Abstract
Background & Aims
Aryl hydrocarbon receptor (AhR) is a liver-enriched xenobiotic receptor that plays important role in detoxification response in liver. This study aimed to investigate how AhR signaling may impact the pathogenesis of alcohol-related liver disease (ALD).
Methods
Chronic alcohol feeding animal studies were conducted with mouse models of hepatocyte-specific AhR knockout (AhRΔhep) and NAD(P)H quinone dehydrogenase 1 (NQO1) overexpression, and dietary supplementation of the AhR ligand indole-3-carbinol. Cell studies were conducted to define the causal role of AhR and NQO1 in regulation of redox balance and apoptosis.
Results
Chronic alcohol consumption induced AhR activation and nuclear enrichment of NQO1 in hepatocytes of both alcoholic hepatitis patients and ALD mice. AhR deficiency exacerbated alcohol-induced liver injury, along with reduction of NQO1. Consistently, in vitro studies demonstrated that NQO1 expression was dependent on AhR. However, alcohol-induced NQO1 nuclear translocation was triggered by decreased cellular oxidized nicotinamide adenine dinucleotide (NAD+)-to-NADH ratio, rather than by AhR activation. Furthermore, both in vitro and in vivo overexpression NQO1 prevented alcohol-induced hepatic NAD+ depletion, thereby enhancing activities of NAD+-dependent enzymes and reversing alcohol-induced liver injury. In addition, therapeutic targeting of AhR in the liver with dietary indole-3-carbinol supplementation efficiently reversed alcoholic liver injury by AhR-NQO1 signaling activation.
Conclusions
This study demonstrated that AhR activation is a protective response to counteract alcohol-induced hepatic NAD+ depletion through induction of NQO1, and targeting the hepatic AhR-NQO1 pathway may serve as a novel therapeutic approach for ALD.
Keywords: AhR, NQO1, NAD+-to-NADH Ratio, Alcohol-Related Liver Disease
Abbreviations used in this paper: 4-HNE, 4-hydroxynonenal; ACD, acetaldehyde; ADH, alcohol dehydrogenase; AF, alcohol-fed; AH, alcoholic hepatitis; AhR, aryl hydrocarbon receptor; ALD, alcohol-related liver disease; ALDH, aldehyde dehydrogenase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; cDNA, complementary DNA; FFA, free fatty acids; I3C, indole-3-carbinol; IF, immunofluorescence; IHC, immunohistochemistry; KD, knockdown; MPO, myeloperoxidase; NAD+, oxidized nicotinamide adenine dinucleotide; NADH, reduced nicotinamide adenine dinucleotide; NQO1, NAD(P)H quinone dehydrogenase 1; NRF2, nuclear factor erythroid 2–related factor 2; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PF, pair-fed; ROS, reactive oxygen species; TG, triglycerides
Graphical abstract
Summary.
The roles of AhR in the pathogenesis of alcohol-related liver disease remains unclear. AhR deficiency disrupted NQO1-mediated regulation of redox balance and exacerbated alcohol-induced liver injury. Activation of AhR-NQO1 signaling efficiently reversed alcohol-induced redox imbalance and liver injury.
The liver is the central organ for metabolizing nutrients and detoxifying toxic substances including xenobiotics.1 Alcohol, as a xenobiotic, is primarily metabolized in the liver, whereas the process of alcohol catabolism generates toxic metabolites, including acetaldehyde (ACD) and reactive oxygen species (ROS). Long-term excessive alcohol consumption produces alcohol-related liver disease (ALD) with a wide spectrum of hepatic lesions, including steatosis, hepatitis, fibrosis or cirrhosis, and hepatocellular carcinoma.2,3 ALD is a leading cause of morbidity and mortality worldwide. It has been estimated that 10%–20% of heavy drinkers will progress to alcoholic cirrhosis, which accounts for more than 40% of all deaths from cirrhosis and for 30% of all hepatocellular carcinomas in the United States.4 Unfortunately, there are currently no Federal Drug Administration–approved pharmacological or nutritional therapies for treating patients with any stage of ALD. Therefore, a better understanding of ALD pathogenesis and identification of therapeutic targets are needed.
Aryl hydrocarbon receptor (AhR) is highly expressed in the hepatocytes and plays a critical role in detoxifying dioxin and xenobiotics.5,6 AhR is a ligand-activated transcriptional factor, and predominantly located in the cytosol that guarded by a chaperone complex (Hsp90/XAP2/p23).7 Upon ligand binding, AhR is translocated into the nucleus and forms a dimer with the AhR nuclear translocator (ARNT), which binds to the xenobiotic/dioxin response elements in the promoter of target genes to initiate gene transcription.8,9 Activated AhR is quickly exported back into the cytosol and degraded by the 26S proteasome,10 thereby preventing AhR from constitutive receptor activity. As a ligand-dependent transcriptional factor, AhR senses a wide range of structurally different exogenous and endogenous molecules and differentially regulates hepatic genes based on the type of ligand.
The first AhR ligand discovered in 1976 is an environmental toxicant, TCDD, which has high affinity to cytosolic AhR in the livers of mice.11 Most early studies on environmental toxicant-induced hepatoxicity demonstrate that AhR activation is a major mechanism mediating toxicant-induced liver injury. Recent studies discovered that many endogenous molecules also serve as AhR ligands. Metabolites from tryptophan, arachidonic acid, and heme have been shown to bind to and activate AhR.6 Activation of AhR by those endogenous ligands leads to regulation of physiological processes. AhR ligands also occur naturally in diet, such as indole-3-carbinol (I3C). Dietary supplementation with I3C has been reported to boost anti-inflammatory function by activating AhR in immune cells.12,13 I3C supplementation also promoted differentiation of intestinal Goblet cells and suppressed lipid accumulation in adipocytes.14,15 A recent study reported that AhR signaling protects against carbon tetrachloride–induced liver fibrosis through preventing activation of hepatic stellate cells and expression of liver fibrogenesis genes in mice.16 These studies suggest that physiological activation of hepatic AhR by endogenous and dietary ligands may result in a protective response.
The effects of alcohol consumption on hepatic AhR are poorly understood. A cell study with mouse hepatic stellate cells showed that acute, but not chronic, alcohol treatment activated AhR expression and upregulated AhR target genes.17 A recent study demonstrated that AhR agonist FICZ (6-formylindolo (3,2-b) carbazole) prevented wild-type mice, but not AhR knockout mice, from alcohol-induced liver injury, suggesting a protective role of AhR against alcohol toxicity.18 However, the underlying mechanisms of AhR-mediated hepatoprotection have not been defined. In this study, we first examined the role of AhR in the pathogenesis of ALD using hepatocyte-specific AhR knockout (AhRΔhep) mice. We also determined a key role of NAD(P)H quinone dehydrogenase (NQO1) in mediating the protective effects of AhR on alcohol-induced hepatic redox imbalance.
Results
Hepatocyte-Specific Knockout of AhR in Mice Exacerbates Alcohol-Induced Liver Injury
To investigate the role of AhR signaling in the pathogenesis of ALD, AhRΔhep mice were generated. Western blot and immunohistochemistry (IHC) showed that chronic alcohol induced AhR nuclear translocation, which was abolished by AhR deletion (Figure 1A). As indicators of liver injury, serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were elevated by alcohol exposure and were significantly exacerbated by AhR deletion (Figure 1B). Histopathological analysis showed that AhR deletion exacerbated alcohol-induced lipid droplet accumulation, hepatocyte degeneration, and inflammatory cell infiltration in the liver (Figure 1C). Biochemical analysis of hepatic triglycerides (TG) and free fatty acids (FFA) showed that alcohol-induced lipid accumulation was worsened by hepatic AhR deletion (Figure 1D). TUNEL assay indicated that alcohol exposure caused apoptotic cell death in alcohol-fed (AF) floxed mice, and more apoptotic cells were observed in the liver of AF AhRΔhep mice (Figure 1E). Moreover, immunostaining of neutrophil marker myeloperoxidase (MPO) showed that alcohol exposure induced more MPO+ neutrophils infiltrated in the liver of AhRΔhep mice compared with the floxed mice (Figure 1F), which was consistent with flow cytometry analysis of CD11b+ Ly6G+ neutrophils (Figure 1G). In accordance, alcohol-induced hepatic expression of Cxcl1, Lcn2, and serum CXCL1 protein were further upregulated by AhR deletion (Figure 1H and I).
Figure 1.
