Highlights
-
•
NUT carcinoma is a rare but aggressive solid tumor that can be diagnosed by presence of the BRD4-NUT fusion.
-
•
This series presents 31 cases of solid tumors that harbor BRD4-NUT but often carry other diagnoses such NSCLC—NOS and NSCLC-SCC.
-
•
Despite lack of PD-L1 expression and a low tumor mutational burden, two index cases responded to either nivolumab or atezolizumab+chemotherapy with partial response or better with 4–5 month duration of response.
-
•
The unexpected response to checkpoint inhibitors could be explained by a very high affinity of the fusion peptide at the junction of BRD4 and NUT to the MHC complex as recently suggested for an exceptional response to an immune checkpoint inhibitor in a fusion bearing low TMB, low PD-L1 expression head and neck carcinoma.
Keywords: NUT midline carcinoma, NUT carcinoma, BRD4-NUT, Checkpoint inhibitor, PD-L1, BRD4
Abstract
Background
The translocation t(15:19) produces the oncogenic BRD4-NUT fusion which is pathognomonic for NUT carcinoma (NC), which is a rare, but extremely aggressive solid tumor. Comprehensive genomic profiling (CGP) by hybrid-capture based next generation sequencing of 186+ genes of a cohort of advanced cancer cases with a variety of initial diagnoses harboring BRD4-NUT may shed further insight into the biology of these tumors and possible options for targeted treatment.
Case presentation
Thirty-one solid tumor cases harboring a BRD4-NUT translocation are described, with only 16% initially diagnosed as NC and the remainder carrying other diagnoses, most commonly NSCLC—NOS (22%) and lung squamous cell carcinoma (NSCLC-SCC) (16%). The cohort was all microsatellite stable and harbored a low Tumor Mutational Burden (TMB, mean 1.7 mut/mb, range 0–4). In two index cases, patients treated with immune checkpoint inhibitors (ICPI) had unexpected partial or better responses of varying duration. Notably, four cases – including the two index cases - were negative for PD-L1 expression. Neo-antigen prediction for BRD4-NUT and then affinity modeling of the peptide-MHC (pMHC) complex for an assessable index case predicted very high affinity binding, both on a ranked (99.9%) and absolute (33 nM) basis.
Conclusions
CGP identifies BRD4-NUT fusions in advanced solid tumors which carry a broad range of initial diagnoses and which should be re-diagnosed as NC per guidelines. A hypothesized mechanism underlying responses to ICPI in the low TMB, PD-L1 negative index cases is the predicted high affinity of the BRD4-NUT fusion peptide to MHC complexes. Further study of pMHC affinity and response to immune checkpoint inhibitors in patients with NC harboring BRD4-NUT is needed to validate this therapeutic hypothesis.
Background
NUT carcinoma (NC), formerly known as NUT midline carcinoma, is an extremely rare and highly aggressive neoplasm with a median survival estimated as less than one year [1]. Morphologically, NC is poorly differentiated and may display squamous features but overall, the appearance can be quite variable.
The seminal study of French demonstrated NC frequently (~60%) harbors a t(15:19) rearrangement encoding the BRD4-NUT fusion and this translocation is essential for the diagnostic criteria for a subset of NC [1,3]. Recent studies have further identified other subsets of NC as harboring other rearrangements of NUTM1 (encoding NUT) fused with other non-BRD4 5′ partners including BRD2, BRD3 and NSD3 together composing approximately 30% of NC cases [2,4]. The diagnostic workup consists of including NC on in the differential diagnosis as based on clinico-pathologic features such as squamous morphology, and then seeking FISH to definitively identify the NUTM1 translocation harbored by a particular case.
Importantly, the development of a monoclonal anti-NUT antibody (C52) for immunohistochemistry (IHC) to detect the strong ectopic expression of NUT associated with the above NUTM1 rearrangements has significantly enhanced the diagnostic screening for NC due to the ease of IHC [5]. However, due to partial uptake of the diagnostic algorithm and associated tools by pathologists, the incidence and prevalence of NC is likely still significantly underestimated [2].
