Abstract
Circular RNAs (circRNAs) have been demonstrated to play critical roles in the initiation and development of breast cancer (BC). This study aimed to uncover the regulatory roles of a novel circRNA, circRPPH1 (hsa_circ_0000514) in BC progression. CircRPPH1, miR-296-5p and FOXP4 levels were determined by qRT-PCR. CircRPPH1 stability was detected in response to ribonuclease (RNase) R digestion and actinomycin D treatment. Cell growth, migration and invasion were evaluated using various functional experiments. Protein levels of proliferating cell nuclear antigen (PCNA), matrix metalloproteinase 9 (MMP-9), hexokinase 2 (HK2) and forkhead box protein 4 (FOXP4) were measured by Western blotting. Metabolic alterations of BC cells were evaluated using commercial kits. The interaction between miR-296-5p and circRPPH1/FOXP4 was assessed using dual-luciferase assay, RNA pull-down, and RNA immunoprecipitation (RIP) assay. The in vivo tumorigenesis was assessed in nude mice. According to the results, up-regulation of circRPPH1 was closely correlated with the poor prognosis of BC patients. Functional experiments showed that knockdown of circRPPH1 repressed BC cell growth, migration, invasion, glycolysis, and in vivo tumor growth. In addition, circRPPH1 could sponge miR-296-5p to enhance FOXP4 expression in BC cells. miR-296-5p inhibition or FOXP4 overexpression restored the malignant properties of circRPPH1-silenced BC cells. Thus, circRPPH1 promoted BC malignant progression through regulating miR-296-5p/FOXP4 axis, indicating a possible novel therapeutic strategy involving circRNA for BC patients.
Keywords: BC, circRPPH1, miR-296-5p, FOXP4
Introduction
Breast cancer (BC), as a heterogeneous malignancy, has been recognized as one of major contributors to tumor-related mortality worldwide [1]. Latest data showed that an estimated 2 million women were diagnosed as BC, which account for about 25% of all women with cancer [1]. Despite the improvement in BC survival attributed to the therapeutic methods, such as radical surgery, postoperative systemic chemotherapy or radiotherapy, easy recurrence and metastasis are the main problems to be urgently solved [2,3]. Hence, it is essential to uncover the pathogenesis of BC to design innovative interventions.
Circular RNAs (circRNAs) are a kind of closed circular structure RNAs without protein-coding ability, which function as critical regulators in multiple physiologic and pathologic processes [4]. The dysregulation of circRNAs has been considered to be involved in the pathogenesis of BC. For instance, circular RNA profiling provides helpful information for early diagnosis and prognosis prediction of BC [5]. Li et al. reported that circ_0104824 was suggested as a therapeutic target for BC patients [6]. Dysregulation of circUSP42 might contribute to the occurrence and development of triple-negative breast cancer [7]. In the online GEO database, circRPPH1 (hsa_circ_0000514) expression is remarkably elevated in BC tissues. However, the biological functions of circRPPH1 in BC remain unclear, which need to be elaborated in detail.
Recently, circRNAs have been identified to play biological functions by sponging microRNAs (miRNAs) [8]. Before the present study, the binding potential between circRPPH1 and miR-296-5p was predicted by several online databases. miR-296-5p has been reported to be down-regulated in BC, thereby promoting the aggressiveness of BC cells [9]. Additionally, Kong et al. demonstrated that miR-296-5p sponged by circPLK1 promoted tumor progression of BC [10]. Forkhead box protein 4 (FOXP4), as a member of FOXP transcription factor family, takes part in tumorigenesis of various cancers [11]. Up-regulation of FOXP4 has been verified to promote metastasis of BC cells [12]. More importantly, FOXP4 was predicted as one of targets of miR-296-5p by online databases. Therefore, the biological functions of circRPPH1, miR-296-5p, and FOXP4 together with their interactions in BC need to be further explored.
This study validated the up-regulation of circRPPH1 in BC, which is closely correlated with poor prognosis of BC patients. circRPPH1 facilitated BC development via sponging miR-296-5p to up-regulate FOXP4. Our findings suggest that circRPPH1 acts as an oncogene and has the potential to be a therapeutic target for BC.
Materials and methods
Clinical sample collection
Seventy pairs of BC tissues and surrounding normal tissues were obtained from the Hospital of Shunyi District, Beijing. All patients did not receive any treatment before surgery and signed informed consents. This study was performed in accordance with the Helsinki Declaration and approved by the Ethics Committee of the Hospital of Shunyi District, Beijing (approval number: 202103).
Cell culture and transfection
T47D, MCF-7, MDA-MB-231, BT549 (BC cell lines), and MCF10A (normal breast epithelial cell line) were purchased from American Type Culture Collection (Manassas, VA, USA). MCF10A cells were cultured in MEGM™ Mammary Epithelial Cell Growth Medium (Lonza, Basel, Switzerland), and BC cells were cultured in Dulbecco’s Modified Eagle’s Medium (BI, Hertzliya Pituach, Israel) or RPMI1640 (BI). All cells were supplemented with 10% fetal bovine serum (FBS; BI) and maintained in an incubator (37°C and 5% CO2). Short hairpin RNAs (shRNAs) specially targeting circRPPH1 transcript but not linear RPPH1 transcript, miR-296-5p mimics, and miR-296-5p inhibitor were obtained from GenePharma (Shanghai, China; sequences are shown in Table 1). The full-length cDNA of FOXP4 was inserted into pcDNA3.1 vector to construct FOXP4 expression plasmid. MCF-7 and MDA-MB-231 cells were transfected with these segments using Lipofectamine 2000 (Thermo Fisher, Franklin Lakes, MA, USA).
Table 1.
