Abstract
Background
Recurrence rates after resection of colorectal cancer liver metastases (CRLM) are high and correlate with worse survival. Postoperative circulating tumour DNA (ctDNA) is a promising prognostic biomarker. Focusing on patients with resected CRLM, this study aimed to evaluate the association between the detection of postoperative ctDNA, pathologic response and recurrence-free survival (RFS).
Methods
Twenty-three patients were selected from an ongoing phase-3 trial who underwent resection of RAS-mutant CRLM after induction systemic treatment. CtDNA analysis was performed by droplet digital PCR using blood samples collected at baseline, before and after resection. Pathologic response of CRLM was determined via the Tumour Regression Grading system.
Findings
With a median follow-up of 19.6 months, the median RFS for patients with detectable (N = 6, [26%]) and undetectable (N = 17, [74%]) postoperative ctDNA was 4.8 versus 12.1 months, respectively. Among 21 patients with available tumour tissue, pathologic response in patients with detectable compared to undetectable postoperative ctDNA was found in one of six (17%) and 15 of 15 (100%) patients, respectively (p < 0.001). In univariable Cox regression analyses both postoperative detectable ctDNA (HR = 3.3, 95%CI = 1.1–9.6, p = 0.03) and pathologic non-response (HR = 4.6, 95%CI = 1.4–15, p = 0.01) were associated with poorer RFS and were strongly correlated (r = 0.88, p < 0.001). After adjusting for clinical characteristics in pairwise multivariable analyses, postoperative ctDNA status remained associated with RFS.
Interpretation
The detection of postoperative ctDNA after secondary resection of CRLM is a promising prognostic factor for RFS and appeared to be highly correlated with pathologic response.
Keywords: Colorectal cancer, Liver metastases, Circulating tumour DNA, Resection, Recurrences
Research in context.
Evidence before this study
Recurrence rates after resection of colorectal liver metastases (CRLM) are high and caused by micro-metastases left in situ after resection. Currently available follow-up methods have limited accuracy for detecting this minimal residual disease (MRD). Studies in patients with stage I-III colorectal cancer demonstrated that postoperative circulating tumour DNA (ctDNA) is a strong independent prognostic biomarker for MRD and recurrence-free survival. Studies investigating postoperative ctDNA in stage IV disease are limited and mostly concern heterogeneous patient groups with both hepatic and extrahepatic disease and varying use of induction systemic treatment.
Added value of this study
This is a proof of concept study reporting on the prognostic value of ctDNA in an upfront carefully selected homogeneous population of patients with RAS mutant initially unresectable CRLM. In addition, this is the first study to analyse the association of postoperative ctDNA detection with pathologic response in patients with metastatic CRC. CtDNA analysis was performed using the relatively fast, inexpensive, and highly sensitive droplet digital PCR to facilitate translation to future clinical practice.
Implications of all the available evidence
The results of this study offer a perspective on the clinical relevance of the assessment of postoperative ctDNA in CRLM patients with a high risk of recurrence. Liquid biopsy ctDNA offers the possibility for longitudinal follow-up, whereas pathologic response can only be assessed after resection. This offers opportunities for the personalisation of postoperative disease management in this common subgroup of patients with metastatic CRC, e.g. by intensifying follow-up or providing adjuvant treatment.
Alt-text: Unlabelled box
1. Introduction
The liver is the primary metastatic site of colorectal cancer (CRC). In patients with metastatic CRC, 70 to 80% have liver metastases [1]. In patients with liver-limited colorectal cancer liver metastases (CRLM), resection offers the only chance for cure or long-term survival [1]. Approximately 20% of patients present with upfront resectable CRLM (primary resectable), and 20–40% of patients with initially unresectable CRLM may convert to resectable disease upon downsizing by induction systemic treatment (secondary resectable) [2]. Nevertheless, reported 3-year recurrence rates for primary and secondary resectable CRLM are up to 60% [3,4] and 80% [4,5], respectively. The majority of recurrences occur within the first two years following resection [4]. Furthermore, over half of the CRLM patients die within five years following resection [4,6]. Pathologic response [7] and early recurrence [8] have been correlated with overall survival in patients with CRLM.
Recurrences are considered to be caused by minimal residual disease (MRD) consisting of micro-metastases left in situ. Currently, available follow-up methods like serum carcinogenic embryonic antigen (CEA) and cross-sectional clinical imaging such as CT- or PET-scans have limited accuracy for detecting MRD due to low sensitivity and specificity [9]. While magnetic resonance imaging (MRI) shows a higher sensitivity compared to CT-scan for detecting small and disappearing metastases in the liver after systemic therapy [10], CT-scan has a higher overall diagnostic accuracy for detecting extrahepatic disease and has clear logistical advantages compared to whole-body MRI. Determining MRD by detecting cell-free circulating tumour DNA (ctDNA) after local treatment of CRLM may offer an alternative approach with important prognostic and therapeutic implications.
Liquid biopsy-derived ctDNA represents a minimally invasive, cancer-specific biomarker with great potential to improve diagnosis and to better determine prognosis, predict drug responsiveness and monitor treatment response [11], [12], [13]. Its short half-life makes ctDNA a dynamic marker indicating the presence of cancer cells and may detect evidence of tumour response or recurrences earlier than imaging and clinical parameters [14,15]. In addition, ctDNA has the potential to provide information about the genomic changes of the tumour [16]. In patients with stage I-III CRC, postoperative ctDNA is a strong independent prognostic biomarker for MRD and recurrence-free survival (RFS) [17], [18], [19]. These data suggest that ctDNA may be a potential marker for selecting early-stage CRC patients for adjuvant systemic therapy [15,[20], [21], [22], [23], [24]]. Compared to other tumour types, patients with metastatic CRC show among the highest levels of detectable ctDNA [24,25]. In unselected patients with metastatic CRC, multiple studies have shown that detectable postoperative ctDNA is also strongly correlated with recurrence rate [26], [27], [28], [29]. However, most of these results were obtained from studies with a small and heterogeneous study population, with limited data on patients with liver-only metastatic disease. Besides, there are no studies involving patients with metastatic CRC that correlated ctDNA results with pathologic response.
The present study makes use of a well-defined selected group of patients participating in a prospective randomised study and aims to determine the prognostic value of postoperative ctDNA for detection of MRD and RFS in patients with CRLM after induction systemic therapy and complete resection of liver metastases. Secondly, the association between postoperative ctDNA detection and pathologic tumour response in liver metastases was evaluated.