Hepatocyte-specific deletion of AhR exacerbates alcohol-induced liver injury. AhRflox/flox (flox) mice and hepatocyte-specific AhR knockout (AhRΔhep) mice were fed control (PF) or alcohol (AF) liquid diet for 8 weeks. (A) Western blot analysis of cytoplasmic and nuclear AhR expression (left), and IHC staining of AhR distribution (right) (scale bar = 20 μm) in the liver of floxed mice and AhRΔhep mice. Arrows indicate nuclear AhR localization. (B) Serum ALT and AST levels. (C) Hematoxylin and eosin staining of liver tissue sections. Arrows indicate hepatocyte degeneration. Arrowheads indicate infiltrated inflammatory cells. Scale bars = 50 μm. (D) Biochemistry analysis of hepatic TG and FFA contents. (E) Apoptotic cells detected by TUNEL assay. Arrows indicate apoptotic cells. (F) MPO immunostaining of liver tissue sections. Arrows indicate MPO+ neutrophils. Arrowheads indicate MPO+ neutrophil cluster. Scale bars = 20 μm. (G) Liver-infiltrated neutrophils (Ly6G+CD11b+) measured by flow cytometry. Gated on singlet 7AAD-CD45+ cells. (H) Cxcl1 messenger RNA (mRNA) expression in the livers of floxed mice and AhRΔhep mice measured by real-time PCR. (I) Serum CXCL1 levels. All data are presented as mean ± SEM. n = 3–5 mice per group. ∗P < .05, ∗∗P < .01. Results were analyzed by one-way analysis of variance followed by Tukey’s multiple comparison test. H&E, hematoxylin and eosin.
AhR Deletion Worsens Alcohol-Reduced Oxidized Nicotinamide Adenine Dinucleotide-to-NADH Ratio in Association With Abrogating NQO1 Induction in the Liver
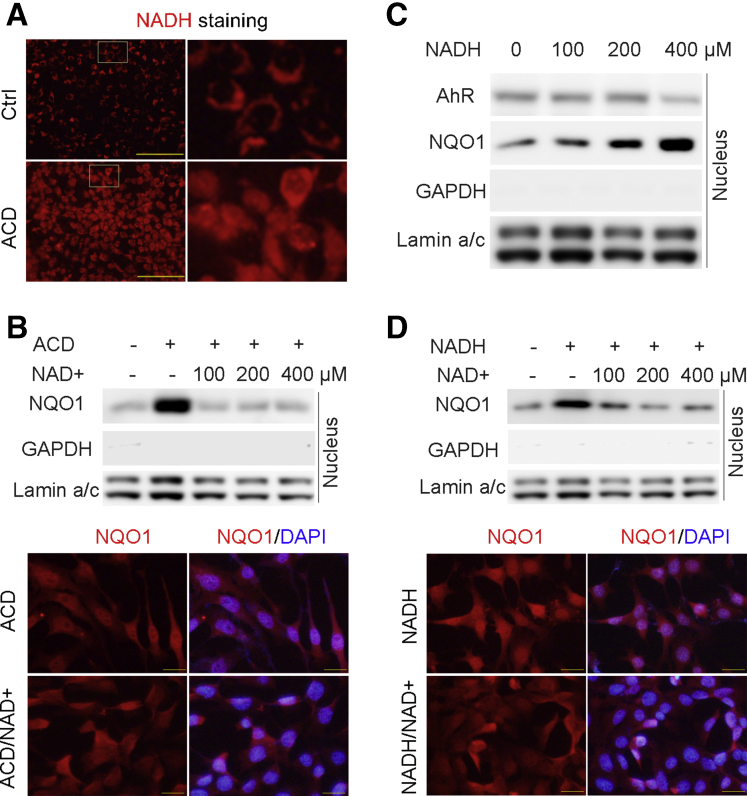
Oxidized nicotinamide adenine dinucleotide (NAD+)-to-NADH ratio reduction is a feature of alcohol metabolism and plays a causal role in alcohol-induced oxidative stress during the development of ALD.19 To understand the mechanisms of how AhR deletion impairs liver function, hepatic NAD+/NADH redox status and oxidative stress were measured. As shown in Figure 2A, alcohol exposure in floxed mice reduced the levels of NAD+ and increased the levels of NADH, leading to a decrease in hepatic NAD+-to-NADH ratio. Unexpectedly, hepatic NAD+-to-NADH ratio was further decreased in AF AhRΔhep mice. Consequently, oxidative stress was assessed by measuring ROS level and lipid peroxidation. Hepatic ROS levels were elevated by alcohol exposure in floxed mice, and a further increase was observed in AF AhRΔhep mice (Figure 2B). Similarly, alcohol-increased serum hydrogen peroxide levels were also exacerbated by AhR deletion (Figure 2C). Consistently, alcohol-induced 4-hydroxynonenal (4-HNE) protein adducts formation was enhanced in AF AhRΔhep mice (Figure 2D). Then, the protein levels of NQO1, an AhR target and NAD+ generator, were examined by Western blot and IHC staining. The data showed that AF floxed mice displayed a significantly increased NQO1 protein levels in the liver, whereas this effect was remarkably abrogated in AF AhRΔhep mice (Figure 2E). Interestingly, IHC showed that alcohol exposure increased NQO1 staining, predominantly in the nuclei, which was attenuated by AhR deletion (Figure 2F). Next, another regulator of NQO1, nuclear factor erythroid 2–related factor 2 (NRF2) was examined by Western blot. The data showed that alcohol exposure reduced NRF2 expression in both AF floxed and AF AhRΔhep mice although it was increased in pair-fed (PF) AhRΔhep mice. Correspondingly, HO-1, a canonical NRF2-regulated gene, was decreased by alcohol feeding along with NRF2 reduction in both AF floxed and AF AhRΔhep mice, and induction in PF AhRΔhep mice (Figure 2E). These data suggest that hepatic NQO1 induction is dependent of AhR, rather than on NRF2, in this alcohol exposure mouse model.
Figure 2.
Hepatocyte-specific AhR deletion aggravates alcohol-induced cellular redox imbalance and oxidative stress in the liver. Liver samples were collected from AhRflox/flox (flox) mice and hepatocyte-specific AhR knockout (AhRΔhep) mice were fed control (PF) or alcohol (AF) liquid diet for 8 weeks. (A) Liver NAD+, NADH, and NAD+-to-NADH ratio were measured by colorimetric assay kit. n = 5 mice per group. (B) Hepatic ROS production was measured by flow cytometry using a fluorescent probe, DCFDA. n = 4 mice per group. (C) Serum hydrogen peroxide levels. n = 7–10 mice per group. (D) IHC staining of hepatic 4-HNE on liver tissue sections. Scale bars = 20 μm. (E) Western blot analysis of hepatic protein levels of NQO1, NRF2, and HO-1. (F) IHC staining of NQO1 on liver tissue sections. Scale bars = 20 μm. All data are presented as mean ± SEM. ∗P < .05, ∗∗P < .01. Different letters indicate statistical significance. Results were analyzed by one-way analysis of variance followed by Tukey’s multiple comparison test.
NQO1 Expression Is Dependent on AhR Activation But Its Nuclear Translocation Is Induced by Reduced NAD+-to-NADH Ratio in Hepa-1c1c7 Cells
Next, to determine whether NQO1 nuclear translocation is dependent on AhR, Hepa-1c1c7 cells and Hapa-1c1c7 cell–derived, AhR-deficient Tao cells were treated with AhR endogenous ligand, FICZ. Western blot and immunofluorescence (IF) microscopy showed that FICZ treatment reduced total protein level of AhR along with AhR nuclear translocation, thereby increased NQO1 expression in Hepa-1c1c7 cells (Figure 3A and B). Regardless of FICZ treatment, Tao cells exhibited a significantly lower abundance of AhR protein, only trace amount of NQO1 was detected (Figure 3A). Furthermore, NRF2 distribution was also analyzed by IF microscopy, and we found that NRF2 was induced by FICZ but was predominantly localized in the cytoplasm (Figure 3C), indicating that NRF2 may not participate in FICZ-induced NQO1 expression. Surprisingly, immunofluorescence (IF) microscopy demonstrated that the NQO1 proteins did not translocated to nucleus, although its expression was upregulated following AhR activation by FICZ (Figure 3D). To further confirm the effects of FICZ on cytoplasmic and nuclear distributions of AhR, NRF2, and NQO1, the cytoplasmic and nuclear fractions were isolated from Hepa-1c1c7 cells. Western blot showed that FICZ treatment dramatically reduced cytoplasmic AhR (Figure 3E) but increased nuclear AhR (Figure 3F) protein levels. However, activation of AhR by FICZ increased cytoplasmic (Figure 3E) but not nuclear NRF2 and NQO1 protein levels ((Figure 3F). These results indicate that AhR activation by FICZ upregulated NQO1 expression but did not induce NQO1 nuclear translocation.
Figure 3.
