Abstract
Aryl hydrocarbon receptor nuclear translocator 2 (ARNT2) is a basic helix-loop-helix (bHLH/PAS) transcription factor involved in the development of paraventricular nucleus of the hypothalamus (PVH) through the heterodimerization with Single-minded 1 (SIM1) (Michaud et al., 2000). Using a Sendai virus-based approach, the four reprogramming factors OCT3/4, SOX2, KLF4 and C-MYC were delivered into Peripheral Blood Mononuclear Cell (PBMCs) from a 14-year-old girl with early onset obesity carrying a de novo variant (p.P130A) in ARNT2. The resulting iPSC line CUIMCi003-A had a normal karyotype, showed pluripotency and three germ layer differentiation capacity in vitro and was heterozygous for the de novo ARNT2 variant.
1. Resource table
| Unique stem cell line identifier | CUIMCi003-A |
| Alternative name(s) of stem cell line | THA20_1 |
| Institution | Columbia University Irving Medical Center (CUIMC) |
| Contact information of distributor | gi2169@cumc.columbia.edu |
| Type of cell line | iPSC |
| Origin | human |
| Additional origin info required for human ESC or iPSC |
Age: 14 Sex: female Ethnicity if known: Caucasian |
| Cell Source | Peripheral Blood Mononuclear Cells (PBMCs) |
| Clonality | Clonal |
| Associated disease | Severe obesity |
| Gene/locus | ARNT2 (p.P130A)/15q25.1 |
| Date archived/stock date | May 2021 |
| Cell line repository/bank | N/A |
| Ethical approval | Columbia University Irving Medical Center Institutional Research Board under Protocol # AAAS4650. This includes approval from the Human Embryonic and Human Pluripotent Stem Cell Research Committee. |
2. Resource utility
CUIMCi003-A is an iPSC line carrying a heterozygous de novo variant (p.P130A) in ARNT2, generated from Peripheral Blood Mononuclear Cells (PBMCs) of a patient with severe early onset obesity. This line can be used for disease modelling and drug screening in weight dysregulation studies. By CRISPR/Cas9 correction, CUIMCi003-A can be used to further investigate the genotype-phenotype correlation.
3. Resource details
ARNT2, a transcription factor required for the development of hypothalamus, is highly expressed in the paraventricular hypothalamus (PVH), one of the key regions for weight regulation (Michaud et al., 2000). In this study, a hiPSC line was established from PBMCs of a 14-year-old girl with severe early onset obesity with a heterozygous missense variant in exon 4 of the ARNT2 (NM_014862.4; c.388C > T) gene. Eight mL of the