The designed sequences for this study
| Name | Sequence (5’-3’) |
|---|---|
| sh-NC | 5’-TTCTCCGAACGTGTCACGT-3’ |
| sh-circRPPH1#1 | 5’-GCGAAGTGAGTTCCCAGAGAA-3’ |
| sh-circRPPH1#2 | 5’-AGCGAAGTGAGTTCCCAGAGA-3’ |
| sh-circRPPH1#3 | 5’-CCAGCGAAGTGAGTTCCCAGA-3’ |
| mimics-NC | UUCUCCGAACGUGUCACGUTT |
| miR-296-5p mimics | UGUCCUAACUCCCCCCCGGGA |
| inhibitor-NC | CAGUACUUUUGUGUAGUACAA |
| miR-296-5p inhibitor | UCCCGGGGGGGAGUUAGGACA |
RNA isolation and quantitative real-time PCR (qRT-PCR)
The nuclear and cytoplasmic fractions were prepared using PARIS kit (Thermo Fisher). Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). The Hairpin-itTM miRNAs qPCR Quantitation Kit (GenePharma) was adopted for evaluating miR-296-5p expression. For measuring circRPPH1 and FOXP4 levels, the First Strand cDNA Synthesis Kit (Sangon, Shanghai, China) was used for cDNA synthesis followed by real-time qRT-PCR using the SYBR Premix Ex Taq™ (Takara, Tokyo, Japan). The 2-ΔΔCt method was used to calculate the gene expression normalized to U6 or GAPDH. The primer sequences are shown in Table 2.
Table 2.
Oligonucleotide primer sets for qPCR
| Name | Sequence (5’-3’) | Length |
|---|---|---|
| circRPPH1 F | AGCTTGGAACAGACTCACGG | 20 |
| circRPPH1 R | AATGGGCGGAGGAGAGTAGT | 20 |
| miR-296-5p F | CGTCTATACAGACCCTGGCTTTTC | 24 |
| miR-296-5p R | CTCAACTGGTGTCGTGGA | 18 |
| ZCCHC3 F | TGGACGTGGAGGACATTGTG | 20 |
| ZCCHC3 R | ATCCCAAACCTGTCGGTCAC | 20 |
| CCAR2 F | CAAACATCCCACACACTTCAC | 21 |
| CCAR2 R | GACCTGGATCCGGCTTGGATG | 21 |
| FOXP4 F | CGACATGATGGTGGAATCTG | 20 |
| FOXP4 R | TGTTTGCTGTCATTGTTCCC | 20 |
| U6 F | TGTTCCACACTACGCAGTCC | 20 |
| U6 R | TTTGTCGTTCCCGTCTCCTG | 20 |
| GAPDH F | GGAGCGAGATCCCTCCAAAAT | 21 |
| GAPDH R | GGCTGTTGTCATACTTCTCATGG | 23 |
Note: ZCCHC3: zinc finger; CCHC-type: containing 3; CCAR2: cell cycle and apoptosis regulator 2; FOXP4: forkhead box protein 4.
Detection of circRPPH1 stability
To evaluate circRPPH1 stability, the BC cells were treated with 2 mg/L actinomycin D for 24 h. The extracted RNA was subjected to 3 U/μg ribonuclease R (RNase R) digestion reaction. Subsequently, qRT-PCR was carried out as described above.
Cell counting kit -8 (CCK-8)
The viability of BC cells was measured using CCK-8 system (MCE, Monmouth Junction, NJ, USA). Briefly, the BC cells with various treatments were added with 10 μL of CCK-8 reagent. After incubation for 3 h, the results were obtained at 450 nm on a microplate reader (Thermo Fisher).
Colony formation assay
Two hundred BC cells in 6-well plates were cultured for 14 d. Thereafter, the plates were immersed in 0.1% crystal violet solution and the images were captured.
Transwell assay
The invasion or migration of BC cells was assessed using transwell plates coated with or without Matrigel (Sigma-Aldrich, Saint Louis, MI, USA). Briefly, the BC cells were seeded into the upper wells without FBS, while the bottom wells were added with culture medium containing 10% FBS. Twenty-four hours later, the invaded or migrated cells were fixed, stained with 0.1% crystal violet solution, and the number of cells were counted under a light microscope.
Determination of glycolysis
The glucose uptake, lactate production and ATP level in BC cells were assessed using the commercial Glucose Colorimetric/Fluorometric Assay Kit, Lactate Assay Kit, and ATP Assay Kit (Sigma-Aldrich) following the manufacturer’s instructions, respectively.
Western blotting analysis
RIPA lysis buffer (Thermo Fisher) was used for total protein isolation. The protein samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by transferring onto the polyvinylidene fluoride membranes. The membranes were incubated with the primary antibodies against FOXP4 (1:1000, PA5-49682, Thermo Fisher), proliferating cell nuclear antigen (PCNA; 1:500, PA5-27214, Thermo Fisher), matrix metalloproteinase (MMP) 9 (1:1,000, MA5-32705, Thermo Fisher), hexokinase 2 (HK2; 1:1,000, MA5-14849, Thermo Fisher), and β-actin (1:5000, bs-0061R, Bioss, Beijing, China). The secondary antibody Goat Anti-rabbit IgG/HRP (1:1000, bs-0295G-HRP, Bioss) was applied. The protein bands were visualized using EnlightTM-Plus (Engreen Biosystem, Birkenhead, AKL, New Zealand).
Tumor formation in nude mice
Twelve six-week-old female nude mice were purchased from Slac Jingda Laboratory Animal Co., Ltd., (Hunan, China) and randomly divided into sh-NC and sh-circRPPH1 groups (n=6 per group). The mice were subcutaneously injected with 1×107 stably transfected MDA-MB-231 cells. Every seven days after the injection, the length and width of transplanted tumors were measured. Tumor volume = (width)2 × length/2. On the 28th day, the xenograft tumors were removed and weighed.
Prediction analysis
The potential target miRNAs of circRPPH1 were predicted using Circatlas (http://circatlas.biols.ac.cn/), CircBank (http://www.circbank.cn/), and StarBase (http://starbase.sysu.edu.cn/index.php) databases. To predict the target genes of miR-296-5p, RNA22 (http://www.mybiosoftware.com/rna22-v2-microrna-target-detection.html), Targetscan (http://www.targetscan.org/vert_71/), and PicTar (https://pictar.mdc-berlin.de/) databases were adopted.
Dual-luciferase reporter assay
The 3’ untranslated regions (3’UTR) sequence of circRPPH1/FOXP4 (circRPPH1/FOXP4-WT) or a mutant sequence (circRPPH1/FOXP4-MUT) was inserted into pmirGLO vector (Promega, Madison, WI, USA). MCF-7 and MDA-MB-231 cells were co-transfected with circRPPH1/FOXP4-WT or circRPPH1/FOXP4-MUT plasmid together with miR-296-5p mimics or mimics NC using Lipofectamine 2000. The dual luciferase activities were detected using a Dual Luciferase Reporter Gene Assay Kit (Yeasen, Shanghai, China).
