PubMed] [Google Scholar]
- 11. Murphy MM, Simons JP, Ng SC, McDade TP, Smith JK, Shah SA, et al. Racial Differences in Cancer Specialist Consultation, Treatment, and Outcomes for Locoregional Pancreatic Adenocarcinoma. Ann Surg Oncol (2009) 16(11):2968–77. 10.1245/s10434-009-0656-5 [DOI] [PubMed] [Google Scholar]
- 12. DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, et al. Cancer Statistics for African Americans, 2016: Progress and Opportunities in Reducing Racial Disparities. CA Cancer J Clin (2016) 66(4):290–308. 10.3322/caac.21340 [DOI] [PubMed] [Google Scholar]
- 13. Kaissis G, Braren R. Pancreatic Cancer Detection and Characterization-State of the Art Cross-Sectional Imaging and Imaging Data Analysis. Transl Gastroenterol Hepatol (2019) 4:35. 10.21037/tgh.2019.05.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: The Process and the Challenges. Magn Reson Imaging (2012) 30(9):1234–48. 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coroller TP, Grossmann P, Hou Y, Rios Velazquez E, Leijenaar RT, Hermann G, et al. Ct-Based Radiomic Signature Predicts Distant Metastasis in Lung Adenocarcinoma. Radiother Oncol (2015) 114(3):345–50. 10.1016/j.radonc.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grove O, Berglund AE, Schabath MB, Aerts HJ, Dekker A, Wang H, et al. Quantitative Computed Tomographic Descriptors Associate Tumor Shape Complexity and Intratumor Heterogeneity With Prognosis in Lung Adenocarcinoma. PloS One (2015) 10(3):e0118261. 10.1371/journal.pone.0118261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leijenaar RT, Carvalho S, Velazquez ER, van Elmpt WJ, Parmar C, Hoekstra OS, et al. Stability of Fdg-Pet Radiomics Features: An Integrated Analysis of Test-Retest and Inter-Observer Variability. Acta Oncol (2013) 52(7):1391–7. 10.3109/0284186x.2013.812798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balagurunathan Y, Kumar V, Gu Y, Kim J, Wang H, Liu Y, et al. Test-Retest Reproducibility Analysis of Lung Ct Image Features. J Digit Imaging (2014) 27(6):805–23. 10.1007/s10278-014-9716-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balagurunathan Y, Gu Y, Wang H, Kumar V, Grove O, Hawkins S, et al. Reproducibility and Prognosis of Quantitative Features Extracted From Ct Images. Transl Oncol (2014) 7(1):72–87. 10.1593/tlo.13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gatenby RA, Grove O, Gillies RJ. Quantitative Imaging in Cancer Evolution and Ecology. Radiology (2013) 269(1):8–15. 10.1148/radiol.13122697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou M, Hall L, Goldgof D, Russo R, Balagurunathan Y, Gillies R, et al. Radiologically Defined Ecological Dynamics and Clinical Outcomes in Glioblastoma Multiforme: Preliminary Results. Transl Oncol (2014) 7(1):5–13. 10.1593/tlo.13730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Permuth JB, Choi J, Balarunathan Y, Kim J, Chen DT, Chen L, et al. Combining Radiomic Features With a Mirna Classifier May Improve Prediction of Malignant Pathology for Pancreatic Intraductal Papillary Mucinous Neoplasms. Oncotarget (2016) 7(52):85785–97. 10.18632/oncotarget.11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Permuth JB, Choi JW, Chen DT, Jiang K, DeNicola G, Li JN, et al. A Pilot Study of Radiologic Measures of Abdominal Adiposity: Weighty Contributors to Early Pancreatic Carcinogenesis Worth Evaluating? Cancer Biol Med (2017) 14(1):66–73. 