Abstract
Objective: To investigate the effect of miR-132-3p and HAVCR1/kidney injury molecule (KIM)-1 on sepsis-induced acute kidney injury (AKI) in mice. Methods: One hundred C57BL/6 mice were divided into five groups with 20 mice in each group: the normal group (normal mice), the model group (mice with sepsis), the miR-132-3p mimic group (miR-132-3p overexpression), the oe-HAVCR1/KIM-1 group (HAVCR1/KIM-1 overexpression), and the miR-132-3p mimic + oe-HAVCR1/KIM-1 group. Dual-luciferase reporter assay was performed to verify the targeting relationship between miR-132-3p and HAVCR1/KIM-1. The expressions of miR-132-3p and HAVCR1/KIM-1 in mice’ kidneys, the levels of renal function markers, the expressions of apoptosis-associated proteins, the renal cell apoptosis rate, and the inflammatory factors in serum were all examined. Results: We found that miR-132-3p can target HAVCR1/KIM-1 and regulate its expression. Compared with the normal mice, the septic mice exhibited lower miR-132-3p level and higher HAVCR1/KIM-1 level (both P<0.05). Moreover, the septic mice had higher levels of cleaved caspase-3, Bax, blood urea nitrogen, creatinine, tumor necrosis factor-α, interleukin-1β, and interleukin-6, higher renal cell apoptosis rate, and lower Bcl-2 level than the normal mice (all P<0.05). MiR-132-3p overexpression could improve the renal function of the mice with sepsis and inhibit renal cell apoptosis and inflammatory progression, whereas HAVCR1/KIM1 overexpression exhibited an opposite effect and could block the renal protective effects of miR-132-3p overexpression on the septic mice. Conclusion: MiR-132-3p overexpression can inhibit renal cell apoptosis and inflammatory progression via suppressing HAVCR1/KIM-1 expression, thereby exert renal protective effects on mice with sepsis.
Keywords: HAVCR1/KIM-1, mir-132-3p, renal function, sepsis-induced acute kidney injury
Introduction
Sepsis is a type of systemic inflammatory response caused by viral, fungal, or bacterial infection. The disease can lead to multiple organ injuries or even organ failure and is a major cause of death among critically ill patients [1-4]. Currently, there is still no effective targeted approach for treating sepsis, and the medical expenses for the treatment of this disease can produce heavy financial burdens to both patients’ families and the society [5,6].
Acute kidney injury (AKI) is one of the major and most severe complications of sepsis and can double the mortality of patients [7,8]. The pathogenesis of AKI involves renal dysfunction, cell damage, and inflammatory infiltration [9-11]. The role of microRNA (miRNA) in sepsis-induced AKI remains unclear. Through searching the literature, we found that miR-132-3p can suppress the inflammation in synovitis as well as promote hippocampal neuron survival and inhibit its apoptosis [12,13]. Other studies have documented that in tuberous sclerosis complex, miR-132-3p can suppress BCL2L11 and regulate cell proliferation and apoptosis [14]. Nevertheless, there have been no reports on the regulatory mechanism of miR-132-3p in AKI. After using the bioinformatics web-based tool, we found that miR-132-3p can bind to HAVCR1/KIM-1. HAVCR1/kidney injury molecule (KIM)-1, as a member of the TIM gene family, is expressed in kidney and has been used as a biomarker of AKI at early stage clinically [15-18]. We speculated that miR-132-3p may play a role in sepsis-induced AKI via HAVCR1/KIM-1.
In this study, we created a mouse model of sepsis-induced AKI by injecting lipopolysaccharide (LPS) to investigate the expression of miR-132-3p in sepsis-induced AKI and its regulatory mechanism.
Materials and methods
Model creation and grouping
One hundred healthy specific pathogen-free C57BL/6 mice were selected for the study (8 weeks of age, 18.63±2.06 g, supplier: Guangdong Medical Laboratory Animal Center, China). The mice were raised at 37°C under a 12-hour light/dark cycle, fed with a normal diet, and given free access to water. The mice were raised for one week to adapt to the housing environment before the subsequent experiments were carried out. All the animal experiments were approved by the Ethics Committee of our hospital.
The mice were divided into the following groups of 20 mice each: the normal group (normal mice), the model group (model), the miR-132-3p mimic group (miR-139-3p overexpression), the oe-HAVCR1/KIM-1 group (HAVCR1/KIM-1 overexpression), and the miR-132-3p mimic + oe-HAVCR1/KIM-1 group (overexpression of both miR-139-3p and HAVCR1/KIM-1). The miR-132-3p mimic and HAVCR/KIM-1 lentiviral vector were purchased from Jrdun Biotechnology, Shanghai, China. The mice in each group were intraperitoneally injected with 1.65 mg/kg corresponding agents. Fifteen minutes after the injection, all the mice except for those in the normal group were given intraperitoneal injections of 10 mg/kg LPS, while the mice in the normal group were administered with same volume of normal saline. After 18 hours, the mice were intraperitoneally injected with pentobarbital sodium solution (0.3%, 0.2 mL/10 g), and their blood was separated by inferior vena cava puncture. The kidneys were collected after the mice were sacrificed [19].
