Abstract
Background: Long non-coding RNA (lncRNA) has gradually received widespread attention due to its role in regulating tumor progression. However, in renal cell cancer (RCC), the exact function of lncRNA LINC00671 remains uncertain. Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was utilized for detecting LINC00671 and miR-221-5p expressions in RCC tissues and cell lines. Western blotting technique was utilized for detecting the expressions of epithelial-mesenchymal transition (EMT)-associated proteins (E-cadherin and N-cadherin) and suppressor of cytokine signaling 1 (SOCS1). The correlation between clinicopathological features and LINC00671 expression was also evaluated. RCC cell multiplication, migration and invasion were measured by CCK-8, EdU and Transwell assays, respectively. The targeted relationships between LINC00671 as well as the SOCS1 3’UTR and miR-221-5p were verified by RNA immunoprecipitation (RIP) and luciferase reporter gene assay. Results: LINC00671 expression in RCC tissues and cells was significantly reduced. Patients with low LINC00671 expression had relatively shorter disease-free survival and overall survival. Moreover, LINC00671 expression was linked to lymph node metastasis, tumor stage, and tumor size. In Caki-1 and 769-P cell lines, LINC00671 overexpression restrained the multiplication, migration, invasion, as well as the EMT process of RCC cells in vitro. In terms of mechanism, miR-221-5p was identified as a target of LINC00671, and LINC00671 could up-regulate SOCS1 by repressing miR-221-5p. Conclusion: LINC00671 regulates the miR-221-5p/SOCS1 axis as a tumor suppressor in RCC.
Keywords: RCC, LINC00671, MiR-221-5p, SOCS1
Introduction
Renal cell cancer (RCC) origins from renal tubular epithelial cells [1]. Statistically there emerge nearly 295,000 new RCC cases globally every year, with about 134,000 cases killed [2]. RCCs have four subtypes: chromophobe RCC, papillary RCC, clear cell RCC (ccRCC) and renal oncocytoma [3]. A majority of the patients with RCC are insensitive to radiotherapy and chemotherapy, and the prognosis outcome of patients with metastatic RCC remains unsatisfactory, and their five-year survival rate is only about 10% [4,5]. Hence, to explore the effective therapy targets for RCC, it is imperative to figure out the molecular mechanism underlying RCC progression.
Known as a category of transcripts, long non-coding RNAs (lncRNAs) are characterized by over 200 nucleotides and having no protein-coding function [6]. A growing number of studies in recent years show that lncRNAs partake in different biological processes, i.e., cell growth, migration, apoptosis, invasion, autophagy, etc., and is pivotal in the progression of human cancers, such as RCC [7-11]. For instance, LINC00673 facilitates cancer cell proliferation and invasion via suppressing KLF4 expression through the interaction with DNMT1 and EZH2 in gastric cancer [9]. LINC00346 promotes the pathogenesis of pancreatic ductal adenocarcinoma by activating c-Myc [10]. In RCC, the over-expression of LINC-PINT is associated with unfavorable prognosis of the patients, and its over-expression induces cell cycle progression, promotes ccRCC cell proliferation, and inhibits apoptosis [11]. In RCC, lncRNA RP11-436H11.5 is highly expressed, and it enhances BCL-W expression through modulating miR-335-5p expression, and enhances the malignant biological behaviors of RCC cells [12]. A recent study shows that LINC00671 is a novel prognostic marker for pancreatic cancer (PC) and plays a cancersuppressing role in PC [13]. Nonetheless, LINC00671’s function in RCC progression is yet to be studied.
Recognized as a category of highly conserved endogenous non-coding RNA, microRNAs (miRNAs) contain 18-25 nucleotides. It binds directly to the target mRNA 3’-untranslated region (3’-UTR), triggering mRNA translation inhibition and degradation [14]. The studies in recent have shown that multiple miRNAs contribute to regulating RCC progression [15,16]. For instance, miR-21 facilitates RCC cells’ proliferation and reduces their apoptosis by activating the mTOR-STAT3 signaling pathway [17]. MiR-181a overexpression boosts cell multiplication and cell cycle progression, and down-regulates KLF6 in ccRCC cells to inhibit apoptosis [18]. MiR-221-5p plays a tumor-suppressing role in colorectal carcinoma and prostate carcinoma [19-22]. In addition, miR-221-5p targets suppressor of cytokine signaling 1 (SOCS1) in prostate cancer cells, and regulates the MAPK/ERK signaling pathway [22].
This work was intended to explore LINC00671’s functions and potential mechanisms in RCC. It was discovered that LINC00671 expression was notably reduced in RCC tissues. Functional experiments showed that LINC00671 repressed RCC cells’ proliferation, migration, invasion. Mechanistically, we found that LINC00671 inhibited RCC progression via targeting miR-221-5p to enhance SOCS1 expression.
Materials and methods
Tissue sample
53 pairs of RCC samples as well as corresponding normal kidney tissue samples were acquired from patients who undergone nephrectomy in Shanghai Seventh People’s Hospital. After being frozen with liquid nitrogen, the samples were preserved at -196°C. The current study obtained the approval of the Ethics Committee of Shanghai Seventh People’s Hospital. Additionally, the patients enrolled signed an informed consent form before surgery. The patients’ clinicopathological features are summarized in Table 1.
Table 1.
Correlation of LINC00671 expression with clinicopathologic characteristics of RCC patients
| Clinicopathologic features | Cases n=53 | LINC000671 expression | P-value | |
|---|---|---|---|---|
|
| ||||
| High (n=26) | Low (n=27) | |||
| Age (years) | ||||
| < 60 | 36 | 16 | 20 | 0.328 |
| ≥ 60 | 17 | 10 | 7 | |
| Gender | ||||
| Male | 38 | 21 | 16 | 0.125 |
| Female | 15 | 5 | 10 | |
| Fuhrman grade | ||||
| Grade 1+2 | 41 | 22 | 19 | 0.215 |
| Grade 3+4 | 12 | 4 | 8 | |
| Tumor size (cm) | ||||
| ≤ 7 | 28 | 18 | 10 | 0.018* |
| > 7 | 25 | 8 | 17 | |
| Tumor stage | ||||
| pT1 | 30 | 10 | 20 | 0.009** |
| pT1+T32 | 23 | 16 | 7 | |
| Lymph node metastasis | ||||
| Yes | 21 | 6 | 15 | 0.012* |
| No | 32 | 21 | 12 | |
P < 0.05 was considered to be statistically significant;
P < 0.01 was considered to be statistically significant.
