Abstract
Purpose: This study was designed to investigate the effect of micro RNA-targeted regulation of FGFR1 on the proliferation and apoptosis of triple-negative breast cancer (TNBC) cells. Methods: TNBC (MAD-MB-231), three types of breast cancer (MCF10A, MCF7, ZR751) cell lines, and normal breast tissue cell lines were extracted. Real-time PCR was used to detect the expression of miRNA-136 in different types of breast cells. The MAD-MB-231 cell lines were transfected with miRNA-136 mimic by lipofection. The effects of miRNA-136 on FGFR1 expression and apoptosis rate of MAD-MB-231 cell lines were determined using western blotting. Results: miRNA-136 expression in TNBC cells was lower than that of controls, and was negatively correlated with TNM staging. miRNA-136 expression in MCF10A, MCF7, ZR751, and MAD-MB-231 cell lines was gradually decreased, and MCF10A expression in the other three cell lines was significantly higher than that of MAD-MB-231 cell line (P<0.05). Transfection with miRNA-136 significantly reduced the proliferation rate of MAD-MB-231, and a higher concentration and longer duration exhibited a more pronounced inhibitory effect on proliferation (P<0.05). Transfection with miRNA-136 significantly reduced FGFR1 expression in the MAD-MB-231 cell lines, without significantly affecting apoptosis. Conclusion: miRNA-136 shows a very low expression level in TNBC cells. Transfection with miRNA-136 can significantly inhibit the proliferation of TNBC cells by external transfection, and has little effect on cell apoptosis. This may be related to miRNA-mediated changes in FGFR1 protein expression.
Keywords: Micro RNA, FGFR1, triple-negative breast cancer, cell proliferation and apoptosis, impact analysis
Introduction
Breast cancer is the most common non-skin cancer in women. The number of new cases worldwide reached 1.38 million in 2013, accounting for about 23% of cancers in women, and 460,000 people died of breast cancer that year, accounting for 14% of all cancer deaths in women [1,2]. In China, breast cancer has been ranked first among female non-skin cancer for the past 20 years [3]. Triple-negative breast cancer is the most aggressive type of breast cancer with a heterogeneous nature, and is clinically manifested as no expression of estrogen receptor, progesterone receptor, or human epidermal growth factor receptor 2 and the malignant cells are extremely aggressive, and patients often have poor prognosis [4]. Triple-negative breast cancer (TNBC) most frequently occurs in women under 40 years of age, and both progression-free survival and overall survival rate are low, with the 5-year survival of only 77%, which decreases to 14% if it progresses to an advanced stage [5]. Early diagnosis and prevention of distant metastases are crucial prerequisites for improving the prognosis of TNBC.
MicroRNA (microRNA or miRNA) is a non-coding RNA, about 22nt in length, widely found in viruses and humans. miRNA can combine with mRNA, block the expression of protein-coding genes, and then regulate the biological processes of the organism by inhibiting the synthesis of specific proteins [6]. Some evidence indicates that miRNAs influence the progression of breast cancer in a variety of pathways, including inhibition of estrogen receptors, proliferation of breast cancer stem cells, and regulation of expression of tumor-associated proteins [7]. Overexpression of miR-21 exists in breast, colon, and lung cancers, and miR-21 may promote the development of breast cancer [8]. miRNA expression may also play an important role in prognosis and treatment of breast cancer [9]. A study on the prognosis of 219 breast cancer patients indicated that miR-210 was associated with the HIF-1α signaling pathway, and that miR-210 could be an independent risk factor for prognosis of breast cancer [10].
This study aimed to investigate the effects of miRNA-136 on the proliferation and apoptosis of TNBC cells through targeted regulation of FGFR1 expression, so as to lay a theoretical foundation for improving the prognosis and survival of patients with TNBC.
Materials and methods
Clinical data
Tumor tissues were obtained from 40 patients with TNBC, and distal paracancerous tissues at more than 5 cm from the tumor edge were collected as controls, from January 2017 to January 2019 in our hospital. This study was approved by ethics committee of the Affiliated Hospital of Liaoning University of Traditional Chinese Medicine. All patients signed informed consent. The clinical data of enrolled breast cancer patients are listed in Table 1.