Clinical evidence for the efficacy of treatment regimens for advanced NC is limited to retrospective case series, which has demonstrated some survival benefit for cytotoxic chemotherapy [2], [6]. Chief among possible targeted therapies for NC are BET (Bromodomain and Extra-terminal) inhibitors, of which MK-8628 have shown a high response rate but limited duration of response in a compassionate use study of four NC patients [7].
We report here a cohort of 31 solid tumor cases carrying NC and other initial diagnoses that harbor BRD4-NUT fusions as identified by clinical comprehensive genomic profiling (CGP) and highlight two index patients that unexpectedly benefitted from PD-1 and PD-L1 inhibition.
Case series
We reviewed a large database of advanced cancer patients assayed with DNA only NGC-based CGP baiting for BRD4 and identified BRD4-NUT fusions in 0.02% (31/181,838) of cases. The original diagnoses of these cases were NC (16%, 5/31), NSCLC—NOS (22%, 7/31), NSCLC-SCC (16%, 5/31), adnexal carcinoma of the skin (6.5%, 2/31) as well as other diagnoses for single cases (3% each) (Table 1). This cohort had a median age of 43 years (range 21–81) and were 14 females and 17 males. 60% had BRD4-NUT as the only functional alteration and the overall series had a mean of less than one (<1) other alteration per case (Supplemental Table 1). The co-occurring alterations were predominantly point mutations, rarely copy number alterations, and no other genomic rearrangements were observed. Consistent with the paucity of point mutations, the median TMB of cases in this series was 1.7 mut/mb and all cases were microsatellite stable (MSS) as well. None (0%) of the BRD4-NUT fusion positive cases harbored evidence of HPV infection (presence of virion DNA). Two cases in these series were known to be treated with immune checkpoint inhibitors (ICPI) and are highlighted below due to the unexpectedly efficacious responses relative to an absence of known predictive biomarkers.
Table 1.
Frequency of BRD4-NUT fusions in advanced cancer cases.
| Disease | Disease Count | Count | % of BRD4-NUT | % of Disease |
|---|---|---|---|---|
| lung non-small cell lung carcinoma (nsclc-nos) | 4996 | 7 | 22.58% | 0.14% |
| lung squamous cell carcinoma (nsclc-scc) | 4895 | 5 | 16.13% | 0.10% |
| nut midline carcinoma | 15 | 5 | 16.13% | 33.33% |