Cell lines and cell culture
From the American Type Culture Collection (ATCC; Rockville, MD, USA), five human RCC cell lines, including 769-P, 786-O, A498, ACHN and Caki-1, and normal human renal cell line HK-2 were bought. They were incubated in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Grand Island, NY, USA) with 100 U/ml penicillin and 0.1 mg/ml streptomycin (Hyclone, Logan, UT, USA) and 10% (v/v) heat-inactivated fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA). All cells were kept in 5% CO2 at 37°C in an incubator.
Transfection
NC mimics and miR-221-5p mimics were bought from GenePharma Co., Ltd. (Shanghai, China). Ribobio Co., Ltd. (Guangzhou, China) provided plasmids: pcDNA3.1-LINC00671 (pc-LINC00671), pcDNA3.1-SOCS1 (pc-SOCS1), and pcDNA3.1-NC (pc-NC). The cells during logarithmic growth were collected and transferred into 6-well plates (2 × 106 cells/well). 12 h later, following the manufacturer’s protocol, the miRNAs and plasmids were transfected into Caki-1 and 769-P cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR) assay
TRIzol reagent (Invitrogen, Waltham, MA, USA) was utilized for the extraction of total RNA. PrimeScript™ RT reagent kit and One Step PrimeScript miRNA cDNA synthesis kit (Takara, Dalian, China) were adopted in reverse transcription. On the Applied Biosystems 7500 detecting system (Applied Biosystems, Foster City, CA, USA), the SYBR Premix Ex Taq™ Kit (Takara, Dalian, China) was employed for conducting qRT-PCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) acted as the internal reference to quantify the expressions of LINC00671 and SOCS1, and U6 functioned as the internal reference for quantifying miR-221-5p. Below are the primer sequences (F for forward; R for reverse): LINC00671: F: 5’-ATGGGAAACTGGCCAGATCA-3’, R: 5’-TTCTCTGGCATTCCTCCTCC-3’; SOCS1: F: 5’-CTGCGGCTTCTATTGGGGAC-3’, R: 5’-AAAAGGCAGTCGAAGGTCTCG-3’; miR-221-5p: F: 5’-ACCTGGCATACAATGTAGATTT-3’, R: universal primers; U6: F: 5’-CTCGCTTCGGCAGCACA-3’, R: 5’-AACGCTTCACGAATTTGCGT-3’; GAPDH: F: 5’-ACCATCTTCCAGGAGCGAGA-3’, R: 5’-GACTCCACACCCACTACTCAGC-3’.
Cell counting kit-8 (CCK-8) assay
CCK-8 (Dojindo Molecular Technologies, Inc., Kyushu, Japan) was conducted for evaluating RCC cell multiplication. After transfection, cells were inoculated at 2 × 103 cells/well into 96-well plates. CCK8 solution (10 μL) was loaded into each well after culturing the cells for 24, 48, 72 and 96 h, respectively. After that, these cells were cultured for another 1 h. Subsequently, the optical density (OD) at 450 nm was measured.
EdU analysis
RCC cells were inoculated into 24-well plates at 5 × 105 cells/well. Subsequently, according to the instruction of EdU kit (Beyotime, Shanghai, China), each well was added with EdU solution. Then after incubation for 4 h, the cells were rinsed by PBS. 4% paraformaldehyde was utilized to fix cells. Following the addition of Apollo solution, in the dark the cells were incubated for 30 min, and the permeability of the cells was increased by the addition of 0.5% Triton X-100. Afterwards, cells were incubated with Hoechest 33342. Eventually, a fluorescence microscope was adopted for observing the cells. The EdU-positive cell rate was calculated as red fluorescence-labeled cell counts/blue fluorescence-labeled cell counts × 100%.
Transwell migration and invasion assays
RCC cell invasion and migration were examined using Transwell chambers (Corning, NY, USA). RCC cells were re-suspended in FBS-free medium, transferred into the top chamber, and medium containing 20% FBS was supplemented to the bottom chambers. For migration assay, no Matrigel was added, while a layer of Matrigel was used to cover the filter to mimic extracellular matrix in invasion assay. The Transwell chambers were kept in the incubator for 24 h, and cotton swabs were employed for wiping the cells remaining on the membrane’s upper surface. Then 95% ethanol was employed for fixing the cells and 0.5% crystal violet was employed for staining the migrated and invaded cells. After staining, PBS was used for washing membranes, and the migrated and invaded cells were counted under a microscope.
Subcellular fractionation
To determine the cellular localization of LINC000671, the isolation of cytoplasmic and nuclear RNA from Caki-1 and 769-P cells was performed using PARISTM Kit (Ambion, Austin, TX, USA). Then the expression of RNA in the cytoplasmic and nuclear fraction was detected by qRT-PCR. GAPDH and U6 served as the cytoplasmic control and the nuclear control, respectively.
RNA immunoprecipitation (RIP) analysis
Magna RIP™ RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) was utilized for conducting RIP assay. To put it briefly, RIP buffer was utilized for lysing RCC cells. Cell extracts and magnetic beads coupled with anti-Ago2 antibody (ab32381, 1:1000, Abcam, Shanghai, China) or control IgG were incubated together. The harvested samples were subsequently processed with proteinase K, and then total RNAs were extracted for qRT-PCR analysis.
Luciferase reporter gene assay
The dual-luciferase reporter assay system (Promega, Madison, WI, USA) was adopted for performing luciferase reporter gene assay. The fragments of LINC00671 and SOCS1 3’-UTR mutant type (Mut) or wild type (Wt) carrying miR-221-5p binding sites, were cloned into the luciferase reporter vector pGL3 (Promega, Madison, WI, USA) to establish LINC000671-mut, LINC000671-wt, SOCS1-mut and SOCS1-wt, respectively. The above plasmids and miR-221-5p mimics or the negative control (NC mimics) were co-transfected into Caki-1 and 769-P cells, respectively, through Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The luciferase activity was detected at 48 h after transfection following the manufacturer’s instruction.