Table 1.
Clinical data
| Baseline data | ||
|---|---|---|
| Average age | 39.19±2.22 | |
| Tumor size | ≤2 cm | 13 |
| >2 cm | 27 | |
| TNM Stage | I | 10 |
| II | 14 | |
| III | 10 | |
| IV | 6 | |
Cell lines and grouping
Breast cancer cell lines MCF10A, MCF7, and ZR751, were purchased from Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, and used as the control group, and the TNBC cell line MDA-MB-231 was selected as the study cell line. All cell lines were cultured in DMEM/F12 medium at 37°C, 5% CO2, and cells in the logarithmic growth were used for model establishment.
RNA extraction and measurement
MCF10A, MCF7, ZR751, and MDA-MB-231 cell lines in the logarithmic growth phase were washed with D-Hanks and treated with lysing solution. Cells were pipetted up and down until fully lysed. After centrifugation, the supernatant was removed, followed by another centrifugation with isopropanol. The white RNA deposited in the bottom of the tube was measured for RNA concentration using a spectrophotometer.
miRNA transfection
The TNBC cell line MDA-MB-231, which was in a logarithmic phase, was divided into five groups, namely a blank control group (cells without transfection), a negative control group (transfected with a blank plasmid), a miRNA-136 mimic group (transfected with an overexpression of miRNA-136), a miRNA-136 inhibitor group (transfected with a miRNA-136 inhibitor), and miRNA-136 inhibitor + siFGFR1 group (transfection of miRNA-136 and inactivation of the FGFR1 gene); and the effect of miRNA-136 transfection on miRNA-136 as well as cell viability was assessed.
Protein expression
The expression of FGFR1 and caspase-1 protein was detected by western blotting using BCA kit (Shanghai Ruiji Biotechnology Development Co., Ltd.). The specific procedure was as follows: A sample of 30 μg was selected for electrophoresis using 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The protein was transferred to polyvinylidene fluoride (PVDF) membrane and sealed with 5% BSA at room temperature for 1 h. Rabbit anti-human antibody of 1:1500 was added and kept overnight at 4°C, followed by rinsing the membrane with TBST 3 times. ECL luminescent agent was added, and X-ray film was used for exposure and development. The operation was strictly in accordance with kit instructions. Each index was measured three times, and the average value was taken as the final result.
Apoptosis rate
Flow cytometry was used to detect the apoptosis rate of the five cell groups. The specific formula is cell survival rate = (OD value of study group - OD value of blank control group)/OD value of blank control group.
Statistical methods
The collected data were entered into SPSS22.0 software. The measured data were expressed as (x̅ ± s), and comparisons between groups and within groups were performed by independent samples t-test. Counted data were expressed as [n (%)]. Comparisons between groups and within groups were performed by chi-square test. The ANVOA test was used for the comparison of multiple time-points within groups. P<0.05 indicated a significant difference [11].
Results
Differences in miRNA-136 expression levels in TNBC and paracancerous tissues
The level of miRNA-136 in the MDA-MB-231 cell lines was significantly lower than that of the paracancerous tissues (P<0.05). The MDA-MB-231 cell lines were categorized as Stage I, Stage II and Stage III according to the TNM staging. With an increase of TNM staging, the expression level of miRNA-136 showed a gradually decreasing trend. The expression of miRNA-136 in Phase III cell lines was significantly lower than that of Phase I cell lines (P<0.05) (Figure 1).
Figure 1.