Dual-luciferase reporter (DLR) assay
The targeting relationship of miR-132-3p with HAVCR1/KIM-1 was analyzed through a bioinformatics website (www.targetscan.org) and verified by DLR assay. HAVCR1/KIM-1 dual-luciferase reporter vectors with and without mutant miR-132-3p binding site were constructed and names as PGL3-HAVCR1/KIM-1-mut and PGL3-HAVCR1/KIM-1 wt, respectively. The reporter plasmids were transfected into HEK 293T cell with renilla plasmid and miR-132-3p mimic or NC plasmids. After 24 h of transfection, DLR assay was conducted according to the manufacturer’s instructions of the kit (Promega, USA) to measure the luciferase activity. The cells from each group were first lysed and centrifuged at 12,000 rpm for 1 min followed by removal of precipitation and collecting the supernatant. The lysed cells were collected in Eppendorf tubes, and 100 μL of firefly luciferase working solution was added to every 10 μL of the samples. After measuring the activity of firefly luciferase, 100 μL of renilla luciferase working solution was used for measuring renilla luciferase activity. Relative luciferase activity = firefly luciferase activity/renilla luciferase activity.
qRT-PCR
The kidney tissues from each group were placed in liquid nitrogen and ground to fine powder. The high purity RNAs were isolated according to the manufacturer’s protocol of the RNA extraction kit (D203-01, GenStar Biosolutions, Beijing, China). The RNAs from each group were reversely transcribed to cDNAs according to the instructions of TaqMan™ MicroRNA Reverse Transcription Kit (4366596, Thermo Fisher Scientific, MA, UK) with the following parameters: 42°C 30-50 min and 85°C 5 s. The primers for miR-132-3p, U6, HAVCR1/KIM-1, and GAPDH were designed by our lab and synthesized by Sangon Biotech, China (Table 1). The expressions of miR-132-3p and HAVCR1/KIM-1 were normalized to U6 and GAPDH, respectively. qRT-PCR was conducted with ABI PRISM® 7300 (Thermo Fisher Scientific, USA) in accordance with the instructions of the kit (SYBR® Premix Ex TaqTM II, RR820A, Xingzhi Biotech, Jiangsu, China). The reaction volume was 50 μL including 25 μL of SYBR® Premix EX TaqTM II (2×), 2 μL of PCR forward primer, 2 μL of reverse primer, 1 μL of ROX reference dye (50×), 4 μL of DNA template, and 16 μL of double-distilled water. The temperature profile of PCR were: 95°C for 10 min (pre-denaturation), 95°C for 15 s (denaturation), and 60°C for 30 s (annealing). The cycles repeated for 40 times followed by extension at 72°C for 1 min. Relative expression level of the target gene was calculated using the 2-ΔΔCt method and experiment was repeated three times. ΔΔCt = (the average Ct value of the target gene in the study group - the average Ct value of the house-keeping gene in the study group) - (the average Ct value of the target gene in the control group - the average Ct value of the house-keeping gene in the control group).
Table 1.
Primer sequences used for qRT-PCR
| Gene | Primer sequence (5’-3’) |
|---|---|
| miR-132-3p | Forward: GCGCGCGTAACAGTCTACAGG |
| Reverse: GTCGTATCCAGTGCAGGGTCC-3 | |
| U6 | Forward: CTCGCTTCGGCAGCACATATACT |
| Reverse: CGCTTCACGAATTTGCGTGT | |
| HAVCR1/KIM-1 | Forward: ACATATCGTGGAATCACAACGAC |
| Reverse: ACTGCTCTTCTGATAGGTGACA | |
| GAPDH | Forward: CCACATCGCTCAGACACCAT |
| Reverse: ACCAGGCGCCCAATACG |
Western blot
The total protein from the kidney tissues was extracted using the radioimmunoprecipitation assay (RIPA) kit (R0010, Solarbio, China). The transfected cells were washed in pre-cooled phosphate buffered saline (PBS) three times and the protein lysis buffer (60% RIPA + 39% SDS + 1% protease inhibitor) was added. Next, the samples were collected into Eppendorf tubes and lysed on ice for 30 min followed by centrifugation at 13,500 rpm at 4°C for 30 min. The supernatant was collected and placed in an icebox (Xiamen Qibing Cold Chain, Fujian, China). The protein concentration was measured using the BCA kit (Jining Shiye, Shanghai, China), and 10% separating gel and 5% stacking gel was prepared using the SDS-PAGE gel preparation kit (Ruji Biotech, Shanghai, China). After running PAGE for 90 min at 100 V, the proteins were transferred to a nitrocellulose membrane by wet-transfer and blocked in 5% BSA at room temperature for 1 h. Subsequently, the samples were incubated with rabbit antibodies to HAVCR1/KIM-1 (ab47635, Abcam, UK), cleaved caspase-3 (ab49822, 1:500, Abcam, UK), Bax (ab32503, 1:1,000, Abcam, UK), Bcl-2 (ab196495, 1:500, Abcam, UK), and GAPDH (ab181602, 1:3,000, Abcam, UK) at 4°C overnight followed by wash in PBS five times (5 min per wash). The membrane was treated with horseradish peroxidase-labeled goat anti-rabbit IgG (1:5,000, ZSGB-Bio, China) and then reacted with ECL (ECL808-25, Biomiga, USA) for 1 min at room temperature. After removing the liquid, the membrane was covered with plastic wrap for X-ray (36209ES01, Qcbio Science & Technologies, Shanghai, China). GAPDH was the internal control for the protein expressions of HAVCR1/KIM-1, cleaved caspase-3, Bax, and Bcl-2. Image J software was used to evaluate the density of the protein. The relative protein expression level was calculated as the grayscale value of the target protein band/the grayscale value of the GAPDH band.