Western blotting
RIPA lysis buffer (Beyotime, Shanghai, China) containing protease inhibitors was utilized for total protein extraction from cells. After denaturation, 8% SDS-PAGE was utilized for separating protein samples, which were then transferred onto the PVDF membrane (Millipore, Billerica, MA, USA). After being blocked with 5% skim milk, the membranes, together with primary antibodies: anti-E-cadherin (ab40772, 1:1000, Abcam, Shanghai, China), anti-GAPDH (ab8245, 1:2000, Abcam, Shanghai, China), anti-SOCS1 (ab3691, 1:1000, Abcam, Shanghai, China), and anti-N-cadherin (ab202030, 1:1000, Abcam, Shanghai, China), were incubated at 4°C for 10 h. Next, the PVDF membranes were rinsed with TBST twice, and incubated with anti-mouse or anti-rabbit horseradish peroxidase-coupled secondary antibodies for 1 h at room temperature, followed by washing membranes three times with TBST. Finally, electrochemical luminescence (ECL) kit (Beyotime, Shanghai, China) was employed for developing the protein bands.
Statistical analysis
All experiments were conducted in triplicate. SPSS Statistics 19 (SPSS Inc., Chicago, IL, USA) was the statistical analysis tool in this study. The results were expressed as mean ± standard deviation (SD). Moreover, two-tailded Student’s t-test was utilized to perform the comparison between two groups, and the comparison among multiple groups was conducted with one-way ANOVA. χ2 test was carried out for the determination of the correlation between LINC00671 expression and RCC patients’ clinical/pathological features. Spearman’s correlation analysis was used for evaluating the correlation between LINC00671 and miR-221-5p expression or SOCS1 expression in RCC tissues. The difference was of statistical significance when P < 0.05.
Results
LINC00671 expression was reduced in RCC tissues
Firstly, LINC00671 expression in 53 patients’ RCC samples and corresponding para-cancerous tissues was detected. The qRT-PCR displayed that LINC00671 was down-regulated in RCC tissues in comparison to para-cancerous tissues (Figure 1A). What’s more, LINC00671 expression in normal human renal cell line HK-2 and five RCC cell lines (769-P, Caki-1, 786-O, ACHN and A498) was detected. As shown, LINC00671 expression in RCC cell lines was markedly lower than HK-2 (Figure 1B). We then searched Gene Expression Profiling Interactive Analysis (GEIPA) database. The results revealed that LINC00671 expression was notably reduced in renal papillary cell carcinoma (KIRP) in comparison with normal tissues, but its expression was not changed in renal clear cell carcinoma (KIRC) (Figure 1C). Furthermore, the disease-free survival and overall survival of RCC patients with high LINC00671 expression were longer than those of patients with low LINC00671 expression (Figure 1D and 1E), and the expression of LINC00671 was decreased with the increase of clinical stage (Figure 1F). We then studied the correlation between the clinicopathological features of patients and LINC00671 expression. All subjects were divided into two groups: high (n=26) and low (n=27) LINC00671 expression groups. These findings indicated that reduced LINC00671 expression was connected with lymph node metastasis, higher clinical stage, and larger tumor size (Table 1).
Figure 1.
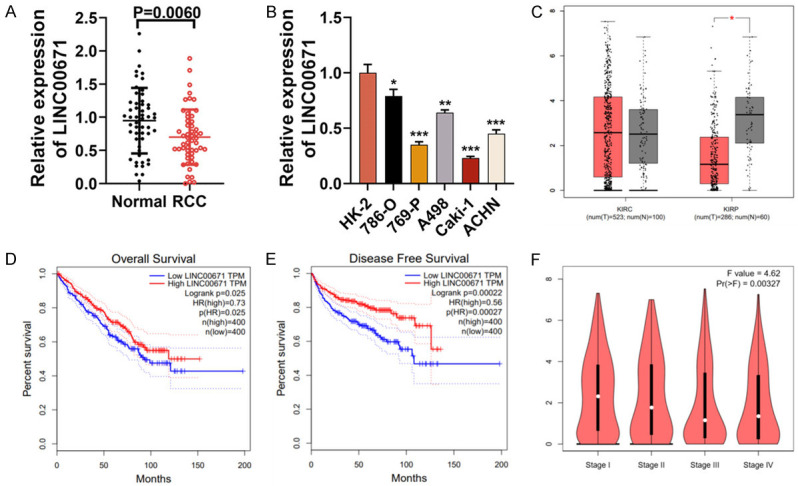
LINC00671 expression was reduced in RCC tissues and LINC00671 was associated with poor clinical outcomes. A. qRT-PCR demonstrated that LINC00671 expression was decreased in RCC samples. B. qRT-PCR implied that LINC00671 expression in five RCC cell lines (Caki-1, 769-P, ACHN, 786-O and A498) was lower than that in human normal kidney cell line HK-2. C. GEPIA RNA-Seq data suggested that in comparison to the adjacent tissues, LINC00671 expression was reduced in RCC tissues. D. GEPIA RNA-Seq data suggested that the overall survival time of patients with high LINC00671 expression in RCC was longer. E. GEPIA RNA-Seq data suggested that the disease-free survival time of patients with high LINC00671 expression in RCC was longer. F. GEPIA RNA-Seq data suggested that LINC00671 expression was decreased with the increase of RCC stage. *P < 0.05, **P < 0.01, and ***P < 0.001.
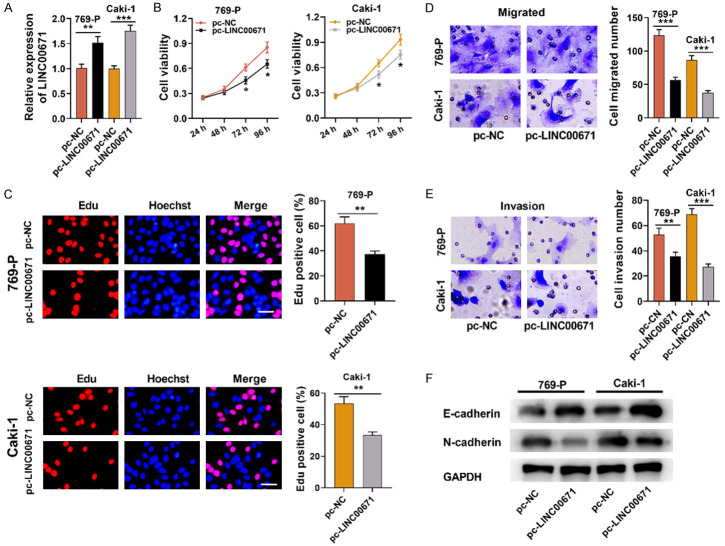
LINC00671 over-expression suppressed RCC cell proliferation, migration, invasion and EMT
To explore LINC00671’s function in RCC progression, we selected Caki-1 and 769-P cells for constructing LINC00671 over-expression cell models (Figure 2A). The effects of LINC00671 over-expression on RCC cell multiplication were assessed by EdU and CCK-8 assays, and it was demonstrated that LINC00671 over-expression significantly suppressed Caki-1 and 769-P cell multiplication (Figure 2B and 2C). Furthermore, Transwell assay indicated that LINC00671 over-expression markedly inhibited Caki-1 and 769-P cell migration and invasion (Figure 2D and 2E). Subsequently, we used Western blot for detecting the expressions of EMT-related proteins and discovered that LINC00671 overexpression remarkably facilitated E-cadherin expression and inhibited N-cadherin expression in both RCC cell lines (Figure 2F).
