Abstract
Metachromatic leukodystrophy (MLD) is a human neurodegenerative disorder characterized by progressive damage on the myelin band in the nervous system. MLD is caused by the impaired function of the lysosomal enzyme Arylsulphatase A (ARSA). The physiopathology mechanisms and the biochemical consequences in the brain of ARSA deficiency are not entirely understood. In recent years, the use of genome-scale metabolic (GEM) models has been explored as a tool for the study of the biochemical alterations in MLD. Previously, we modeled the metabolic consequences of different lysosomal storage diseases using single GEMs. In the case of MLD, using a glia GEM, we previously predicted that the metabolism of glycosphingolipids and neurotransmitters was altered. The results also suggested that mitochondrial metabolism and amino acid transport were the main reactions affected. In this study, we extended the modeling of the metabolic consequences of ARSA deficiency through the integration of neuron and glial cell metabolic models. Cell-specific models were generated from Recon2, and these were used to create a neuron-glial bi-cellular model. We propose a workflow for the integration of this type of model and its subsequent study. The results predicted the impairment pathways involved in the transport of amino acids, lipids metabolism, and catabolism of purines and pyrimidines. The use of this neuron-glial GEM metabolic reconstruction allowed to improve the prediction capacity of the metabolic consequences of ARSA deficiency, which might pave the way for the modeling of the biochemical alterations of other inborn errors of metabolism with central nervous system involvement.
Keywords: Metachromatic leukodystrophy, System biology, Arylsulphatase A, Myelin sheath
Metachromatic leukodystrophy; System biology; Arylsulphatase A; Myelin sheath.
1. Introduction
Myelin sheath is a protein-lipid band that covers neural axons and facilitates the transmission of the nervous impulse. This band is formed by the cell membrane of the oligodendrocytes in the central nervous system (CNS) or Schwann cells in the peripheral nervous system. Myelin membrane is rich in lipids with a composition that varies significantly from other biological membranes. Since myelination requires a high level of lipid synthesis, the integrity of myelin is susceptible to numerous lipid metabolism disorders [1]. One of the few described myelin metabolic pathways is the beginning of the catabolism of the lipids present in the myelin sheath [2]. The lysosomal enzyme arylsulfatase A (ARSA) catalyzes the first step in the degradation of galactosyl-3-sulfate ceramide (cerebroside sulfate or sulfatide), one of the significant membrane lipids of the myelin sheaths [3]. Sulfatide located on the surface of the myelin sheaths loses the sulfate group located in the 3 position by the action of ARSA, releasing galactosylceramide that later is transformed into ceramide by the action of galactocerebrosidase [2]. Deficiency of ARSA leads to the neurodegenerative disease Metachromatic Leukodystrophy (MLD) [4]. A clear genotype-phenotype relationship has not been described for MLD patients, and there is not a direct relationship between the severity of the disease and the residual enzymatic activity [2, 4, 5, 6]. In addition, the physiopathology of the disease cannot be explained exclusively by the lysosomal accumulation of the non-degraded substrate, as evidenced by reports that currently describe new mechanisms of cell damage [6, 7].
Advances in genomics, transcriptomics, proteomics, metabolomics, and other omics sciences, in addition to the increase of the computational power, allowed GEM models to became a useful approach to increase our understanding of several cellular processes [8]. One aspect of the application of GEM is the study of the complexity of the human brain and associated diseases, a field in which we have been working on for several years [9, 10, 11], as well as other authors [8, 12]. One of the most popular and used human GEM reconstruction is Recon2 [13], which has 7440 metabolic reactions, 5063 metabolites, and eight compartments. This model has allowed the construction of different tissue-specific models, including cells from CNS, such as neurons and glial cells from the cortex, hippocampus, and lateral ventricle tissue [14].
Due to the possibility of integration between different Recon2 models, multi-cell-type metabolic networks have modeled the study of brain metabolism. For instance, Lewis et al. [15], modeled the integration of astrocytes, glutamatergic, GABAergic, and cholinergic neurons to study Alzheimer's disease and cholinergic neurotransmission. They also presented a workflow for generating tissue-specific multicellular metabolic models. The integration of these cell models has been used to study mechanisms of neurotransmission or hypoxia [16]. A dynamic model to analyze the interaction between the astrocytes and neurons by employing a different approach was recently reported [6], in which astrocytes were modeled as units without compartmentalization, while the neurons were represented as spot-like stimuli defined with a set of mathematical equations to resemble the neuronal firing behavior. Cakir et al. [17], constructed a central metabolism, lipid metabolism, reactive oxygen species (ROS), detoxification, amino acid metabolism, glutamate-glutamine cycle, and neurotransmitter metabolism for neurons and astrocytes. The integration of neuron and glial cell metabolic reconstruction models has been reported and used to model specific processes such as the role of lactate between neurons and glial cells [18, 19]. The integration of neuron and glial cell models may allow us to increase our understanding of the cellular process involved in the relationship between these cells, and also to model diseases or conditions in which these cells are affected.
Previously, we used a GEM of a glial cell to study the metabolic consequences of ARSA deficiency. The results predicted that the metabolism of glycosphingolipids and neurotransmitters were the main impairment pathways [10]. In this study, we constructed a bi-cellular GEM of glial and neuron cells from the human cortex. This bi-cellular GEM allowed us to understand how ARSA enzyme deficiency alters the metabolic communication between both cell types. The results predict that ARSA deficiency resulted in the impairment of several metabolic pathways in both cell types. ARSA deficiency affected 370 and 293 reactions in the neuron and glial cell models, respectively, which represent about 16 % of the reactions of each model. In addition, the results showed that ARSA deficiency induced changes in the flux distributions of several metabolic pathways in a different way to that observed for the independent cellular models. To the best of our knowledge, this is the first time that the integration of neuron and glial cell metabolic reconstructions has been used to explore the impact of the impairment of lipids metabolism, such as that produced by ARSA deficiency, on cell metabolism. This integration will not only allow us to model, at the cellular level, the alterations observed in MLD but also may serve as a starting point to model other lipid metabolic diseases or inborn errors of metabolism in which neuron and glial cells are affected.