2. Methods
2.1. Patient selection
Patients were selected from the ongoing CAIRO5 randomised phase 3 trial of the Dutch Colorectal Cancer Group (DCCG), in which the currently most effective first-line systemic regimens of chemotherapy plus targeted therapy are being compared in patients with initially unresectable CRLM (registration number: NCT02162563). A total of 564 patients are planned to be enrolled in the CAIRO5 clinical trial based on statistical assumptions previously described [30]. CRLM are deemed initially unresectable after assessment following predefined baseline resectability criteria considering R0-resection cannot be achieved in one procedure with one surgical intervention only. Patients are stratified for RAS and BRAF V600E mutation status and sidedness of primary tumour. Mutation analyses were performed on DNA isolated from the primary tumour for most patients because tissue from metastases was rarely available (91% versus 9%, respectively). Patients are evaluated every two months by an expert panel of liver surgeons and abdominal radiologists for the possibility of local treatment of CRLM following current practice [31]. Patients in whom local treatment of CRLM is achieved continue postoperatively with the preoperative systemic regimen but without the targeted agent for a total duration of pre- and postoperative treatment of six months. After patients signed informed consent, formalin-fixed paraffin-embedded (FFPE) tumour tissue was collected prior to treatment for translational research. In addition, blood samples were collected longitudinally every two months until resection and every three months after resection. For the current observational translational research subgroup analysis patients were selected who were randomised between the start of the study (June 2014) and August 2018, with RAS mutated tumours treated with bevacizumab plus either doublet or triplet chemotherapy, complete (R0/R1) resection of the primary tumour and liver metastases (resection and/or local ablation), and available baseline, pre- and postoperative liquid biopsies. Follow-up was recorded until May 2020. ctDNA analyses were performed on the subset of patients with RAS hotspot mutations, which can be analysed using the relatively fast, inexpensive and highly sensitive ddPCR test. Patients with a first postoperative liquid biopsy drawn after starting adjuvant systemic therapy were excluded to avoid the confounding effect of chemotherapy. After completing systemic treatment, follow-up was performed according to the standard of care, including a three-monthly clinical review, six-monthly serum CEA, and CT imaging.
2.2. Ethics
The medical ethical committee of the Amsterdam Medical Center approved the CAIRO5 study under reference number METC 2014_008, NL47650.018.14, and all patients signed written informed consent for study participation as well as liquid biopsy and tumour tissue collection for translational research.
2.3. Clinicopathological data
Baseline clinicopathological patient characteristics were prospectively collected, such as age, sex, characteristics of the primary tumour (sidedness of the tumour, type of RAS mutation), time to metastases (with metachronous disease defined as a disease-free interval of more than six months after diagnosis of the primary tumour [32]), size and number of metastases, serum CEA levels, clinical risk score (CRS) [33] (low risk 0–2 points and high risk 3–5 points), chemotherapy regimen (doublet or triplet), number of cycles and documented radiologic response according to the RECIST 1.1 criteria, type of local therapies for CRLM, and R-status of resections (R0 or R1).
Pathologic response assessment was done by evaluating hematoxylin- and eosin-stained slides by an independent pathologist blinded for ctDNA outcomes. Pathologic response was scored according to the tumour Regression Grading (TRG) [34]. TRG was graded from 1 to 5, with TRG 4 and 5 indicating no or minor pathological response.
Previous studies have shown that early recurrence after resection of CRLM, defined as recurrence within six to eight months, correlates with prognosis [8,35,36]. Therefore, we defined early recurrence as occurring within eight months of local treatment of CRLM. RFS was calculated from the date of hepatic resection until documented progression or censored on the last clinical visit date. In the case of a two-stage hepatic resection, RFS was calculated from the last surgical procedure.
2.4. Cell-free DNA isolation and quantification
Prior to systemic treatment (baseline), preoperatively, a maximum of 100 days postoperatively, and during follow-up, 10 ml of blood was collected using a cell-stabilising BCT® tube (Streck, La Vista, USA) at the medical centre of inclusion. For analyses, all liquid biopsies were shipped to the Clinical Chemistry laboratory at the Netherlands Cancer Institute (Amsterdam, the Netherlands). Cell-free plasma was collected in a two-step centrifugation process; 10 min at 1.700 g followed by 10 min at 20.000 g before storage at −80 °C. Cell-free DNA (cfDNA) was isolated using the QIAsymphony (Qiagen, Germany) with an elution volume set to 60 µl. The concentration of the cfDNA was measured using the Qubit™ dsDNA High-Sensitivity Assay (TFS, Waltham, USA) and ranged from 0.12 to 60.4 ng/µl.
2.5. Cell-free DNA RAS mutation analyses
KRAS and NRAS mutation analyses using extracted cfDNA from plasma were performed by droplet digital PCR (ddPCR) (Bio-Rad, Hercules, USA). For these analyses, the ddPR™ KRAS G12/G13 (#1863506), ddPCR™ KRAS Q61 (#12001626), ddPCR™ KRAS A146 (#10049550) and the ddPCR™ NRAS Q61 (#12001006) Screening Kits were used according to the manufacturer's instruction making use of 1 µl multiplex assay, 11 µl ddPCR supermix for probes (no dUTP), 9 µl sample and 1 µl H2O. When necessary, samples were diluted to 2 ng/µl. All measurements were performed in duplicate and included a blank (nuclease-free water) and an in-house positive control. Data were analysed using the QuantaSoft™ software version 1.6.6 (Bio-Rad, Hercules, USA). Individual wells with less than 10.000 total events (droplets) were excluded from the analysis, and all results were corrected based on a predefined false-positive rate, based on 60-fold analyses of commercial reference wildtype DNA (Promega; Fitchburg, WI, USA) [37].
2.6. Statistics
Patient and tumour characteristics were summarised as frequency counts and percentages, or as medians and range. Differences between groups were analysed using Pearson's chi-square test and Fisher exact test, as appropriate. Survival data were analysed using the Kaplan-Meier method, and survival curves were compared using the log-rank test. Cox proportional hazards regression analysis was performed to analyse prognostic factors for RFS. Hazard ratios (HRs) and corresponding 95% confidence intervals (95% CI) were estimated. Given the small sample size and the limited number of events available, a maximum of two variables was introduced in multivariable analyses. Given the strong association between ctDNA and pathologic response, they were not analysed together in the same multivariable model. A multivariable Cox regression analysis including more than one covariate together with postoperative ctDNA was performed as sensitivity analysis. Spearman's correlation coefficient was estimated to evaluate the association between pathologic response and postoperative ctDNA status. Analyses were performed using SPSS software version 25 (IBM, New York, USA).
2.7. Role of the funding source
This study was supported by the Dutch Cancer Society (Grant No. 10438) and by a scientific grant from Amgen, The Netherlands. The funders had no role in the design, conduct and submission of the study, nor the decision to submit the manuscript for publication. All authors had full access to all the data in the study and accepted the responsibility to submit for publication.