Figure 2.
LINC00671 overexpression suppressed RCC proliferation, migration, invasion and EMT. A. LINC00671 overexpression cell models were established, and qRT-PCR showed that LINC00671 expression in 769-P and Caki-1 cells was up-regulated after transfection. B, C. CCK-8 and EdU experiments were employed for examining the effect of LINC00671 over-expression on 769-P and Caki-1 cell proliferation. D, E. Transwell assay was employed for examining the effect of LINC00671 over-expression on Caki-1 and 769-P cell migration and invasion. F. Western blot was utilized for examining the effect of LINC00671 over-expression on the expression of EMT-related proteins (E-cadherin and N-cadherin). pc-NC: pcDNA3.1-NC; pc-LINC00671: pcDNA3.1-LINC00671; CCK-8: Cell Counting Kit-8. *P < 0.05, **P < 0.01, and ***P < 0.001.
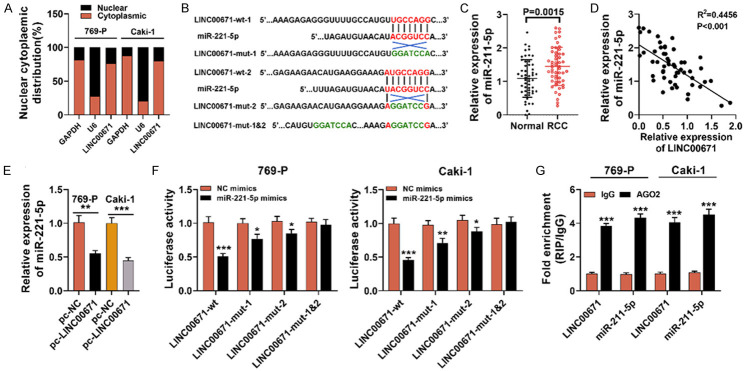
LINC00671 serves as the miR-221-5p target gene in RCC cells
LncRNA can sponge miRNA to act as competitive endogenous RNA (ceRNA) [23]. Based on nuclear-cytoplasmic fractionation, we demonstrated that LINC00671 was located mainly in the cytoplasm of Caki-1 and 69-P cells, suggesting that LINC00671 could likely act as a ceRNA (Figure 3A). According to LncBase Predicted v2 database, LINC000671 had two predicted miR-221-5p binding sites (Figure 3B). It was demonstrated from qRT-PCR that miR-221-5p expression was enhanced in RCC samples (Figure 3C) and it was inversely related to LINC00671 expression (Figure 3D). Meanwhile, qRT-PCR displayed that the expression of miR-221-5p was notably reduced in Caki-1 and 769-P cells with LINC00671 overexpression (Figure 3E). Subsequently, we constructed mutated luciferase reporter vectors LINC00671-mut-1, LINC00671-mut-2, LINC00671-mut-1&2 and wild type luciferase reporter vector LINC00671-wt, and they and miR-211-5p mimics or NC mimics, were co-transfected into 769-P and Caki-1 cell lines respectively. It was discovered that the transfection of miR-221-5p mimics markedly lowered LINC00671-wt’s luciferase activity, LINC00671-mut-1 and LINC00671-mut-2, and had no significant effect on that of LINC00671-mut-1&2 (Figure 3F). RIP was utilized for validating the direct interaction between miR-221-5p and LINC00671 in RCC cells, and it was unveiled that LINC00671 and miR-221-5p were both enriched in the immunoprecipitate containing Ago2 (Figure 3G). The above results indicated that LINC00671 could targetedly inhibit miR-221-5p expression by sponging it in RCC cells.
Figure 3.
miR-221-5p served as a target of LINC00671 in RCC cells. A. Subcellular localization of LINC00671 in 769-P and Caki-1 cells was performed by RNA isolation and qRT-PCR. GAPDH is the cytoplasmic control, and U6 serves as the nuclear control. B. Bioinformatics predicted two binding sites between LINC00671 and miR-221-5p. C. qRT-PCR indicated that miR-221-5p was up-regulated in RCC samples. D. Spearman’s correlation analysis was adopted to examine the correlation of LINC00671 expression with miR-221-5p in RCC samples. E. qRT-PCR displayed that miR-221-5p expression was reduced after LINC00671 is over-expressed in 769-P and Caki-1 cells. F. LINC00671-wt, LINC00671-mut-1, LINC00671-mut-2 or LINC00671-mut-1&2 was co-transfected with miR-221-5p mimics or NC mimics into 769-P and Caki-1 cell lines, and dual-luciferase reporter assay was utilized to validate the predicted binding sites. G. RIP experiment was used for evaluating the direct interaction between LINC00671 and miR-221-5p, and it indicated that LINC00671 and miR-221-5p were immunoprecipitated with anti-AGO2 antibody from lysates of 769-P and Caki-1 cells. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; pc-NC: pcDNA3.1-NC; pc-LINC00671: pcDNA3.1-LINC00671; LINC00671-wt: LINC00671 wild type; LINC00671-mut: LINC00671 mutant type; RIP: RNA immunoprecipitation. *P < 0.05, **P < 0.01, and ***P < 0.001.
MiR-221-5p served as a tumor promoter in RCC cells
Next, we evaluated miR-221-5p’s functions in RCC cells. MiR-221-5p mimics were transfected into cell lines Caki-1 and 769-P respectively. qRT-PCR indicated that the miR-221-5p mimics transfection greatly up-regulated miR-221-5p expression in RCC cells (Figure 4A). EdU and CCK-8 assays indicated that miR-221-5p over-expression enhanced Caki-1 and 769-P cell proliferation (Figure 4B and 4C). Transwell assay displayed that the miR-221-5p mimics transfection facilitated Caki-1 and 769-P cell migration and invasion (Figure 4D and 4E). Additionally, it was discovered that the miR-221-5p mimics transfection increased N-cadherin expression and inhibited E-cadherin expression in both RCC cell lines (Figure 4F).
Figure 4.