RNA immunoprecipitation (RIP)
The EZ-Magna RIP Kit (Millipore, Bedford, MA, USA) was used for assessing the binding between circRPPH1 and miR-296-5p. Briefly, the lysate of BC cells was incubated with the complex of magnetic beads with Ago-2 or IgG antibody at 4°C overnight. Thereafter, the immunoprecipitated RNA was isolated from the magnetic beads using TRIzol reagent. CircRPPH1 and miR-296-5p levels were detected by qRT-PCR.
RNA pull-down
The biotinylated miR-296-5p (bio-miR-296-5p) or bio-NC was transfected into BC cells. Forty-eight hours later, the cells were lysed in lysis buffer, followed by incubation with magnetic beads (Sigma-Aldrich) at 4°C overnight. Then the bound RNA was isolated, and circRPPH1 level was detected by qRT-PCR.
Statistical analysis
Experimental data are presented as mean ± standard deviation (x̅ ± sd). Student’s t test for two groups’ comparison or One-Way ANOVA for multiple group comparison was performed using GraphPad prism 6. P value less than 0.05 was considered as significant difference.
Results
circRPPH1 expression was increased in BC
The differentially expressed circRNAs in BC tissues were obtained from the online GEO datasets (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101123) containing circRNA expression profile (GSE101123). We observed that circRPPH1 was significantly up-regulated in BC tissues (Figure 1A). Furthermore, the up-regulation of circRPPH1 was validated in 70 BC tissues (Figure 1B). Notably, the increased expression of circRPPH1 was associated with poor prognosis of BC patients (Figure 1C). Moreover, the clinical parameters of BC patients with high and low circRPPH1 expression were summarized in Table 3. We found that high circRPPH1 expression was closely correlated with lymph node metastasis and higher TNM stages. Similarly, circRPPH1 level was elevated in a series of BC cells (Figure 1D). MCF-7 and MDA-MB-231 cells with higher circRPPH1 level were selected for the subsequent experiments. Moreover, actinomycin D or RNase R treatment was used to investigate circRPPH1 stability. As detected by qRT-PCR, the linear mRNA level was significantly reduced, whereas the circRPPH1 level in BC cells was not changed in response to actinomycin D or RNase R treatment (Figure 1E, 1F). The above data indicated that circRPPH1 might play a role in BC development.
Figure 1.
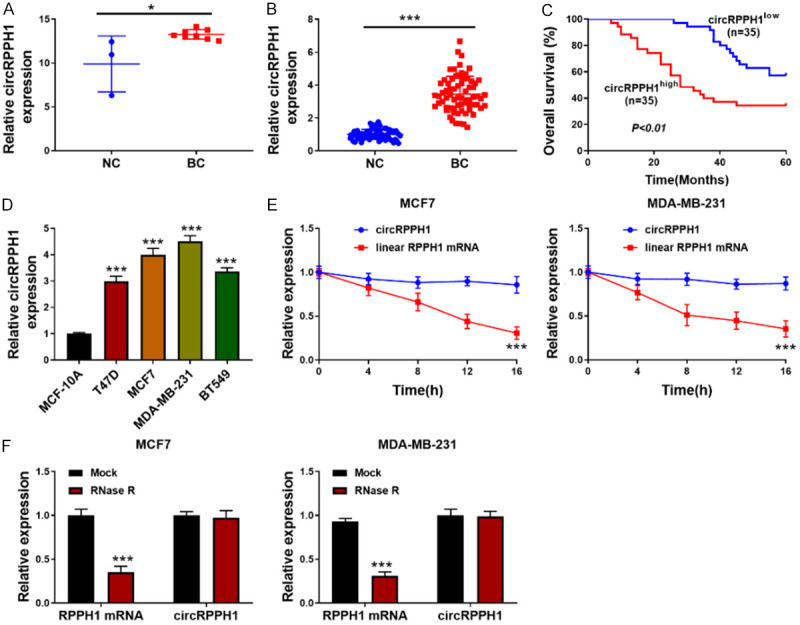
circRPPH1 was up-regulated in BC. A: circRPPH1 was up-regulated in BC tissues from GEO database (GSE101123); B: qRT-PCR for circRPPH1 level in 70 paired BC and surrounding normal tissues; C: The overall survival of BC patients; D: CircRPPH1 level in various BC cells was examined using qRT-PCR; E: BC cells were treated with actinomycin D, and circRPPH1, and its linear mRNA expression was evaluated by qRT-PCR; F: qRT-PCR for circRPPH1 and its linear mRNA expression in BC cells in response to RNase R. Data are shown as mean ± standard deviation, *P<0.05, ***P<0.001.
Table 3.
Correlation between circRPPH1 expression and clinicopathological parameters of BC patients
| Characteristics | Number | circRPPH1 expression | P | |
|---|---|---|---|---|
|
| ||||
| High (n=35) | Low (n=35) | |||
| Age (years) | 0.811 | |||
| <50 | 37 | 19 | 18 | |
| ≥50 | 33 | 16 | 17 | |
| Menopause | 0.212 | |||
| No | 45 | 25 | 20 | |
| Yes | 25 | 10 | 15 | |
| Tumor size (cm) | 0.337 | |||
| ≤2 | 38 | 17 | 21 | |
| >2 | 32 | 18 | 14 | |
| Lymph node metastasis | 0.031* | |||
| Yes | 37 | 23 | 14 | |
| No | 33 | 12 | 21 | |
| TNM stage | 0.008* | |||
| I | 31 | 10 | 21 | |
| II-III | 39 | 25 | 14 | |
Note: BC: Breast cancer.
P<0.05, statistically significant.