Hematoxylin and eosin (H&E) staining
Six paraffin sections of the renal tissues from each group were dewaxed with dimethyl benzene and ethanol and stained with hematoxylin (Solarbio, Beijing, China) for 5 min at room temperature followed by wash in tap water. The sections were then treated with 1% hydrochloric acid for differentiation for 5 s and washed in tap water again. Subsequently, 0.6% aqueous ammonia was added and the sections were rinsed in running water for 15 min before being stained with eosin for 1 min. Next, the sections were dehydrated in a gradient of alcohol, sealed with neutral balsam, and observed under an inverted microscope (Thermo Fisher Scientific, USA).
Detecting cellular apoptosis using the terminal deoxynucleotidyl transferase-dUTP nick end labeling (TUNEL) staining
Three renal sections from each group were collected to examine the renal cell apoptosis. The sections were placed on glass slides and numbered. Next, the sections were dewaxed, hydrated, and washed in PBS twice followed by treatment with permeabilization buffer for 8 min. The sections were washed in PBS twice and were incubated with 50 μL of TUNEL reaction mixture in a humidified box for 1 h at room temperature. The sections were then washed in PBS three times and observed under a fluorescence microscope (Nikon, TE2000, Tokyo, Japan). The TUNEL positive cells were analyzed using the Image-Pro Plus 6.0. Five fields were randomly picked to count the positive cells.
ELISA
Levels of BUN (blood urea nitrogen), serum creatinine (SCr), tumor necrosis factor (TNF)-α, interleukin (IL)-1β, and IL-6 in mice’ serum were detected according to the manufacturer’s instructions of the kits (ELISA kits for SCr and BUN: CST, USA; kits for TNF-α, IL-1β, and IL-6: Abcam, UK). The optical density value of each sample at 450 nm was measured with a spectrometer (Thermo Fisher Scientific, USA) for detecting the levels of these markers.
Statistical analysis
SPSS 21.0 (SPSS Inc., IL, USA) was applied for data analysis. Measurement data are expressed as mean ± standard deviation. Comparison between groups was conducted by one-way analysis of variance and Bonferroni post-hoc test. Statistical significance was indicated by P<0.05.
Results
miR-132-3p can target HAVCR1 and inhibit its expression
The bioinformatics website predicted the pairing of miR-132-3p with HAVCR1/KIM-1 (Figure 1A). Results of DLR assay showed that compared with the cells transfected with miR-132-3p mimic NC, the luciferase activity of the cells co-transfected with miR-132-3p mimic and HAVCR1/KIM-1 wt was much lower (Figure 1B), suggesting that miR-132-3p can target HAVCR1/KIM-1. Meanwhile, qRT-PCR and Western blot were performed to measure the expression levels of miR-132-3p and HAVCR1/KIM-1 (Figure 1C-E). Compared with the normal group, the other groups presented lower miR-132-3p expression levels and higher HAVCR1/KIM-1 mRNA and protein expression levels in the mice’ kidneys (all P<0.05). Compared with the model group, the miR-132-3p mimic group had a higher miR-132-3p expression level and lower HAVCR1/KIM-1 mRNA and protein expression levels (all P<0.05), the oe-HAVCR1 group showed no difference in the miR-132-3p level (P>0.05) but presented higher mRNA and protein expression levels of HAVCR1/KIM (both P<0.05), and the miR-132-3p mimic + oe-HAVCR1/KIM-1 group had significantly higher expression level of miR-132-3p and similar expression level of HAVCR1/KIM. These results reveal that miR-12-3p can bind to HAVCR1/KIM-1 and inhibit its expression.
Figure 1.
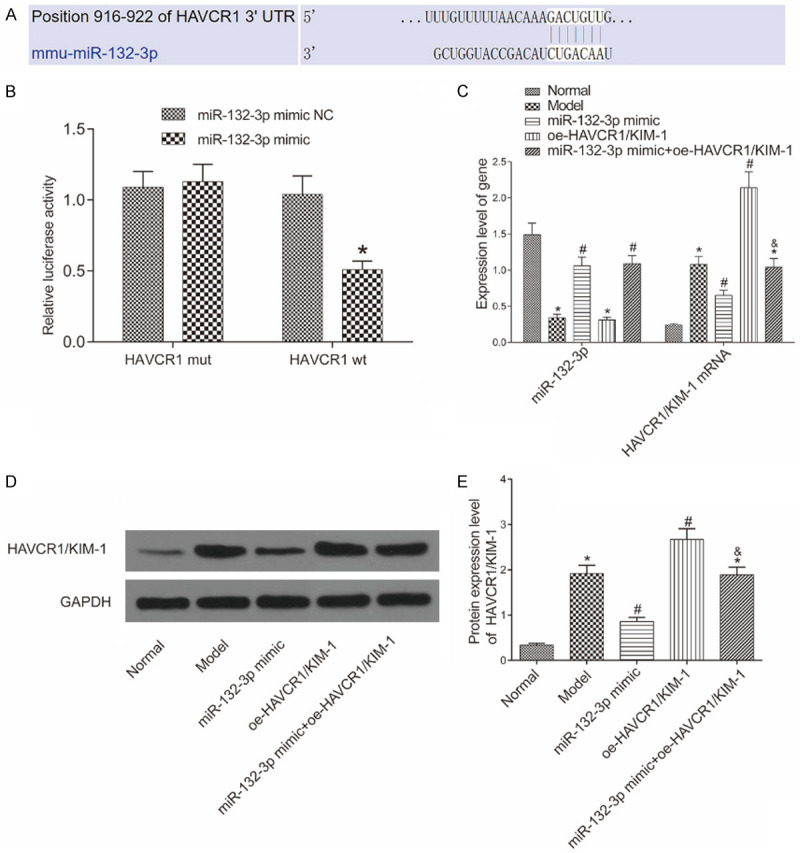
Relationship between miR-132-3p and HAVCR1/KIM-1. A: The pairing of miR-132-3p with HAVCR1/KIM-1 predicted by the bioinformatics website; B: miR-132-3p can target HAVCR1/KIM-1 as verified by dual-luciferase reporter assay. *P<0.05 vs. the miR-132-3p mimic NC group; C: The expression levels of miR-132-3p and HAVCR1/KIM-1 in the mice’ kidneys determined by qRT-PCR (n=6); D: Western blot image; E: The protein expression of HAVCR1/KIM-1 in each group (n=6). *P<0.05 vs. the normal group, #P<0.05 vs. the model group, &P<0.05 vs. the miR-132-3p mimic group.