patient’s peripheral blood were collected into BD Vacutainer cell preparation tube (CPT) with sodium citrate and centrifuged (1800 RCF, 30 min, RT) to extract PBMCs. The isolated cells were grown in Expansion Medium (EM): QBSF-60 (cat# 160204101, Quality Biologicals), antibiotics (Primocin, Invivogen, #ant-pm-1; Pen/Strep, Life Technologies, #15140–155; L-Ascorbic Acid, Sigma, # A4544–25G), and growth factors (EPO, #287-TC-500; IL-3, #203-IL-010/CF; IGF-1, #291-G1–200; SCF, #255-SC-010/CF; R&D Systems) and Dexamethasone (Sigma, #D8893–1MG) for few days. After 12 days, 2.5 × 105 cells were collected and reprogrammed into iPSC using a non-integrating Sendai virus approach (CytoTune™-iPS 2.0 Sendai Reprogramming Kit, cat# A16518, ThermoFisher Scientific), with MOI 5:5:3, KOS:c-myc: KLF4 (Yang et al., 2008–2012) following manufacturer’s instructions, while the remaining cells were banked. At D12 post-transduction, small colonies with iPSC-like morphology appeared and single clone picking was performed. Each clone was further expanded and the established hiPSC line CUIMCi003-A was selected for further characterization (Fig. 1, Tables 1 and 2). This line exhibited a normal stem cell-like morphology (Fig. 1A, 1000 μm) and G-banded karyotype analysis revealed a normal karyotype (46, XX) (tested at passage 6, Fig. 1B). The absence of Sendai viral transgenes in CUIMCi003-A at passage 17 was confirmed by qRT-PCR: the patient PBMCs after Sendai transduction were used as a positive control, while a previously published iPSC line (Patel et al., 2020) and the hESC line H9 (WA09) were used as negative control (Fig. 1C). Immunostaining was performed to assess the expression of the stemness markers Oct 4, Nanog and Sox2 (tested at passage 18, Fig. 1D). The expression of both cell surface (Tra-1–60 and SSEA4) and intracellular stemness markers (Nanog and Oct4) was also quantified by flow cytometry (tested at passage 18, Fig. 1E). The expression of ectoderm (OTX2), mesoderm (Brachyury) and endoderm (SOX17) markers upon differentiation demonstrated the ability of CUIMCi003-A to differentiate in vitro into the three germ-layers (tested at passage 18, Fig. 1F). Short tandem repeat (STR) analysis of the generated CUIMCi003-A line was performed through DNA fingerprint analysis (Cell Line Genetics, Madison, WI). The test revealed that all 15 hiPSC allele loci were consistent with those of the parental PBMCs (data available upon request). Mycoplasma test at passage 20 resulted negative (Supplementary Fig. 1). The de novo (p.P130A) ARNT2 variant was confirmed in CUIMCi003-A by Sanger sequencing (Fig. 1G).