3. Results
3.1. Patient characteristics
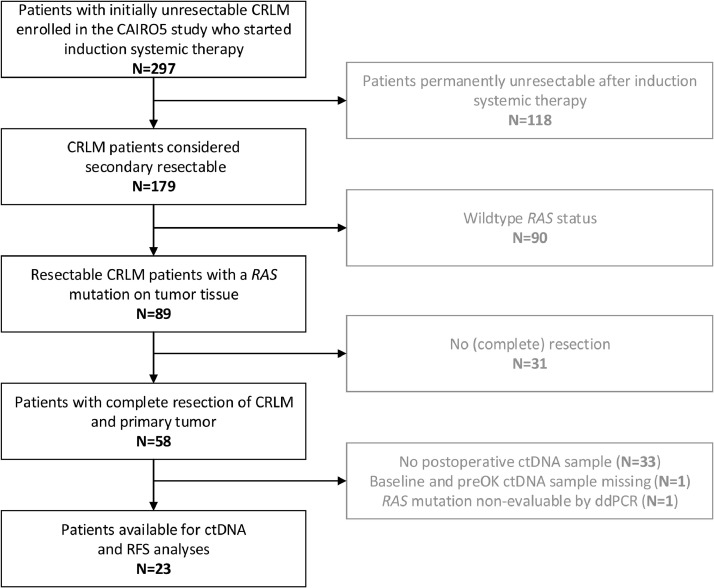
Patient selection and study overview are presented in Fig. 1. Between November 2014 and August 2018, 297 patients with initially unresectable CRLM were enrolled in the CAIRO5 study. According to tumour tissue analyses, fifty-nine patients carried a RAS mutation and achieved a confirmed complete resection of liver metastases and primary tumour after systemic induction therapy. After exclusion of patients with unavailable preoperative and/or postoperative liquid biopsies, a total of 23 patients, one with a NRAS mutation and 22 with a KRAS mutation, were eligible for further ctDNA and RFS analysis. The follow-up was recorded until the 20th of April 2020. The baseline patient characteristics of this cohort are displayed in Table 1 and show synchronous metastases in 19 (83%) patients, with a median number of metastases of eight (range 1–37), and 20 (87%) patients with a high CRS. Ten (44%) patients received doublet chemotherapy (FOLFOX or FOLFIRI) plus bevacizumab, and 13 (57%) patients triplet chemotherapy (FOLFOXIRI) plus bevacizumab.
Fig. 1.
Flowchart of patient selection.
Table 1.
Summary of clinicopathological patient characteristics.
| Clinical characteristics | All patients (N = 23) | |
|---|---|---|
| Age, median (range) | 63 (54–76) | |
| Sex, n (%) | ||
| Male | 15 (65) | |
| Female | 8 (35) | |
| Tumour site, n (%) | ||
| Left colon | 17 (74) | |
| Right colon | 6 (26) | |
| RAS mutation, n (%) | ||
| KRAS mutation | 22 (96) | |
| NRAS mutation | 1 (4) | |
| Source tissue mutation analysis, n (%) | ||
| Primary tumour | 21 (91) | |
| Liver metastases | 2 (9) | |
| Synchronous liver metastases, n (%) | ||
| No | 4 (17) | |
| Yes | 19 (83) | |
| Number of metastases, median (range) | 8 (1–37) | |
| Prior resection of primary tumour, n (%) | ||
| No | 11 (48) | |
| Yes | 12 (52) | |
| CEA, median (range) | 9.5 (1–3469) | |
| Fong risk score, n (%) | ||
| Low (0–2) | 3 (13) | |
| High (3–5) | 20 (87) | |
| Perioperative systemic therapy, n (%) | ||
| Doublet chemotherapy + target therapy | 10 (44) | |
| Triplet chemotherapy + target therapy | 13 (57) | |
| Cycles preoperative therapy, mean (range) | 7.7 (4–13) | |
| Cycles postoperative therapy, mean (range) | 1.9 (0–7) | |
| Best response (RECIST), n (%) | ||
| Partial response | 17 (74) | |
| Stable disease | 5 (22) | |
| Progression of disease | 1 (4) | |
| Type of resection, n (%) | ||
| 1-stage | 19 (83) | |
| 2-stage | 3 (13) | |
| R-status, n (%) | ||
| R0 | 20 (83) | |
| R1 | 3 (13) | |
| Local ablative therapy | 1 (4) | |
| Baseline ctDNA, n (%) | ||
| Undetectable | 2 (9) | |
| Detectable | 18 (78) | |
| Missing baseline sample | 3 (13) | |
| Histopathological response (TRG), n (%) | ||
| Pathologic response (TRG 1–3) | 16 (65) | |
| No pathologic response (TRG 4–5) | 5 (22) | |
| Missing | 2 (9) | |
| Postoperative ctDNA, days after last surgery, median (range) | 38 (1–99) | |
Abbreviations; CEA; carcinogenic embryonic antigen; RECIST; response evaluation criteria in solid tumours; ctDNA; circulating tumour DNA; TRG; tumour regression grade
3.2. Detection of ctDNA at baseline, preoperatively and postoperatively
Within the group of 23 patients, preoperative ctDNA analyses were performed on baseline blood samples in 20 patients (87%) and on preoperative blood samples in 22 patients (96%). Analyses of the postoperative liquid biopsies showed that six (26%) patients had detectable ctDNA compared to 17 (74%) patients with undetectable postoperative ctDNA. Patients with detectable versus undetectable postoperative ctDNA did not differ in baseline characteristics (Supplementary Table 1).