Pathological changes in each group
To investigate the kidney injury in each group, we conducted H&E staining to evaluate the pathological changes in the mice’ kidneys (Figure 2). The kidney structure in the normal group was normal without any noticeable pathological change, whereas the other groups exhibited different degrees of edema and degeneration of renal tubular epithelial cell and noticeable inflammatory cell infiltration in renal interstitium. In comparison with the model group, the miR-132-3p mimic group had less severe edema and degeneration of the renal epithelial cells and a lower number of inflammatory cells in renal interstitium, whereas the oe-HAVCR1/KIM-1 group had more severe edema and necrosis of the epithelial cells and more noticeable inflammatory cell infiltration. No differences were observed in the renal pathological changes between the model group and the miR-132-3p mimic + oe-HAVCR1/KIM-1 group.
Figure 2.
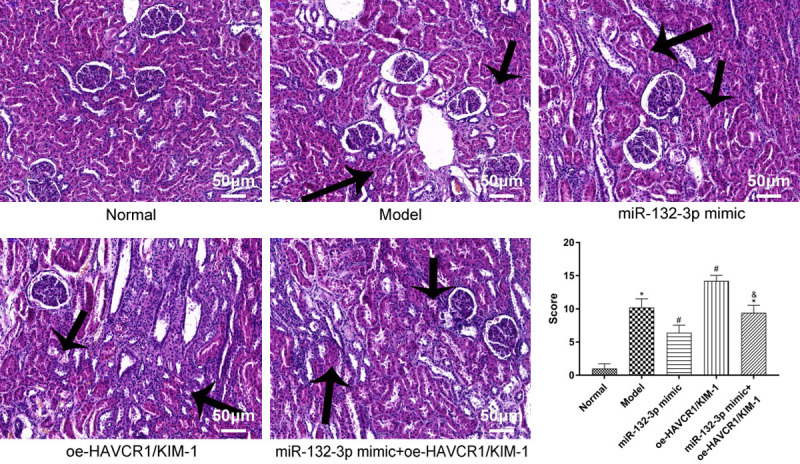
Pathological changes in mice’ renal tissues detected by HE stain (200×). HE: hematoxylin and eosin. Arrows indicate severe edema and degeneration of renal tubular epithelial cell and noticeable inflammatory cell infiltration in renal interstitium. *P<0.05 vs. the normal group, #P<0.05 vs. the model group, &P<0.05 vs. the miR-132-3p mimic group.
Changes in renal function in each group
Levels of renal function markers are presented in Figure 3. Unlike the normal group, the other groups had higher levels of BUN and SCr in the mice’ serum (all P<0.05). Compared with the model group, the levels of BUN and SCr were lower in the miR-132-3p mimic group and higher in the oe-HAVCR1/KIM-1 group (all P<0.05). Moreover, the miR-132-3p mimic + oe-HAVCR1/KIM-1 group had higher BUN and SCr content than the miR-132-3p mimic group (both P<0.05).
Figure 3.
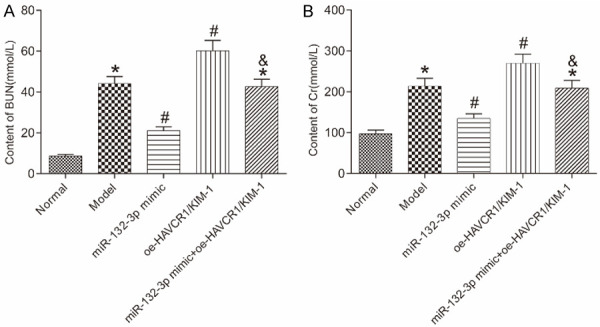
Levels of renal function markers in the mice’s serum in each group detected by ELISA. A: BUN content; B: SCr content. *P<0.05 vs. the normal group; #P<0.05 vs. the model group; &P<0.05 vs. the miR-132-3p mimic group. BUN: blood urea nitrogen; SCr: serum creatinine; ELISA: enzyme-linked immunosorbent assay.
Renal cell apoptosis in each group
One of the main manifestations of AKI is renal cell apoptosis. In this study, we carried out the TUNEL assay to examine the apoptosis in each group (Figure 4A, 4B). The results displayed that in comparison with the normal group, the other groups had higher apoptosis rates (all P<0.05). The renal cell apoptosis rate was lower in the miR-132-3p mimic group and higher in the oe-HAVCR1/KIM-1 group as compared to the model group (both P<0.05). The miR-132-3p mimic + oe-HAVCR1/KIM-1 group had a higher apoptosis rate than the miR-132-3p mimic group (P<0.05). Moreover, the protein expression levels of apoptosis-associated factors, cleaved caspase-3, Bax, and Bcl-2, in the renal tissues were examined by Western blot (Figure 4C, 4D). Compared with the normal group, the model group had higher protein expressions of cleaved caspase-3, Bax, and lower expression of Bcl-2 (all P<0.05). Compared with the model group, the miR-132-3p mimic group had lower protein expression levels of cleaved capsase-3 and Bax and higher Bcl-2 level, whereas the the oe-HAVCR1/KIM-1 group had higher protein expression levels of cleaved capsase-3 and Bax and lower Bcl-2 level (all P<0.05). The miR-132-3p mimic + oe-HAVCR1/KIM-1 group had higher protein expression levels of cleaved caspase-3, Bax and lower level of Bcl-2 than the miR-132-3p mimic group (all P<0.05).