Fig. 1.
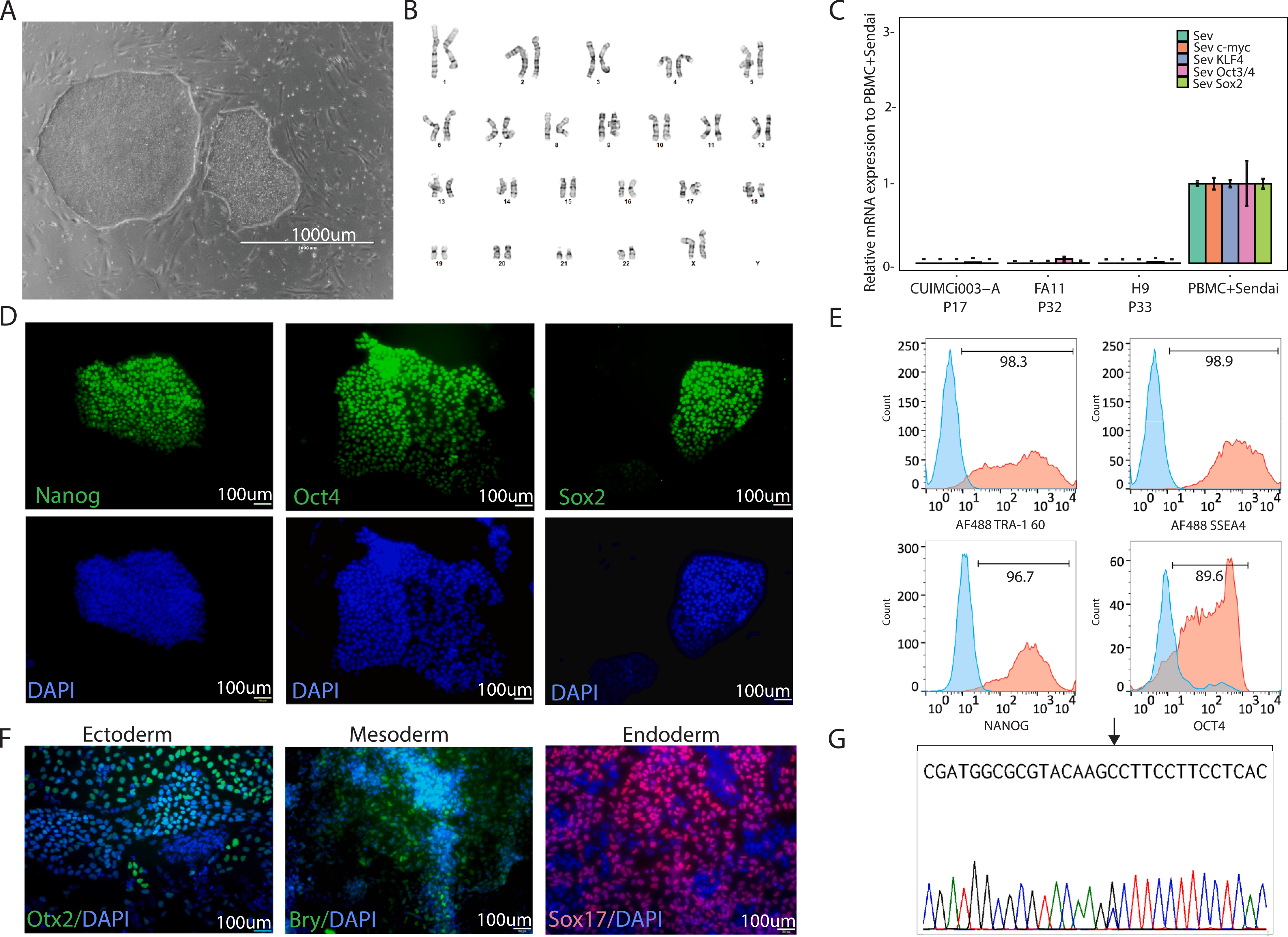
CUIMCi003-A characterization.
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Typical morphology | Fig. 1A |
| Phenotype | Qualitative analysis (Immunocytochemistry) | Assess staining/expression of stemness markers at P18: Nanog, Oct4, Sox2 | Fig. 1D |
| Quantitative analysis (Flow cytometry, RT-qPCR) | Assess % of positive cells or transcripts for antigen & cell surface/intracellular markers at P18: Tra 1–60+: 98.3%, SSEA4+: 98.9%, Nanog+: 96.7%, Oct4+: 89.6% Sendai clearance assessment by qRT-PCR (at P17 for CUIMCi003-A; after transduction for the positive control) |
Fig. 1E Fig. 1C |
|
| Genotype | Karyotype (G-banding) Resolution 450–500 | 46XX Resolution 450–500 at P6 | Fig. 1B |
| Identity | STR analysis | DNA Profiling Performed 16 loci analyzed, all matching donor of origin at P17 | Supplemental Table 1 available with authors |
| Mutation analysis (IF APPLICABLE) | Sequencing | Heterozygous variant | Fig. 1G |
| Southern Blot OR WGS | N/A | ||
| Microbiology and virology | Mycoplasma | PCR, Negative at P18 | Supplemental Fig. 1 |
| Differentiation potential | e.g. Embryoid body formation OR Directed differentiation | Proof of three germ-layers formation: positive OTX2 (ectoderm) staining, positive Brachyury (mesoderm) staining and positive SOX17 (endoderm) staining (at P18) | Fig. 1 F |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping | N/A | N/A |
| HLA tissue typing | N/A | N/A | |
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Stemness Marker | Rabbit anti-OCT4 (Alexa Fluor 488 Conjugate) | 1:50 | Cell Signaling Technology Cat# 5177S, RRID: AB_10693303 |
| Stemness Marker | Rabbit anti-NANOG | 1:400 | Cell Signaling Technology Cat# 4903P, RRID: AB_10559205 |
| Stemness Marker | Mouse anti-SOX2 (Alexa Fluor 488 Conjugate) | 1:50 | Santa Cruz Biotechnology, Cat# sc-365823 RRID: AB_10842165 |
| Stemness Marker (FACS) | Alexa Fluor 488 Mouse anti-Human TRA-1–60 | 1:20 | BD Biosciences, Cat#560173 |
| Stemness Marker (FACS) | Alexa Fluor 488 Mouse anti-SSEA-4 | 1:20 | BD Biosciences, Cat#560308 |
| Stemness Marker (FACS) | Alexa Fluor 488 Mouse anti-Human Nanog | 1:20 | BD Biosciences, Cat#560791 |
| Stemness Marker (FACS) | Oct-4A - Rabbit mAb (Alexa Fluor 488 Conjugate) | 1:20 | Cell Signaling Technology Cat#5177 |