NQO1 is induced in an AhR-dependent manner. Hepa-1c1c7 cells and AhR-deficient Tao cells were treated with or without FICZ for 6 hours, and protein expression and distribution of AhR, NRF2, and NQO1 were analyzed by Western blot and IF. (A) AhR and NQO1 total protein levels in Hepa-1c1c7 cells and Tao cells. (B) IF microscopy of AhR. (C) IF microscopy of NRF2. (D) IF microscopy of NQO1. (E) Cytoplasmic protein levels of AhR, NRF2, and NQO1. (F) Nuclear protein levels of AhR, NRF2, and NQO1. Scale bars = 20 μm.
To further explore the mechanism of how alcohol may induce NQO1 nuclear translocation, Hepa-1c1c7 cells were treated with ethanol. Unexpectedly, IF microscopy showed that ethanol treatment did induced neither AhR nuclear translocation (Figure 4A) nor NQO1 upregulation (Figure 4B). Western blot further confirmed that ethanol did not affect either cytoplasmic or nuclear protein levels of AhR and NQO1. To understand why ethanol failed to activate AhR as observed in the mouse study, the protein levels of ethanol metabolizing enzymes in Hepa-1c1c7 cells were examined. As shown in Figure 4D, Hepa-1c1c7 cells did not express ethanol metabolizing enzymes, including alcohol dehydrogenase (ADH) and CYP2E1, but expressed ALDH2, which catabolizes ACD, the ethanol metabolite. Accordingly, Hepa-1c1c7 cells were treated with ACD. IF microscopy showed that ACD treatment remarkably induced nuclear translocation of both AhR (Figure 4E) and NQO1 (Figure 4F), which was further confirmed by Western blot using isolated cytoplasmic and nuclear fractions (Figure 4G).
Figure 4.
ACD but not alcohol induces AhR activation and NQO1 nuclear translation in Hepa-1c1c7 cells. Hepa-1c1c7 cells were treated with ethanol or ACD for 24 hours, and protein expression and distribution of AhR and NQO1 were analyzed by Western blot and IF. (A) IF microscopy of AhR with ethanol treatment. (B) IF microscopy of NQO1 with ethanol treatment. (C) AhR and NQO1distribution analysis by Western blot. (D) Alcohol metabolized enzymes analysis by Western blot in Hepa-1c1c7 cells. (F) IF microscopy of NQO1 with ACD treatment. Scale bars = 20 μm. (G) Western blot analysis of cytoplasmic and nuclear protein levels of AhR and NQO1.
It is well known that ACD metabolism by ALDH2 generates NADH. To understand why ACD induced NQO1 nuclear translocation, the effect of ACD on cellular NADH levels was measured by a fluorescent NADH probe. As shown in Figure 5A, ACD treatment dramatically increased the cytosolic and nuclear NADH levels. To further determine whether reduced NAD+-to-NADH ratio plays a role in NQO1 nuclear translocation, exogeneous NAD+ was added along with ACD treatment. Western blot and IF microscopy showed that ACD-increased nuclear NQO1 protein levels and localization were abrogated by NAD+ treatment (Figure 5B), indicating that NADH/NAD+ ratio plays crucial role in NQO1 nuclear translocation. To validate this hypothesis. Hepa-1c1c7 cells were treated with NADH with and without NAD+. As shown in Figure 5C, nuclear NQO1 accumulation was increased upon NADH in a dose-dependent manner. Similarly, NADH also increased nuclear NQO1 in a dose-dependent manner, which was blocked by adding NAD+ (Figure 5D). Taken together, these results indicate that reduction of cellular NAD+-to-NADH ratio serves as a trigger of ACD-induced NQO1 nuclear translocation, following AhR-dependent NQO1 induction.
Figure 5.
Decreased NAD+-to-NADH ratio induces NQO1 nuclear translocation. NQO1 nuclear translocation was determined by IF microscopy and Western blot in ACD, exogenous NADH, or NAD+-treated Hepa-1c1c7 cells. (A) NADH staining of ACD-treated Hepa-1c1c7 cells using a fluorescence probe. Scale bars = 50 μm. (B) NQO1 distribution was analyzed by Western blot and IF after ACD with or without NAD+ treatment. Scale bars = 20 μm. (C) Western blot analysis of nuclear AhR and NQO1 in NADH-treated Hepa-1c1c7 cells. (D) NQO1 distribution was analyzed by Western blot and IF after NADH or NADH plus NAD+ treatment. Scale bars = 20 μm.
NQO1 Mediates the Protective Effect of AhR Against ACD-Induced Apoptosis in Hepa-1c1c7 Cells and Tao Cells
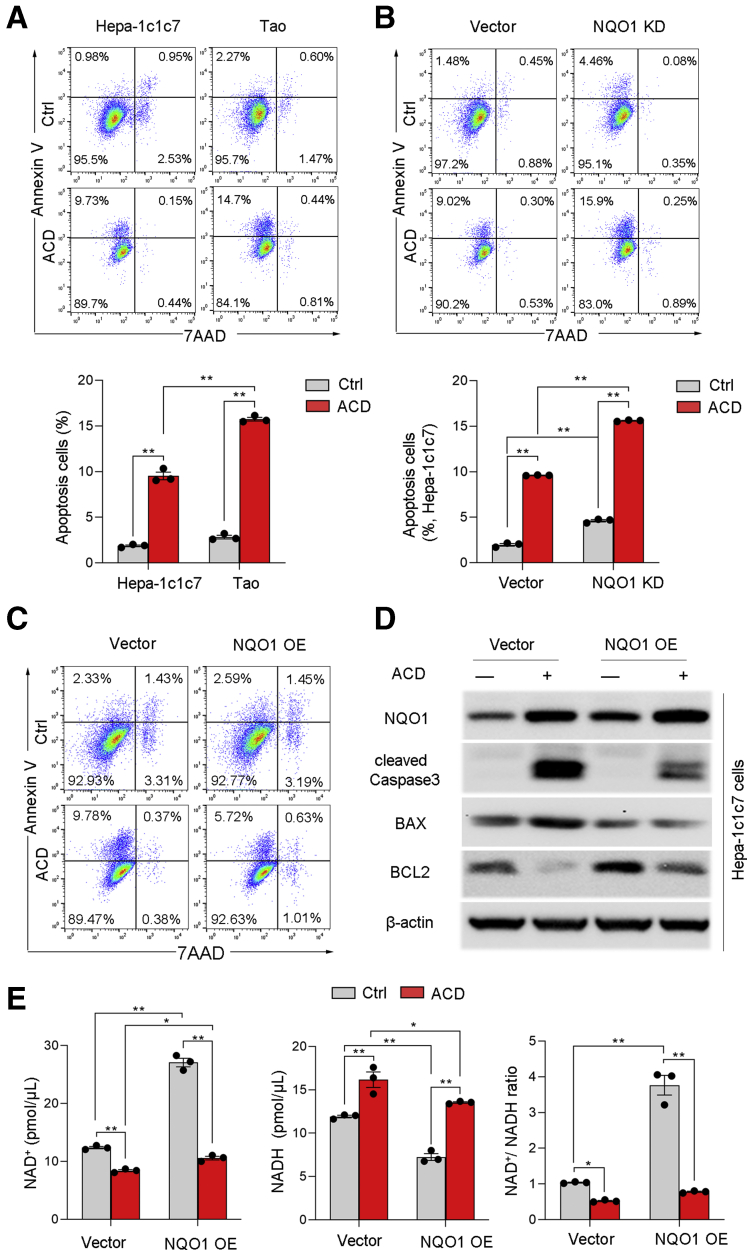
To determine the cytoprotective effect of AhR activation, Hepa-1c1c7 cells and AhR-deficient Tao cells were treated with ACD. As shown in Figure 6A, ACD treatment induced apoptosis (Annexin V–positive cells) in both cell types. However, Tao cells exhibited much higher frequencies of Annexin V–positive cells than Hepa-1c1c7 cells, suggesting an antiapoptotic role of AhR. To define whether NQO1 may mediates the antiapoptotic role of AhR, Hepa-1c1c7 cells with NQO1 knockdown (KD) were treated with ACD. As shown in Figure 6B, compared with control Hepa-1c1c7 cells, NQO1 KD significantly increased the proportion of Annexin V–positive cells, indicating that NQO1 critically mediates the antiapoptotic role of AhR. To further assess the role of NQO1 in protection against ACD toxicity, 2 stable NQO1-overexpressing cell lines were created using Hepa-1c1c7 cells and Tao cells, respectively. NQO1 overexpression significantly reduced ACD-induced Annexin V–positive cells (Figure 6C), and attenuated ACD-induced caspase-3 activation and BAX upregulation (Figure 6D), while antiapoptotic protein BCL-2 was upregulated by NQO1 overexpression. Moreover, quantitative assay of NAD+ and NADH levels demonstrated that NQO1 protects cells from ACD-induced apoptosis in association with increased cellular NAD+ and reduced cellular NADH levels, although this effect was diminished after ACD treatment (Figure 6E).
Figure 6.