Figure 4.
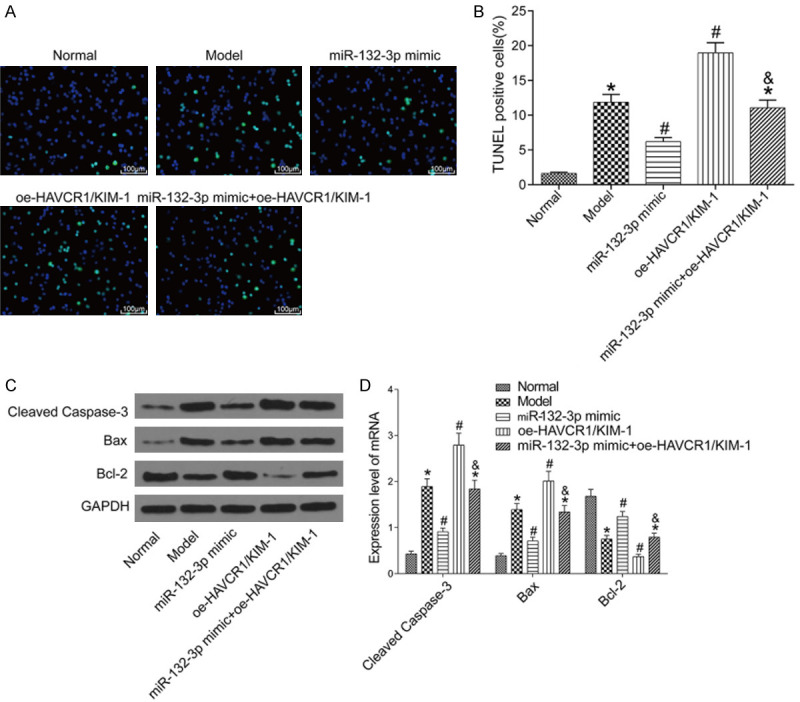
Renal cell apoptosis in each group. A: TUNEL stain (200×); B: Renal cell apoptosis rate in each group; C: Western blot image; D: The protein expressions of apoptosis-associated factors in the renal tissues of each group (n=6). *P<0.05 vs. the normal group; #P<0.05 vs. the model group; &P<0.05 vs. the miR-132-3p mimic group. TUNEL: terminal deoxynucleotidyl transferase-dUTP nick end labeling.
Levels of inflammatory factors in each group
ELISA was performed to measure the levels of inflammatory markers, TNF-α, IL-1β, and IL-6, in the mice’ serum in each group (Figure 5). Unlike the normal group, the other groups had higher levels of TNF-α, IL-1β, and IL-6 (all P<0.05). Compared with the model group, the levels of TNF-α, IL-1β, and IL-6 were lower in the miR-132-3p mimic group and higher in the oe-HAVCR1/KIM-1 group (all P<0.05). The miR-132-3p mimic + oe-HAVCR1/KIM-1 group had higher levels of TNF-α, IL-1β, and IL-6 than the miR-132-3p mimic group (all P<0.05).
Figure 5.
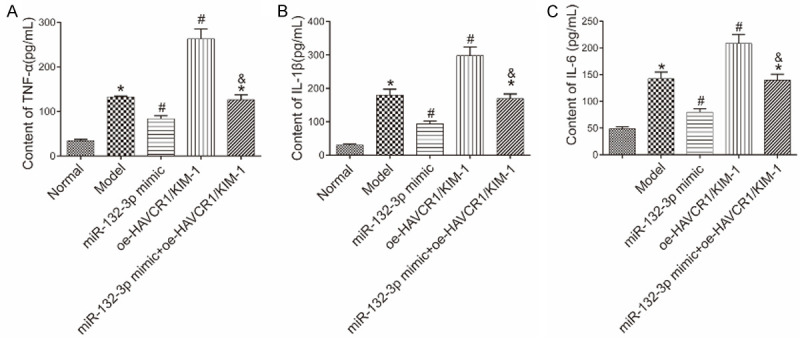
Levels of inflammatory factors in the mice’ serum in each group. A: TNF-α content; B: IL-1β content; C: IL-6 content. *P<0.05 vs. the normal group; #P<0.05 vs. the model group; &P<0.05 vs. the miR-132-3p mimic group. TNF: tumor necrosis factor; IL: interleukin.
Discussion
Sepsis can cause multiple organ injury in a short period of time, leading to organ failure and even death of patients [20]. AKI is one of the main complications of this disease. So far, there has been no effective method for treating sepsis-induced organ damage clinically [21].