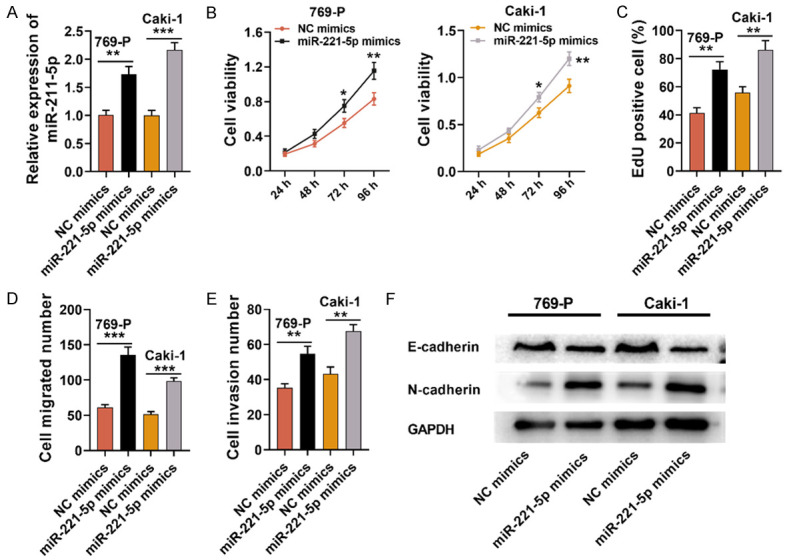
MiR-221-5p served as a tumor promoter in RCC cells. A. MiR-221-5p mimics or NC mimics were transfected into cell lines 769-P and Caki-1, and qRT-PCR was utilized for detecting miR-221-5p expression. B, C. EdU and CCK-8 experiments were used for examining the effect of miR-221-5p over-expression on Caki-1 and 769-P cell proliferation. D, E. Transwell assay was employed to detect the effect of miR-221-5p over-expression on Caki-1 and 769-P cell invasion and migration. F. Western blot was utilized for testing the effect of miR-221-5p over-expression on the expressions of EMT-related proteins (N-cadherin and E-cadherin). NC: negative control; CCK-8: Cell Counting Kit-8. *P < 0.05, **P < 0.01, and ***P < 0.001.
MiR-221-5p targeted SOCS1 and inhibits its expression
To delve into the downstream mechanism of miR-221-5p in RCC progression, TargetScan database was employed to perform bioinformatics analysis, and it was manifested that SOCS1 was a potential miR-221-5p target (Figure 5A). It was indicated by dual-luciferase reporter assay that the luciferase activity was remarkably lowered in Caki-1 and 769-P cells co-transfected with SOCS1-wt and miR-221-5p mimics, while co-transfection with miR-221-5p mimics and SOCS1-mut caused no luciferase activity change (Figure 5B). Moreover, SOCS1 expression in RCC samples was markedly higher as opposed to para-cancerous tissues (Figure 5C), and SOCS1 and LINC00671 expression were positively correlated in RCC samples (Figure 5D). In addition, Western blot and qRT-PCR indicated that SOCS1 mRNA and protein expressions were markedly lowered in Caki-1 and 769-P cells overexpressing miR-221-5p (Figure 5E and 5F). So in conclusion, SOCS1 was the miR-221-5p direct target, and its expression can be suppressed by miR-221-5p in RCC.
Figure 5.
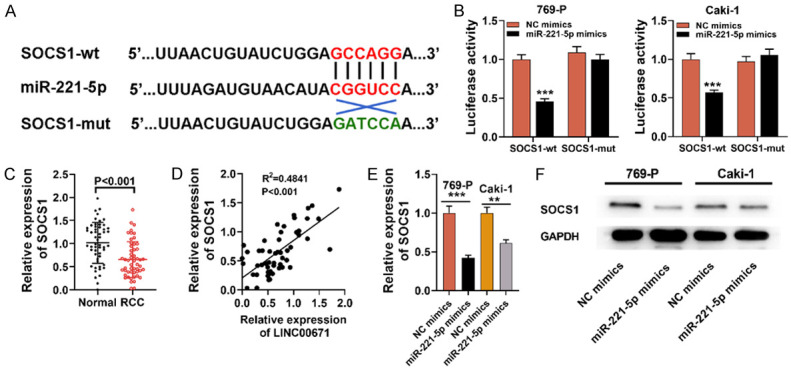
MiR-221-5p targeted SOCS1 and inhibited its expression. A. Bioinformatics was utilized to predict the targeting site between miR-221-5p and SOCS1 3’-UTR. B. SOCS1-wt or SOCS1-mut was co-transfected with miR-221-5p mimics or NC mimics into 769-P and Caki-1 cell lines, and dual-luciferase reporter assay was utilized for validating the predicted binding site. C. qRT-PCR suggested that SOCS1 expression in RCC samples was down-regulated. D. Spearman’s correlation analysis was adopted to examine the correlation of LINC00671 expression with SOCS1 expression in RCC samples. E. qRT-PCR suggested that SOCS1 expression was decreased in 769-P and Caki-1 cells transfected by miR-221-5p mimics. F. Western blot was utilized for detecting SOCS1 expression in Caki-1 and 769-P cells transfected by miR-221-5p mimics or NC mimics. SOCS1-wt: SOCS1 3’-UTR wild type; SOCS1-mut: SOCS1 3’-UTR mutant type; NC: negative control. **P < 0.01, and ***P < 0.001.
MiR-211-5p promoted RCC progression by repressing SOCS1
To further explore whether SOCS1 mediated miR-221-5p’s biological effects on RCC cells, we transfected SOCS1 over-expression plasmids into cell lines Caki-1 and 769-P transfected with miR-221-5p mimics (Figure 6A and 6B). Subsequently, CCK-8, EdU and Transwell assays, and Western blotting were utilized for detecting RCC cell multiplication, migration, invasion and EMT, and it was suggested that SOCS1 overexpression counteracted the promoting impact of miR-221-5p over-expression on the malignant biological behaviors of RCC cells (Figure 6C-G). In conclusion, miR-221-5p promoted RCC progression by inhibiting SOCS1 expression.
Figure 6.