3.3. Association of ctDNA detection with recurrence of disease
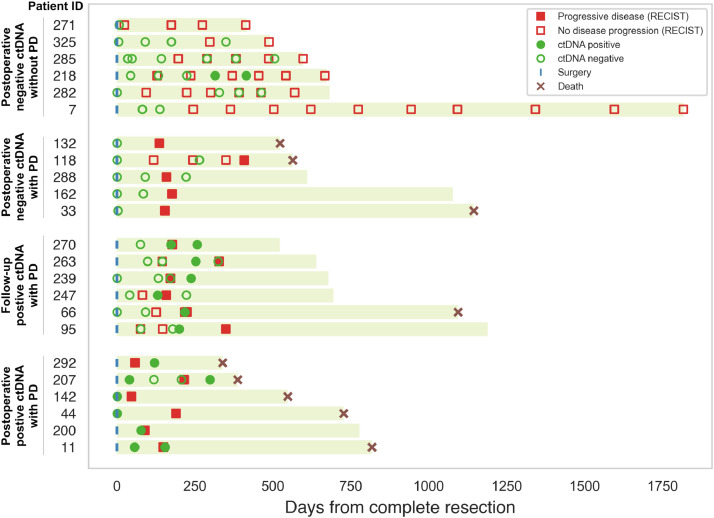
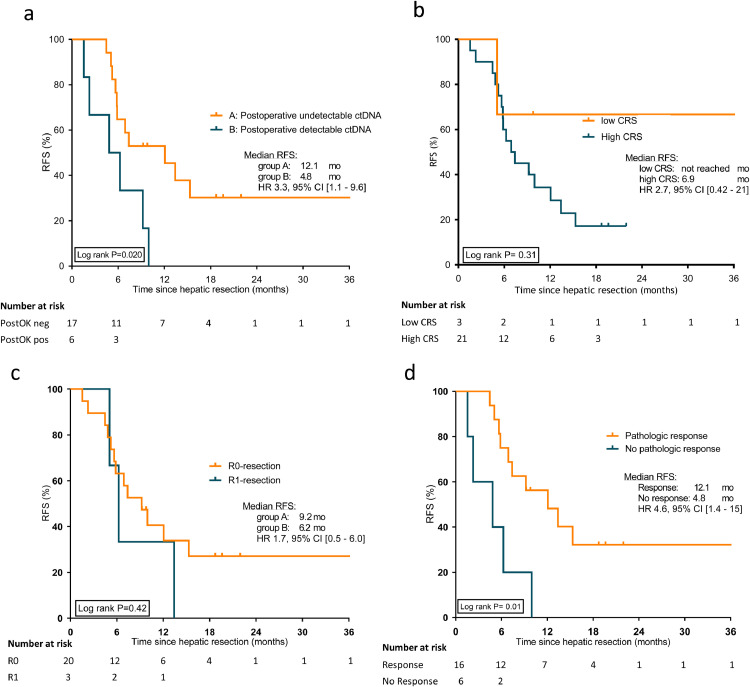
At a median follow-up of 19.6 months (range 1.5 – 60 months), 17 patients (74%) had recurrence of disease, with 12 patients (52%) showing early disease recurrence (≤ eight months), see Table 2. In nine patients (53%), the first recurrence occurred at an extrahepatic site. In patients with postoperatively detectable ctDNA compared to undetectable ctDNA, early disease recurrence was observed in four (67%) patients versus eight (47%) patients, respectively. However, this was not significant (Pearson's chi-squared test, p = 0.41, specificity 81% and sensitivity 33%). Fig. 2 presents the postoperative ctDNA status and lead-time to recurrence detected by ctDNA and imaging studies for all 23 patients. A detailed overview of both pre- and postoperative ctDNA detection per patient is presented in Supplementary Figure 1. In analysing the performance of ctDNA in the detection of MRD, we found that six patients (100%) with postoperative detectable ctDNA and 11 patients (65%) with undetectable postoperative ctDNA had a recurrence during follow-up. For a total of 15 patients, serum CEA was determined within 100 days following resection. Of patients with serum CEA levels within the normal range (N = 14) versus elevated (> 5 ng/ml) (N = 1), 11 (79%) and one (100%) patient developed recurrences during follow-up, respectively. Postoperative ctDNA detection was significantly associated with poorer RFS, with a median RFS for patients with postoperative undetectable versus detectable ctDNA of 12.1 and 4.8 months, respectively (HR 3.3, 95%CI 1.1–9.6, log-rank p = 0.03), see Fig. 3.
Table 2.
Follow-up and recurrence-free survival for patients with postoperative undetectable and postoperative detectable ctDNA.
| All patients (N = 23) | Postoperative undetectable ctDNA (N = 17) | Postoperative detectable ctDNA (N = 6) | |||
|---|---|---|---|---|---|
| Median follow-up, months (95% CI) | 19.6 (17.8 – 21.4) | ||||
| Median RFS, months | 7.4 | 12.1 | 4.8 | ||
| Number of patients with recurrence, n (%) | 17 (74) | 11 (65) | 6 (100) | ||
| Early recurrence (≤ 8 months), n (%) | |||||
| No | 11 (48) | 9 (53) | 2 (33) | ||
| Yes | 12 (52) | 8 (47) | 4 (67) | ||
| Site of recurrence | |||||
| Liver | 8 (47) | 6 (55) | 2 (33) | ||
| Extrahepatic | 9 (53) | 5 (45) | 4 (67) | ||
| No recurrence | 6 | 6 | – | ||
Abbreviations; ctDNA; circulating tumour DNA; RFS; recurrence-free survival
Fig. 2.
Overview of surveillance for disease recurrence in 23 patients with colorectal liver metastases (CRLM) after complete resection following induction systemic treatment. Clinical response evaluation is depicted until progression of disease (PD), where all liquid biopsy ctDNA ddPCR analysis results are showed. A distinction was made between four groups; patients with postoperative positive ctDNA with PD, patients with follow-up positive ctDNA with PD, patients with postoperative negative ctDNA with PD, and patients with postoperative negative ctDNA without PD.
Fig. 3.
Kaplan-Meier curves showing recurrence-free survival according to: (a) postoperative ctDNA mutation status (undetectable versus detectable), (b) Fong clinical risk score (low versus high) (c) resection margin (R0 versus R1), and (d) pathologic response (TRG 1–3 versus TRG 4–5).
3.4. Postoperative ctDNA detection and association with pathologic response
For one patient only local ablative therapy was executed, and for one patient no HE-slides were available. Therefore, pathologic response was assessed on resected tissue from liver metastases of 21 (91%) patients using Slide Score [38]. In patients with liver metastases available for pathologic response assessment, major pathologic response (TRG 1 or 2), partial (TRG 3), and no pathologic response (TRG 4 or 5) was scored in 10 (48%), six (29%), and five (24%) patients, respectively. Postoperative ctDNA status was strongly correlated with pathologic response (TRG 1–3) (Spearman's correlation, r = 0.88, p < 0.001). All patients (N = 15, 100%) with undetectable ctDNA had partial or major pathologic response compared to only one (17%) patient with detectable ctDNA (Pearson's Chi-squared test, p < 0.001).