The pathogenesis of sepsis-induced AKI remains unclear. It is believed that the major etiological factors include cell damage, worsening renal function, and inflammatory cell infiltration in renal interstitium [22-25]. Inflammatory cell infiltration is the major event for AKI at an early stage. The infiltration can be observed around blood capillaries of renal interstitium in patients with sepsis, and during this process, there is a marked increase in the levels of pro-inflammatory factors, TNF-α, IL-1β, and IL-6 [25,26]. The expression of these factors can act on renal cells to activate chemotactic factors and white blood cells, and the activated white blood cell can further adhere to the surface of the renal cells, penetrate the cells, and cause kidney damage [27,28]. Moreover, the development of inflammation can damage the renal cell function and cause cellular apoptosis, thus aggravating AKI [29,30]. In the present study, we created a mouse model of sepsis-induced AKI by injecting LPS and this model showed evident renal function damage, elevated renal cell apoptosis rate, and increased levels of TNF-α, IL-1β, and IL-6 in serum.
miRNA, a type of non-coding RNA, can regulate all kinds of life activities in mammals. In this study, a down-regulation of miR-132-3p expression was observed in the renal tissues of mice with sepsis-induced AKI, whereas miR-132-3p overexpression improved the renal function, inhibited the renal cell apoptosis, and lowered content of TNF-α, IL-1β, and IL-6 in the mice’ serum. Thus, we speculated that in the mouse model of sepsis-induced AKI, miR-132-3p may promote the expression of anti-apoptotic factor Bcl-2 via inhibition of the expressions of pro-apoptotic factors, cleaved caspase-3 and Bax, to reduce the renal cell apoptosis, and miR-132-3p may suppress the inflammatory progression through inhibiting the expressions of pro-inflammatory factors, thereby protecting the renal function of the mice and ameliorating the kidney injury in the septic mice. However, the link between HAVCR1/KIM-1 in the apoptotic signaling and the inflammatory pathway remains unclear, and other mechanisms may also be involved in this regulation. In this study, we only analyzed the levels of inflammatory markers TNF-α, IL-1β, and IL-6 in serum and didn’t investigate these levels in kidney tissues, thus more studies need to be carried out in the future for further verification.
Currently, there has been no study to elucidate the regulatory mechanism of miR-132-3p in sepsis-induced AKI and the investigation of this miRNA is now mainly in the area of oncology since it is found that miR-132-3p can regulate the apoptosis of tumor cells via regulating caspase-3 activity [31,32]. In chronic neuropathic pain, miR-132-3p can ameliorate nervous system diseases by inhibiting the inflammatory progression [33,34]. In this study, we found that miR-132-3p could regulate the expression of HAVCR1/KIM-1, a factor related to kidney injury, and the overexpression of HAVCR1/KIM-1 could block the repairing effect of miR-132-3p on sepsis-induced AKI mouse model. KIM-1, coded by the HAVCR1 gene, is a type I transmembrane glycoprotein [35]. Since HAVCR1/KIM-1 expression is very low in normal kidneys but is significantly elevated when the kidney is damaged, HAVCR1/KIM is now deemed as one of the main marker genes in kidney injury [36]. Some researchers believed that the persistent expression of HAVCR1/KIM-1 can cause the occurrence of chronic kidney disease. They have reported that HAVCR1/KIM-1 can regulate the inflammatory progression in the kidney by affecting the immunoregulation in the body and can further aggravate the kidney injury by damaging the nephron structure and function in mice [37,38]. Moreover, some studies have indicated that the persistent expression of HAVCR1/KIM-1 can activate the mTOR signaling pathway, leading to renal interstitial fibrosis and renal cell apoptosis [39,40]. Together, our findings support that miR-132-3p can inhibit the expression of downstream gene HAVCR1/KIM-1 to decrease renal cell apoptosis and inhibit inflammatory factor expression, thereby ameliorating kidney injury.
However, since miRNA regulates life activities via targeting multiple genes, whether miR-132-3p can regulate AKI through other pathways needs to be further investigated. In our study, the miR-132-3p mimic group had more edema renal tubular epithelial cells and inflammatory infiltration, but less severe degeneration or necrosis of renal tubular epithelial cells compared to the model group. The worsened edema and inflammatory infiltration may be due to that the intraperitoneal injection of the miR-132-3p cross-reacted with LPS, thus resulting in acute inflammatory and renal pathology in the mice. Moreover, bioinformatics database indicate that miR-132-3p can target casp6 and Bcl-2, thus the effect of miR-132-3p on apoptotic gene might be independent of the effect of HAVCR1/KIM-1 on apoptotic gene. The mechanism of the regulatory axis of HAVCR1/KIM-1/miR-132-3p and the apoptosis need to be further investigated.
In summary, we have shown that miR-132-3p can regulate sepsis-induced AKI in mice by targeting HAVCR1/KIM-1. The finding may help to further explain the pathogenesis of sepsis-induced AKI and provide some theoretical basis for the clinical treatment of this disease.
Disclosure of conflict of interest
None.