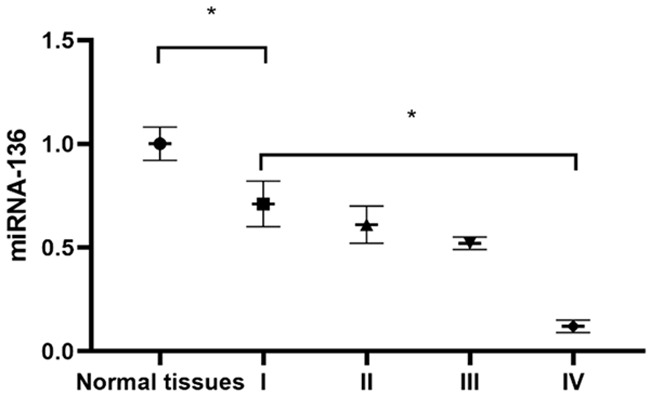
miRNA-136 expression level in TNBC and paracancerous tissues. *P<0.05.
Differences in miRNA-136 expression levels in breast cancer tissues
MCF10A cell lines had the highest miRNA-136 expression level, followed by MCF7, ZR751, and MAD-MB-231. The expression of miRNA-136 in the MCF10A cell lines was significantly higher than that of the MAD-MB-231 cell line (P<0.05) (Figure 2).
Figure 2.
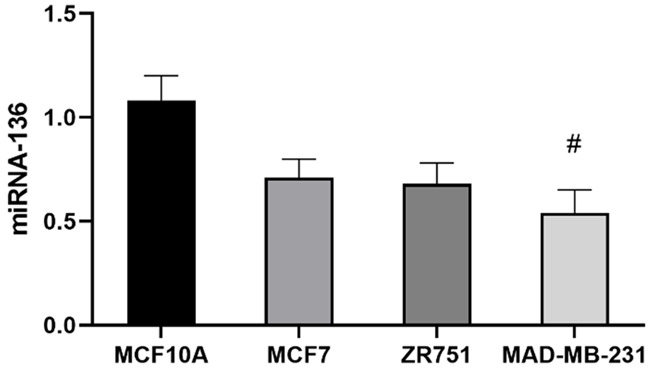
Differences in miRNA-136 expression levels in different breast cancer tissues. #P<0.05 compared with the MCF10A cell line.
Differences in miRNA-136 expression after transfection of MAD-MB-231 cell lines
The miRNA-136 levels exhibited no significant difference among the blank control, negative control, miRNA-136 mimic, miRNA-136 inhibitor, and miRNA-136 inhibitor + siFGFR1 groups (P>0.05). miRNA-136 expression in the miRNA-136 mimic group was significantly higher than the rest of the groups (P<0.05) (Figure 3).
Figure 3.
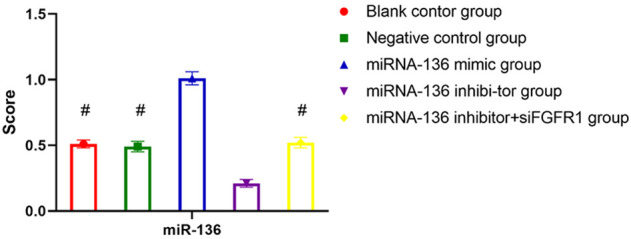
Differences in miRNA-136 expression after transfection of the MAD-MB-231 cell lines. #P<0.05 compared with the miR-136 mimic group.
Effect of miRNA-136 transfection on proliferative activity of MAD-MB-231 cell lines
After digestion and passaging, the well-grown MAD-MB-231 cell lines were inoculated in culture plates at 37°C and 5% CO2 for 12 h, and were treated with 0.1 mM, 0.5 mM, 1.0 mM and 2.0 mM of miRNA-136, respectively. The results showed that different concentrations of miRNA-136 led to different survival rates. Overall, with an increase of miRNA-136 concentration and the extension of the intervention time, the survival rate of MAD-MB-231 cells was decreased accordingly, showing a negative correlation. A higher concentration of miRNA-136 indicated greater damage to the cells (Figure 4).
Figure 4.