Overexpression (OE) of NQO1 ameliorates ACD-induced apoptosis in Hepa-1c1c7 cells. Hepa1c1c7 cells, AhR-deficient TAO cells, and NQO1 KD or OE Hepa-1c1c7 cells were treated with or without ACD at 100 μmol/L for 24 hours, and apoptosis, NAD+ levels, NADH levels, and NAD+-to-NADH ratio were measured. (A) Flow cytometry analysis of apoptosis in Hepa-1c1c7 cells and Tao cells. n = 3 per group. (B) ACD-induced apoptosis was analyzed by flow cytometry in vector control cells and NQO1 KD cells. n = 3 per group. (C) ACD-induced apoptosis was analyzed by flow cytometry in vector control cells and NQO1 OE cells. (D) Western blot analysis of apoptosis related proteins in NQO1 OE cells with or without ACD treatment. (E) NAD+, NADH, and NAD+-to-NADH ratio were measured by colorimetric assay kit. n = 3 per group. All data are presented as mean ± SEM. ∗P < .05, ∗∗P < .01. Results were analyzed by one-way analysis of variance followed by Tukey’s multiple comparison test.
In addition, NQO1 overexpression also attenuated ACD-induced mitochondrial potential disruption (Figure 7A) and ROS overgeneration (Figure 7B). Similarly, NQO1 overexpression in Tao cells also protected the cells from ACD-induced oxidative stress (Figure 7C) and apoptosis (Figure 7D and E).
Figure 7.
NQO1 overexpression (OE) prevents cells from ACD-induced mitochondria potential change, oxidative stress, and apoptosis in Hepa-1c1c7 cells and Tao cells. (A) Mitochondrial membrane potential assay in Hepa-1c1c7 cells by using a fluorescent probe, TMRE. (B) Cellular ROS staining by DCFDA in Hepa-1c1c7 cells with or without NQO1 OE. (C) Cellular ROS staining by DCFDA in Tao cells with or without NQO1 OE. (D) ACD-induced apoptosis was measured by flow cytometry in Tao cells with or without NQO1 OE. (E) Western blot analysis for ACD-induced apoptosis related protein in Tao cells with or without NQO1 OE. Scale bars = 20 μm. All data are presented as mean ± SEM. ∗∗P < .01. Results were analyzed by one-way analysis of variance followed by Tukey’s multiple comparison test.
Hepatic NQO1 Overexpression in Mice Protects Against Alcohol-Induced Apoptosis via Interaction With SIRT1
To test whether overexpression of NQO1 in the liver of mice may counter alcohol-induced redox imbalance and thereby alleviating the development of ALD, hepatocyte-targeted gene delivery of NQO1 in mice was achieved by injection of AAV8-TBG-m-NQO1. NQO1 gene delivery significantly increased total hepatic protein levels of NQO1, with an enriched distribution in the nuclei of hepatocytes (Figure 8A and 8B). NQO1 overexpression increased hepatic NAD+ and NAD+-to-NADH ratios and decreased hepatic NADH levels (Figure 8C). In addition to correcting NAD+-to-NADH ratio, NQO1 overexpression also increased its binding to SIRT1 (sirtuin 1) and increased SIRT1 activity (Figure 8D and 8E). Along with increased SIRT1 activity, hepatic total lysine acetylation levels (Figure 8F) and FoxO1 (forkhead box O1) acetylation levels (Figure 8G) were significantly decreased by NQO1 overexpression. Consequently, NQO1 overexpression reduced alcohol-upregulated pro-apoptotic proteins (Figure 8H) and TUNEL-positive cells (Figure 8I).
Figure 8.
Hepatocyte-specific NQO1 overexpression improves chronic alcohol-induced apoptosis. C57BL/6J mice without or with hepatocyte-specific NQO1 gene delivery (AF/NQO1) were fed alcohol liquid diet (AF) for 8 weeks. Hepatic NQO1 protein expression (left) and distribution (right) were measured by (A) Western blot and (B) IHC staining (scale bars = 20 μm), respectively. (C) Hepatic NAD+, NADH, and NAD+-to-NADH ratio were measured by colorimetric assay kit. n = 5–6 mice per group. (D) Immunoprecipitation analysis of SIRT1 and NQO1 interaction and (E) SIRT1 enzyme activities measurement. n = 3 mice per group. (F) IHC staining of acetylated lysine. Scale bars = 20 μm. (G) Immunoprecipitation analysis of acetylated FoxO1 level. (H) FoxO1-targeted apoptotic protein expression including BIM and PUMA were analyzed by Western blot. (I) Hepatic apoptosis was analyzed by TUNEL staining. Scale bars = 50 μm. Arrows indicate apoptotic cells. All data are presented as mean ± SEM. ∗P < .05, ∗∗P < .01. Results were analyzed by independent-samples t test.
NQO1 overexpression also attenuated alcohol-induced ROS overgeneration and lipid peroxidation (Figure 9A), and increased ADH and ALDH activities, leading to a reduction of serum ethanol levels (Figure 9B). Alcohol-induced liver damage was improved by NQO1 overexpression, as indicated by reduced serum ALT and AST levels and histopathological alterations, including reduced lipid droplet accumulation and hepatocyte degeneration (Figure 9C). NQO1 overexpression also reduced hepatic neutrophil infiltration (Figure 9D) as well as reduced Cxcl1 and Lcn2 expression (Figure 9E).
Figure 9.
Hepatocyte-specific NQO1 overexpression attenuates alcohol-induced oxidative stress, liver injury, and inflammation. (A) Hepatic ROS production and 4-HNE levels were measured by flow cytometry and IHC staining (scale bars = 50 μm), respectively. n = 4 mice per group. (B) ADH, ALDH enzyme activities, and serum ethanol levels. n = 3 mice per group. (C) Serum ALT and AST levels and liver hematoxylin and eosin (H&E) staining. n = 4–7 mice per group. Arrows indicate hepatocyte degeneration. Arrowheads indicate infiltrated inflammatory cells. (D) Flow cytometry analysis of hepatic infiltration of neutrophils (Ly6G+CD11b+). n = 3 mice per group. (E) Cxcl1 and Lcn2 messenger RNA (mRNA) expression was measured by real-time PCR. n = 3–4 mice per group. All data are presented as mean ± SEM. ∗P < .05, ∗∗P < .01. Results were analyzed by independent-samples t test.
Dietary I3C Supplementation Activates Hepatic AhR-NQO1 Signaling Pathway and Reverses Alcohol-Induced Liver Injury
To evaluate the therapeutic potential of AhR activation, dietary AhR ligand, I3C, was supplemented to the liquid diets, starting from the sixth week in an 8-week alcohol feeding protocol. IHC showed that dietary I3C supplementation increased nuclear AhR and NQO1 protein levels (Figure 10A) in the liver, indicating an activation of AhR-NQO1 signaling pathway. Meanwhile, increased hepatic NAD+ and reduced NADH levels were detected, which led to an elevation of NAD+-to-NADH ratio (Figure 10B). Consequently, I3C administration improved alcohol-induced oxidative stress by reducing hepatic ROS generation (Figure 10C). Furthermore, I3C reversed alcohol-induced liver injury, as indicated by reduced serum ALT and AST levels, and improved histopathological alterations (Figure 10D and E). I3C also increased the binding of NQO1 with SIRT1 and increased SIRT1 activities (Figure 10F), which further led to FoxO1 targeting cleaved caspase-3, and PUMA (p53 upregulated modulator of apoptosis) expression decreased (Figure 10G) reversal of alcohol-induced hepatic apoptosis. Dietary I3C supplementation also alleviated neutrophil infiltration in the liver (Figure 10H) as well as alcohol-induced Cxcl1 and Lcn2 upregulation (Figure 10I).
Figure 10.
Dietary I3C supplementation activates AhR-NQO1 signaling and reverses alcohol-induced liver injury. C57BL/6J mice were an fed alcohol liquid diet (AF) without or with dietary I3C supplementation (AF/I3C) at 5 mg/10 mL, 3 times a week, starting from the sixth week in an 8-week feeding protocol. (A) IHC staining of AhR and NQO1. Scale bars = 20 μm. (B) Hepatic NAD+, NADH, and NAD+-to-NADH ratio. n = 4–5 mice per group. (C) Hepatic ROS production measured by flow cytometry using the fluorescent probe, DCFDA. n = 4 mice per group. (D) Serum ALT and AST levels. n = 5–10 mice per group. (E) Liver histopathology (hematoxylin and eosin [H&E] staining). Arrows indicate hepatocyte degeneration. Arrowheads indicate infiltrated inflammatory cells. Scale bars = 50 μm. (F) Immunoprecipitation analysis of SIRT1 and NQO1 interaction, and SIRT1 enzyme activities measurement. n = 3 mice per group. (G) Western blot analysis of cleaved caspase 3 and PUMA. (H) Flow cytometry analysis of hepatic infiltration of neutrophils (Ly6G+CD11b+). n = 3 mice per group. (I) Real-time PCR analysis of Cxcl1 and Lcn2 messenger RNA (mRNA) levels. n = 3–4 mice per group. All data are presented as mean ± SEM. ∗P < .05, ∗∗P < .01. Results were analyzed by independent-samples t test.