References
- 1.Hu QY, Ren JN, Ren HJ, Wu J, Wu XW, Liu S, Wang GF, Gu GS, Guo K, Li JS. Urinary mitochondrial DNA identifies renal dysfunction and mitochondrial damage in sepsis-induced acute kidney injury. Oxid Med Cell Longev. 2018;2018:8074936. doi: 10.1155/2018/8074936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho AY, Yoon HJ, Lee KY, Sun IO. Clinical characteristics of sepsis-induced acute kidney injury in patients undergoing continuous renal replacement therapy. Ren Fail. 2018;40:403–409. doi: 10.1080/0886022X.2018.1489288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheng B, Zhao L, Zang XF, Zhen J, Chen W. miR-375 ameliorates sepsis by downregulating miR-21 level via inhibiting JAK2-STAT3 signaling. Biomed Pharmacother. 2017;86:254–261. doi: 10.1016/j.biopha.2016.11.147. [DOI] [PubMed] [Google Scholar]
- 4.Li XH, Li YS, Shen K, Li HQ, Bai JW. The protective effect of ticagrelor on renal function in a mouse model of sepsis-induced acute kidney injury. Platelets. 2019;30:199–205. doi: 10.1080/09537104.2017.1392499. [DOI] [PubMed] [Google Scholar]
- 5.Holt BG, White JJ, Kuthiala A, Fall P, Szerlip HM. Sustained low-efficiency daily dialysis with hemofiltration for acute kidney injury in the presence of sepsis. Clin Nephrol. 2008;69:40–46. doi: 10.5414/cnp69040. [DOI] [PubMed] [Google Scholar]
- 6.Hu JF, Wang HX, Li HH, Hu J, Yu Y, Gao Q. Inhibition of ALDH2 expression aggravates renal injury in a rat sepsis syndrome model. Exp Ther Med. 2017;14:2249–2254. doi: 10.3892/etm.2017.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sunahara S, Watanabe E, Hatano M, Swanson PE, Oami T, Fujimura L, Teratake Y, Shimazui T, Lee C, Oda S. Influence of autophagy on acute kidney injury in a murine cecal ligation and puncture sepsis model. Sci Rep. 2018;8:1050. doi: 10.1038/s41598-018-19350-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai ZY, Sheng ZX, Yao H. Pachymic acid ameliorates sepsis-induced acute kidney injury by suppressing inflammation and activating the Nrf2/HO-1 pathway in rats. Eur Rev Med Pharmacol Sci. 2017;21:1924–1931. [PubMed] [Google Scholar]
- 9.Matejovic M, Valesova L, Benes J, Sykora R, Hrstka R, Chvojka J. Molecular differences in susceptibility of the kidney to sepsis-induced kidney injury. BMC Nephrol. 2017;18:183. doi: 10.1186/s12882-017-0602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Qiu JL, Chen B, Lin YP, Chen YL, Xie GJ, Qiu JM, Tong HS, Jiang DX. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-kappaB pathway. Int Immunopharmacol. 2018;59:252–260. doi: 10.1016/j.intimp.2018.03.023. [DOI] [PubMed] [Google Scholar]
- 11.Pei Z, Deng S, Xie D, Lv M, Guo W, Liu D, Zheng Z, Long X. Protective role of fenofibrate in sepsis-induced acute kidney injury in BALB/c mice. RSC Adv. 2018;8:28510–28517. doi: 10.1039/c8ra00488a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandes JG, Borrego A, Jensen JR, Cabrera WHK, Correa MA, Starobinas N, Ribeiro OG, Ibañez OM, De Franco M. miRNA expression and interaction with genes involved in susceptibility to pristane-induced arthritis. J Immunol Res. 2018;2018:1928405. doi: 10.1155/2018/1928405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majer A, Medina SJ, Niu YL, Abrenica B, Manguiat KJ, Frost KL, Philipson CS, Sorensen DL, Booth SA. Early mechanisms of pathobiology are revealed by transcriptional temporal dynamics in hippocampal ca1 neurons of prion infected mice. PLoS Pathog. 2012;8:e1003002. doi: 10.1371/journal.ppat.1003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doi K, Hu X, Yuen PS, Leelahavanichkul A, Yasuda H, Kim SM, Schnermann J, Jonassen TE, Frøkiaer J, Nielsen S, Star RA. AP214, an analogue of alpha-melanocyte-stimulating hormone, ameliorates sepsis-induced acute kidney injury and mortality. Kidney Int. 2008;73:1266–1274. doi: 10.1038/ki.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopes JA, Fernandes P, Jorge S, Resina C, Santos C, Pereira A, Neves J, Antunes F, Gomes da Costa A. Long-term risk of mortality after acute kidney injury in patients with sepsis: a contemporary analysis. BMC Nephrol. 2010;11:9. doi: 10.1186/1471-2369-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu YX, Wang H, Sun RH, Ni Y, Ma LL, Xv F, Hu XP, Jiang LZ, Wu AP, Chen X, Chen MH, Liu JQ, Han F. Urinary netrin-1 and KIM-1 as early biomarkers for septic acute kidney injury. Ren Fail. 2014;36:1559–1563. doi: 10.3109/0886022X.2014.949764. [DOI] [PubMed] [Google Scholar]
- 18.Zhao XH, Liu BH, Zhang W, Zhang L, Ji GH. Early diagnostic value of serum procalcitonin combined with kidney injury molecule-1 for children sepsis complicated with acute kidney injury. Doctor. 2017;2:53–54. 66. [Google Scholar]
- 19.Dong W, Li ZL, Chen YH, Zhang L, Ye ZM, Liang HB, Li RZ, Xu LX, Zhang B, Liu SX, Wang WD, Li CL, Shi W, Liang XL. Necrostatin-1 attenuates sepsis-associated acute kidney injury by promoting autophagosome elimination in renal tubular epithelial cells. Mol Med Rep. 2018;17:3194–3199. doi: 10.3892/mmr.2017.8214. [DOI] [PubMed] [Google Scholar]
- 20.Wieczorek A, Lipińska-Gediga M, Siebert A, Patrza D. Results of kidney transplant from donors with sepsis-induced acute kidney injury. Transplantation. 2017;101:S63. [Google Scholar]
- 21.Lin L, Huang L, School G, University GM, Nephrology D. Starting time of continuous renal replacement therapy based on the serum cystatin C level in patients with sepsis-induced acute kidney injury. Chin Gen Pract. 2017 [Google Scholar]