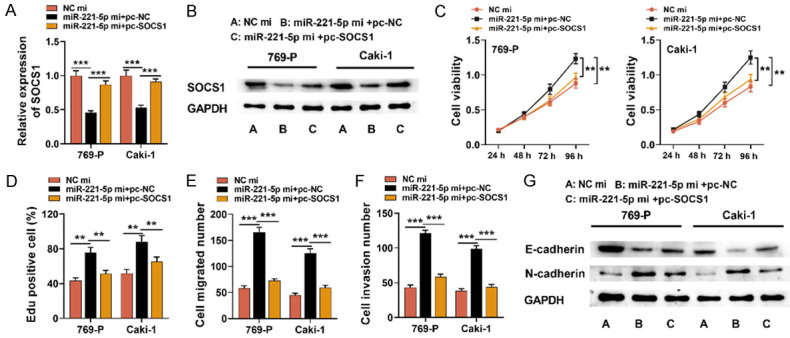
SOCS1 over-expression inhibited the promotion on RCC progression induced by miR-221-5p. A, B. SOCS1 overexpression plasmid was transfected into Caki-1 and 769-P cells with miR-221-5p over-expression, and qRT-PCR and Western blot suggested that SOCS1 mRNA and protein expressions were up-regulated. C, D. CCK-8 and EdU experiments were employed for the detection of the effect of SOCS1 over-expression on Caki-1 and 769-P cell proliferation. E, F. Transwell assay was utilized for detecting the effect of SOCS1 over-expression on 769-P and Caki-1 cell invasion and migration. G. Western blot was utilized to test the expressions of EMT-related proteins (N-cadherin and E-cadherin). NC mi: NC mimics; miR-221-5p mi: miR-221-5p mimics; pc-NC: pcDNA3.1-NC; pc-SOCS1: pcDNA3.1-SOCS1; CCK-8: Cell Counting Kit-8. **P < 0.01, ***P < 0.001.
LINC00671 suppressed tumor development in RCC via miR-221-5p/SOCS1 axis
To further clarify whether LINC00671 regulated RCC progression through the miR-221-5p/SOCS1 axis, miR-221-5p mimics were transfected into Caki-1 and 769-P cells with LINC00671 over-expression (Figure 7A). Western blot and qRT-PCR indicated that miR-221-5p over-expression dramatically reduced the expression of SOCS1 compared with RCC cells with LINC00671 (Figure 7B and 7C). Next, CCK-8, EdU and Transwell assay and Western blot revealed that miR-221-5p over-expression not only partially counteracted the effects that LINC00671 over-expression had on RCC cell multiplication, migration and invasion, but also significantly restored the EMT process (Figure 7D-H). Therefore, it was confirmed that LINC00671 inhibited RCC development through modulating the miR-221-5p/SOCS1 axis.
Figure 7.
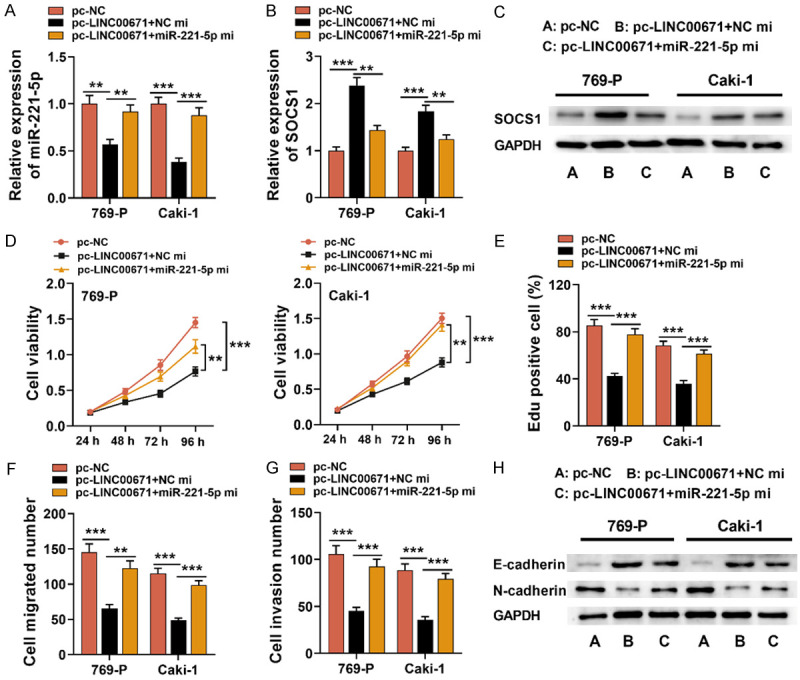
LINC00671 suppressed RCC progression via regulating miR-221-5p/SOCS1 axis. A-C. MiR-221-5p mimics were transfected into Caki-1 and 769-P cell lines with LINC00671 over-expression, and qRT-PCR and Western blot were used to test the expressions of miR-221-5p and SOCS1 after transfection. D, E. CCK-8 and EdU experiments were used for the detection of the proliferation of 769-P and Caki-1 cells. F, G. Transwell assay was employed for the detection of Caki-1 and 769-P cell migration and invasion. H. Western blot was utilized to detect the expressions of EMT-related proteins (N-cadherin and E-cadherin). pc-NC: pcDNA3.1-NC; pc-LINC00671: pcDNA3.1-LINC00671; NC mi: NC mimics; miR-221-5p mi: miR-221-5p mimics; CCK-8: Cell Counting Kit-8. **P < 0.01, ***P < 0.001.
Discussion
Accumulating studies show that lncRNA contributes to tumorigenesis and cancer progression via modulating diverse key biological processes [24-27]. Many aberrantly expressed lncRNAs are reportedly related to the malignant biological behaviors of RCC cells [28,29]. LINC00511, as a ceRNA, regulates cyclin D1 expression by sponging miR-625 in RCC to facilitate RCC progression [30]. LncRNA ADAMTS9-AS2 suppresses RCC development and reverses chemoresistance of RCC cells through regulating miR-27a-3p/FOXO1 expression [31]. It is found that LINC00671 is lowly expressed in PC tissues and the serum of the patients, and the survival rate of patients with high LINC00671 expression is notably improved [13]. Nevertheless, previously, the biological function and expression characteristics of LINC00671 in RCC remain unclear. The present work indicated that in RCC tissues and cell lines, LINC00671 expression was reduced. The under-expression of LINC00671 was strongly related to lymph node metastasis, higher tumor stage, and larger tumor size in patients with RCC. Importantly, the data from The Cancer Genome Atlas (TCGA) suggested, the prognosis of RCC patients with highly expressed LINC00671 was better than those with lowly expressed LINC00671. Functional experiments displayed that LINC00671 overexpression could block RCC cell multiplication, migration, invasion and EMT. Collectively, for the first time, our work suggested that LINC00671 was a novel tumor suppressor in RCC.