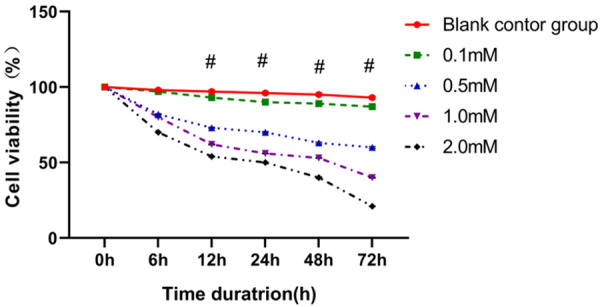
Effect of miRNA-136 transfection on the proliferative activity of the MAD-MB-231 cell lines. Different concentrations of miRNA-136 and different intervention times significantly affected the proliferative activity of MAD-MB-231 cell lines.
Effect of miRNA-136 transfection on protein expression of MAD-MB-231 cell lines
At 12 h after transfection, FGFR1 levels showed no significant difference among blank control, negative control, and miRNA-136 inhibitor + siFGFR1 groups (P>0.05). FGFR1 expression in the miRNA-136 mimic group was significantly lower than that of other groups (P<0.05) (Figures 5 and 6).
Figure 5.
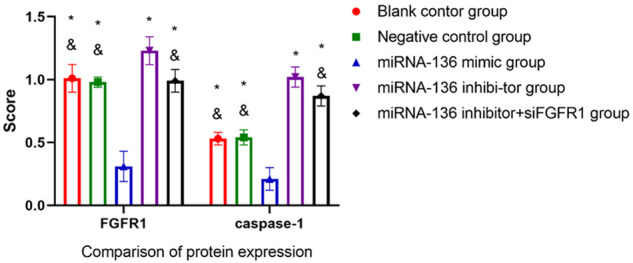
Effect of miRNA-136 transfection on protein expression in MAD-MB-231 cell lines. *P<0.05 compared to the miRNA-136 mimic group; #P<0.05 compared to the miRNA-136 inhibitor group.
Figure 6.
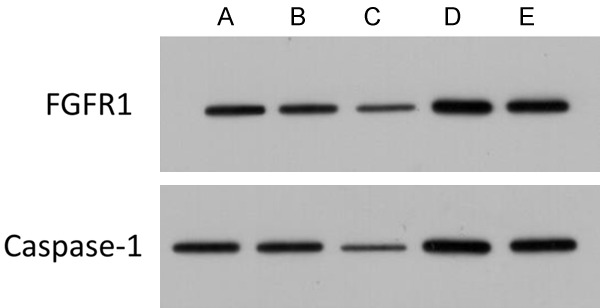
Effect of miRNA-136 transfection on protein expression of MAD-MB-231 cell lines. A: Blank control group; B: Negative control group; C: miRNA-136 mimic group; D: miRNA-136 inhibitor group; E: miRNA-136 inhibitor + siFGFR1 group.
Effect of miRNA-136 transfection on apoptosis rate of MAD-MB-231 cell lines
Transfection with miRNA-136 did not significantly affect the apoptosis rate of MAD-MB-231 cell lines (P>0.05) (Figure 7).
Figure 7.
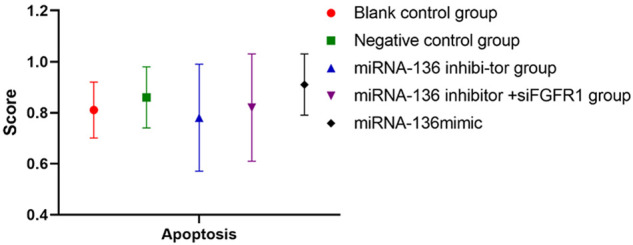
Effect of miRNA-136 transfection on apoptosis of MAD-MB-231 cell lines. Transfection with miRNA-136 produced some changes in the apoptosis rate of MAD-MB-231 cells, but the differences between groups were not significant (P>0.05).
Discussion
TNBC is a special subtype of breast cancer, negative for estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 [12]. The prevalence of TNBC varies by region and ethnicity. However, it generally accounts for about 10%-20% of all breast cancers [13]. TNBC is most commonly diagnosed in women under 45 years of age, and is characterized by high malignancy, young onset, strong aggressiveness, and rapid progression [14].