Silencing of circRPPH1 repressed BC cell proliferation, metastasis and glycolysis
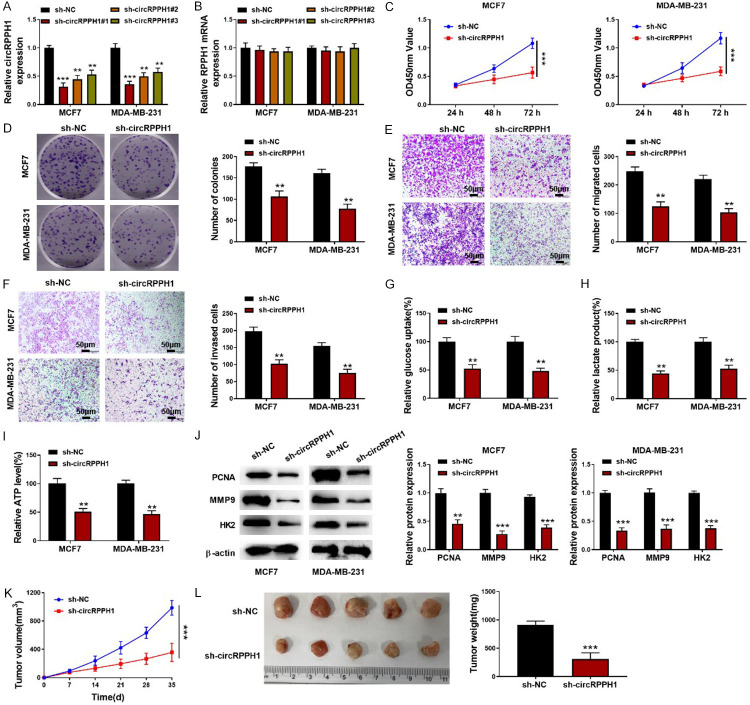
Next, three shRNAs targeting circRPPH1 were transfected into BC cells. The silencing efficiency was validated by qRT-PCR (Figure 2A). Among three shRNAs, the sh-circRPPH1 #1 exhibited the highest efficiency and therefore was selected in the subsequent experiments. As presented in Figure 2B, the linear mRNA level was not affected by shRNAs targeting circRPPH1. In addition, MCF-7 and MDA-MB-231 cells were transfected with sh- circRPPH1 #1. We observed that circRPPH1 depletion suppressed the growth of BC cells (Figure 2C, 2D). The migration and invasion were strikingly inhibited in circRPPH1-silenced BC cells (Figure 2E, 2F). Additionally, down-regulation of circRPPH1 remarkably repressed glycolysis of BC cells (Figure 2G-I). Western blotting assay revealed that the protein levels of PCNA, MMP9, and HK2 were decreased after silencing of circRPPH1 (Figure 2J). As shown in Figure 2K, 2L, a significant decrease in tumor volume and weight was observed in circRPPH1-silenced group in comparison to sh-NC group. These results revealed that circRPPH1 knockdown inhibited proliferation, metastasis and glycolysis of BC cells.
Figure 2.
Knockdown of circRPPH1 restrained growth, metastasis and glycolysis of BC cells. A, B: Expression of circRPPH1 and its linear mRNA was detected by qRT-PCR after transfection with shRNAs targeting circRPPH1; C, D: CCK-8 and colony formation assay for the growth of BC cells; E, F: Transwell assays for evaluating BC cell migration and invasion (200×); G-I: The glycolysis of BC cells was assessed using commercial kits; J: Western blotting for determining PCNA, MMP9, and HK2 protein abundance; K, L: The tumor volume and weight were detected. Data are shown as mean ± standard deviation, **P<0.01, ***P<0.001.
CircRPPH1 directly bond to miR-296-5p
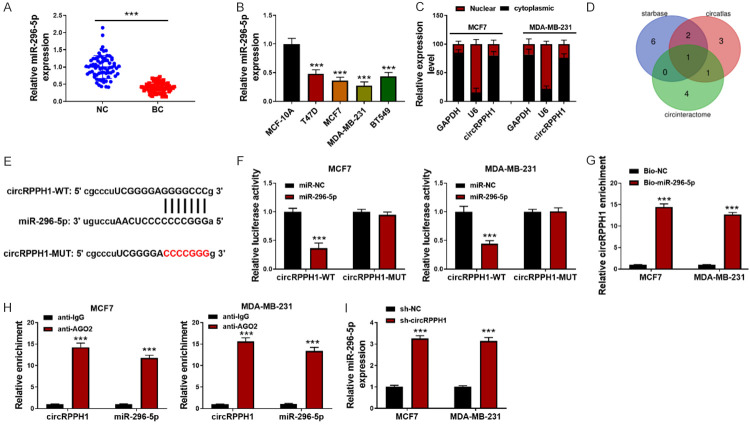
As assessed by qRT-PCR, a lower miR-296-5p expression was found in 70 BC tissues in comparison with surrounding normal tissues (Figure 3A). Moreover, down-regulation of miR-296-5p was validated in multiple BC cells (Figure 3B). To assess the cellular distribution of circRPPH1 in BC cells, the nuclear and cytoplasmic fractions were prepared. As illustrated in Figure 3C, circRPPH1 was mostly located in the cytoplasm of BC cells. To predict the target miRNAs of circRPPH1, we overlapped the results of Circatlas, CircBank, and StarBase databases and miR-296-5p was found to be the only candidate (Figure 3D). The complementary sites between circRPPH1 and miR-296-5p are shown in Figure 3E. A reduced luciferase activity was observed after co-transfection with miR-296-5p mimics and circRPPH1-WT in MCF-7 and MDA-MB-231 cells, which was abolished in circRPPH1-MUT group (Figure 3F). BC cells were transfected with bio-miR-296-5p or bio-NC. RNA pull-down experiment showed that use of biotin-labeled miR-296-5p probe led to increased enrichment of circRPPH1 in BC cells (Figure 3G). In addition, interaction between circRPPH1 and miR-296-5p was further confirmed by RIP assay (Figure 3H). Moreover, miR-296-5p level was remarkably enhanced after silencing of circRPPH1 (Figure 3I). Collectively, miR-296-5p was demonstrated to be the target gene of circRPPH1.
Figure 3.
CircRPPH1 directly interacts with miR-296-5p. A: qRT-PCR for miR-296-5p level in 70 paired BC and surrounding normal tissues; B: miR-296-5p expression in multiple BC cells was assessed by qRT-PCR; C: qRT-PCR for assessing expression of circRPPH1 in the nuclear and cytoplasmic fractions of BC cells; D: MiR-296-5p was predicted as a target of circRPPH1 by Circatlas, CircBank, and StarBase databases; E: The putative binding sites between circRPPH1 and miR-296-5p are shown; F: The direct binding between circRPPH1 and miR-296-5p was confirmed by dual luciferase assay; G: RNA pull-down assay; H: RIP assay; I: qRT-PCR for miR-296-5p expression in BC cells after silencing of circRPPH1. Data are shown as mean ± standard deviation, ***P<0.001.