2. Methods
For the construction of the bi-cellular model, we used the previously constructed glial (Biomodels Database number: MODEL1310110064) and neuron cortex (Biomodel Database number: MODEL1310110033) metabolic reconstructions [14], which were originally derived from expression data from the Human Protein Atlas [20, 21]. We share the code and the model (xls and xml format) in the following link: https://github.com/dasalazarb/Neuro-Glia_GEM. The following procedure describes the steps used to reconstruct the glia-neuron bi-cellular model:
Step 1
Individualization of models.
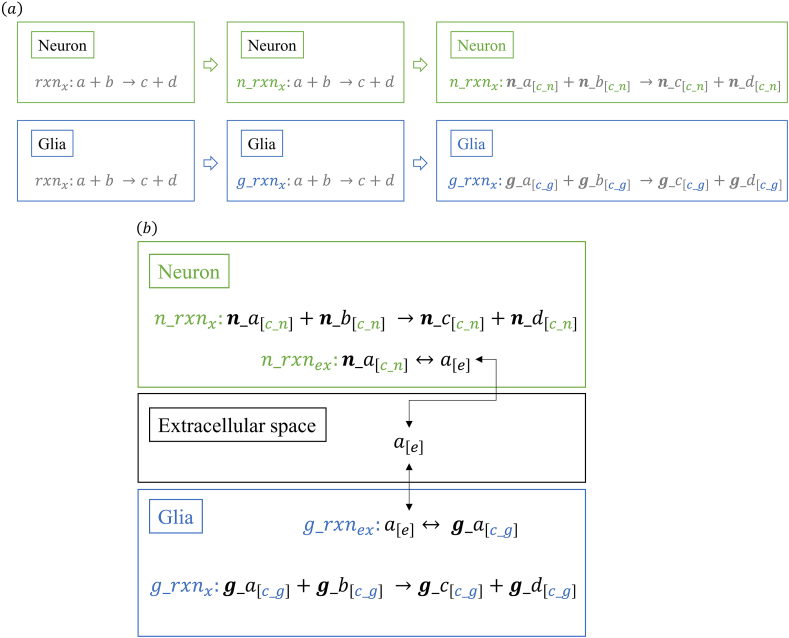
Recon2 model has a standard annotation to describe all the derived cell tissue models [22]. Based on that, the nomenclature of models was individualized by adding the prefixes ‘n_’ and ‘g_’ to the reactions, compartments, and metabolites of the neuron and glia metabolic reconstruction, respectively. In this sense, for an rxnx reaction, which is equal in both models, specific reactions ‘n_rxnx’ and ‘g_rxnx’ were created within the neuron and glia models, respectively. In addition, the compartmentalization of metabolites in each model was also differentiated. For instance, a metabolite x[c], which is allocated in the cytosol in both models, was changed to x [c_n] and x[c_g] within the neuron and glia models, respectively (Figure 1a). Finally, we differentiated between names of metabolites using the prefix ‘g_’ for glial but not for neuron metabolites because once the glia metabolites have been differentiated, there is no need to change the neuron metabolites. For example, for an ‘x’ metabolite, the new nomenclature was ‘g_x’ and ‘x’ (Figure 1a).
Step 2
Interaction between models.
The coupling of metabolic models is based on the metabolites that are shared (i.e., consumed and produced) by glia and neuron cortex models [23]. In this sense, to integrate both metabolic reconstructions, we took all the exchange metabolites from the neuron and glia model as metabolites that allow communication between the cells. The nomenclature of extracellular metabolites and transport reactions allowed the creation of a common space for both cells, i.e., an extracellular compartment. For instance, for two hypothetical extracellular transport reactions as ‘n_rxnx: a[c_n] ↔ a[e]’ and ‘g_rxnx: a[e] ↔ g_a[c_g]’, the common metabolite is ‘a[e]’. In this sense, the glia and neuron models were able to share metabolites in the extracellular compartment, producing the interaction of the models (Figure 1b).
Step 3
Testing the mathematical bi-cellular model.
The integration of the two metabolic reconstructions represents the union of the corresponding stoichiometric matrices which allows the use of a Flux Balance Analysis (FBA) to solve the same optimization problem used for the single metabolic models [24]. The optimization problem consists of a stoichiometric matrix (S), of size m∗n, where m corresponds to metabolites and n corresponds to reactions, linked by stoichiometric coefficients, in which a negative coefficient means consumption of a metabolite in a reaction and a positive coefficient means production. The optimization problem indicated that in a steady-state exist a vector of fluxes () that optimize the objective function (, where c is a vector of weights, indicating how much each reaction contributes to the objective function) to a maximum or minimum. In addition, the fluxes () are subject to constraints (mass or thermodynamic) that limit the solutions of the problem. In a mathematical language, it is defined as:
Maximize/minimize
Subject to
and lower bound ≤ ≤ upper bound
When the optimization problem is feasible, a flux distribution () is obtained, which shows the contribution of each reaction to the objective function. For reading, debugging, joining the models, and FBA computation, we used COBRA (COnstraint-Based Reconstruction and Analysis) Toolbox [25, 26] implemented in MATLAB software.