MicroRNAs are single-stranded RNAs with powerful regulatory abilities to control gene expression. miRNAs affect the expression of specific proteins by targeting the expression of RNAs, thus regulating metabolic function [15]. The role of miRNAs in malignant tumors has been gradually recognized. A study found that miRNAs can negatively regulate protein expression through specific binding to certain untranslated regions, thus affecting the proliferation and metastasis of rectal cancer cells, and it is believed that miRNA levels can be used to assess the condition of patients and predict prognosis [16]. Research on gastric cancer has found that specific miRNAs play a vital role in cell proliferation, differentiation, apoptosis, gene regulation and even tumorigenesis. Different clinical outcomes showed significant differences in miRNA expression between gastric cancer patients, suggesting the potential of miRNA in prognostic assessment of patients with gastric cancer [17].
In this study, we investigated the effect of miRNA on the proliferation and apoptosis of TNBC cells, and initially investigated the regulatory mechanism. Results showed that the expression of miRNA-136 was lower in TNBC tissues compared to paracancerous tissues. It has been noted that miRNAs can promote or inhibit the expression of target genes by specifically recognizing and binding target gene mRNAs [18]. miRNA-136 is a new miRNA discovered in recent years, but there are few studies on the expression and mechanism of miRNA-136 with regard to TNBC [19]. Results obtained in this study indicated that miRNA-136 was a tumor suppressor miRNA, and a decrease of miRNA-136 expression with higher TNM staging also confirmed our speculation. There were also some differences in miRNA-136 expression among MCF10A, MCF7, ZR751, and MAD-MB-231 cell lines, and it has been noted that MCF10A is a minimally-invasive breast cancer line, while MAD-MB-231 is a cell line with high invasiveness [20,21], suggesting that a higher invasiveness correlates with lower miRNA-136 expression. This is also evidenced by the lowest miRNA-136 level being in tissues with TNM stage IV. However, the mechanism remains to be further validated.
There was no sigificant difference in miR-136 levels among blank control, negative control, and miR-136 inhibitor + siFGFR1 groups, and miR-136 expression in the miR-136 mimic group was significantly higher than that of other groups. It was found that different transfection media resulted in differences in the expression of FGFR1, a gene that has been widely demonstrated to be associated with malignant tumors [22,23]. A study of endometrial cancer found significant differences in FGFR1 expression in three types of tissues: normal endometrium, atypical hyperplastic endometrium, and endometrial cancer, with FGFR1 expression significantly higher in endometrial cancer. FGFR1 expression was also positively correlated with the differentiation of endometrial cancer [24,25]. Caspase-1 is a regulatory protein in tumor development, and the results of an animal study on breast cancer showed that caspase-1 could exert inhibitory effects on tumor by regulating the development of specific cells in peripheral tissues, suggesting that caspase-1 may be a target for cancer therapy [26]. The expression differences of miRNA-136 and FGFR1 in different transfected cell lines suggest that FGFR1 is a target of miRNA-136 and can be used to regulate the proliferation of TNBC cell lines. This is also confirmed by the changes in the survival rate of MAD-MB-231 cells under different concentrations of miRNA-136 and durations of intervention. miRNA-136 had no significant effect on the apoptotic process of the TNBC cell line, which was different from the findings of other studies and may be related to the short intervention time.
In summary, miRNA-136 showed a markedly low expression level in TNBC. miRNA-136 significantly inhibited the proliferation of TNBC cells by external transfection, but had no significant effect on apoptosis, and the mechanism may be related to the possible involvement of miRNA in the regulation of FGFR1 protein expression. The limitation of this study is that only one miRNA was explored, thus future studies are needed.
Disclosure of conflict of interest
None.