CircRPPH1 affected BC cell proliferation, metastasis and glycolysis via regulating miR-296-5p
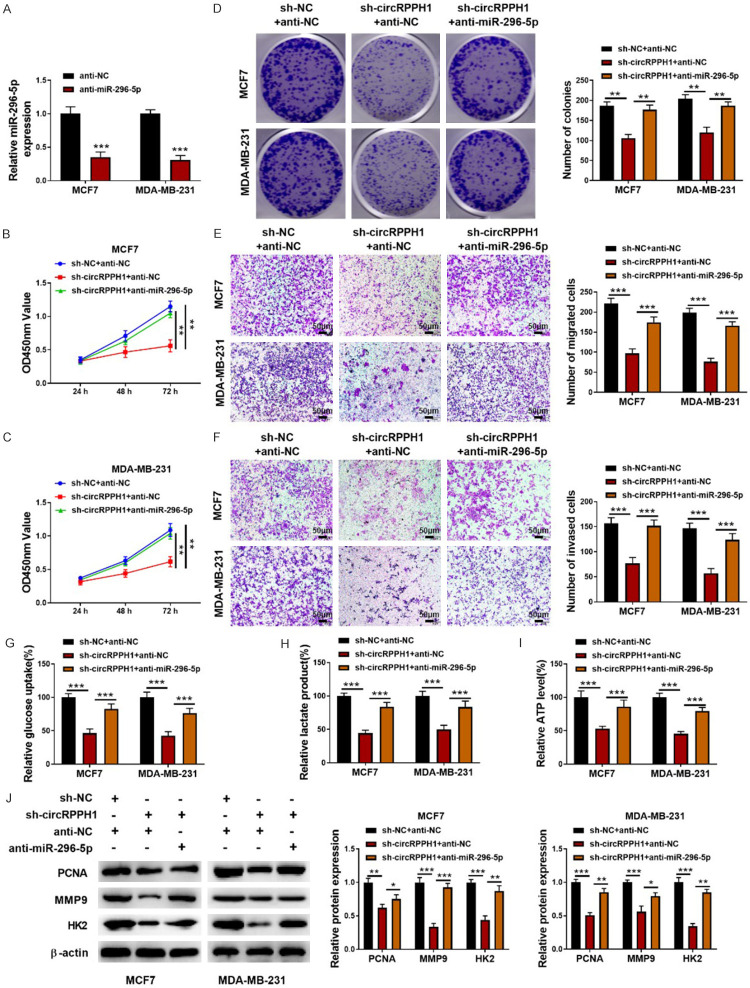
The involvement of miR-295-5p in circRPPH1-mediated biological function was determined using rescue experiments. For this purpose, MCF-7 and MDA-MB-231 cells were transfected with sh-circRPPH1 together with or without miR-295-5p inhibitor. The inhibition of miR-295-5p expression after transfection with miR-295-5p inhibitor was validated (Figure 4A). Functional experiments demonstrated that sh-circRPPH1-mediated suppression in proliferation and metastasis was reversed by miR-295-5p inhibitor (Figure 4B-F). Besides, the reduced glucose uptake, lactate production, and ATP level in circRPPH1-depleted BC cells were restored by the inhibition of miR-295-5p (Figure 4G-I). In addition, miR-295-5p inhibitor effectively counteracted the decreased protein levels of PCNA, MMP9, and HK2 induced by circRPPH1 suppression (Figure 4J). Therefore, circRPPH1 affected the malignant phenotypes of BC cells by targeting miR-295-5p.
Figure 4.
miR-296-5p abolished the regulation of circRPPH1 in BC cells. A: Anti-miR-295-5p-mediated decreased miR-295-5p expression was validated by qRT-PCR; B, C: CCK8 assay for the proliferation of BC cells from different groups; D: The growth of BC cells was measured by colony formation assay; E, F: Transwell assays for the migration and invasion of BC cells (200x); G-I: The glycolysis of BC cells with various treatments was determined using commercial kits; J: Western blotting for PCNA, MMP9, and HK2 protein abundance. Data are shown as mean ± standard deviation, *P<0.05, **P<0.01, ***P<0.001.
FOXP4 was a target gene of miR-296-5p
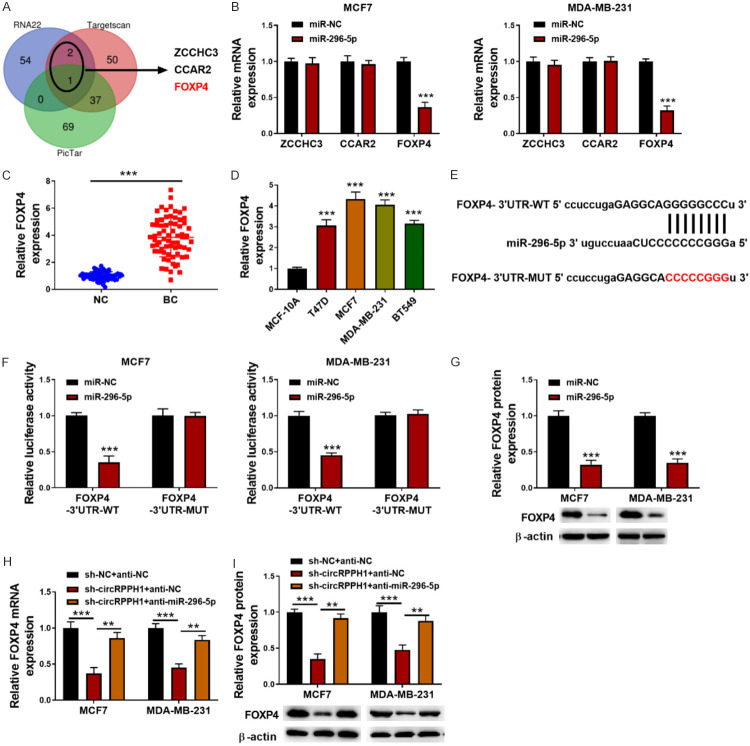
To explore the potential target genes of miR-296-5p, RNA22, Targetscan and Pictar databases were adopted. By overlapping the results, ZCCHC3, CCAR2, and FOXP4 were displayed as the candidate genes (Figure 5A). Further experiments indicated that miR-296-5p overexpression strikingly down-regulated FOXP4 mRNA level, whereas ZCCHC3 and CCAR2 mRNA levels were not affected (Figure 5B). Thus, FOXP4 was selected as the target of miR-296-5p. As shown in Figure 5C, 5D, the mRNA level of FOXP4 was up-regulated in 70 BC tissues and a series of BC cells. miR-296-5p and FOXP4 binding sites are shown in Figure 5E. miR-296-5p mimics obviously reduced the luciferase activity of FOXP4-WT in MCF-7 and MDA-MB-231 cells, but not that of FOXP4-MUT (Figure 5F). Besides, enforced expression of miR-296-5p reduced FOXP4 protein level (Figure 5G). Furthermore, sh-circRPPH1 with or without miR-295-5p inhibitor was transfected into MCF-7 and MDA-MB-231 cells. The down-regulated mRNA and protein levels of FOXP4 in circRPPH1-silenced BC cells were counteracted by miR-296-5p inhibitor (Figure 5H, 5I). These findings indicated that circRPPH1 sequestered miR-296-5p to suppress its inhibition of FOXP4 expression.