Activation of AhR-NQO1 Pathway in the Liver of Patients With Alcoholic Hepatitis
To determine whether alcohol consumption in human also activates AhR-NQO1 signaling pathway, IHC was conducted with liver tissue samples from normal subjects and patients with severe alcoholic hepatitis (AH). Compared with the uniform staining in the normal liver tissue sections, enriched nuclear staining of both AhR and NQO1 were observed in some hepatocytes in the liver of AH patients (Figure 11A). While both the AhR- and NQO1-positive hepatocytes exhibited normal cell size, most of the negative hepatocytes displayed enlarged cell size.
Figure 11.
AhR-NQO1 signaling is activated in liver in AH patients, and a working model of AhR-NQO1 signaling in ALD. (A) Immunohistochemistry staining of AhR and NQO1 in normal liver and AH specimens. (B) A working model of AhR-NQO1 signaling in ALD. Alcohol metabolism in hepatocytes reduces NAD+-to-NADH ratio and induces AhR-NQO1 signaling activation. Increased NADH induces NQO1 nuclear translocation. Converting NADH to NAD+ by NQO1 restores cellular redox balance and reactivates NAD+-dependent enzymes such as SIRT1. Dietary I3C supplementation activates AhR-NQO1 signaling and thereby reverses alcohol-induced liver injury.
Discussion
The role of AhR has been investigated in diverse liver diseases, including toxic liver injury, nonalcoholic steatohepatitis, fibrosis, and autoimmune hepatitis, during the past 4 decades.20 Of note, among of these disease models, AhR signaling activation exhibited a contradictory cellular and pathological consequence. In a mouse model of TCDD-induced liver fibrosis, AhR deletion prevented the development of liver fibrosis,21 suggesting a pathophysiologic role of AhR in toxic fibrosis. However, another study reported that AhR-null mice displayed reduced liver size and exhibited spontaneous liver fibrosis,22 indicating a physiological role of AhR. Studies on the role of AhR in liver fibrogenesis demonstrated that the nontoxic AhR ligand, ITE, prevents CCl4-induced liver fibrosis, whereas TCDD causes typical hepatotoxicity.16 A recent study demonstrated that treatments with probiotics and AhR agonist FICZ reduced alcohol-induced liver lesions, but the beneficial effects were abrogated by whole-body deletion of AhR.18 These studies indicate that activation of AhR by different type of ligands may lead to differential regulation of cellular pathways and generate contrary consequences. In this study, we showed that chronic alcohol exposure activated hepatic AhR, and hepatocyte-specific deletion of AhR exacerbated alcohol-induced liver injury, indicating a protective role of AhR in ALD. Dietary I3C supplementation study further demonstrated that activation of AhR alleviated alcohol-induced liver injury. Moreover, NQO1 as one of AhR targets involved in protective role in ALD through modulation of alcohol-reduced NAD+-to-NADH ratio (Figure 11B). Thus, the results from our present study may help to establish AhR and its target NQO1 as novel therapeutic and preventive targets for ALD.
Oxidative stress and inflammatory responses have been recognized as a central mechanism in the pathogenesis of alcoholic liver injury.23 Previous studies have shown that AhR participates in antioxidant responses through activation of NRF2.24 NRF2 is a master regulator of antioxidant response. It regulates a variety of antioxidant enzymes, such as NQO1 and HO-1 by binding to its cognate binding sites, known as antioxidant response elements after nuclear translocation. Interestingly, NQO1 is also considered as an AhR target gene, which contain AhR binding sites known as xenobiotic response element binding sites.25 In this study, we found that alcohol-induced NQO1 expression is dependent on AhR, as AhR deletion decreased alcohol-induced NQO1 expression without affecting HO-1 and NRF2 expression. HO-1, as a canonical NRF2 regulated gene, exhibited a similar expression pattern with NRF2 in ALD mice. Our in vitro study further demonstrated that FICZ, an endogenous AhR ligand, strongly induced AhR translocation to the nucleus and increased NQO1 expression. In fact, FICZ also induced NRF2 expression but without nuclear translocation, indicating that NQO1 expression is dependent on AhR, but not on NRF2, in our model system. In line with NQO1 suppression, AhR knockout exacerbated hepatic lipid peroxidation and ROS overgeneration in both the liver and blood. Hepatocyte-specific overexpression of NQO1 in mice confirmed that NQO1 protected against chronic alcohol exposure–induced hepatic ROS overgeneration and lipid peroxidation as well as against liver injury. A recent study demonstrated that hepatic NQO1 promoted IκBζ (inhibitor of kappa B zeta) degradation and subsequently attenuated expression of IκBζ target genes, indicating a potential role of NQO1 in inflammatory responses.26 Thus, IκBζ target genes, including Cxcl1 and Lcn2, were measured in our chronic ALD models. A negative correlation between NQO1 and Cxcl1/Lcn2 was observed, the messenger RNA levels of Cxcl1 and Lcn2 were significantly increased following NQO1 reduction by AhR knockout and decreased by NQO1 overexpression. These results suggest that induction of NQO1 is a critical downstream event mediating the hepatoprotective response of AhR signaling against alcohol-induced oxidative stress and inflammation.
In addition to its antioxidative effect, NQO1 also has other functions. One of its emerging roles is the function as a powerful generator of NAD+.27 Alcohol metabolism is well known to consume NAD+ and generate NADH, leading to a reduction of cellular NAD+-to-NADH ratio, a fundamental cellular disorder in the pathogenesis of ALD.28 In contract to ADH and ALDH, NQO1 consumes NADH and produces NAD+. Thus, NQO1 induction may resolve alcohol-induced decrease in cellular NAD+-to-NADH ratio. Indeed, our study showed that chronic alcohol exposure upregulated NQO1 in association with reduction of NAD+-to-NADH ratio. Along with reduction of NQO1 in AhR knockout mice, we observed a further decrease in hepatic NAD+-to-NADH ratio, which suggests that activation of the AhR-NQO1 signaling pathway is an adaptive response to alcohol-induced decrease in hepatic NAD+-to-NADH ratio. Furthermore, upregulation of hepatic NQO1 by AAV gene delivery or activation of AhR by dietary I3C improved chronic alcohol exposure–induced decrease in hepatic NAD+-to-NADH ratio along with amelioration of ALD. Furthermore, serum ethanol levels correlatively elevated in association with a reduction of hepatic NAD+-to-NADH ratio in AF AhRΔhep mice and declined in association with increased hepatic NAD+-to-NADH ratio in AF/NQO1 mice or AF/I3C mice.
Another interesting finding from this study is that NQO1 translocated to the nuclei under a condition of cellular reduced NAD+-to-NADH ratio. NQO1 is historically regarded as a cytosolic enzyme,29 which contains NADH binding site at its C-terminal domain.30 Thus, high levels of NADH in the cytosol may mediate NQO1 translocation. It is well known that high levels of NADH were produced by consuming NAD+ during alcohol/ACD detoxification in hepatocytes. Indeed, ACD-induced high levels of nuclear NADH were observed in Hepa-1c1c7 cells by using a fluorescence NADH imaging probe in present study. Meanwhile, NQO1 nuclear translocation was also clearly observed after ACD treatment. Furthermore, our in vitro studies demonstrated that decreased NAD+-to-NADH ratio by adding exogenous NADH induces NQO1 nuclear translocation, which was blocked by treatment with exogenous NAD+. These results indicate that decreased NAD+-to-NADH ratio serves as an actual trigger for NQO1 nuclear translocation in liver.
Modulation of exogenous NAD+ biosynthesis has shown therapeutic benefits against tissue damage in animal models of muscle degeneration,31 small intestine injury,32 and liver fibrosis.33 Similarly, several studies also showed that increased cellular NAD+ levels by activation of NQO1 resulted in beneficial effects on acute kidney injury,34 lung fibrosis,35 and cardiac dysfunction.36 In this study, we found that NQO1 upregulation by AAV gene delivery and I3C administration increased hepatic SIRT1 activity and reduced FoxO1 acetylation. As a deacetylase, SIRT1 requires NAD+ as a substrate to receive the acetyl group in order to function properly.37, 38, 39, 40, 41 Our results demonstrated that NAD+ is the molecule linking AhR-NQO1 signaling with SIRT1-FoxO1 signaling in protection against alcohol-induced apoptosis in the liver. These results suggest that NQO1 plays a critical role in liver function and correction of alcohol-reduced NAD+-to-NADH ratio is an important mechanism underlying the protective role of AhR-NQO1 signaling pathway.