References
- 1.Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zangouri VM, Akrami MM, Tahmasebi SM, Talei AM, Ghaeini Hesarooeih AM. Medullary breast carcinoma and invasive ductal carcinoma: a review study. Iran J Med Sci. 2018;43:365–371. [PMC free article] [PubMed] [Google Scholar]
- 3.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, Pasetto A, Langhan M, Shelton T, Prickett T, Gartner J, Jia L, Trebska-McGowan K, Somerville RP, Robbins PF, Rosenberg SA, Goff SL, Feldman SA. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med. 2018;24:724–730. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charmet R, Duffy S, Keshavarzi S, Gyorgy B, Marre M, Rossing P, McKnight AJ, Maxwell AP, Ahluwalia TVS, Paterson AD, Trégouët DA, Hadjadj S. Novel risk genes identified in a genome-wide association study for coronary artery disease in patients with type 1 diabetes. Cardiovasc Diabetol. 2018;17:61. doi: 10.1186/s12933-018-0705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rados DV, Pinto LC, Leitão CB, Gross JL. Screening for coronary artery disease in patients with type 2 diabetes: a meta-analysis and trial sequential analysis. BMJ Open. 2017;7:e015089. doi: 10.1136/bmjopen-2016-015089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morbach C, Wagner M, Güntner S, Malsch C, Oezkur M, Wood D, Kotseva K, Leyh R, Ertl G, Karmann W, Heuschmann PU, Störk S. Heart failure in patients with coronary heart disease: prevalence, characteristics and guideline implementation - results from the German EuroAspire IV cohort. BMC Cardiovasc Disord. 2017;17:108. doi: 10.1186/s12872-017-0543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgeois-Daigneault MC, Roy DG, Aitken AS, El Sayes N, Martin NT, Varette O, Falls T, St-Germain LE, Pelin A, Lichty BD, Stojdl DF, Ungerechts G, Diallo JS, Bell JC. Neoadjuvant oncolytic virotherapy before surgery sensitizes triple-negative breast cancer to immune checkpoint therapy. Sci Transl Med. 2018;10:eaao1641. doi: 10.1126/scitranslmed.aao1641. [DOI] [PubMed] [Google Scholar]
- 8.Feng A, Yuan X, Li X. MicroRNA-345 inhibits metastasis and epithelial-mesenchymal transition of gastric cancer by targeting FOXQ1. Oncol Rep. 2017;38:2752–2760. doi: 10.3892/or.2017.6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naga Prasad SV, Gupta MK, Duan ZH, Surampudi VS, Liu CG, Kotwal A, Moravec CS, Starling RC, Perez DM, Sen S, Wu Q, Plow EF, Karnik S. A unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. PLoS One. 2017;12:e0170456. doi: 10.1371/journal.pone.0170456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuzaki K, Fujita K, Jingushi K, Kawashima A, Ujike T, Nagahara A, Ueda Y, Tanigawa G, Yoshioka I, Ueda K, Hanayama R, Uemura M, Miyagawa Y, Tsujikawa K, Nonomura N. MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget. 2017;8:24668–24678. doi: 10.18632/oncotarget.14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park S, Eom K, Kim J, Bang H, Wang HY, Ahn S, Kim G, Jang H, Kim S, Lee D, Park KH, Lee H. MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer. 2017;17:658. doi: 10.1186/s12885-017-3642-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi B, Deng W, Long X, Zhao R, Wang Y, Chen W, Xu G, Sheng J, Wang D, Cao S. miR-21 increases c-kit(+) cardiac stem cell proliferation in vitro through PTEN/PI3K/Akt signaling. PeerJ. 2017;5:e2859. doi: 10.7717/peerj.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Gao Y, Duan L, Wei S, Liu J, Tian L, Quan J, Zhang Q, Liu J, Yang J. Metformin ameliorates skeletal muscle insulin resistance by inhibiting miR-21 expression in a high-fat dietary rat model. Oncotarget. 2017;8:98029–98039. doi: 10.18632/oncotarget.20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z, He Y, Li D, Fang X, Shang T, Zhang H, Zheng X. Long noncoding RNA MEG3 suppressed endothelial cell proliferation and migration through regulating miR-21. Am J Transl Res. 2017;9:3326–3335. [PMC free article] [PubMed] [Google Scholar]