Step 4
Manual curation and annotation of the model
The glia and neuron metabolic reconstructions were manually curated by adding reactions to complete the metabolic pathways corresponding to the sulfatide degradation from the myelin band, which correspond to the beginning of the myelin degradation within a cortical neuron and glia cells [27]. Hence, the glia model is not only representing the oligodendrocyte metabolism but also the interaction of astrocyte-oligodendrocyte since these cells are gap junction-coupled in the central nervous system [28]. In addition, the network formed by these cells favors the regulation of ions, also providing nutrients for axonal metabolism [27]. This bi-cellular model does not contain an axon compartment but is intended to stimulate the metabolism and interaction of glia and neuron cells. Although the metabolism of these cells is not fully understood, certain metabolic features are known, such as the presence of a high pentose phosphate activity or the anaplerotic replenishment of the tricarboxylic acid (TCA) cycle with the recycling of pyruvate [29]. Metabolic pathways of glycolysis, TCA cycle, pentose phosphate, and oxidative phosphorylation were completed within the glia and neuron models. Synthesis and degradation pathways of glycogen were added to the glia model [30, 31, 32], while the reactions that were not well annotated in both models were completed based on the information from Recon2. The medium conditions used and the reactions added in simulations are described in Supplementary File 1.
Step 5
Context-specific bi-cellular model simulation
To study the effect of model integration, we performed a FBA for the bi-cellular model and each cellular model separately and compared the flow distributions obtained. In all scenarios, the objective function was ATP synthesis, taking into account that we did not intend to simulate any scenario related to other processes such as synapses or neuronal excitation. Since the solution of FBA is not unique, we performed a random sampling of the flow distribution in the space of possible solutions defined by the stoichiometric matrix using the Hit and Run algorithm implemented in COBRA [29, 33]. From a sampling of 5,000 points within this space, for each reaction, the mean and standard deviation were determined, and a Z-score where the difference between the means in each of the conditions divided by the standard deviation of this difference, was calculated as reported by Bordel et al. [33], which quantifies the significance of the change in each flux between the evaluated conditions [33]. A t-test was carried out in R, and the interpretation analysis was performed using score with a p-value < 0.05.
For modeling the ARSA deficiency, as a first approximation to a pathological state, and an initial use of a bi-cellular model designed, the lower and upper flux limits for the reaction catalyzed by ARSA enzyme (EC 3.1.6.8) were set to zero. The ATP synthesis was selected as the objective function to model an excited neuron that releases neurotransmitters and therefore presents a highly oxidative metabolism. In addition, the biological scenario was the neuronal excitation, where sufficient energy is required (ATP synthesis optimization).
Figure 1.
Construction of metabolic bi-cellular model. (a) Individualization of the models into compartments. (b) Adding of the extracellular reactions for communication between models.
3. Results
3.1. Metabolic construction and integration of models
The coupling of metabolic reconstructions of glia and neuron cortex cells might allow us to understand how these cells communicate and how an enzyme deficiency can alter the homeostasis between both cells [34]. Although other metabolic models of integration of these cell types have been reported, the number of studied pathways is reduced. Therefore, it is not possible to have a broad picture of the interaction between these two cells [35]. Although the model was manually debugged, expression data obtained from different omics could enrich the present model [36] and allows further predictions. For blocked reactions, orphan metabolites, and dead-end metabolites, we used the memote tool [37] which reports in detail the composition of a GEM to determine errors and ensure that the model meets the minimum requirements for use. We found 227 blocked reactions, 16 orphan-metabolites, and 8 dead-end metabolites (Supplementary File 1). These reactions correspond to the degradation of keratan, heparan, and chondroitin sulfates, in addition to exchange and transport reactions.
The bi-cellular GEM consisted of two types of brain cells, neurons, and glial cells. The glial cell within the model was adapted to resemble the astrocyte-oligodendrocyte network, which is the main actor of neuron maintenance [38]. The bi-cellular model consisted of three large compartments: neuron, glial, and extracellular compartment; with the latter being responsible for metabolites exchange (i.e. communication) between the two cell models (Supplementary File 2 in formats .xls and .xml). The model has a total of 3845 reactions (1911 neuron, 1615 glia, and 319 extracellular reactions). The neuron and glia models were compartmentalized individually in seven sub-compartments (cytosol [c], mitochondria [m], endoplasmic reticulum [r], peroxisome [x], nucleus [N], lysosome [l] and Golgi [g]). In total, 2460 metabolites were found in the bi-cellular model, with 1192, 939, and 329 metabolites present in neuron, glial, and extracellular compartments, respectively (Supplementary File 2 in formats .xls and .xml).
We initially tested the hypothesis that the integration of neuron and glial cell models will affect the reaction fluxes of each model. For this purpose, we compared the FBA results of independent (i.e. neuron and glial cell models) and the bi-cellular models (Supplementary File 3). The results show that the integration of the models affected 34 % and 39 % of the reactions in the neuron and glial cell compartments, respectively (Table 1). In this sense, these results suggest that neuron and glial cell compartments, in the bi-cellular model, were connected through the extracellular compartment by the exchange of metabolites.
Table 1.