| skin adnexal carcinoma | 135 | 2 | 6.45% | 1.48% |
| lung small cell undifferentiated carcinoma (sclc) | 1723 | 1 | 3.23% | 0.06% |
| nasopharynx and paranasal sinuses squamous cell carcinoma (scc) | 92 | 1 | 3.23% | 1.09% |
| unknown primary (nos) | 817 | 1 | 3.23% | 0.12% |
| testis germ cell tumor (non-seminoma) | 108 | 1 | 3.23% | 0.93% |
| salivary gland carcinoma (nos) | 294 | 1 | 3.23% | 0.34% |
| unknown primary melanoma | 2590 | 1 | 3.23% | 0.04% |
| nasopharynx and paranasal sinuses undifferentiated carcinoma | 177 | 1 | 3.23% | 0.56% |
| trachea squamous cell carcinoma (scc) | 6 | 1 | 3.23% | 16.67% |
| breast carcinoma (nos) | 9459 | 1 | 3.23% | 0.01% |
| unknown primary germ cell tumor | 28 | 1 | 3.23% | 3.57% |
| head and neck squamous cell carcinoma (hnscc) | 2245 | 1 | 3.23% | 0.04% |
| thymus carcinoma | 294 | 1 | 3.23% | 0.34% |
| All | 181,838 | 31 | 100.00% | 0.02% |
Case presentation: index patient #1
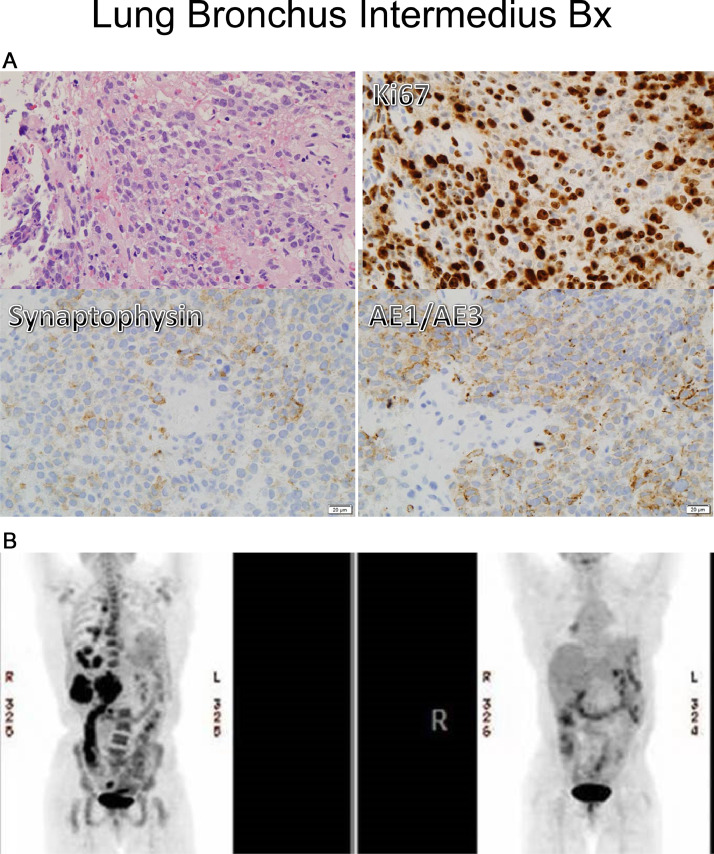
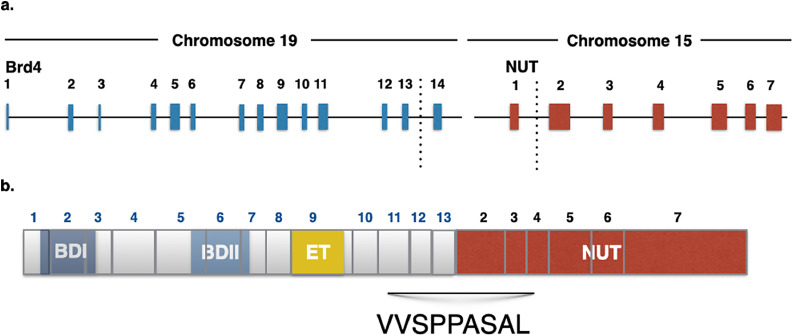
A 52-year-old African American never-smoker female was diagnosed with Stage IV non-small cell lung cancer with high-grade neuroendocrine features, metastatic to lung, lymph nodes, bones, and liver (Fig. 1). As small cell lung carcinoma (SCLC) was in the differential diagnosis, limited molecular testing was performed for TP53 and RB1, both of which were wild-type. The patient experienced disease progression on cisplatin and etoposide, irinotecan, and docetaxel. Pathologic examination of a repeat biopsy showed both squamous and neuroendocrine markers and the histologic diagnosis was changed to NSCLC-SCC. This new biopsy was assayed by comprehensive genomic profiling (CGP) and shown to harbor BRD4-NUTM1 (BRD4 exons 1–12 to NUTM1 exons 2–7, t(15:19) (Fig. 3). A repeat biopsy 15 months later re-confirmed the translocation and identified an additional truncating mutation in BCOR (BCOR 5756*). TMB was 0.8 mutations per megabase (mut/mb) with status of MSS. PD-L1 immunohistochemical (IHC) staining was negative by two assays (Ventana SP142 and Dako 22C3).
Fig. 1.
First index patient. A) Pathologic examination showed small blue cells that stained positive for KI67 (top right), but negative for synaptophysin and Keratin AE1/3 (lower left and right). B) PET imaging of index case #1. Pre-treatment (top panel left) and two months nivolumab (top panel right).