- 15.Li WJ, Yin RX, Cao XL, Chen WX, Huang F, Wu JZ. DOCK7-ANGPTL3 SNPs and their haplotypes with serum lipid levels and the risk of coronary artery disease and ischemic stroke. Lipids Health Dis. 2018;17:30. doi: 10.1186/s12944-018-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burzotta F, Lassen JF, Banning AP, Lefèvre T, Hildick-Smith D, Chieffo A, Darremont O, Pan M, Chatzizisis YS, Albiero R, Louvard Y, Stankovic G. Percutaneous coronary intervention in left main coronary artery disease: the 13th consensus document from the European Bifurcation Club. EuroIntervention. 2018;14:112–120. doi: 10.4244/EIJ-D-18-00357. [DOI] [PubMed] [Google Scholar]
- 17.Bechthold A, Boeing H, Schwedhelm C, Hoffmann G, Knüppel S, Iqbal K, De Henauw S, Michels N, Devleesschauwer B, Schlesinger S, Schwingshackl L. Food groups and risk of coronary heart disease, stroke and heart failure: a systematic review and dose-response meta-analysis of prospective studies. Crit Rev Food Sci Nutr. 2019;59:1071–1090. doi: 10.1080/10408398.2017.1392288. [DOI] [PubMed] [Google Scholar]
- 18.Kwong JC, Schwartz KL, Campitelli MA, Chung H, Crowcroft NS, Karnauchow T, Katz K, Ko DT, McGeer AJ, McNally D, Richardson DC, Rosella LC, Simor A, Smieja M, Zahariadis G, Gubbay JB. Acute myocardial infarction after laboratory-confirmed influenza infection. N Engl J Med. 2018;378:345–353. doi: 10.1056/NEJMoa1702090. [DOI] [PubMed] [Google Scholar]
- 19.Chen Q, Qiu F, Zhou K, Matlock HG, Takahashi Y, Rajala RVS, Yang Y, Moran E, Ma JX. Pathogenic role of microRNA-21 in diabetic retinopathy through downregulation of PPARα. Diabetes. 2017;66:1671–1682. doi: 10.2337/db16-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo L, Xu K, Yan H, Feng H, Wang T, Chai L, Xu G. MicroRNA expression signature and the therapeutic effect of the microRNA-21 antagomir in hypertrophic scarring. Mol Med Rep. 2017;15:1211–1221. doi: 10.3892/mmr.2017.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J, Kuang TH, Chen J, Yang JW, Liu YX. The diagnostic and prognostic values of microRNA-21 in patients with gastric cancer: a meta-analysis. Eur Rev Med Pharmacol Sci. 2017;21:120–130. [PubMed] [Google Scholar]
- 22.Wen Q, Liu Y, Lyu H, Xu X, Wu Q, Liu N, Yin Q, Li J, Sheng X. Long noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int J Gynecol Cancer. 2017;27:1096–1108. doi: 10.1097/IGC.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Wang J, Ma H, Xiao Z, Dong X. MicroRNA-21 inhibits mitochondria-mediated apoptosis in keloid. Oncotarget. 2017;8:92914–92925. doi: 10.18632/oncotarget.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Cai Z, Hong G, Zheng G, Huang Y, Zhang S, Dai J. MicroRNA21 increases cell viability and suppresses cellular apoptosis in nonsmall cell lung cancer by regulating the PI3K/Akt signaling pathway. Mol Med Rep. 2017;16:6506–6511. doi: 10.3892/mmr.2017.7440. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Chen J, Gu Y, Shen W. MicroRNA-21 functions as an oncogene and promotes cell proliferation and invasion via TIMP3 in renal cancer. Eur Rev Med Pharmacol Sci. 2017;21:4566–4576. [PubMed] [Google Scholar]
- 26.Tseng HH, Tseng YK, You JJ, Kang BH, Wang TH, Yang CM, Chen HC, Liou HH, Liu PF, Ger LP, Tsai KW. Next-generation sequencing for microRNA profiling: microRNA-21-3p promotes oral cancer metastasis. Anticancer Res. 2017;37:1059–1066. doi: 10.21873/anticanres.11417. [DOI] [PubMed] [Google Scholar]