Comparison between independent and bi-cellular model fluxes. The FBA obtained for the individual models was compared with the FBA of the bi-cellular model. The fluxes of the reactions changed concerning the bi-cellular model.
| Neuron | Glial | ||
|---|---|---|---|
| Total reactions in bi-cellular model | 1906 | 1609 | |
| Number of reactions with different flux in the bi-cellular model | Higher | 270 | 251 |
| Lower | 258 | 220 | |
| Number of on/off reactions in bi-cellular but not in the individual model | On | 127 | 116 |
| Off | 124 | 158 | |
| Number of reactions with different value between model bicellular and individual model | 652 | 629 | |
From this comparison, we found that 160 metabolic pathways were altered in the bi-cellular model, with 76, 83, and 1 pathways present in the glial, neuron, and extracellular compartments, respectively. The structure of these pathways is listed in Supplementary File 4. The sphingolipids metabolism is one of the pathways that was different between the neuron and glial cell models, which is an expected result due to the role of sphingolipids in the synthesis of the axon by glial cells [39]. The neuron contains 44 sphingolipid reactions, including synthesis of sphinganine, phytosphingosine, sphingosine, ceramide, glucosylceramide, sulfatides, galactosylceramide, and digalactosylceramide sulfate. On the other hand, the glia contains 11 reactions, including the synthesis of phosphoethanolamine, sphingosine, phytosphingosine, sphinganine, ceramide, galactosylceramide, sulfatide, and digalactosylceramide sulfate [10].
3.2. Modeling of ARSA deficiency
Previously, we predicted that ARSA deficiency (i.e a total flux blockage of the reaction catalyzed by ARSA) in a glia GEM, affected amino acids transport and neurotransmitters metabolism, without any effect in the sphingolipids pathway [10]. As a continuation, in this study, we analyzed the metabolic consequences of ARSA deficiency in the neuron-glia bi-cellular GEM. It is important to mention that this bi-cellular model predicts the relationship between a glial cell (cerebral cortex glial cell) and a neuron in an excited state (cerebral cortex neuron cell), in the complete absence of ARSA. Therefore, this model partially represents the real condition of a MLD patient, not only because of the cell types present but also because the total absence of enzyme activity is not a common feature of this disorder.
The results predicted that within the bi-cellular model, ARSA deficiency resulted in the impairment of 370 and 293 reactions in the neuron and glial compartments, respectively. These impaired reactions represent about 16 % of the total reaction present in each compartment (Table 2). We found changes in different metabolisms such as amino acids, carbohydrates, and mitochondrial pathways among others might suggest that the functional alteration of ARSA in the bi-cellular model, significantly impairs the cell homeostasis.
Table 2.
Characteristics of the affected reactions in the ARSA deficient bi-cellular model compared to ARSA activated bi-cellular model. The number of affected reactions is expressed as a percentage of the total number of reactions for each compartment.
| Cell Type | Neuron | Glial |
|---|---|---|
| Total reactions | 1906 | 1609 |
| Number of reactions with different flux (p-value > 0.05) | 17.0 % | 16.5 % |
| Percentage of increased reactions in deficiency ARSA model vs ARSA activated model | 41.7 % | 51.5 % |
| Percentage of decreased reactions in deficiency ARSA model vs ARSA activated model | 46.2 % | 42.3 % |
| Percentage of reactions with inverted flux | 12.1 % | 6.2 % |
To continue the analysis of ARSA deficiency within the bi-cellular model, we considered that the biological function of a reaction is preserved when the reaction conserves the original direction (i.e., A → B). On the other hand, when the direction of a reaction is inverted (i.e., A ← B) the biological function may be affected, which might be reflected in the disease phenotype. In this sense, since most of the affected reactions (90,4 %) maintained the original direction, these results may suggest that the cells within the model (i.e., neuron and glia) may undergo metabolic changes to adjust their metabolism and retain the functionality and exchange between them. From a physiological point of view, it is usual that in neurodegenerative disorders, the disease onset is followed by a symptomatology stabilization period [4, 6, 40]. This period could be associated with a nervous system adjustment to the new myelin condition to conserve the functionality as much as possible.
3.3. Metabolic changes in glial and neuron compartments
For the glial cell compartment within the bi-cellular model, the ARSA deficiency reduced the flux of 72 reactions compared to a model with normal ARSA activity (Table 3). These were mainly grouped within the following pathways: glycolysis/gluconeogenesis, lysine degradation, fatty acid degradation, tyrosine metabolism, and metabolism of xenobiotics by cytochrome P450. On the other hand, the ARSA deficiency increased the flux of 44 reactions compared to a model with normal ARSA activity (Table 4). These reactions were associated with several pathways, such as pyrimidines, purines, lysine, carbon, β-alanine, and pyruvate metabolism (Table 4) [41, 42, 43, 44, 45, 46, 47].
Table 3.
Metabolic pathways in glia cell compartment with reactions that had lower flux values in the ARSA deficient bi-cellular model compared to the model with normal ARSA activity. The adjusted p-value corresponds to Fisher's exact test for genes.
| Metabolic Pathway | Count | % | Adjusted p-value |
|---|---|---|---|
| Glycolysis/Gluconeogenesis | 20 | 29,4 | 6,4E-23 |
| Lysine degradation | 16 | 23,5 | 3,2E-18 |
| Fatty acid degradation | 13 | 19,1 | 1,1E-14 |
| Tyrosine metabolism | 11 | 16,2 | 2,3E-12 |
| Metabolism of xenobiotics by cytochrome P450 | 12 | 17,6 | 2,7E-10 |
Table 4.