Fig. 3.
Schematic of BRD4-NUT translocation A) Genomic structure of translocation in index case #1 B) Predicted protein domain structure of BRD4 NUT fusion.
Treatment with the anti-PD-1 ICPI nivolumab was initiated, and the patient had a near-complete response as assessed on metabolic imaging at the two-month mark (Fig. 1B). Imaging at four months demonstrated progression at two skeletal foci, a rib and left iliac bone lesion, which were each irradiated. Subsequently, the patient received additional treatment including the nivolumab-ipilumumab combination on which she rapidly progressed, and thereafter transitioned to hospice.
Case presentation: index patient #2
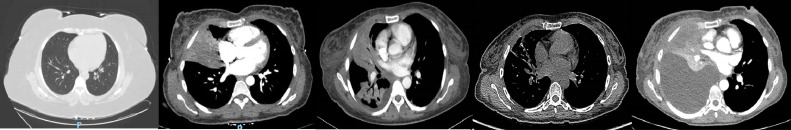
A 39-year-old previously healthy Caucasian female with a 13 pack-year smoking history and a family history of lung cancer (maternal grandmother) presented with a 5.5 × 7 cm right middle lobe tumor protruding from the chest wall (Fig. 2A). Pathologic examination of a core needle biopsy showed a poorly differentiated squamous cell carcinoma of uncertain anatomic origin, positive for CK7, p63 and CK5/6, and negative for TTF1, synaptophysin and chromogranin. Ki67 staining was up to 70%. Staining for PD-L1 was rated at 1% and EGFR, ALK, and ROS1 were wild-type by limited molecular testing. CGP was performed and a BRD4-NUTM1 fusion was identified, as well as TMB of 1 mut/mb and MSS status along with CEBPA and AKT3 alterations.
Fig. 2.
Second index patient. A to E Left to Right (A)-Baseline before diagnosis, (B)-At diagnosis, (C)-Partial response to Cisplatin and Etoposide, (D)-Response to Carboplatin-Paclitaxel and Atezolizumab, (E)-Disease progression.
Imaging by FDG-18 PET-CT indicated locally advanced disease (Fig. 2B) with supraclavicular involvement, stage IIIA. The patient was treated with combined chemo-radiation therapy of 4 cycles of cisplatin and etoposide with 60 GY chest radiation. A partial response was noted (Fig. 2C). Subsequently, a 5 cm consolidation developed at the right middle lobe, with multiple pleural plaques and malignant pleural effusion.
Atezolizumab with carboplatin-paclitaxel was started, as based on safety data presented in the IMpower150 trial and palliative RT 8 Gy to chest(5). The patient had a clinical and radiographic response to treatment (Fig. 2D). Disease progression was observed five months after initiating the combined immunotherapy-chemotherapy regimen (Fig. 2E) and the patient passed away thereafter.
Fusion peptide: MHC analysis
We modeled the neoantigenicity of the BRD4-NUT fusion and HLA type of index case #1 to assess the affinity of the fusion peptide at the junction of BRD4-NUT to patient-specific MHC using IEDB NetMHCpan with methodology akin to a previous stud [8,9]. The neoantigenic sequence flanking the breakpoint was included as input and binding affinities for all the patient-specific HLA-A/B/C alleles were assessed. For this patient, HLA-C*16:01 was predicted to bind the neopeptide of the BRD4-NUT observed here: the fusion sequence VVSPP|ASAL (5 amino acids of BRD4 exon 13 and 4 amino acids of NUTM1 exon 2). This peptide/HLA pair is predicted to have a strong binding affinity of 33 nM, which is ranked as >99.9% compared to a set of random peptides.