In summary, the present study provides the first evidence showing hepatic AhR activation and NQO1 nuclear enrichment in both ALD patients and mouse model of alcohol abuse. Hepatocyte-specific deletion of AhR in mice exacerbated chronic alcohol exposure–induced cellular redox imbalance and liver injury by downregulation of NQO1. Cell studies revealed that a reduced NAD+-to-NADH ratio serves as an inducer of NQO1 nuclear translocation. Hepatocyte-specific gene delivery of NQO1 in mice increased nuclear NQO1 and ameliorated alcoholic liver injury through increasing NAD+-to-NADH ratio and restoring NAD+-dependent enzyme activities. Dietary I3C supplementation activated hepatic AhR-NQO1 signaling and reversed chronic alcohol exposure–induced liver injury. Taken together, these results demonstrate that AhR signaling drives a protective response against alcohol induced hepatotoxicity and NQO1 plays a critical role in mediating the protective effect of AhR. This study also supports that dietary AhR ligands may serve as an attractive intervention for ALD.
Materials and Methods
Human Liver Samples
Liver explant specimens from patients with severe AH and wedge biopsies from the donor livers (normal control) were collected at Johns Hopkins University under the support of National Institute on Alcohol Abuse and Alcoholism–funded Clinical Resource for Alcoholic Hepatitis Investigations (R24AA025017). Collecting tissues from explanted livers or biopsies from donor livers had been approved by Institutional Review Boards at Johns Hopkins Medical Institutions (IRB00107893, IRB00021325).
Mice
C57BL/6J wild-type, AhR floxed mice (AhRflox/flox, stock no. 006203), and Albumin-Cre mice (Alb-Cre) (stock no. 003574) were purchased from the Jackson Laboratory (Bar Harbor, ME). AhRflox/flox mice were bred with Alb-Cre mice to generate mice with hepatocyte-specific knockout of AhR (AhRΔhep). Sibling littermates of AhRflox/flox mice (flox) were used as control animals for the AhRΔhep mice (AhRΔhep). For hepatocyte-specific NQO1 overexpression, C57/BL6J mice at 10 weeks old were injected via the retro orbital sinus with recombinant AAV8 gene transfer vectors (5 × 1011 GC/per mouse) bearing mouse NQO1 complementary DNA (cDNA) under the control of hepatocyte-specific promoter (TBG). Mice injected with AAV8-TBG-null vectors were served as control (AAV8-Null). Mice were handled and all experiments were performed in accordance with the protocol approved by the North Carolina Research Campus Institutional Animal Care and Use Committee.
Chronic Alcohol Feeding and Treatments
Male mice at 12 weeks old were fed an alcohol-containing Lieber-DeCarli liquid diet (AF) or an isocaloric control liquid diet (PF) for 8 weeks as described previously.42 All ingredients used in the liquid diets were obtained from Dyets (Bethlehem, PA) except for ethanol and I3C (Sigma-Aldrich, St Louis, MO). To test the therapeutic potential of the dietary AhR ligand, mice were fed with liquid diet added with I3C at a dosage of 5 mg/10 mL, 3 times a week, starting from the sixth week in an 8-week feeding protocol.
Cell Culture and Treatments
Hepa-1c1c7 mouse hepatoma cells and Tao cells (American Type Culture Collection, Rockville, MD) were seeded at a density of 1 × 104 cells/cm2 in 25-cm2 sealable flasks (Thermo Fisher Scientific, Rockford, IL). After reaching 80% confluence, cells were treated with FICZ at 0–200 nmol/L or ethanol at 50–200 mmol/L for 6 hours and treated with ACD at 100 μmol/L, NADH at 100–400 μmol/L, or NAD+ at 100–400 μmol/L (Sigma-Aldrich) for 24 hours.
Establishment of Stable NQO1 Overexpressing/Knockdown Cell Lines
In a 6-well tissue culture plate, 1.5 × 105 cells were seeded in 3 mL of standard growth medium per well. After reaching to 60% confluency, cells were transfected with NQO1 lentiviral activation particles (SC-421913-LAC; Santa Cruz Biotechnology, Dallas, TX) or control lentiviral activation particles (SC-437282; Santa Cruz Biotechnology) following the manufacturer’s guidelines. For NQO1 knockdown, cells were transfected with NQO1 Double Nickase Plasmid (SC-421913; Santa Cruz Biotechnology) or control plasmid (SC-437281; Santa Cruz Biotechnology) following the manufacturer’s guidelines. After 24 hours of transfection, cells were selected with puromycin at 1–3 μg/mL. The stable cell lines were established and confirmed by Western blot analysis.
Serum ALT, AST, and CXCL1 Levels
Plasma ALT and AST levels were colorimetrically measured by Infinity ALT Reagent and Infinity AST Reagent (Thermo Fisher Scientific), respectively. For serum CXCL1, levels were measured by mouse CXCL1/KC quantikine enzyme-linked immunosorbent assay kit (R&D Systems, Minneapolis, MN)
Liver TG and FFA Levels
The hepatic levels of TG and FFA were measured with colorimetric assay kits (BioVision, Milpitas, CA). Briefly, lipids were extracted using chloroform/methanol (2:1), vacuumed, and redissolved in 5% Triton X-100/methyl alcohol mixture (1:1 vol/vol), then FFA and TG contents were determined according to the manufacturer’s instructions.
Flow Cytometry
The frequency of liver neutrophils, the mean fluorescence intensity of DCFDA, and the Annexin V–positive cells were determined by flow cytometry using a fluorescence-activated cell sorting Melody analyzer (BD Biosciences, San Jose, CA), and data were analyzed with FlowJo 10.1 software (Tree Star, Ashland, OR).
Histopathology and Immunohistochemistry
Histopathology staining was performed as previously described.43,44 Briefly, liver tissues were fixed in 10% formalin and processed for paraffin embedding. Paraffin sections were cut in 5 μm and processed with hematoxylin and eosin staining. For IHC staining, hepatic acetylated lysine, AhR, NQO1, and 4-HNE were detected. Liver tissue paraffin sections were incubated with 3% hydrogen peroxide for 10 minutes to inactivate endogenous peroxidases. The endogenous mouse IgG was blocked by incubation with a mouse-to-mouse blocking reagent (ScyTek Laboratories, Logan, UT). Tissue sections were then incubated with a monoclonal antibody at 4°C overnight, followed by incubation with EnVision+ Labelled Polymer-HRP-conjugated anti-mouse IgG (DAKO, Carpinteria, CA) at room temperature for 30 minutes. Diaminobenzidine was used as horseradish peroxidase substrate for visualization.
Western Blot and Immunoprecipitation
Whole protein lysates of the liver and Hepa-1c1c7 cells were extracted using lysis buffer supplemented with the protease inhibitor and phosphatase inhibitor (Sigma-Aldrich). Nuclear proteins were extracted using a commercial kit followed by manufacturer’s protocol (Boster Biological Technology, Pleasanton, CA). Aliquots containing 40 μg of proteins were loaded onto 10%–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, transblotted onto polyvinylidene difluoride membrane, blocked with 5% nonfat milk or 5% bovine serum albumin in Tris-buffered saline solution with 0.1% Tween 20 for 30 minutes at room temperature, and incubated with NQO1, β-actin, GAPDH (Abcam, Cambridge, MA), acetylated lysine, FoxO1, BIM, BAX, PUMA, cleaved caspase3, BCL2, SIRT1, Lamin a/c (Cell Signaling Technology, Danvers, MA), AhR (Enzo Life Sciences, Farmingdale, NY), and HO-1 (Boster Biological Technology), respectively. Membranes were washed and incubated with secondary antibodies (Thermo Fisher Scientific). The bound complexes were detected via enhanced chemiluminescence (GE Healthcare, Piscataway, NJ). Protein bands were quantified, and the ratio to β-actin or GAPDH was calculated as fold changes, setting the values of PF or normal control at 1.
For immunoprecipitation studies, total protein (1000 μg) was incubated in 20-μL protein A/G plus agarose beads (Santa Cruz Biotechnology) on a rocker for 1 hour at 4°C and then was centrifuged at 14,000 g for 30 seconds at 4°C to remove the protein A/G. The supernatant was collected and incubated with 2.5-μg FoxO1 or SIRT1 antibody for overnight at 4°C on a rocker, and then 20-μL protein A/G plus agarose beads was added, followed by incubation overnight with mixing, and then was centrifuged at 14,000 g for 30 seconds at 4°C. After washing 3 times with cold phosphate-buffered saline (PBS), pellet was resuspended in 60 μL of electrophoresis sample buffer and boiled for 5 minutes, and sodium dodecyl sulfate polyacrylamide gel electrophoresis was performed following Western blotting procedures.