Figure 5.
miR-296-5p directly targets FOXP4 in BC cells. A: ZCCHC3, FOXP4, and CCAR2 were predicted as target genes of miR-296-5p using RNA22, Targetscan, and Pictar databases; B: qRT-PCR for the mRNA expression of ZCCHC3, FOXP4, and CCAR2 in miR-296-5p-overexpressed BC cells; C: The mRNA expression of FOXP4 in 70 paired BC and surrounding normal tissues was determined by qRT-PCR; D: FOXP4 mRNA expression in various BC cells was detected by qRT-PCR; E: The putative binding sites between FOXP4 and miR-296-5p are shown; F: Dual luciferase assay for evaluating the interaction between FOXP4 and miR-296-5p; G: Western blotting for detecting FOXP4 protein abundance in miR-296-5p-overexpressed BC cells; H, I: FOXP4 expression in BC cells from different groups was assessed by qRT-PCR and Western blotting, respectively. Data are shown as mean ± standard deviation, **P<0.01, ***P<0.001.
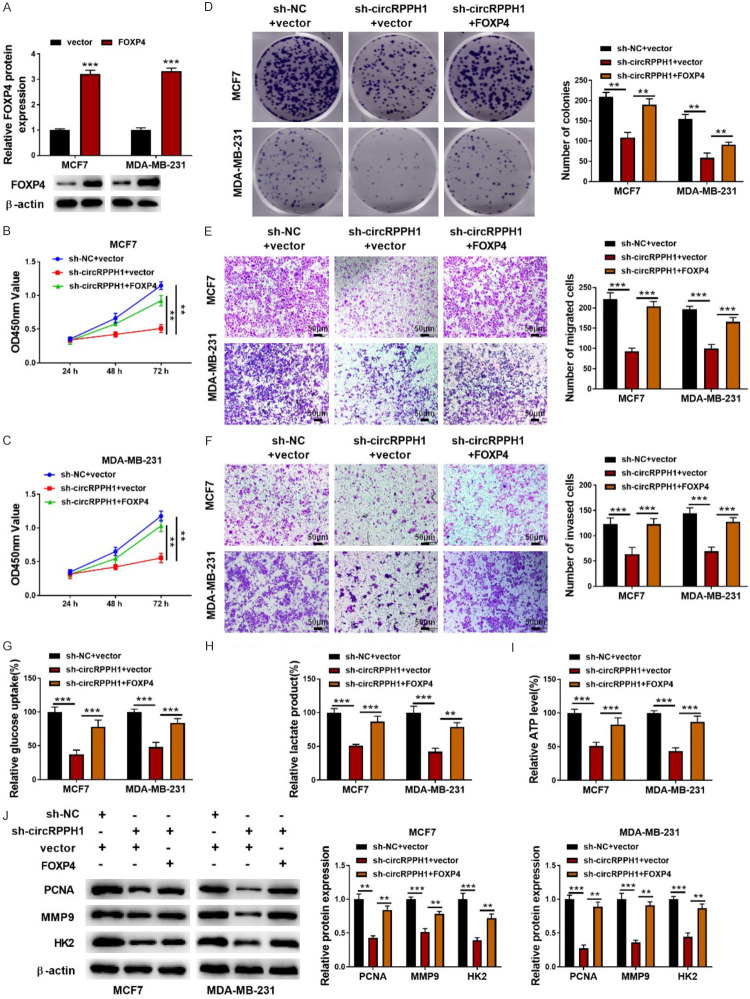
FOXP4 abolished the inhibition of circRPPH1 knockdown on BC cell proliferation, metastasis and glycolysis
To determine whether circRPPH1 regulated BC progression via miR-296-5p/FOXP4 axis, BC cells transfected with sh-circRPPH1 were further transfected with vector or FOXP4 overexpression plasmid. As presented in Figure 6A, overexpression of FOXP4 in BC cells after transfection with FOXP4 plasmid was verified by Western blotting. Overexpression of FOXP4 counteracted sh-circRPPH1-mediated repression in growth, migration, and invasion of BC cells (Figure 6B-F). Likewise, circRPPH1 knockdown-induced glycolysis inhibition was abrogated by FOXP4 overexpression as evidenced by enhanced glucose uptake, lactate production, and ATP level (Figure 6G-I). Western blotting analysis demonstrated that the decreased levels of PCNA, MMP9, and HK2 in circRPPH1-silenced MCF7 and MDA-MB-231 cells were reversed by FOXP4 overexpression (Figure 6J). Therefore, FOXP4 was involved in circRPPH1-mediated BC progression.
Figure 6.
FOXP4 weakened circRPPH1 knockdown-mediated suppression of BC cell growth, metastasis and glycolysis. A: Overexpression of FOXP4 was confirmed by Western blotting; B-D: CCK8 and colony formation assay for determining the growth of BC cells; E, F: Transwell assays for evaluating BC cell migratory and invasive abilities (200×); G-I: The glycolysis of BC cells was assessed using commercial kits; J: Western blotting for PCNA, MMP9, and HK2 protein abundance. Data are shown as mean ± standard deviation, **P<0.01, ***P<0.001.