3.5. Postoperative ctDNA and pathologic non-response are associated with poor RFS
Univariable survival analysis showed detectable postoperative ctDNA (HR 3.3, 95%CI 1.1–9.6, log-rank p = 0.03) and pathologic non-response (TRG 4–5) (HR 4.6, 95%CI 1.4–15, log-rank p = 0.01) to be associated with poorer RFS (see Table 3). After adjusting postoperative ctDNA for age, sex, Fong CRS, radiological response, sidedness and R-status in separate pairwise multivariable analyses, detectable postoperative ctDNA remained significantly associated with poorer RFS. The association between postoperative ctDNA and RFS remained strong in the sensitivity analysis adjusting for all the aforementioned variables simultaneously in a multivariable model (HR 4.1, 95%CI 1.19–14.47, log-rank p = 0.026). No indications of an association between RECIST response or non-response and pathologic response (Fisher's Exact, p = 0.761), detection of postoperative ctDNA (Fisher's Exact, p = 0.083), or recurrence of disease (Fisher's Exact, p = 0.217) was found.
Table 3.
Cox regression univariable recurrence-free survival analysis by clinicopathological variables and postoperative ctDNA status.
| Variable | Number patients | Event RFS | Univariable analysis |
|||
|---|---|---|---|---|---|---|
| n (%) | n | HR | 95% CI | Log-rank P | ||
| Age, years | ||||||
| < 60 | 8 (35) | 6 | ||||
| > 60 | 15 (65) | 11 | 1.2 | 0.4 – 3.1 | 0.78 | |
| Sex | ||||||
| Male | 15 (65) | 11 | ||||
| Female | 8 (35) | 6 | 0.98 | 0.4 – 3.7 | 0.98 | |
| Sidedness primary tumour | ||||||
| Left | 17 (74) | 13 | ||||
| Right | 6 (26) | 4 | 1.2 | 0.4 – 3.8 | 0.74 | |
| Clinical risk score* | ||||||
| Low | 3 (13) | 1 | ||||
| High | 20 (87) | 16 | 2.7 | 0.4 – 21 | 0.33 | |
| Postoperative serum CEA | ||||||
| Normal | 15 (94) | 10 | ||||
| Elevated (> 5 ng/ml) | 1 (6) | 1 | 0.9 | 0.1 – 6.8 | 0.90 | |
| Resection status | ||||||
| R0-resection | 19 (86) | 13 | ||||
| R1-resection | 3 (14) | 3 | 1.7 | 0.5 – 6.0 | 0.43 | |
| Radiological response on induction treatment | ||||||
| Response | 17 (74) | 14 | ||||
| No Response | 6 (26) | 3 | 2.5 | 0.7 – 8.7 | 0.16 | |
| Tumour regression grade | ||||||
| Response (TRG 1–3) | 16 (76) | 10 | ||||
| No response (TRG 4–5) | 5 (24) | 5 | 4.6 | 1.4 – 15 | 0.01 | |
| Postoperative ctDNA status | ||||||
| Undetectable | 17 (74) | 11 | ||||
| Detectable | 6 (26) | 6 | 3.3 | 1.1 – 9.6 | 0.03 | |
Abbreviations; RFS; recurrence-free survival; CEA; carcinogenic embryonic antigen; TRG; tumour regression grade; ctDNA; circulating tumour DNA; *Clinical risk groups are classified according to Fong
4. Discussion
This study analysed the association between postoperative ctDNA and both pathologic response and RFS in patients with initially unresectable CRLM after radical resection of both CRLM and primary tumour. The results indicate that postoperative ctDNA analysis within a high-risk cohort may potentially identify patients with a higher risk of disease recurrence after secondary resection. In addition, postoperative ctDNA showed a strong association with pathologic response on systemic therapy as assessed by the tumour regression grade and is an independent prognostic factor for RFS.
Liquid biopsies are a rich source of minimal invasive biomarkers such as circulating tumour cells (CTCs) and ctDNA, which have the potential to be applied for the clinical management of patients with CRC [39]. In this study we focused on the analysis of ctDNA, considering the higher detection rate of ctDNA compared to CTCs in patients with metastatic CRC [40]. Limited data is available on the value of ctDNA in patients with CRLM [40], [41], [42]. Narayan et al. showed an association of preoperative ctDNA with overall survival in patients with upfront resectable CRLM [41]. The PRODIGE-14 METHEP-2 trial showed in initially unresectable CRLM patients that preoperative ctDNA levels correlate with R0/R1 resections and overall survival [40]. The trial of He et al. involving twenty CRLM patients, not clearly defined as initially resectable or unresectable and with approximately 50% receiving neo-adjuvant systemic therapy, demonstrated a prolonged RFS for patients with low preoperative ctDNA [42]. Further studies in patients with resected CRLM concerned heterogeneous populations in terms of CRC stage among the whole population, presence of extrahepatic metastases, first presentation and relapse of CRLM [27,28], inclusion of both radical and non-radical resections [28], primary and secondary resectable CRLM and types of local therapy [29,42].
To our knowledge, this is the first study to analyse the association of postoperative ctDNA detection and pathologic response in resected liver metastases in CRLM patients. Pathologic response is a well-known independent prognostic factor for overall survival in patients with CRLM [7] and can therefore be used as an early surrogate marker for survival. Our results show a strong association between postoperative ctDNA status and pathologic response. After adjusting for clinical characteristics, both postoperative ctDNA and pathologic response were independent prognostic factors for RFS in separately conducted pairwise multivariable analysis. The added value of ctDNA compared to pathologic response is the ability to perform serial ctDNA analyses in longitudinal follow-up, whereas pathologic response is only possible after resection. Additionally, ctDNA is analysed by a simple blood draw while pathologic response requires tumour tissue. These factors combined with the results of this study might have clinically relevant implications since ctDNA could be used as a surrogate marker for pathologic response and clinical outcome in metastatic CRC patients without available tumour tissue after systemic therapy, such as patients treated with local ablative therapy only or patients on palliative systemic therapy.
The promising monitoring and prognostic value of ctDNA have raised major interest in ctDNA driven adjuvant trials [43]. Adjuvant systemic therapy in CRLM patients has failed to show a 5 year survival benefit [6]. However, this study concerned relatively low-risk CRLM patients (with four or fewer metastases), and retrospective studies suggest that an adequate selection of patients with a high risk of recurrence could help select the patients who might benefit from adjuvant treatment [44,45]. Our results show that postoperative ctDNA status is an independent prognostic factor for RFS and might be a promising biomarker in future trials to select very high-risk CRLM patients for adjuvant trials or otherwise for individualised therapy.