Immunofluorescence Microscopy
Cells were fixed with 4% formaldehyde for 15 minutes at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 10 minutes and incubated for 30 minutes in 10% normal donkey serum. Then cells were incubated with primary antibodies in PBS containing 1% bovine serum albumin, 0.1% cold fish skin gelatin, 0.1% Triton X-100, and 0.05% sodium azide (primary antibody dilution buffer) overnight at 4°C. The next day, cells were incubated with Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific) in PBS for 1 hour at room temperature and then 5 minutes to DAPI (Life Technologies, Carlsbad, CA) in PBS.
To determine hepatic neutrophil infiltration, cryostat sections of mouse liver were incubated with anti-MPO (LSBio, Seattle, WA), followed by Alexa Fluor 594-conjugated donkey anti-rat IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). The nuclei were counterstained by DAPI (Life Technologies).
Immunofluorescence Staining
For mitochondrial membrane potential staining by TMRE (tetramethylrhodamine, ethyl ester) mitochondrial kit (Abcam), cellular ROS staining by DCFDA (Invitrogen, Carlsbad, CA), and NADH staining by JZL1707 NAD(P)H sensor (AAT Bioquest, Sunnyvale, CA) according to manufacturer's instructions, respectively.
RNA Isolation and Real-Time Polymerase Chain Reaction
Total RNA was isolated using TRIzol Reagent (Invitrogen) from mouse liver tissue. The cDNA was total RNA by cDNA Synthesis kit (Takara Bio, Tokyo, Japan). The resulting cDNA was subjected to real-time polymerase chain reaction (PCR) analysis with gene-specific primers in the presence of SYBR Green Supermix kit (Takara) on a Vii7 system (ABI, Carlsbad, CA). Data were analyzed using the 2−ΔΔCt threshold cycle method. Primers used for real-time PCR are depicted in Table 1.The messenger RNA levels of genes were normalized to that of RPS17 rRNA and expressed as relative to the control.
Table 1.
Overview of Real-Time PCR Primers and Sequences
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| AhR | CCATCCCCGCTGAAGGAATTA | CTGCCAGTCTCTGATTTGTGC |
| Cxcl1 | CCAGAGCTTGAAGGTGTTGC | AAGCCTCGCGACCATTCTTG |
| Lcn2 | TCCTCAGGTACAGAGCTACAA | GCTCCTTGGTTCTTCCATACA |
| Nqo1 | CATTGCAGTGGTTTGGGGTG | TCTGGAAAGGACCGTTGTCG |
| RPS17 | GGAGATCGCCATTATCCCCA | ATCTCCTTGGTGTCGGGATC |
PCR, polymerase chain reaction.
Statistical Analysis
Results were analyzed using the independent-samples t test or one-way analysis of variance followed by Tukey’s multiple comparison. Data were expressed as mean ± SEM. In all tests, P < .05 was taken as significant.
Acknowledgments
The authors thank Dr Zhaoli Sun at Johns Hopkins University School of Medicine for help with human samples collecting. The authors also thank Tianjiao Li, PhD candidate at the University of North Carolina at Greensboro, for discussion and comments during manuscript preparation.
CRediT Authorship Contributions
Zhanxiang Zhou, PhD (Conceptualization: Equal; Funding acquisition: Lead; Projectadministration: Lead; Supervision: Lead; Writing – review & editing: Lead)
Haibo Dong (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead;Software: Lead; Writing – original draft: Lead)
Liuyi Hao (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal;Writing – review & editing: Supporting)
Wenliang Zhang (Data curation: Supporting; Resources: Lead)
Wei Zhong (Conceptualization: Supporting; Project administration: Equal; Writing –review & editing: Supporting)
Wei Guo (Conceptualization: Supporting; Data curation: Supporting; Writing – review &editing: Supporting)
Ruichao Yue (Conceptualization: Supporting; Data curation: Supporting; Writing –review & editing: Supporting)
Xinguo Sun (Methodology: Equal; Resources: Equal)
Footnotes
Funding This research was supported by the National Institutes of Health grants R01AA018844 (to Zhanxiang Zhou) and R01AA020212 (to Zhanxiang Zhou).
Conflicts of interest All authors report no conflict of interest.
References
- 1.Vondracek J., Machala M. Environmental ligands of the aryl hydrocarbon receptor and their effects in models of adult liver progenitor cells. Stem Cells Int 2016. 2016 doi: 10.1155/2016/4326194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Shea R.S., Dasarathy S., McCullough A.J. Practice Guideline Committee of the American Association for the Study of Liver Disease, Practice Parameters Committee of the American College of Gastroenterology. Alcoholic liver disease. Hepatology. 2010;51:307–328. doi: 10.1002/hep.23258. [DOI] [PubMed] [Google Scholar]
- 3.Warren K.R., Murray M.M. Alcoholic liver disease and pancreatitis: global health problems being addressed by the US National Institute on Alcohol Abuse and Alcoholism. J Gastroenterol Hepatol. 2013;28(Suppl 1):4–6. doi: 10.1111/jgh.12246. [DOI] [PubMed] [Google Scholar]
- 4.Rehm J., Samokhvalov A.V., Shield K.D. Global burden of alcoholic liver diseases. J Hepatol. 2013;59:160–168. doi: 10.1016/j.jhep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Lafita-Navarro M.C., Kim M., Borenstein-Auerbach N., Venkateswaran N., Hao Y.H., Ray R., Brabletz T., Scaglioni P.P., Shay J.W., Conacci-Sorrell M. The aryl hydrocarbon receptor regulates nucleolar activity and protein synthesis in MYC-expressing cells. Genes Dev. 2018;32:1303–1308. doi: 10.1101/gad.313007.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen L.P., Bradfield C.A. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21:102–106. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kudo I., Hosaka M., Haga A., Tsuji N., Nagata Y., Okada H., Fukuda K., Kakizaki Y., Okamoto T., Grave E., Itoh H. The regulation mechanisms of AhR by molecular chaperone complex. J Biochem. 2018;163:223–232. doi: 10.1093/jb/mvx074. [DOI] [PubMed] [Google Scholar]
- 8.Kazlauskas A., Poellinger L., Pongratz I. Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (aryl hydrocarbon) receptor. J Biol Chem. 1999;274:13519–13524. doi: 10.1074/jbc.274.19.13519. [DOI] [PubMed] [Google Scholar]
- 9.Hord N.G., Perdew G.H. Physicochemical and immunocytochemical analysis of the aryl hydrocarbon receptor nuclear translocator: characterization of two monoclonal antibodies to the aryl hydrocarbon receptor nuclear translocator. Mol Pharmacol. 1994;46:618–626. [PubMed] [Google Scholar]
- 10.Pollenz R.S. The mechanism of AH receptor protein down-regulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem Biol Interact. 2002;141:41–61. doi: 10.1016/s0009-2797(02)00065-0. [DOI] [PubMed] [Google Scholar]
- 11.Beischlag T.V., Luis Morales J., Hollingshead B.D., Perdew G.H. The aryl hydrocarbon receptor complex and the control of gene expression. Crit Rev Eukaryot Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson J.M., Shepherd D.M. Dietary ligands of the aryl hydrocarbon receptor induce anti-inflammatory and immunoregulatory effects on murine dendritic cells. Toxicol Sci. 2011;124:327–338. doi: 10.1093/toxsci/kfr249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh N.P., Singh U.P., Rouse M., Zhang J., Chatterjee S., Nagarkatti P.S., Nagarkatti M. Dietary indoles suppress delayed-type hypersensitivity by inducing a switch from proinflammatory Th17 cells to anti-inflammatory regulatory T cells through regulation of MicroRNA. J Immunol. 2016;196:1108–1122. doi: 10.4049/jimmunol.1501727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J.H., Lee J.M., Lee E.J., Hwang W.B., Kim D.J. Indole-3-carbinol promotes goblet-cell differentiation regulating wnt and notch signaling pathways AhR-dependently. Mol Cells. 2018;41:290–300. doi: 10.14348/molcells.2018.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M.L., Lin S.H., Hou Y.Y., Chen Y.H. Suppression of lipid accumulation by indole-3-carbinol is associated with increased expression of the aryl hydrocarbon receptor and CYP1B1 proteins in adipocytes and with decreased adipocyte-stimulated endothelial tube formation. Int J Mol Sci. 2016;17:1256. doi: 10.3390/ijms17081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan J., Tung H.C., Li S., Niu Y., Garbacz W.G., Lu P., Bi Y., Li Y., He J., Xu M., Ren S., Monga S.P., Schwabe R.F., Yang D., Xie W. Aryl hydrocarbon receptor signaling prevents activation of hepatic stellate cells and liver fibrogenesis in mice. Gastroenterology. 2019;157:793–806.e14. doi: 10.1053/j.gastro.2019.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H.F., Lin X.H., Yang H., Zhou L.C., Guo Y.L., Barnett J.V., Guo Z.M. Regulation of the activity and expression of aryl hydrocarbon receptor by ethanol in mouse hepatic stellate cells. Alcohol Clin Exp Res. 2012;36:1873–1881. doi: 10.1111/j.1530-0277.2012.01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wrzosek L., Ciocan D., Hugot C., Spatz M., Dupeux M., Houron C., Lievin-Le Moal V., Puchois V., Ferrere G., Trainel N., Mercier-Nome F., Durand S., Kroemer G., Voican C.S., Emond P., Straube M., Sokol H., Perlemuter G., Cassard A.M. Microbiota tryptophan metabolism induces aryl hydrocarbon receptor activation and improves alcohol-induced liver injury. Gut. 2020 Oct 1 doi: 10.1136/gutjnl-2020-321565. [E-pub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Osna N.A., Donohue T.M., Jr., Kharbanda K.K. Alcoholic liver disease: pathogenesis and current management. Alcohol Res. 2017;38:147–161. [PMC free article] [PubMed] [Google Scholar]