| Stemness Marker – Isotype (FACS) | Alexa Fluor488 Mouse IgG1 κ Isotype Control | 1:20 | BD, Biosciences, Cat#557702 |
| Stemness Marker – Isotype (FACS) | Rabbit mAb IgG - Isotype Control (Alexa Fluor488 Conjugate) | 1:20 | Cell Signaling Technology, Cat#2975S |
| Differentiation Markers | Goat anti-OTX2 | 1:20 | R&D Systems Cat# AF1979, RRID: AB 2157172 |
| Differentiation Markers | Goat anti-Brachyury | 1:20 | R&D Systems Cat# AF2085, RRID: AB_2200235 |
| Differentiation Markers | Goat anti-SOX17 (NL557 Conjugate) | 1:10 | R&D Systems Cat# NL1924R, RRID: AB_2195645 |
| Secondary antibody | Rabbit anti-Goat IgG Alexa Fluor 488 | 1:1000 | Thermo Fisher Scientific Cat# A27012, RRID: AB_2536077 |
| Secondary antibody | Goat anti-Rabbit IgG Alexa Fluor 488 | 1:1000 | Thermo Fisher Scientific Cat# A11008, RRID: AB_143165 |
| Secondary antibody (FACS) | Goat anti mouse IgG1-PE | 1:500 | Molecular Probes Cat# P21129, RRID: AB_2539816 |
| Secondary antibody (FACS) | Goat anti mouse IgM-488 | 1:500 | Molecular Probes Cat# A21042, RRID: AB_141357 |
| Primers qPCR | Target | Forward/Reverse primer (5′–3′) | |
| Sendai virus detection | SeV | GGATCACTAGGTGATATCGAGC ACCAGACAAGAGTTTAAGAGATATGTATC |
|
| Transgene detection | KOS | ATGCACCGCTACGACGTGAGCGC ACCTTGACAATCCTGATGTGG |
|
| Transgene detection | Klf4 | TTCCTGCATGCCAGAGGAGCCC AATGTATCGAAGGTGCTCAA |
|
| Transgene detection | c-Myc | TAACTGACTAGCAGGCTTGTCG TCCACATACAGTCCTGGATGATGATG |
|
| Housekeeping gene | 18S | Hs99999901s1, Applied Biosystem | |
| Primers mutational screening | Target | Forward/Reverse primer (5′–3′) | |
| ARNT2 | ARNT2 | GGTGTTAGCCCCTAGTTCCTGG TGGCTTCATTCCTTCCTCAACC |
|
4. Materials and methods
4.1. iPSC maintenance
niPSCs were grown on feeders (γMEF, Life Technologies, cat #A34181) in DMEM/Ham’s F-12 (Corning) supplemented with 20% KO-SR, 1x non-essential amino acids, 1x Penicillin-Streptomycin, 1x glutamine, 1x β-Mercaptoethanol (all Life Technologies) and 10 ng/ml FGF2 (R&D Systems). Cells were fed daily and split 1:6 when they reached 80% confluency using EDTA. Established iPSCs were maintained in feeder-free condition using Matrigel (Corning) coated plates and mTeSR1 or mTeSR Plus medium (Stem Cell Technologies). All cells were kept in an incubator at 37 °C, 5% CO2, 20% O2.
4.2. Karyotyping
CUIMCi003-A cells at passage 6 were examined by standard G-banding analysis (Cell Line Genetics, Madison, WI) on twenty-G banded metaphase cells at 450–500 band resolution.