10.20892/j.issn.2095-3941.2017.0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polk SL, Choi JW, McGettigan MJ, Rose T, Ahmed A, Kim J, et al. Multiphase Computed Tomography Radiomics of Pancreatic Intraductal Papillary Mucinous Neoplasms to Predict Malignancy. World J Gastroenterol (2020) 26(24):3458–71. 10.3748/wjg.v26.i24.3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith AD, Gray MR, Del Campo SM, Shlapak D, Ganeshan B, Zhang X, et al. Predicting Overall Survival in Patients With Metastatic Melanoma on Antiangiogenic Therapy and Recist Stable Disease on Initial Posttherapy Images Using Ct Texture Analysis. AJR Am J Roentgenol (2015) 205(3):W283–93. 10.2214/ajr.15.14315 [DOI] [PubMed] [Google Scholar]
- 26. Skogen K, Ganeshan B, Good C, Critchley G, Miles K. Measurements of Heterogeneity in Gliomas on Computed Tomography Relationship to Tumour Grade. J Neurooncol (2013) 111(2):213–9. 10.1007/s11060-012-1010-5 [DOI] [PubMed] [Google Scholar]
- 27. Ganeshan B, Skogen K, Pressney I, Coutroubis D, Miles K. Tumour Heterogeneity in Oesophageal Cancer Assessed by Ct Texture Analysis: Preliminary Evidence of an Association With Tumour Metabolism, Stage, and Survival. Clin Radiol (2012) 67(2):157–64. 10.1016/j.crad.2011.08.012 [DOI] [PubMed] [Google Scholar]
- 28. Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S, Miles K. Tumour Heterogeneity in Non-Small Cell Lung Carcinoma Assessed by Ct Texture Analysis: A Potential Marker of Survival. Eur Radiol (2012) 22(4):796–802. 10.1007/s00330-011-2319-8 [DOI] [PubMed] [Google Scholar]
- 29. Ganeshan B, Goh V, Mandeville HC, Ng QS, Hoskin PJ, Miles KA. Non-Small Cell Lung Cancer: Histopathologic Correlates for Texture Parameters at Ct. Radiology (2013) 266(1):326–36. 10.1148/radiol.12112428 [DOI] [PubMed] [Google Scholar]
- 30. Andersen MB, Harders SW, Ganeshan B, Thygesen J, Torp Madsen HH, Rasmussen F. Ct Texture Analysis Can Help Differentiate Between Malignant and Benign Lymph Nodes in the Mediastinum in Patients Suspected for Lung Cancer. Acta Radiol (2015) 57(6):669–76. 10.1177/0284185115598808 [DOI] [PubMed] [Google Scholar]
- 31. Hanania AN, Bantis LE, Feng Z, Wang H, Tamm EP, Katz MH, et al. Quantitative Imaging to Evaluate Malignant Potential of Ipmns. Oncotarget (2016) 7(52):85776–84. 10.18632/oncotarget.11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Attiyeh MA, Chakraborty J, McIntyre CA, Kappagantula R, Chou Y, Askan G, et al. Ct Radiomics Associations With Genotype and Stromal Content in Pancreatic Ductal Adenocarcinoma. Abdom Radiol (NY) (2019) 44(9):3148–57. 10.1007/s00261-019-02112-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carmicheal J, Patel A, Dalal V, Atri P, Dhaliwal AS, Wittel UA, et al. Elevating Pancreatic Cystic Lesion Stratification: Current and Future Pancreatic Cancer Biomarker(s). Biochim Biophys Acta Rev Cancer (2020) 1873(1):188318. 10.1016/j.bbcan.2019.188318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chakraborty J, Midya A, Gazit L, Attiyeh M, Langdon-Embry L, Allen PJ, et al. Ct Radiomics to Predict High-Risk Intraductal Papillary Mucinous Neoplasms of the Pancreas. Med Phys (2018) 45(11):5019–29. 10.1002/mp.13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chu LC, Park S, Kawamoto S, Fouladi DF, Shayesteh S, Zinreich ES, et al. Utility of Ct Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma From Normal Pancreatic Tissue. AJR Am J Roentgenol (2019) 213(2):349–57. 10.2214/ajr.18.20901 [DOI] [PubMed] [Google Scholar]