Liquid biopsy ctDNA is a promising biomarker to optimize strategies for monitoring disease recurrence after resection of CRLM. Early detection of a recurrence limited to the liver might offer an opportunity for repeated local treatments with curative intent. Further studies are needed to determine if patients with detectable postoperative ctDNA have clinical benefit from intensified follow-up strategies, like more frequent evaluations or additional imaging methods such as MRI or PET-CT, resulting in better survival outcomes than the current standard of care follow-up strategies. With the additional advantage of liquid biopsies providing the ability for longitudinal monitoring of disease recurrence, having less burden to patients and lower costs than radiological imaging, ctDNA is an interesting biomarker to investigate in future prospective trials. Furthermore, combining radiologic and ctDNA assessments might also help interpret indeterminate radiological findings such as nonspecific liver or lung nodules. Currently, serum CEA is used after resection of CRLM to monitor disease recurrence. However, serum CEA has a low sensitivity and specificity, which might be explained by expression in both neoplastic and normal cells [45], [46], [47]. Liquid biopsy ctDNA was shown to perform better [41,48] with higher sensitivity compared to serum CEA, 100% versus 56% (p = 0.01) [26]. In our population with high-risk CRLM patients, we confirmed that ctDNA is a stronger prognostic marker for RFS than CEA. Secondly, in pairwise multivariable analysis with other potential clinicopathological risk factors for disease recurrence (e.g. CEA, CRS, R-status), we found indications that postoperative ctDNA status was an independent prognostic factor for RFS in patients with secondary resection of CRLM.
An ideal test to diagnose MRD after resection, and further tailor adjuvant systemic treatment, has high sensitivity and specificity [49]. Previously, postoperative ctDNA in metastatic CRC was shown to have high specificity but relatively low sensitivity, since a considerable number of patients with undetectable postoperative ctDNA still developed a recurrence [27], [28], [29]. Similarly, in our study investigating a homogeneous group of CRLM patients, we found a high specificity, where all patients with postoperative detectable ctDNA had a recurrence during follow-up, but lower sensitivity, since 65% of the patients with undetectable postoperative ctDNA also developed a recurrence. A possible factor contributing to our study's sensitivity is the use of ddPCR as a hotspot detection method (detection of one mutation). Our study focused on patients whose RAS mutation status was determined as part of the clinical diagnostic workflow, to establish their eligibility for anti-EGFR treatment. Methodologically, ddPCR-based assays for detecting ctDNA hotspot mutations have high sensitivity and are relatively cheap [37]. This ensures more widespread applicability in daily clinical practice as compared to NGS analyses of gene panels and rendered ddPCR a logical choice for detecting ctDNA in this subset of patients in the present study. Another explanation for the phenomenon of undetectable postoperative ctDNA in patients with MRD leading to recurrence might be the use of preoperative systemic therapy in all patients in our study. This could have (temporarily) reduced the proliferation and apoptosis of minimal residual tumour cells postoperatively, thereby reducing the shedding of ctDNA [49]. Also the time window from postoperative blood draw till disease recurrence might have been too long. Lastly, the site of recurrence might have an impact on ctDNA detection in the circulation [25]. Future studies should determine the optimal time window for the sampling of ctDNA after surgery. Liquid biopsy cfDNA levels after tissue damage resulting from the surgery itself can be elevated up to four weeks, which may result in masking ctDNA with false-negative outcomes. It has been recommended that a second blood sample, collected after four weeks, is analysed for patients with postoperative undetectable ctDNA [50].
Limitations of our study include the small sample size, in part caused by the exclusion of patients with missing postoperative blood samples. The challenging logistics of blood sampling for translational research are well established [40]. Also, the sample size was limited to patients with a known RAS mutation, present in only 40–56% of patients with metastatic CRC [51], [52], [53]. A strength of our study is the homogeneous study population relative to other studies assessing the value of postoperative ctDNA in CRLM patients [28,29,41,42]. Integrating clinical, pathological and molecular markers can help to improve and customise therapy.
In conclusion, the detection of postoperative ctDNA is a promising prognostic factor for disease recurrence and median RFS in patients after secondary resection of RAS mutated colorectal cancer liver-only metastases. In addition, postoperative ctDNA showed a strong association with pathologic response. Further analysis with a bigger sample size would be needed to confirm these promising findings.
Contributors
Study concept and design: KB, IvtE, CJAP, RJAF
Financial support: GAM, CJAP, RJAF
Data collection: KB, IvtE, CM, PMDvD, AK, RJS
Statistical analyses: KB, IvtE, MLY
Data interpretation: KB, IvtE, CJAP, RJAF
Manuscript writing: KB, IvtE, CJAP, RJAF
Critical revision of the manuscript: All authors
Data sharing statement
This translational research study makes use of patients who participate in the currently ongoing CAIRO5 clinical trial, however, does not report clinical trial data. Therefore, except for the information presented in this study no additional individual participant data is currently available. Other available documents are the CAIRO5 study protocol and informed consent form. These are accessible via: https://dccg.nl/trial/cairo5
Declaration of Competing Interest
C.J.A.P. has an advisory role for Nordic Pharma. This funding is not related to the current research.
G.A.M. reports non-financial support from Exact Sciences, non-financial support from Sysmex, non-financial support from Sentinel CH. SpA, non-financial support from Personal Genome Diagnostics (PGDX), other from Hartwig Medical Foundation, grants from CZ (OWM Centrale Zorgverzekeraars groep Zorgverzekeraar u.a), other from Royal Philips, other from GlaxoSmithKline, other from Keosys SARL, other from Open Clinica LLC, other from Roche Diagnostics Nederland BV, other from The Hyve BV, other from Open Text, other from SURFSara BV, other from Vancis BV, other from CSC Computer Sciences BV, outside the submitted work; In addition, G.A.M. has several patents pending.
R.J.A.F. reports grants and non-financial support from Personal Genome Diagnostics, grants from MERCK BV, non-financial support from Pacific Biosciences, non-financial support from Cergentis BV, outside the submitted work; In addition, R.J.A.F. has several patents pending.
The remaining authors declare no potential conflicts of interest.
Acknowledgements
This study was supported by scientific grants from the Dutch Cancer Society (Grant No. 10438) and Amgen, The Netherlands. We thank Mirthe Lanfermeijer, Dorothé Linders, Kalpana Ramkisoensing, Margriet Lemmens, Anne Bolijn and Marianne Tijssen for laboratory assistance. We thank all participating hospitals and their research teams involved in the CAIRO5 study. We would like to acknowledge the NKI-AVL Core Facility Molecular Pathology & Biobanking (CFMPB) for supplying NKI-AVL Biobank material.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103498.