Metabolic pathways in neuron cell compartment with reactions that had higher flux values in ARSA deficient bi-cellular model compared to the model with normal ARSA activity. The adjusted p-value corresponds to Fisher's exact test for genes.
| Metabolic Pathway | Count | % | Adjusted p-value |
|---|---|---|---|
| Pyrimidine metabolism | 10 | 30,3 | 5,5E-9 |
| Purine metabolism | 10 | 30,3 | 3,8E-7 |
| Lysine degradation | 7 | 21,2 | 5,7E-7 |
| Carbon metabolism | 7 | 21,2 | 4,8E-5 |
| beta-Alanine metabolism | 5 | 15,2 | 4,5E-5 |
| Pyruvate metabolism | 5 | 15,2 | 1,1E-4 |
The most affected in the ARSA deficient bi-cellular model were glycolysis/gluconeogenesis, the metabolism of purine, pyrimidine, pyruvate, citric acid and beta-alanine, fatty acids degradation, and the metabolism of some amino acids (Leu, Val, Iso, Gly, Ser, Thr, Lys, and Tyr).
In the neuron compartment within the bi-cellular model, 35 reactions had lower flux values (Table 5); while 52 reactions had higher flux values (Table 6) in ARSA deficient model compared to the model with normal ARSA activity. The latter compromises several pathways including pyruvate, citrate cycle, glycolysis, and fatty acids.
Table 5.
Metabolic pathways in neuron cell compartment with reactions that had lower flux values in ARSA deficient bi-cellular model compared to the model with normal ARSA activity. The adjusted p-value corresponds to Fisher's exact test for genes.
| Metabolic Pathway | Count | % | Adjusted p-value |
|---|---|---|---|
| Fatty acid degradation | 12 | 24,0 | 2,8E-14 |
| PPAR signaling-pathway | 12 | 24,0 | 5,3E-12 |
| Fatty acid metabolism | 11 | 22,0 | 4,3E-12 |
Table 6.
Metabolic pathways in neuron cell compartment with reactions that had higher flux values in ARSA deficient bi-cellular model compared to the model with normal ARSA activity. The adjusted p-value corresponds to Fisher's exact test for genes.
| Metabolic Pathway | Count | % | Adjusted p-value |
|---|---|---|---|
| Carbon metabolism | 15 | 38,5 | 6,1E-15 |
| Pyruvate metabolism | 11 | 28,2 | 8,3E-14 |
| Glynlysis Gluconeogenesis | 11 | 28,2 | 1,8E-11 |
| Citrate cycle (TCA cam) | 9 | 23,1 | 2,3E-11 |
| Glyosylphosphatidylinositol (GPO-anchor biosynthesis | 6 | 15,4 | 2,0E-6 |
The model predicted that oxidoreductases have the highest number of affected reactions in both types of cells (Table 7). Noteworthy, it was not observed an important effect on the lysosomal hydrolases after ARSA knock-out (i.e., 15 % and 1 % of the glycosphingolipids reactions in the neuron and glial compartments, respectively), an unexpected result due to the impairment of the lysosomal metabolism induced by the glycosphingolipids accumulation.
Table 7.
Enzyme type affected in the ARSA deficient bi-cellular model.
| Compartment | Neuron (%) | Glial (%) |
|---|---|---|
| Oxidoreductases | 45 | 37 |
| Transferases | 48 | 35 |
| Hydrolases | 16 | 18 |
| Lyases | 4 | 5 |
| Isomerases | 2 | 1 |
| Ligases | 1 | 3 |
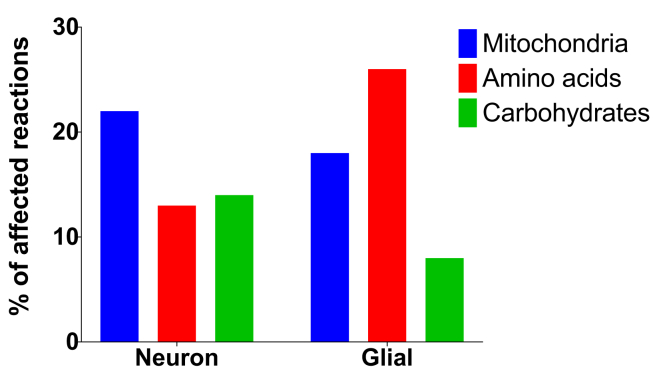
When the nature of the affected reactions was explored within bi-cellular model, we observed that the main affected reactions were related to amino acids transport, mitochondrial metabolism, and carbohydrate metabolism (Figure 2).
Figure 2.
Type of reactions affected in the neuron and glial cell compartments within the bi-cellular model.
4. Discussion
In this paper, we described the generation of a GEM produced by the integration of metabolic models of glial and neuron cells. Only a few bioinformatics studies have described the integration of the metabolic models of these two cell types. For instance, Cakir et. al., modeled the glutamate/glutamine/GABA cycle fluxes using minimization of metabolic adjustments, an alternative method to FBA, that allowed to find sub-optimal flux distributions after a gene knock-out [17]. In 2014, Sertbas et. al., extend this model to include metabolic pathways such as glycolysis, amino acid, and inositol, and modeled six diseases based on transcriptional data. Noteworthy, they found disease-specific metabolites and processes, such as the alteration of the sphingolipid metabolism in Parkinson's disease [48].
The bi-cellular GEM generated in this study integrates three compartments that are part of the neuronal synapse, i.e. neuron, glial, and the extracellular space. The latter compartment allows the communication between glial and neuron cells, which is required for the regulation of neurotrophic factors and the control of neurotransmitters, metabolites, and ions involved in neurotransmission, among others [49]. Our model comprises different metabolic pathways that are important for cell survival such as carbon and tricarboxylic acid cycles, as well as specialized ones such as those related to sphingolipids. Despite that the glial cell model was obtained from transcriptome data, this model is not distinguished as a specific glial cell (i.e. astrocyte, microglia, or oligodendrocyte), and by incorporating sphingolipid and ceramide metabolisms we could evaluate the behavior of ARSA enzyme deficiency. Our model was able to optimize the target function of producing ATP in the electron transport chain in the mitochondria, showing a significant metabolic change when ARSA knocked out. However, it is important to note that the present model corresponds to a complete absence of ARSA activity, which is a rare finding in MLD patients.