MiRNA can bind directly to the target mRNA 3’UTR and induce mRNA degradation or translation inhibition, thereby contributing to various pathological and physiological processes [14,15,32]. A lot of studies have confirmed that miRNAs are possible targets for RCC treatment [33]. The biological functions of miR-221-5p in different cancers are distinct [19-22,34]. In RCC, miR-221-5p is proved to be an oncogenic miRNA: its expression is enhanced in RCC tissues, and it facilitates carcinoma cell multiplication and migration, and suppresses apoptosis in 786-O and ACHN cell lines [34]. This work also displayed that miR-221-5p was abnormally up-regulated in RCC tissues, and its mimics promoted the malignant biological behaviors of other two RCC cell lines Caki-1 and 69-P, which are consistent with the previous report [34]. Additionally, in this work, we predicted and verified that LINC00671 could targetedly suppress miR-221-5p expression, and this partly explained the mechanism by which miR-221-5p was up-regulated in RCC.
SOCS1 is a key inhibitor of MAPK/ERK signal transduction, and it contributes to inhibiting cell proliferation via arresting cell cycle and promoting apoptosis [35,36]. SOCS1 plays a tumor-suppressing role in multiple cancers, including gastric carcinoma, prostate carcinoma and multiple myeloma [37-39]. In RCC, SOCS1 is also a tumor suppressor. Ubiquitination and proteasome degradation of SOCS1 is crucial for the metastasis of RCC cells mediated by von Hippel-Lindau (VHL), and SOCS1 facilitates nuclear redistribution and K63-ubiquitylation of VHL in response to DNA double-strand breaks to maintain the high genomic instability of RCC cells [40,41]. It is reported that miR-221-5p specifically targets SOCS1 to accelerate prostate carcinoma cell growth and migration in vivo and in vitro [22]. In this study, bioinformatics prediction was conducted, and dual-luciferase reporter assay was utilized for validating that miR-221-5p could target the 3’-UTR of SOCS1 in RCC cells. It was discovered that miR-221-5p in RCC cells could suppress SOCS1 mRNA and protein expressions, conforming to the previous report [22]. Importantly, SOCS1 was also proved to be up-regulated by LINC00671, and SOCS1 over-expression reversed the promotional impact of miR-221-5p over-expression on the malignant biological behaviors of RCC. Collectively, it was revealed that LINC00671 played a cancer-suppressing role by regulating the miR-221-5p/SOCS1 axis.
To sum up, the present work demonstrates that LINC00671 expression is markedly reduced in RCC, and that its down-regulation is closely related to unfavorable clinicopathologic characteristics and adverse prognosis. It is also confirmed that LINC00671 inhibits RCC cell multiplication, migration and invasion, and EMT process via the miR-221-5p/SOCS1 axis. Collectively, LINC00671 can function as a favorable biomarker and new treatment target for RCC.
Acknowledgements
We thank Hubei Yican Health Industry Co., Ltd. for its linguistic assistance during the preparation of this manuscript. This work was funded by Talents Training Program of the Seventh People’s Hospital, Shanghai University of Traditional Chinese Medicine (Grant No. BDX202001).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009–17027. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Cao Q, Bai P. Role of autophagy in renal cancer. J Cancer. 2019;10:2501–2509. doi: 10.7150/jca.29285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motzer RJ, Molina AM. Targeting renal cell carcinoma. J. Clin. Oncol. 2009;27:3274–3276. doi: 10.1200/JCO.2009.21.8461. [DOI] [PubMed] [Google Scholar]
- 6.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu YB, Tu YN. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. 2017;8:95542–95553. doi: 10.18632/oncotarget.20980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng WX, He RZ, Zhang Z, Yang L, Mo YY. LINC00346 promotes pancreatic cancer progression through the CTCF-mediated Myc transcription. Oncogene. 2019;38:6770–6780. doi: 10.1038/s41388-019-0918-z. [DOI] [PubMed] [Google Scholar]
- 11.Duan J, Ma X, Shi J, Xuan Y, Wang H, Li P, Zhang Y, Fan Y, Gong H, Ma X, Pang Y, Wang L, Yan Y, Zhang X. Long noncoding RNA LINC-PINT promotes proliferation through EZH2 and predicts poor prognosis in clear cell renal cell carcinoma. Onco Targets Ther. 2019;12:4729–4740. doi: 10.2147/OTT.S202938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Jin W, Song Y, Fei X. LncRNA RP11-436H11.5, functioning as a competitive endogenous RNA, upregulates BCL-W expression by sponging miR-335-5p and promotes proliferation and invasion in renal cell carcinoma. Mol Cancer. 2017;16:166–176. doi: 10.1186/s12943-017-0735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res. 2018;10:2648–2658. [PMC free article] [PubMed] [Google Scholar]
- 14.Gu L, Li H, Chen L, Ma X, Gao Y, Li X, Zhang Y, Fan Y, Zhang X. MicroRNAs as prognostic molecular signatures in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget. 2015;6:32545–32560. doi: 10.18632/oncotarget.5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He YH, Chen C, Shi Z. The biological roles and clinical implications of microRNAs in clear cell renal cell carcinoma. J Cell Physiol. 2018;233:4458–4465. doi: 10.1002/jcp.26347. [DOI] [PubMed] [Google Scholar]