Appendix. Supplementary materials
References
- 1.Adam R., Kitano Y. Multidisciplinary approach of liver metastases from colorectal cancer. Ann Gastroenterol Surg. 2019;3(1):50–56. doi: 10.1002/ags3.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolhuis K., Kos M., Van Oijen M.G.H., Swijnenburg R.J., Punt C.J.A. Conversion strategies with chemotherapy plus targeted agents for colorectal cancer liver-only metastases: a systematic review. Eur J Cancer. 2020;141:225–238. doi: 10.1016/j.ejca.2020.09.037. [DOI] [PubMed] [Google Scholar]
- 3.Hallet J., Sa Cunha A., Adam R., Goere D., Bachellier P., Azoulay D. Factors influencing recurrence following initial hepatectomy for colorectal liver metastases. Br J Surg. 2016;103(10):1366–1376. doi: 10.1002/bjs.10191. [DOI] [PubMed] [Google Scholar]
- 4.Angelsen J.H., Viste A., Loes I.M., Eide G.E., Hoem D., Sorbye H. Predictive factors for time to recurrence, treatment and post-recurrence survival in patients with initially resected colorectal liver metastases. World J Surg Oncol. 2015;13:328. doi: 10.1186/s12957-015-0738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mima K., Beppu T., Chikamoto A., Miyamoto Y., Nakagawa S., Kuroki H. Hepatic resection combined with radiofrequency ablation for initially unresectable colorectal liver metastases after effective chemotherapy is a safe procedure with a low incidence of local recurrence. Int J Clin Oncol. 2013;18(5):847–855. doi: 10.1007/s10147-012-0471-z. [DOI] [PubMed] [Google Scholar]
- 6.Nordlinger B., Sorbye H., Glimelius B., Poston G.J., Schlag P.M., Rougier P. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14(12):1208–1215. doi: 10.1016/S1470-2045(13)70447-9. [DOI] [PubMed] [Google Scholar]
- 7.Blazer D.G., Kishi Y., Maru D.M., Kopetz S., Chun Y.S., Overman M.J. Pathologic response to preoperative chemotherapy: a new outcome end point after resection of hepatic colorectal metastases. J Clin Oncol. 2008;26(33):5344–5351. doi: 10.1200/JCO.2008.17.5299. [DOI] [PubMed] [Google Scholar]
- 8.Imai K., Allard M.A., Benitez C.C., Vibert E., Sa Cunha A., Cherqui D. Early recurrence after hepatectomy for colorectal liver metastases: what optimal definition and what predictive factors? Oncologist. 2016;21(7):887–894. doi: 10.1634/theoncologist.2015-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao M., Gibbs P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow-up for patients with early-stage colon cancer. J Clin Oncol. 2009;27(36):e279–e280. doi: 10.1200/JCO.2009.25.6156. author reply e81. [DOI] [PubMed] [Google Scholar]
- 10.Barimani D., Kauppila J.H., Sturesson C., Sparrelid E. Imaging in disappearing colorectal liver metastases and their accuracy: a systematic review. World J Surg Oncol. 2020;18(1):264. doi: 10.1186/s12957-020-02037-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cristiano S., Leal A., Phallen J., Fiksel J., Adleff V., Bruhm D.C. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570(7761):385–389. doi: 10.1038/s41586-019-1272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phallen J., Sausen M., Adleff V., Leal A., Hruban C., White J. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9(403) doi: 10.1126/scitranslmed.aan2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan J.C.M., Massie C., Garcia-Corbacho J., Mouliere F., Brenton J.D., Caldas C. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17(4):223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 14.Tie J., Kinde I., Wang Y., Wong H.L., Roebert J., Christie M. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26(8):1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Li L., Cohen J.D., Kinde I., Ptak J., Popoli M. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol. 2019;5(8):1118–1123. doi: 10.1001/jamaoncol.2019.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai H.X., Lee A.M., Yang L., Zhang P., Davatzikos C., Maris J.M. Imaging genomics in cancer research: limitations and promises. Br J Radiol. 2016;89(1061) doi: 10.1259/bjr.20151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R., Zhao A., Cao N., Li Z., Zhang G., Liu F. The value of circulation tumor DNA in predicting postoperative recurrence of colorectal cancer: a meta-analysis. Int J Colorectal Dis. 2020;35(8):1463–1475. doi: 10.1007/s00384-020-03667-y. [DOI] [PubMed] [Google Scholar]
- 18.Tie J., Cohen J.D., Lo S.N., Wang Y., Li L., Christie M. Prognostic significance of postsurgery circulating tumor DNA in nonmetastatic colorectal cancer: individual patient pooled analysis of three cohort studies. Int J Cancer. 2020;148(4):1014–1026. doi: 10.1002/ijc.33312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinert T., Henriksen T.V., Christensen E., Sharma S., Salari R., Sethi H. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5(8):1124–1131. doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tie J., Wang Y., Tomasetti C., Li L., Springer S., Kinde I. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346):346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarazona N., Gimeno-Valiente F., Gambardella V., Zuniga S., Rentero-Garrido P., Huerta M. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30(11):1804–1812. doi: 10.1093/annonc/mdz390. [DOI] [PubMed] [Google Scholar]
- 22.Fleming C.A., O’Leary D.P., Wang J., Redmond H.P. Association of observed perioperative cell-free DNA dynamics with early recurrence in patients with colon cancer. JAMA Surg. 2019;155(2):168–170. doi: 10.1001/jamasurg.2019.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tie J., Cohen J.D., Wang Y., Li L., Christie M., Simons K. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019;68(4):663–671. doi: 10.1136/gutjnl-2017-315852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidal J., Muinelo L., Dalmases A., Jones F., Edelstein D., Iglesias M. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28(6):1325–1332. doi: 10.1093/annonc/mdx125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scholer L.V., Reinert T., Orntoft M.W., Kassentoft C.G., Arnadottir S.S., Vang S. Clinical Implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23(18):5437–5445. doi: 10.1158/1078-0432.CCR-17-0510. [DOI] [PubMed] [Google Scholar]
- 28.Benesova L., Halkova T., Ptackova R., Semyakina A., Menclova K., Pudil J. Significance of postoperative follow-up of patients with metastatic colorectal cancer using circulating tumor DNA. World J Gastroenterol. 2019;25(48):6939–6948. doi: 10.3748/wjg.v25.i48.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boysen A.K., Pallisgaard N., Andersen C.S.A., Spindler K.G. Circulating tumor DNA as a marker of minimal residual disease following local treatment of metastases from colorectal cancer. Acta Oncol. 2020;59(12):1424–1429. doi: 10.1080/0284186X.2020.1806357. [DOI] [PubMed] [Google Scholar]