4.3. Quantitative real-time polymerase chain reaction (qRT-PCR)
At passage 17, 1 × 106 cells were collected and 1 μg of RNA was extracted using RNeasy Mini Kit (Qiagen, Cat # 74104). Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher, #4387406). The qRT-PCR was conducted on Applied Biosystems QuantStudio Flex 7 using the TaqMan® iPSC Sendai Detection Kit (ThermoFisher, #A13640). Primer sequences are listed in Table 2. PBMCs from the same cell line after Sendai transduction were used as a positive control for the expression of exogenous genes (=1 in the relative quantification). The iPSC line CUIMCi002-A (passage 32) and the hESC line H9 (WA09, passage 33) were used as negative control.
4.4. Immunostaining
At passage 18, iPSCs were fixed in 4% PFA (Santa Cruz Biotechnologies) for 10 min at room temperature (RT), washed twice with 1x PBS, then permeabilized for 30 min at RT with 0.1% Triton X-100. Cells were then incubated with Protein Block (Agilent Dako, Santa Clara, CA) for 30' at RT followed by primary antibodies for markers of stemness or differentiation (Table 2) in 1x PBS + 1% BSA over-night at RT. After washing twice, when required, cells were incubated with secondary antibody (Table 2) in 1x PBS + 1% BSA for 1hr at RT, followed by washing with 1x PBS + 1% BSA. Nuclei were stained with DAPI and cells visualized under an Olympus IX73 inverted microscope connected to a XM10 monochrome camera (Olympus, Tokyo, Japan).
4.5. Flow cytometry
Cell were dissociated by accutase treatment for 5 min at 37 °C, then fixed with 4% PFA for 15 min at RT. For intracellular staining, cells were permeabilized in methanol. Cells were then incubated for 15 min at RT with directly conjugated antibodies against OCT-3/4, NANOG, TRA-1–60 and SSEA4 (Table 2). Flow cytometry was performed on S3e (Bio-Rad). Data were analyzed using FlowJo™ Software (FlowJo 10.7.1. Ashland, OR: Becton, Dickinson and Company).
4.6. In vitro germ layer differentiation
To assess the 3-germ layer differentiation capability of CUIMCi003-A cells in vitro, the Human Pluripotent Stem Cell Functional Identification kit (R&D Systems, #SC027) was used according to manufacturer’s instructions.
4.7. Mycoplasma detection
Absence of mycoplasma contamination was confirmed using e-Myco™ plus Mycoplasma PCR Detection Kit (Intron, Burlington, MA, #25234) according to manufacturer’s instructions
4.8. Mutation analysis
Genomic DNA was extracted and purified from CUIMCi003-A cells by DNase blood and tissue kit (Qiagen, #69504). Genotyping of the heterozygous P130A variant in the ARNT2 gene was performed by Sanger sequencing (Genewiz). Primers are listed in Table 2.
4.9. Short tandem repeat profiling
THA20_1 is the donor PBMC line for the iPSC line CUIMCi003-A. iPSC were tested at P17.
Supplementary Material
Acknowledgements
The authors would like to thank the patients who generously donated their blood for the research conducted; Michael Kissner, Director of Operations, and the personnel of the Columbia Stem Cell Initiative Flow Core; Cecilia Sena, research coordinator.
5. Funding sources
This research was funded in part by Pilot and Feasibility Grant, New York Obesity and Nutrition Research Center, NIH-NIDDK, P30 DK026687-39, and Louis V. Gerstner Clinical Scholar Program at Columbia University Irving Medical Center.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2021.102432.
References
- Michaud JL, DeRossi C, May NR, Holdener BC, Fan C-M, 2000. ARNT2 acts as the dimerization partner of SIM1 for the development of the hypothalamus. Mech. Dev. 90 (2), 253–261. [DOI] [PubMed] [Google Scholar]
- Yang et al. iPSC Reprogramming from Human Peripheral Blood Using Sendai Virus Mediated Gene Transfer StemBook. Cambridge (MA): Harvard Stem Cell Institute; 2008–2012. Jun 10. [PubMed] [Google Scholar]
- Patel A, Garcia Diaz A, Moore JC, Sirabella D, Corneo B, 2020. Establishment and characterization of two iPSC lines derived from healthy controls. Stem Cell Res. 47, 101926. 10.1016/j.scr.2020.101926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