The design of the bi-cellular model using neuron and glia GEM permitted improvement in the modeling of ARSA deficiency in the CNS. It was predicted that the knock-out of ARSA in glial cells led to the downregulation of several metabolic pathways including glycolysis/gluconeogenesis, the degradation of fatty acids, and the metabolism of several amino acids. The impairment of β-oxidation of fatty acids compromises the entire oxidation cycle. Although acetyl-CoA is obtained from both mitochondrial and extramitochondrial sources, the low biological activity of β-oxidation enzymes may lead to the low production of acetyl-CoA whose ultimate purpose is the generation of ketones and energy. Thus, the metabolic impairment of these enzymes may lead to a decrease in energy production from fatty acids. Similarly, the model predicted is a significant compromise in the metabolism of several amino acids, such as Gly, Ser, Thr, Lis, and Tyr, which are directly involved in the metabolism of neurotransmitters, and maybe the result of a blockage in sulfatide catabolism present in the myelin band.
On the other hand, the knock-out of ARSA in glial cells led to the up-regulation of a diversity of metabolic pathways, including glycolysis/gluconeogenesis, the degradation of valine, leucine, and isoleucine, and the metabolism of purine, pyrimidine, pyruvate, citric acid, and β-alanine. The model also predicted the up-regulation of other important pathways such as the synthesis and hydrolysis of carnosine and homocarnosine, which could have the ability to protect neuron cells against ischemic injury and oxidative stress, as well as to increase their resistance to functional exhaustion [50, 51]. It has been postulated that carnosine and homocarnosine protection mechanism lies in the anti-glycating and antioxidant activities of these molecules [52, 53], as well as on a membrane-protecting effect [54].
In neuron cells on their part, the knock-out of ARSA in neuron cells led to a down-regulation of several metabolic pathways, including lipolysis/lipogenesis, transport, storage, DNA synthesis, and glycolysis. The main impairment occurred in lipolysis since the four main enzymes that catalyze the initial step of β-oxidation of fatty acids were inhibited. This involves both linear, or branched-chain fatty acids with short, medium, long or very-long chains, long-chain dicarboxylic acids, and poly-unsaturated fatty acids. The clinical impact of these deficiencies has been evidenced using brain images showing progressive white matter demyelination [55, 56].
On the other hand, it was predicted that in neuron cells, up-regulated metabolic pathways mainly involved the metabolism of nucleotides, directly affecting the catabolism of purines and pyrimidines and therefore the production of ATP for energy use. In addition to the up-regulation of pyruvate metabolism, it was also observed the impairment of glycolysis and gluconeogenesis pathways, which have a positive feedback effect within each other, possibly seeking to compensate for the lack of energy caused by reduction of ATP production and in general of triphosphate nucleotides. Clinically it is to be expected an important impact on organs that depend directly on glucose as an energy source, such as the brain, kidney, testicles, and erythrocytes; as well as in organs with high energy demand such as muscle, heart, and liver.
This bi-cellular model agreed with the previous prediction using the glia GEM, strongly suggesting that ARSA deficiency affects metabolic pathways involved in the transport of amino acids, which could lead to activation of neuroprotective mechanisms in response to the metabolic blockage. In addition, the use of this bi-cellular model allowed us to discard the previous prediction of the impairment in the metabolism of several neurotransmitters as a consequence of ARSA deficiency [10]. The bi-cellular model did not predict any alteration in acetylcholine, dopamine, GABA, glycine, glutamate, epinephrine, norepinephrine, and choline, which supports the concept that the neurological compromise in MLD is a result of the distortion of the myelin band caused by the accumulation of non-degraded sulfatide rather that a change in neurotransmitters metabolism. However, the limitations of our model are related to its debugging process, as this may lead to imbalances in the reactions used. For example, we found blocked reactions in glycosaminoglycan degradation metabolism, which are incomplete in the original models. In summary, the use of a neuron-glia metabolic reconstruction allowed to improve the prediction capacity of the model to understand the metabolic consequences of ARSA deficiency. This bi-cellular model may pave the way for modeling the biochemical alterations of other inborn errors of metabolism with CNS involvement.
Declarations
Author contribution statement
Echeverri-Peña Olga Y.: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Salazar-Barreto Diego A: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Rodríguez-Lopez Alexander, González Janneth, Barrera Luis A: Analyzed and interpreted the data; Wrote the paper.
Alméciga-Díaz Carlos J.: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Verano-Guevara Cristian H: Performed the experiments; Analyzed and interpreted the data.
Funding statement
Olga Y. Echeverri-Peña and Luis A. Barrera were supported by the grant number SIAP 4197, Pontificia Universidad Javeriana.
Data availability statement
Data will be made available on request.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Olga Y. Echeverri-Peña, Email: oyecheve@javeriana.edu.co.