Discussion
BC has been considered as one of the key reasons for cancer-related death worldwide. To improve the therapeutic effectiveness of BC, it is crucial to better understand its pathogenesis. In this study, a novel circRNA, circRPPH1, was identified to be up-regulated in BC tissues and cells. One mechanism is that circRPPH1 enhanced FOXP4 expression through sponging miR-296-5p. Functional experiments demonstrated that circRPPH1 promoted the growth, metastasis and glycolysis of BC cells via regulating miR-296-5p/FOXP4 axis.
Growing evidence has demonstrated that circRNAs participate in the complicated pathogenesis of BC [13,14]. Even so, the biological functions and mechanisms of circRNAs in BC are still far from clear. By the selection of differentially expressed circRNA from the online GEO database (GSE101123), circRPPH1 was focused on in the present study. Our further results validated that circRPPH1 was remarkably up-regulated in BC cells and tissues, which was closely correlated with lymph node metastasis and TMN stages. Besides, functional experiments demonstrated that depletion of circRPPH1 repressed the growth and metastasis of BC cells. Moreover, the glycolysis of BC cells was further investigated. Metabolic alteration has been widely recognized as a notable feature of cancer cells, by which glycolysis provides cancer cells with energy needed, even under normoxia condition [15,16]. Cancer cells are prone to produce ATP through transforming glucose into lactate by the glycolysis pathway, which may result in increased glucose uptake and lactate production [17]. In the present study, circRPPH1 knockout significantly restrained the glycolysis of BC cells. These data revealed that circRPPH1 functioned as a cancer-promoting gene in BC.
It has been widely recognized that circRNAs affect tumor progression through regulating gene expression via sequestering miRNAs [18]. For instance, hsa_circ_0001785 delayed the progression of BC via sponging miR-942 to increase SOCS3 expression [19]. Whereas, cirCHIPK3 functioned as a promotor in BC development by sequestering miR-193a to enhance HMGB1 expression [20]. In this study, we inferred a predicted binding between circRPPH1 and miR-296-5p. The anti-tumor effects of miR-296-5p have been documented in various human cancers. For example, miR-296-5p suppressed esophageal squamous cell carcinoma cell metastasis by decreasing STAT3 expression [21]. Shi et al. found that miR-296-5p restrained the stemness potency of liver tumor cells by modulating Brg1/Sall4 pathway [22]. Besides, miR-296-5p level was verified to be decreased in BC, and enforced expression of miR-296-5p could repress in vivo tumor growth [23]. In this study, the interaction between circRPPH1 and miR-296-5p was confirmed. Furthermore, inhibition of miR-296-5p could abolish sh-circRPPH1-mediated anti-tumor effects on BC. Collectively, these data demonstrated that circRPPH1 conferred the malignant properties of BC cells via sponging miR-296-5p.
Finally, the target gene of circRPPH1/miR-296-5p axis was investigated. miRNAs perform their roles in various biological processes via regulating the expression of target genes [24]. In this study, FOXP4 was predicted as a target gene of miR-296-5p by RNA22, Targetscan, and Pictar databases, which was further confirmed by dual luciferase assay. It has been revealed that up-regulation of FOXP4 could facilitate the malignant progression of multiple human cancers, including liver cancer, lung cancer, and prostate cancer [25-27]. Notably, FOXP4 was up-regulated in BC, which contributed to the malignant development of BC [12]. In this work, FOXP4 was dramatically down-regulated by miR-296-5p overexpression. Compelling evidence suggests that circRNAs can post-transcriptionally modulate gene expression through functioning as ceRNAs to competitively sponge their targeted miRNAs [28]. A recent study by Li et al. reported that hsa_circ_0001785 enhanced SOCS3 expression via sponging miR-942, which prevented the progression of BC [19]. Jiang et al. demonstrated that circ_002178 depletion restrained BC progression via sequestering miR-1258 to attenuate its inhibitory effect on KDM7A expression [29]. Consistently, our data indicated that circRPPH1 depletion suppressed FOXP4 expression, which was remarkably reversed by transfection with miR-296-5p inhibitor. Furthermore, FOXP4 inhibition counteracted the anti-cancer effect of sh-circRPPH1 on BC cells. Therefore, these findings suggested that circRPPH1 contributed to the malignant development of BC via sequestering miR-296-5p to enhance FOXP4 expression.
In this study, the differential expression of circRPPH1, miR-296-5p and FOXP4 was examined across various sub-types of BC cells, including MCF-7 and T47D (luminal A-type BC cells), MDA-MB-231 and BT549 (triple-negative BC cells). Moreover, the function of circRPPH1/miR-296-5p/FOXP4 axis was explored in MCF-7 and MDA-MB-231 cells, two different sub-types of BC. Therefore, circRPPH1/miR-296-5p/FOXP4 axis had a uniform function in different sub-types of BC cells.
Taken together, circRPPH1 was up-regulated in BC, which was closely correlated with the malignant progression of BC patients. Moreover, we found that circRPPH1 silencing restrained BC cell growth, metastasis, and glycolysis by modulating the miR-296-5p/FOXP4 axis. Collectively, circRPPH1 may be an effective therapeutic target for multiple sub-types of BC.
Disclosure of conflict of interest
None.