- 22.Yasuda H, Sekine K, Abe T, Suzaki S, Katsumi A, Harada N, Higashi H, Kishihara Y, Suzuki H, Takebayashi T. Comparison of two polysulfone membranes for continuous renal replacement therapy for sepsis: a prospective cross-over study. Renal Replacement Therapy. 2018;4:6. [Google Scholar]
- 23.Hinkelbein J, Böhm L, Braunecker S, Adler C, Robertis ED, Cirillo F. Decreased tissue COX5B expression and mitochondrial dysfunction during sepsis-induced kidney injury in rats. Oxid Med Cell Longev. 2017;2017:8498510. doi: 10.1155/2017/8498510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronco C, Bellomo R, Kellum JA. Acute kidney injury in sepsis. Intensive Care Med. 2017;43:1–13. doi: 10.1007/s00134-017-4755-7. [DOI] [PubMed] [Google Scholar]
- 25.Ge QM, Huang CM, Zhu XY, Bian F, Pan SM. Differentially expressed miRNAs in sepsis-induced acute kidney injury target oxidative stress and mitochondrial dysfunction pathways. PLoS One. 2017;12:e0173292. doi: 10.1371/journal.pone.0173292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fani F, Regolisti G, Delsante M, Cantaluppi V, Castellano G, Gesualdo L, Villa G, Fiaccadori E. Recent advances in the pathogenetic mechanisms of sepsis-associated acute kidney injury. J Nephrol. 2018;31:351–359. doi: 10.1007/s40620-017-0452-4. [DOI] [PubMed] [Google Scholar]
- 27.Nusshag C, Weigand MA, Zeier M, Morath C, Brenner T. Issues of acute kidney injury staging and management in sepsis and critical illness: a narrative review. Int J Mol Sci. 2017;18:1387. doi: 10.3390/ijms18071387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin YX, Zhang HL, Yu BL, Shi SH. Effects of Xuebijing injection for patients with sepsis-induced acute kidney injury after wenchuan earthquake. Altern Ther Health Med. 2017;23:36–42. [PubMed] [Google Scholar]
- 29.Liu C, Hu J, Mao Z, Kang HJ, Liu H, Fu WL, Lv YF, Zhou FH. Acute kidney injury and inflammatory response of sepsis following cecal ligation and puncture in d-galactose-induced aging rats. Clin Interv Aging. 2017;12:593–602. doi: 10.2147/CIA.S132277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valley TS, Nallamothu BK, Heung M, Iwashyna TJ, Cooke CR. Hospital variation in renal replacement therapy for sepsis in the united states. Crit Care Med. 2018;46:e158–e165. doi: 10.1097/CCM.0000000000002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qu XC, Chen ZQ, Fan DW, Sun CG, Zeng Y. MiR-132-3p regulates the osteogenic differentiation of thoracic ligamentum flavum cells by inhibiting multiple osteogenesis-related genes. Int J Mol Sci. 2016;17:1370. doi: 10.3390/ijms17081370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li G, Liu KY, Du XH. Long non-coding RNA TUG1 promotes proliferation and inhibits apoptosis of osteosarcoma cells by sponging mir-132-3p and upregulating sox4 expression. Yonsei Med J. 2018;59:226–235. doi: 10.3349/ymj.2018.59.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu LP, Liao Q, Chen XG, Xu L, Zeng X, Lv ZY, Sun X, Zhen HQ, Wu ZD. Dynamic expression of miR-132, miR-212, and miR-146 in the brain of different hosts infected with angiostrongylus cantonensis. Parasitol Res. 2014;113:91–99. doi: 10.1007/s00436-013-3630-x. [DOI] [PubMed] [Google Scholar]
- 34.Leinders M, Üçeyler N, Pritchard RA, Sommer C, Sorkin LS. Increased miR-132-3p expression is associated with chronic neuropathic pain. Exp Neurol. 2016;283:276–286. doi: 10.1016/j.expneurol.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun IO, Shin SH, Cho AY, Yoon HJ, Chang MY, Lee KY. Clinical significance of NGAL and KIM-1 for acute kidney injury in patients with scrub typhus. PLoS One. 2017;12:e0175890. doi: 10.1371/journal.pone.0175890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goli F, Karimi J, Khodadadi I, Tayebinia H, Kheiripour N, Hashemnia M, Rahimi R. Silymarin attenuates ELMO-1 and KIM-1 expression and oxidative stress in the kidney of rats with type 2 diabetes. Indian J Clin Biochem. 2019;34:172–179. doi: 10.1007/s12291-018-0735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JY, Ismail OZ, Zhang XZ, Haig A, Lian DM, Gunaratnam L. Donor kidney injury molecule-1 promotes graft recovery by regulating systemic necroinflammation. Am J Transplant. 2018;18:2021–2028. doi: 10.1111/ajt.14745. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Li Q, Fang M, Ma Y, Liu N, Yan X, Zhou J, Li F. The kidney injury induced by short-term PM2.5 exposure and the prophylactic treatment of essential oils in BALB/c mice. Oxid Med Cell Longev. 2018;2018:9098627. doi: 10.1155/2018/9098627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim JY, Erikson DW, Burghardt RC, Spencer TE, Wu GY, Bayless KJ, Johnson GA, Bazer FW. Secreted phosphoprotein 1 binds integrins to initiate multiple cell signaling pathways, including FRAP1/mTOR, to support attachment and force-generated migration of trophectoderm cells. Matrix Biol. 2010;29:369–382. doi: 10.1016/j.matbio.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Sivertsson E, Friederich-Persson M, Öberg CM, Fasching A, Hansell P, Rippe B, Palm F. Inhibition of mammalian target of rapamycin decreases intrarenal oxygen availability and alters glomerular permeability. Am J Physiol Renal Physiol. 2018;314:F864–F872. doi: 10.1152/ajprenal.00033.2017. [DOI] [PubMed] [Google Scholar]