- 30.Huiskens J., van Gulik T.M., van Lienden K.P., Engelbrecht M.R., Meijer G.A., van Grieken N.C. Treatment strategies in colorectal cancer patients with initially unresectable liver-only metastases, a study protocol of the randomised phase 3 CAIRO5 study of the Dutch Colorectal Cancer Group (DCCG) BMC Cancer. 2015;15:365. doi: 10.1186/s12885-015-1323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Cutsem E., Cervantes A., Adam R., Sobrero A., Van Krieken J.H., Aderka D. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 32.Mekenkamp L.J., Koopman M., Teerenstra S., van Krieken J.H., Mol L., Nagtegaal I.D. Clinicopathological features and outcome in advanced colorectal cancer patients with synchronous vs metachronous metastases. Br J Cancer. 2010;103(2):159–164. doi: 10.1038/sj.bjc.6605737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fong Y., Fortner J., Sun R.L., Brennan M.F., Blumgart L.H. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230(3):309–318. doi: 10.1097/00000658-199909000-00004. discussion 18-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubbia-Brandt L., Giostra E., Brezault C., Roth A.D., Andres A., Audard V. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neo-adjuvant chemotherapy followed by liver surgery. Ann Oncol. 2007;18(2):299–304. doi: 10.1093/annonc/mdl386. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi S., Konishi M., Nakagohri T., Gotohda N., Saito N., Kinoshita T. Short time to recurrence after hepatic resection correlates with poor prognosis in colorectal hepatic metastasis. Jpn J Clin Oncol. 2006;36(6):368–375. doi: 10.1093/jjco/hyl027. [DOI] [PubMed] [Google Scholar]
- 36.Vigano L., Capussotti L., Lapointe R., Barroso E., Hubert C., Giuliante F. Early recurrence after liver resection for colorectal metastases: risk factors, prognosis, and treatment. A LiverMetSurvey-based study of 6,025 patients. Ann Surg Oncol. 2014;21(4):1276–1286. doi: 10.1245/s10434-013-3421-8. [DOI] [PubMed] [Google Scholar]
- 37.Vessies D.C.L., Greuter M.J.E., van Rooijen K.L., Linders T.C., Lanfermeijer M., Ramkisoensing K.L. Performance of four platforms for KRAS mutation detection in plasma cell-free DNA: ddPCR, Idylla, COBAS z480 and BEAMing. Sci Rep. 2020;10(1):8122. doi: 10.1038/s41598-020-64822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slide score. [https://www.slidescore.com], 2020.
- 39.Marcuello M., Vymetalkova V., Neves R.P.L., Duran-Sanchon S., Vedeld H.M., Tham E. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol Asp Med. 2019;69:107–122. doi: 10.1016/j.mam.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 40.Bidard F.C., Kiavue N., Ychou M., Cabel L., Stern M.H., Madic J. Circulating tumor cells and circulating tumor DNA detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the unicancer prodige-14 trial. Cells. 2019;8(6) doi: 10.3390/cells8060516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narayan R.R., Goldman D.A., Gonen M., Reichel J., Huberman K.H., Raj S. Peripheral circulating tumor DNA detection predicts poor outcomes after liver resection for metastatic colorectal cancer. Ann Surg Oncol. 2019;26(6):1824–1832. doi: 10.1245/s10434-019-07201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Y., Ma X., Chen K., Liu F., Cai S., Han-Zhang H. Perioperative circulating tumor DNA in colorectal liver metastases: concordance with metastatic tissue and predictive value for tumor burden and prognosis. Cancer Manag Res. 2020;12:1621–1630. doi: 10.2147/CMAR.S240869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schraa S.J., van Rooijen K.L., van der Kruijssen D.E.W., Rubio Alarcon C., Phallen J., Sausen M. Circulating tumor DNA guided adjuvant chemotherapy in stage II colon cancer (MEDOCC-CrEATE): study protocol for a trial within a cohort study. BMC Cancer. 2020;20(1):790. doi: 10.1186/s12885-020-07252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahbari N.N., Reissfelder C., Schulze-Bergkamen H., Jager D., Buchler M.W., Weitz J. Adjuvant therapy after resection of colorectal liver metastases: the predictive value of the MSKCC clinical risk score in the era of modern chemotherapy. BMC Cancer. 2014;14:174. doi: 10.1186/1471-2407-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirokawa F., Hayashi M., Miyamoto Y., Asakuma M., Shimizu T., Komeda K. Reconsideration of the indications for adjuvant chemotherapy for liver metastases from colorectal cancer after initial hepatectomy. Ann Surg Oncol. 2014;21(1):139–146. doi: 10.1245/s10434-013-3310-1. [DOI] [PubMed] [Google Scholar]
- 46.Benson A.B., Desch C.E., Flynn P.J., Krause C., Loprinzi C.L., Minsky B.D. 2000 update of American society of clinical oncology colorectal cancer surveillance guidelines. J Clin Oncol. 2000;18(20):3586–3588. doi: 10.1200/JCO.2000.18.20.3586. [DOI] [PubMed] [Google Scholar]
- 47.Ayez N., Lalmahomed Z.S., van der Pool A.E., Vergouwe Y., van Montfort K., de Jonge J. Is the clinical risk score for patients with colorectal liver metastases still useable in the era of effective neoadjuvant chemotherapy? Ann Surg Oncol. 2011;18(10):2757–2763. doi: 10.1245/s10434-011-1819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong R., Tie J., Lee M., Cohen J., Wang Y., Li L. The potential role of circulating tumor DNA (ctDNA) in the further investigation of colorectal cancer patients with nonspecific findings on standard investigations. Int J Cancer. 2019;145(2):540–547. doi: 10.1002/ijc.32117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coakley M., Garcia-Murillas I., Turner N.C. Molecular residual disease and adjuvant trial design in solid tumors. Clin Cancer Res. 2019;25(20):6026–6034. doi: 10.1158/1078-0432.CCR-19-0152. [DOI] [PubMed] [Google Scholar]
- 50.Henriksen T.V., Reinert T., Christensen E., Sethi H., Birkenkamp-Demtroder K., Gogenur M. The effect of surgical trauma on circulating free DNA levels in cancer patients - implications for studies of circulating tumor DNA. Mol Oncol. 2020 doi: 10.1002/1878-0261.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peeters M., Kafatos G., Taylor A., Gastanaga V.M., Oliner K.S., Hechmati G. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: a pooled analysis of randomised controlled trials. Eur J Cancer. 2015;51(13):1704–1713. doi: 10.1016/j.ejca.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 52.Hsu H.C., Thiam T.K., Lu Y.J., Yeh C.Y., Tsai W.S., You J.F. Mutations of KRAS/NRAS/BRAF predict cetuximab resistance in metastatic colorectal cancer patients. Oncotarget. 2016;7(16):22257–22270. doi: 10.18632/oncotarget.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Therkildsen C., Bergmann T.K., Henrichsen-Schnack T., Ladelund S., Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: a systematic review and meta-analysis. Acta Oncol. 2014;53(7):852–864. doi: 10.3109/0284186X.2014.895036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.