- 20.Tian J., Feng Y., Fu H., Xie H.Q., Jiang J.X., Zhao B. The aryl hydrocarbon receptor: a key bridging molecule of external and internal chemical signals. Environ Sci Technol. 2015;49:9518–9531. doi: 10.1021/acs.est.5b00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierre S., Chevallier A., Teixeira-Clerc F., Ambolet-Camoit A., Bui L.C., Bats A.S., Fournet J.C., Fernandez-Salguero P., Aggerbeck M., Lotersztajn S., Barouki R., Coumoul X. Aryl hydrocarbon receptor-dependent induction of liver fibrosis by dioxin. Toxicol Sci. 2014;137:114–124. doi: 10.1093/toxsci/kft236. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Salguero P., Pineau T., Hilbert D.M., McPhail T., Lee S.S., Kimura S., Nebert D.W., Rudikoff S., Ward J.M., Gonzalez F.J. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 23.Ambade A., Mandrekar P. Oxidative stress and inflammation: essential partners in alcoholic liver disease. Int J Hepatol. 2012;2012:853175. doi: 10.1155/2012/853175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeager R.L., Reisman S.A., Aleksunes L.M., Klaassen C.D. Introducing the "TCDD-inducible AhR-Nrf2 gene battery. Toxicol Sci. 2009;111:238–246. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietrich C. Antioxidant functions of the aryl hydrocarbon receptor. Stem Cells Int. 2016;2016:7943495. doi: 10.1155/2016/7943495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y., Feng D., Hwang S., Mackowiak B., Wang X., Xiang X., Rodrigues R.M., Fu Y., Ma J., Ren T., Ait-Ahmed Y., Xu M., Liangpunsakul S., Gao B. Interleukin-20 exacerbates acute hepatitis and bacterial infection by downregulating IkappaBzeta target genes in hepatocytes. J Hepatol. 2021 Feb 18 doi: 10.1016/j.jhep.2021.02.004. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross D., Siegel D. Functions of NQO1 in cellular protection and CoQ10 metabolism and its potential role as a redox sensitive molecular switch. Front Physiol. 2017;8:595. doi: 10.3389/fphys.2017.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comporti M., Signorini C., Leoncini S., Gardi C., Ciccoli L., Giardini A., Vecchio D., Arezzini B. Ethanol-induced oxidative stress: basic knowledge. Genes Nutr. 2010;5:101–109. doi: 10.1007/s12263-009-0159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernster L., Danielson L., Ljunggren M. DT diaphorase. I. Purification from the soluble fraction of rat-liver cytoplasm, and properties. Biochim Biophys Acta. 1962;58:171–188. doi: 10.1016/0006-3002(62)90997-6. [DOI] [PubMed] [Google Scholar]
- 30.Medina-Carmona E., Neira J.L., Salido E., Fuchs J.E., Palomino-Morales R., Timson D.J., Pey A.L. Site-to-site interdomain communication may mediate different loss-of-function mechanisms in a cancer-associated NQO1 polymorphism. Sci Rep. 2017;7:44532. doi: 10.1038/srep44532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goody M.F., Kelly M.W., Reynolds C.J., Khalil A., Crawford B.D., Henry C.A. NAD+ biosynthesis ameliorates a zebrafish model of muscular dystrophy. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandit A., Kim H.J., Oh G.S., Shen A., Lee S.B., Khadka D., Lee S., Shim H., Yang S.H., Cho E.Y., Kwon K.B., Kwak T.H., Choe S.K., Park R., So H.S. Dunnione ameliorates cisplatin-induced small intestinal damage by modulating NAD(+) metabolism. Biochem Biophys Res Commun. 2015;467:697–703. doi: 10.1016/j.bbrc.2015.10.081. [DOI] [PubMed] [Google Scholar]
- 33.Pham T.X., Bae M., Kim M.B., Lee Y., Hu S., Kang H., Park Y.K., Lee J.Y. Nicotinamide riboside, an NAD+ precursor, attenuates the development of liver fibrosis in a diet-induced mouse model of liver fibrosis. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2451–2463. doi: 10.1016/j.bbadis.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh G.S., Kim H.J., Choi J.H., Shen A., Choe S.K., Karna A., Lee S.H., Jo H.J., Yang S.H., Kwak T.H., Lee C.H., Park R., So H.S. Pharmacological activation of NQO1 increases NAD(+) levels and attenuates cisplatin-mediated acute kidney injury in mice. Kidney Int. 2014;85:547–560. doi: 10.1038/ki.2013.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh G.S., Lee S.B., Karna A., Kim H.J., Shen A., Pandit A., Lee S., Yang S.H., So H.S. Increased cellular NAD(+) level through NQO1 enzymatic action has protective effects on bleomycin-induced lung fibrosis in mice. Tuberc Respir Dis (Seoul) 2016;79:257–266. doi: 10.4046/trd.2016.79.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khadka D., Kim H.J., Oh G.S., Shen A., Lee S., Lee S.B., Sharma S., Kim S.Y., Pandit A., Choe S.K., Kwak T.H., Yang S.H., Sim H., Eom G.H., Park R., So H.S. Augmentation of NAD(+) levels by enzymatic action of NAD(P)H quinone oxidoreductase 1 attenuates adriamycin-induced cardiac dysfunction in mice. J Mol Cell Cardiol. 2018;124:45–57. doi: 10.1016/j.yjmcc.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Canto C., Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H., Xing R., Cheng X., Li Q., Liu F., Ye H., Zhao M., Wang H., Wang G., Hao H. De-novo NAD+ synthesis regulates SIRT1-FOXO1 apoptotic pathway in response to NQO1 substrates in lung cancer cells. Oncotarget. 2016;7:62503–62519. doi: 10.18632/oncotarget.11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H.Y., Li Q.R., Cheng X.F., Wang G.J., Hao H.P. NAMPT inhibition synergizes with NQO1-targeting agents in inducing apoptotic cell death in non-small cell lung cancer cells. Chin J Nat Med. 2016;14:582–589. doi: 10.1016/S1875-5364(16)30068-1. [DOI] [PubMed] [Google Scholar]
- 40.Gu X., Han D., Chen W., Zhang L., Lin Q., Gao J., Fanning S., Han B. SIRT1-mediated FoxOs pathways protect against apoptosis by promoting autophagy in osteoblast-like MC3T3-E1 cells exposed to sodium fluoride. Oncotarget. 2016;7:65218–65230. doi: 10.18632/oncotarget.11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo G., Jian Z., Zhu Y., Zhu Y., Chen B., Ma R., Tang F., Xiao Y. Sirt1 promotes autophagy and inhibits apoptosis to protect cardiomyocytes from hypoxic stress. Int J Mol Med. 2019;43:2033–2043. doi: 10.3892/ijmm.2019.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong W., Zhang W., Li Q., Xie G., Sun Q., Sun X., Tan X., Sun X., Jia W., Zhou Z. Pharmacological activation of aldehyde dehydrogenase 2 by Alda-1 reverses alcohol-induced hepatic steatosis and cell death in mice. J Hepatol. 2015;62:1375–1381. doi: 10.1016/j.jhep.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Q., Zhong W., Zhang W., Zhou Z. Defect of mitochondrial respiratory chain is a mechanism of ROS overproduction in a rat model of alcoholic liver disease: role of zinc deficiency. Am J Physiol Gastrointest Liver Physiol. 2016;310 doi: 10.1152/ajpgi.00270.2015. G205–G204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Q., Zhang W., Zhong W., Sun X., Zhou Z. Pharmacological inhibition of NOX4 ameliorates alcohol-induced liver injury in mice through improving oxidative stress and mitochondrial function. Biochim Biophys Acta Gen Subj. 2017;1861:2912–2921. doi: 10.1016/j.bbagen.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]