Abbreviations
- circRNAs
circular RNAs
- BC
breast cancer
- RNase
ribonuclease
- miRNAs
microRNAs
- FOXP4
forkhead box protein 4
- FBS
fetal bovine serum
- CCK-8
cell counting kit -8
- MMP
matrix metalloproteinase
- HK2
hexokinase 2
- PCNA
proliferating cell nuclear antigen
- 3’UTR
3’ untranslated regions
- ZCCHC3
zinc finger CCHC-type containing 3
- CCAR2
cell cycle and apoptosis regulator
- RIP
RNA immunoprecipitation
- shRNA
short hairpin RNA
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Clarke M, Collins R, Darby S, Davies C, Elphinstone P, Evans V, Godwin J, Gray R, Hicks C, James S, MacKinnon E, McGale P, McHugh T, Peto R, Taylor C, Wang Y. Effects of radiotherapy and of differences in the extent of surgery for early breast cancer on local recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;366:2087–2106. doi: 10.1016/S0140-6736(05)67887-7. [DOI] [PubMed] [Google Scholar]
- 3.Harjes U. Breast cancer: staying silent. Nat Rev Cancer. 2018;18:136. doi: 10.1038/nrc.2018.17. [DOI] [PubMed] [Google Scholar]
- 4.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 5.Ma Y, Niu X, Yan S, Liu Y, Dong R, Li Y. Circular RNA profiling facilitates the diagnosis and prognostic monitoring of breast cancer: a pair-wise meta-analysis. J Clin Lab Anal. 2021:e23575. doi: 10.1002/jcla.23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Ma F, Wu L, Zhang X, Tian J, Li J, Cao J, Ma Y, Zhang L, Wang L. Identification of Hsa_circ_0104824 as a potential biomarkers for breast cancer. Technol Cancer Res Treat. 2020;19:1533033820960745. doi: 10.1177/1533033820960745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Shen W, Xu J, Gong B, Gao B, Zhu J. circUSP42 is downregulated in triple-negative breast cancer and associated with poor prognosis. Technol Cancer Res Treat. 2020;19:1533033820950827. doi: 10.1177/1533033820950827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinami R, Buemi V, Sestito R, Zappone A, Ciani Y, Mano M, Petti E, Sacconi A, Blandino G, Giacca M, Piazza S, Benetti R, Schoeftner S. Epigenetic silencing of mir-296 and mir-512 ensures htert dependent apoptosis protection and telomere maintenance in basal-type breast cancer cells. Oncotarget. 2017;8:95674–95691. doi: 10.18632/oncotarget.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong Y, Yang L, Wei W, Lyu N, Zou Y, Gao G, Ou X, Xie X, Tang H. CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics. 2019;11:1163–1176. doi: 10.2217/epi-2019-0093. [DOI] [PubMed] [Google Scholar]
- 11.Myatt SS, Lam EW. The emerging roles of forkhead box proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 12.Ma T, Zhang J. Upregulation of FOXP4 in breast cancer promotes migration and invasion through facilitating EMT. Cancer Manag Res. 2019;11:2783–2793. doi: 10.2147/CMAR.S191641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Jiang B, He Z, Zhu H, He R, Fan S, Wu X, Xie L, He X. circIQCH sponges mir-145 to promote breast cancer progression by upregulating DNMT3A expression. Aging. 2020;12:15532–15545. doi: 10.18632/aging.103746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Liu F, Ma H, Cui X, Yang S, Qin R. circCDYL acts as a tumor suppressor in triple negative breast cancer by sponging mir-190a-3p and upregulating TP53INP1. Clin Breast Cancer. 2020;20:422–430. doi: 10.1016/j.clbc.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Hsu PP, Sabatini DM. Cancer cell metabolism: warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Cao L, Wu J, Qu X, Sheng J, Cui M, Liu S, Huang X, Xiang Y, Li B, Zhang X, Cui R. Glycometabolic rearrangements--aerobic glycolysis in pancreatic cancer: causes, characteristics and clinical applications. J Exp Clin Cancer Res. 2020;39:267. doi: 10.1186/s13046-020-01765-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shankaraiah RC, Veronese A, Sabbioni S, Negrini M. Non-coding RNAs in the reprogramming of glucose metabolism in cancer. Cancer Lett. 2018;419:167–174. doi: 10.1016/j.canlet.2018.01.048. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol. 2015;8:30. doi: 10.1186/s13045-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Zheng J, Lin W, Weng J, Hong W, Zou J, Zhang T, Ye C, Chen Y. Circular RNA hsa_circ_0001785 inhibits the proliferation, migration and invasion of breast cancer cells in vitro and in vivo by sponging mir-942 to upregulate SOCS3. Cell Cycle. 2020;19:2811–2825. doi: 10.1080/15384101.2020.1824717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen ZG, Zhao HJ, Lin L, Liu JB, Bai JZ, Wang GS. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thorac Cancer. 2020;11:2660–2671. doi: 10.1111/1759-7714.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang ZZ, Luo YR, Du J, Yu Y, Yang XZ, Cui YJ, Jin XF. MiR-296-5p inhibits cell invasion and migration of esophageal squamous cell carcinoma by downregulating STAT3 signaling. Eur Rev Med Pharmacol Sci. 2019;23:5206–5214. doi: 10.26355/eurrev_201906_18185. [DOI] [PubMed] [Google Scholar]
- 22.Shi DM, Shi XL, Xing KL, Zhou HX, Lu LL, Wu WZ. miR-296-5p suppresses stem cell potency of hepatocellular carcinoma cells via regulating Brg1/Sall4 axis. Cell Signal. 2020;72:109650. doi: 10.1016/j.cellsig.2020.109650. [DOI] [PubMed] [Google Scholar]
- 23.Savi F, Forno I, Faversani A, Luciani A, Caldiera S, Gatti S, Foa P, Ricca D, Bulfamante G, Vaira V, Bosari S. miR-296/scribble axis is deregulated in human breast cancer and mir-296 restoration reduces tumour growth in vivo. Clin Sci. 2014;127:233–242. doi: 10.1042/CS20130580. [DOI] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G, Zhang G. Upregulation of FoxP4 in HCC promotes migration and invasion through regulation of EMT. Oncol Lett. 2019;17:3944–3951. doi: 10.3892/ol.2019.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang T, Li H, Thakur A, Chen T, Xue J, Li D, Chen M. FOXP4 modulates tumor growth and independently associates with mir-138 in non-small cell lung cancer cells. Tumour Biol. 2015;36:8185–8191. doi: 10.1007/s13277-015-3498-8. [DOI] [PubMed] [Google Scholar]
- 27.Huang C, Deng H, Wang Y, Jiang H, Xu R, Zhu X, Huang Z, Zhao X. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23:6112–6119. doi: 10.1111/jcmm.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Yang X, Shi C, Zhou Z. Hsa_circ_002178 promotes the growth and migration of breast cancer cells and maintains cancer stem-like cell properties through regulating miR-1258/KDM7A axis. Cell Transplant. 2020;29:963689720960174. doi: 10.1177/0963689720960174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]