- 17.Liang T, Hu XY, Li YH, Tian BQ, Li ZW, Fu Q. MicroRNA-21 regulates the proliferation, differentiation, and apoptosis of human renal cell carcinoma cells by the mTOR-STAT3 signaling pathway. Oncol Res. 2016;24:371–380. doi: 10.3727/096504016X14685034103356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lei Z, Ma X, Li H, Zhang Y, Gao Y, Fan Y, Li X, Chen L, Xie Y, Chen J, Wu S, Tang L, Zhang X. Up-regulation of miR-181a in clear cell renal cell carcinoma is associated with lower KLF6 expression, enhanced cell proliferation, accelerated cell cycle transition, and diminished apoptosis. Urol Oncol. 2018;36:93.e23–93.e37. doi: 10.1016/j.urolonc.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 19.Song J, Ouyang Y, Che J, Li X, Zhao Y, Yang K, Zhao X, Chen Y, Fan C, Yuan W. Potential value of miR-221/222 as diagnostic, prognostic, and therapeutic biomarkers for diseases. Front Immunol. 2017;8:56–54. doi: 10.3389/fimmu.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan K, Xie K, Fox J, Zeng H, Gao H, Huang C, Wu M. Decreased levels of miR-224 and the passenger strand of miR-221 increase MBD2, suppressing maspin and promoting colorectal tumor growth and metastasis in mice. Gastroenterology. 2013;145:853–864. e859. doi: 10.1053/j.gastro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiener M, Chen L, Krebs M, Grosjean J, Klima I, Kalogirou C, Riedmiller H, Kneitz B, Thalmann GN, Snaar-Jagalska E, Spahn M, Kruithof-de Julio M, Zoni E. miR-221-5p regulates proliferation and migration in human prostate cancer cells and reduces tumor growth in vivo. BMC Cancer. 2019;19:627–643. doi: 10.1186/s12885-019-5819-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao N, Ma G, Zhang J, Zhu W. miR-221-5p enhances cell proliferation and metastasis through post-transcriptional regulation of SOCS1 in human prostate cancer. BMC Urol. 2018;18:14–22. doi: 10.1186/s12894-018-0325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:1310–1335. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang LN, Zhu XQ, Song XS, Xu Y. Long noncoding RNA lung cancer associated transcript 1 promotes proliferation and invasion of clear cell renal cell carcinoma cells by negatively regulating miR-495-3p. J Cell Biochem. 2018;119:7599–7609. doi: 10.1002/jcb.27099. [DOI] [PubMed] [Google Scholar]
- 25.He H, Wang N, Yi X, Tang C, Wang D. Long non-coding RNA H19 regulates E2F1 expression by competitively sponging endogenous miR-29a-3p in clear cell renal cell carcinoma. Cell Biosci. 2017;7:65–76. doi: 10.1186/s13578-017-0193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN, Klibanski A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70:2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 28.Zhang M, Lu W, Huang Y, Shi J, Wu X, Zhang X, Jiang R, Cai Z, Wu S. Downregulation of the long noncoding RNA TUG1 inhibits the proliferation, migration, invasion and promotes apoptosis of renal cell carcinoma. J Mol Histol. 2016;47:421–428. doi: 10.1007/s10735-016-9683-2. [DOI] [PubMed] [Google Scholar]
- 29.Xiong J, Liu Y, Luo S, Jiang L, Zeng Y, Chen Z, Shi X, Lv B, Tang W. High expression of the long non-coding RNA HEIRCC promotes Renal Cell Carcinoma metastasis by inducing epithelial-mesenchymal transition. Oncotarget. 2017;8:6555–6563. doi: 10.18632/oncotarget.14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng H, Huang C, Wang Y, Jiang H, Peng S, Zhao X. LINC00511 promotes the malignant phenotype of clear cell renal cell carcinoma by sponging microRNA-625 and thereby increasing cyclin D1 expression. Aging (Albany NY) 2019;11:5975–5991. doi: 10.18632/aging.102156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song EL, Xing L, Wang L, Song WT, Li DB, Wang Y, Gu YW, Liu MM, Ni WJ, Zhang P, Ma X, Zhang X, Yao J, Chen Y, An RH. LncRNA ADAMTS9-AS2 inhibits cell proliferation and decreases chemoresistance in clear cell renal cell carcinoma via the miR-27a-3p/FOXO1 axis. Aging (Albany NY) 2019;11:5705–5725. doi: 10.18632/aging.102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 33.Boguslawska J, Poplawski P, Alseekh S, Koblowska M, Iwanicka-Nowicka R, Rybicka B, Kedzierska H, Gluchowska K, Hanusek K, Tanski Z, Fernie AR, Piekielko-Witkowska A. MicroRNA-mediated metabolic reprograming in renal cancer. Cancers (Basel) 2019;11:1825–1845. doi: 10.3390/cancers11121825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Wang Y, Li W, Yu S, Wen Z, Chen Z, Lin F. miR-221-5p acts as an oncogene and predicts worse survival in patients of renal cell cancer. Biomed Pharmacother. 2019;119:109406. doi: 10.1016/j.biopha.2019.109406. [DOI] [PubMed] [Google Scholar]
- 35.Zhou H, Miki R, Eeva M, Fike FM, Seligson D, Yang L, Yoshimura A, Teitell MA, Jamieson CA, Cacalano NA. Reciprocal regulation of SOCS1 and SOCS3 enhances resistance to ionizing radiation in glioblastoma multiforme. Clin Cancer Res. 2007;13:2344–2353. doi: 10.1158/1078-0432.CCR-06-2303. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Metze D, Nashan D, Muller-Tidow C, Serve HL, Poremba C, Luger TA, Bohm M. Expression of SOCS-1, suppressor of cytokine signalling-1, in human melanoma. J Invest Dermatol. 2004;123:737–745. doi: 10.1111/j.0022-202X.2004.23408.x. [DOI] [PubMed] [Google Scholar]
- 37.Fukushima N, Sato N, Sahin F, Su GH, Hruban RH, Goggins M. Aberrant methylation of suppressor of cytokine signalling-1 (SOCS-1) gene in pancreatic ductal neoplasms. Br J Cancer. 2003;89:338–343. doi: 10.1038/sj.bjc.6601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.To KF, Chan MW, Leung WK, Ng EK, Yu J, Bai AH, Lo AW, Chu SH, Tong JH, Lo KW, Sung JJ, Chan FK. Constitutional activation of IL-6-mediated JAK/STAT pathway through hypermethylation of SOCS-1 in human gastric cancer cell line. Br J Cancer. 2004;91:1335–1341. doi: 10.1038/sj.bjc.6602133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galm O, Yoshikawa H, Esteller M, Osieka R, Herman JG. SOCS-1, a negative regulator of cytokine signaling, is frequently silenced by methylation in multiple myeloma. Blood. 2003;101:2784–2788. doi: 10.1182/blood-2002-06-1735. [DOI] [PubMed] [Google Scholar]
- 40.Wu KL, Miao H, Khan S. JAK kinases promote invasiveness in VHL-mediated renal cell carcinoma by a suppressor of cytokine signaling-regulated, HIF-independent mechanism. Am J Physiol Renal Physiol. 2007;293:F1836–1846. doi: 10.1152/ajprenal.00096.2007. [DOI] [PubMed] [Google Scholar]
- 41.Metcalf JL, Bradshaw PS, Komosa M, Greer SN, Stephen Meyn M, Ohh M. K63-ubiquitylation of VHL by SOCS1 mediates DNA double-strand break repair. Oncogene. 2014;33:1055–1065. doi: 10.1038/onc.2013.22. [DOI] [PubMed] [Google Scholar]