Carlos J. Alméciga-Díaz, Email: cjalmeciga@javeriana.edu.co.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Li Z.-H. Progress in metabolism and function of myelin lipids. Acta Physiol. Sin. 2017;69:817–829. [PubMed] [Google Scholar]
- 2.Kohlschütter A. Chapter 164 - lysosomal leukodystrophies: krabbe disease and metachromatic leukodystrophy. In: Olivier Dulac M.L., Harvey B.S., editors. Handbook of Clinical Neurology. Elsevier; 2013. pp. 1611–1618. [DOI] [PubMed] [Google Scholar]
- 3.Yu R.K., Nakatani Y., Yanagisawa M. The role of glycosphingolipid metabolism in the developing brain. J. Lipid Res. 2009;50(Suppl):S440–S445. doi: 10.1194/jlr.R800028-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gieselmann V., Ingeborg K.-M. Metachromatic leukodystrophy. In: Beaudet A.L., editor. The Online Metabolic and Molecular Bases of Inherited Disease. The McGraw-Hill Companies, Inc.; New York, NY: 2014. [Google Scholar]
- 5.Deconinck N. Metachromatic leukodystrophy without arylsulfatase A deficiency: a new case of saposin-B deficiency. Eur. J. Paediatr. Neurol. 2008;12(1):46–50. doi: 10.1016/j.ejpn.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Tan M.A.F. Biochemical profiling to predict disease severity in metachromatic leukodystrophy. Mol. Genet. Metabol. 2010;99(2):142–148. doi: 10.1016/j.ymgme.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Goodrum J.F. Fatty acids from degenerating myelin lipids are conserved and reutilized for myelin synthesis during regeneration in peripheral nerve. J. Neurochem. 1995;65(4):1752–1759. doi: 10.1046/j.1471-4159.1995.65041752.x. [DOI] [PubMed] [Google Scholar]
- 8.Cook D.J., Nielsen J. Genome-scale metabolic models applied to human health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017;9(6) doi: 10.1002/wsbm.1393. [DOI] [PubMed] [Google Scholar]
- 9.Salazar D.A. Systems biology study of mucopolysaccharidosis using a human metabolic reconstruction network. Mol. Genet. Metabol. 2016;117(2):129–139. doi: 10.1016/j.ymgme.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Olga Y.E. Understanding the metabolic consequences of human arylsulfatase A deficiency through a computational systems biology study. Cent. Nerv. Syst. Agents Med. Chem. 2016;16:1–6. [PubMed] [Google Scholar]
- 11.Olarte-Avellaneda S. Computational analysis of human N-acetylgalactosamine-6-sulfate sulfatase enzyme: an update in genotype-phenotype correlation for Morquio A. Mol. Biol. Rep. 2014;41(11):7073–7088. doi: 10.1007/s11033-014-3383-3. [DOI] [PubMed] [Google Scholar]
- 12.Wade J.J. Bidirectional coupling between astrocytes and neurons mediates learning and dynamic coordination in the brain: a multiple modeling approach. PloS One. 2011;6(12) doi: 10.1371/journal.pone.0029445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swainston N. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiele I. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013;31(5) doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis N.E. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat. Biotechnol. 2010;28(12):1279–1285. doi: 10.1038/nbt.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen E.P., Song R.S., Chen X. Mathematical model of hypoxia and tumor signaling interplay reveals the importance of hypoxia and cell-to-cell variability in tumor growth inhibition. BMC Bioinf. 2019;20(1):507. doi: 10.1186/s12859-019-3098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cakir T. Reconstruction and flux analysis of coupling between metabolic pathways of astrocytes and neurons: application to cerebral hypoxia. Theor. Biol. Med. Model. 2007;4:48. doi: 10.1186/1742-4682-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aubert A., Costalat R. Compartmentalization of brain energy metabolism between glia and neurons: insights from mathematical modeling. GLIA. 2007;55(12):1272–1279. doi: 10.1002/glia.20360. [DOI] [PubMed] [Google Scholar]
- 19.Jolivet R. Comment on recent modeling studies of astrocyte-neuron metabolic interactions. J. Cerebr. Blood Flow Metabol. 2010;30(12):1982–1986. doi: 10.1038/jcbfm.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlén M. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 21.Uhlen M. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 22.Pfau T., Pacheco M.P., Sauter T. Towards improved genome-scale metabolic network reconstructions: unification, transcript specificity and beyond. Briefings Bioinf. 2016;17(6):1060–1069. doi: 10.1093/bib/bbv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bordbar A. A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst. Biol. 2011;5:180. doi: 10.1186/1752-0509-5-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bordbar A. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014;15(2):107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 25.Schellenberger J. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 2011;6(9):1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonora M. Molecular mechanisms of cell death: central implication of ATP synthase in mitochondrial permeability transition. Oncogene. 2015;34(12):1608. doi: 10.1038/onc.2014.462. [DOI] [PubMed] [Google Scholar]
- 27.Saab A.S., Tzvetanova I.D., Nave K.A. The role of myelin and oligodendrocytes in axonal energy metabolism. Curr. Opin. Neurobiol. 2013;23(6):1065–1072. doi: 10.1016/j.conb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Tress O. Panglial gap junctional communication is essential for maintenance of myelin in the CNS. J. Neurosci. 2012;32(22):7499–7518. doi: 10.1523/JNEUROSCI.0392-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amaral A.I. Characterization of glucose-related metabolic pathways in differentiated rat oligodendrocyte lineage cells. GLIA. 2016;64(1):21–34. doi: 10.1002/glia.22900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown A.M., Ransom B.R. Astrocyte glycogen and brain energy metabolism. Glia. 2007;55(12):1263–1271. doi: 10.1002/glia.20557. [DOI] [PubMed] [Google Scholar]