- 36. Harrington KA, Williams TL, Lawrence SA, Chakraborty J, Al Efishat MA, Attiyeh MA, et al. Multimodal Radiomics and Cyst Fluid Inflammatory Markers Model to Predict Preoperative Risk in Intraductal Papillary Mucinous Neoplasms. J Med Imaging (Bellingham) (2020) 7(3):31507. 10.1117/1.Jmi.7.3.031507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nasief H, Zheng C, Schott D, Hall W, Tsai S, Erickson B, et al. A Machine Learning Based Delta-Radiomics Process for Early Prediction of Treatment Response of Pancreatic Cancer. NPJ Precis Oncol (2019) 3:25. 10.1038/s41698-019-0096-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen X, Yang F, Yang P, Yang M, Xu L, Zhuo J, et al. A Contrast-Enhanced Computed Tomography Based Radiomics Approach for Preoperative Differentiation of Pancreatic Cystic Neoplasm Subtypes: A Feasibility Study. Front Oncol (2020) 10:248. 10.3389/fonc.2020.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Al-Hawary MM, Francis IR, Chari ST, Fishman EK, Hough DM, Lu DS, et al. Pancreatic Ductal Adenocarcinoma Radiology Reporting Template: Consensus Statement of the Society of Abdominal Radiology and the American Pancreatic Association. Gastroenterology (2014) 146(1):291–304 e1. 10.1053/j.gastro.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 40. Mackin D, Fave X, Zhang L, Yang J, Jones AK, Ng CS, et al. Harmonizing the Pixel Size in Retrospective Computed Tomography Radiomics Studies. PloS One (2017) 12(9):e0178524. 10.1371/journal.pone.0178524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shafiq-Ul-Hassan M, Zhang GG, Latifi K, Ullah G, Hunt DC, Balagurunathan Y, et al. Intrinsic Dependencies of Ct Radiomic Features on Voxel Size and Number of Gray Levels. Med Phys (2017) 44(3):1050–62. 10.1002/mp.12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B-Methodological (1995) 57(1):289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 43. Permuth-Wey J, Chen DT, Fulp WJ, Yoder SJ, Zhang Y, Georgeades C, et al. Plasma Micrornas as Novel Biomarkers for Patients With Intraductal Papillary Mucinous Neoplasms of the Pancreas. Cancer Prev Res (Phila) (2015) 8(9):826–34. 10.1158/1940-6207.capr-15-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zaid M, Widmann L, Dai A, Sun K, Zhang J, Zhao J, et al. Predictive Modeling for Voxel-Based Quantification of Imaging-Based Subtypes of Pancreatic Ductal Adenocarcinoma (Pdac): A Multi-Institutional Study. Cancers (Basel) (2020) 12(12):596931. 10.3390/cancers12123656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brouwer A, De Laere B, Peeters D, Peeters M, Salgado R, Dirix L, et al. Evaluation and Consequences of Heterogeneity in the Circulating Tumor Cell Compartment. Oncotarget (2016) 7(30):48625–43. 10.18632/oncotarget.8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Caswell DR, Swanton C. The Role of Tumour Heterogeneity and Clonal Cooperativity in Metastasis, Immune Evasion and Clinical Outcome. BMC Med (2017) 15(1):133. 10.1186/s12916-017-0900-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dagogo-Jack I, Shaw AT. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat Rev Clin Oncol (2018) 15(2):81–94. 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
- 48. Marusyk A, Almendro V, Polyak K. Intra-Tumour Heterogeneity: A Looking Glass for Cancer? Nat Rev Cancer (2012) 12(5):323–34. 10.1038/nrc3261 [DOI] [PubMed] [Google Scholar]
- 49. Permuth JB, Dezsi KB, Vyas S, Ali KN, Basinski TL, Utuama OA, et al. The Florida Pancreas Collaborative Next-Generation Biobank: Infrastructure to Reduce Disparities and Improve Survival for a Diverse Cohort of Patients With Pancreatic Cancer. Cancers (Basel) (2021) 13(4):1–24. 10.3390/cancers13040809 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.