- 31.Rich L.R., Harris W., Brown A.M. The role of brain glycogen in supporting physiological function. Front. Neurosci. 2019;13:1176. doi: 10.3389/fnins.2019.01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson R.A. Physiologic coupling of glial glycogen metabolism to neuronal activity in brain. Can. J. Physiol. Pharmacol. 1992;70(Suppl):S138–S144. doi: 10.1139/y92-255. [DOI] [PubMed] [Google Scholar]
- 33.Bordel S., Agren R., Nielsen J. Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS Comput. Biol. 2010;6(7) doi: 10.1371/journal.pcbi.1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben Achour S., Pascual O. Astrocyte–neuron communication: functional consequences. Neurochem. Res. 2012;37(11):2464–2473. doi: 10.1007/s11064-012-0807-0. [DOI] [PubMed] [Google Scholar]
- 35.Cakir T., Tacer C.S., Ulgen K.O. Metabolic pathway analysis of enzyme-deficient human red blood cells. Biosystems. 2004;78 doi: 10.1016/j.biosystems.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Blazier A., Papin J. Integration of expression data in genome-scale metabolic network reconstructions. Front. Physiol. 2012;3(299) doi: 10.3389/fphys.2012.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieven C. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 2020;38(3):272–276. doi: 10.1038/s41587-020-0446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allaman I., Bélanger M., Magistretti P.J. Astrocyte–neuron metabolic relationships: for better and for worse. Trends Neurosci. 2011;34(2):76–87. doi: 10.1016/j.tins.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Yamashita T. Interruption of ganglioside synthesis produces central nervous system degeneration and altered axon–glial interactions. Proc. Natl. Acad. Sci. U. S. A. 2005;102(8):2725–2730. doi: 10.1073/pnas.0407785102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rappard D.F., Boelens J.J., Wolf N.I. Metachromatic leukodystrophy: disease spectrum and approaches for treatment. Best Pract. Res. Clin. Endocrinol. Metabol. 2015;29(2):261–273. doi: 10.1016/j.beem.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Marchitti S.A., Deitrich R.A., Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol. Rev. 2007;59(2):125–150. doi: 10.1124/pr.59.2.1. [DOI] [PubMed] [Google Scholar]
- 42.Elpeleg O., Mandel H., Saada A. Depletion of the other genome-mitochondrial DNA depletion syndromes in humans. J. Mol. Med. (Berl.) 2002;80(7):389–396. doi: 10.1007/s00109-002-0343-5. [DOI] [PubMed] [Google Scholar]
- 43.Nakai M. Effects of ligation of the ductus deferens on the fowl epididymal region. Nihon Juigaku Zasshi. 1989;51(3):521–529. doi: 10.1292/jvms1939.51.521. [DOI] [PubMed] [Google Scholar]
- 44.Toepfer C.N. SarcTrack. Circ. Res. 2019;124(8):1172–1183. doi: 10.1161/CIRCRESAHA.118.314505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkes H.A. Overexpression of acyl-CoA synthetase-1 increases lipid deposition in hepatic (HepG2) cells and rodent liver in vivo. Am. J. Physiol. Endocrinol. Metab. 2006;291(4):E737–E744. doi: 10.1152/ajpendo.00112.2006. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y. SIRT3 and SIRT5 regulate the enzyme activity and cardiolipin binding of very long-chain acyl-CoA dehydrogenase. PloS One. 2015;10(3) doi: 10.1371/journal.pone.0122297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cairella M., Volpari B. Clinical observations on the anti-inflammatory activity of the association of benzydamine and tetracycline. Clin. Ter. 1969;51(5):439–451. [PubMed] [Google Scholar]
- 48.Sertbas M., Ulgen K., Cakir T. Systematic analysis of transcription-level effects of neurodegenerative diseases on human brain metabolism by a newly reconstructed brain-specific metabolic network. FEBS Open Bio. 2014;4:542–553. doi: 10.1016/j.fob.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Luca C. Neurons, glia, extracellular matrix and neurovascular unit: a systems biology approach to the complexity of synaptic plasticity in health and disease. Int. J. Mol. Sci. 2020;21(4) doi: 10.3390/ijms21041539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoen P. Serum carnosinase activity in plasma and serum: validation of a method and values in cardiopulmonary bypass surgery. Clin. Chem. 2003;49(11):1930–1932. doi: 10.1373/clinchem.2003.019398. [DOI] [PubMed] [Google Scholar]
- 51.Drozak J. Molecular identification of carnosine synthase as ATP-grasp domain-containing protein 1 (ATPGD1) J. Biol. Chem. 2010;285(13):9346–9356. doi: 10.1074/jbc.M109.095505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Preston J.E. Toxic effects of beta-amyloid(25-35) on immortalised rat brain endothelial cell: protection by carnosine, homocarnosine and beta-alanine. Neurosci. Lett. 1998;242(2):105–108. doi: 10.1016/s0304-3940(98)00058-5. [DOI] [PubMed] [Google Scholar]
- 53.Tabakman R., Lazarovici P., Kohen R. Neuroprotective effects of carnosine and homocarnosine on pheochromocytoma PC12 cells exposed to ischemia. J. Neurosci. Res. 2002;68(4):463–469. doi: 10.1002/jnr.10228. [DOI] [PubMed] [Google Scholar]
- 54.Alabovsky V.V. Effect of histidine-containing dipeptides on isolated heart under ischemia/reperfusion. Biochemistry (Mosc.) 1997;62(1):77–87. [PubMed] [Google Scholar]
- 55.Ferdinandusse S. Adult peroxisomal acyl-coenzyme A oxidase deficiency with cerebellar and brainstem atrophy. J. Neurol. Neurosurg. Psychiatry. 2010;81(3):310–312. doi: 10.1136/jnnp.2009.176255. [DOI] [PubMed] [Google Scholar]
- 56.Alfardan J. Characterization of new ACADSB gene sequence mutations and clinical implications in patients with 2-methylbutyrylglycinuria identified by newborn screening. Mol. Genet. Metabol. 2010;100(4):333–338. doi: 10.1016/j.ymgme.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.