To examine how frequently this specific BRD4:NUT fusion of index case #1 is predicted to bind each HLA type, we ran binding predictions for the VVSPP|ASAL neoantigen for all HLA-A/B/C types available in the NetMHCPan 4.0 database (2915). 3.5% (103/2915) of HLA types were predicted to strongly bind the fusion neoantigen (predicted affinity <50 nM). All the alleles predicted to strongly bind this sequence impacted HLA-C and these strong binders clustered into only three HLA allele groups (2-digit): HLA-C*03, HLA-C*12, and HLA-C*16. To understand how frequently these alleles are represented in the population, we examined a cohort of 4147 cancer patients with HLA allele predictions available (predicted using OptiType); 32.7% (1357/4147) of patients harbored at least one of the strong-binding HLA alleles above [10].
For the second index case, the fusion sequence could not be definitively determined, as the BRD4 breakpoint identified by CGP was in exon 13, and the NUTM1 breakpoint was 2.5 KB 5′ of NUTM1.
Discussion
The cases in this cohort are defined by the presence of BRD4-NUT, which is now accepted as diagnostic for NC [11]. For example in the first index case, consideration of an NC diagnosis came only on identification of the BRD4-NUT fusion after an initial diagnostic workup that did not include NUT IHC. Similarly, the other cases in this series should be re-evaluated for a diagnosis of NC, including the cases of cutaneous specimens harboring BRD4-NUT as these may be metastatic NC [12].
Shifting focus to the prognostic implications of this rediagnosis, NC carries an extremely poor prognosis with median overall survival reported as 5 months in a recent systematic review of 119 patients [1]. Chemotherapy does positively impact survival, but long-term control is still needed. Given the great need for efficacious therapy, bromodomain inhibitors of BRD4 are being developed as therapy for NC patients and other diseases. These inhibitors are referred to as the BET inhibitors, referring to the overarching Bromodomain and Extra terminal (BET) family of proteins, all of which have two bromodomains, an extra terminal domain (ET), and a C-terminal domain (CTD), of which BRD4 is the canonical member. The BET inhibitors in clinical development include BMS-986,158, TEN-010, Birabresib (MK-8628/OTX015), and molibresib (GSK525762) as well as others. At present, these inhibitors have modest response rates and duration of responses in early clinical trials for confirmed NC patients. The birabresib phase Ib trial includes responses for 3/10 patents in the receiving continuous dosing of 80 mg or higher [13]. In the molibresib phase I, 2/19 NC patients had confirmed partial responses (PR), and another two had unconfirmed PRs [14]. Notably amongst all these cases, a NC patient harboring BRD3-NUT receiving BMS-986,158 had 16% tumor regression that persisted for 279 days [15].
The responses presented in this report suggest that immunotherapy should be investigated as an additional therapeutic avenue for NC patients. The responses reported here are modest in duration relative to the response of checkpoint inhibitors in other tumor types, but nonetheless striking in that a response occurred given the status of the predictive biomarkers of PD-L1 expression, TMB, Microsatellite stability in these NC cases are all negatively correlated with ICPI response [16], [17], [18], [19], [20], [21]. Interestingly, a near CR in patient 1 did not lead to a durable response, unlike deep response correlated with durability in other types. One limitation of the assessment of response for index patient 2, the response cannot be formally solely attributed to the PD-L1 inhibitor atezolizumab given the combination chemotherapy regimen administered.
We hypothesized that the fusion peptide from BRD4-NUT had a high affinity to the MHC complex, as this affinity is predicted to correlate with immune response. A recent Nature Medicine study demonstrated that neoantigens derived from gene fusions can evoke potent a host cell cytotoxic T-cell response, both as assessed experimentally in an index case, and then as suggested by in silico analysis of a larger cohort of cases [21]. This seminal study described a patient with head and neck cancer that was low TMB, PD-L1, and MSS, but had an exceptional response to an immune checkpoint inhibitor. The authors attribute this response to the potency of the fusion neoantigen in the context of MHC binding in evoking a T-Cell response as experimentally assessed. This relationship between fusion neoantigen and HLA binding was supported by an in-silico analysis of fusion neoantigens and pMHC affinity of other head and neck cancer cases and immune checkpoint inhibitor responses. In analogy to this seminal study, we sought to investigate the NC cases here for their peptide MHC affinity.
For the first index case, [22] the junctional BRD4-NUT neopeptide was modeled with an HLA binding affinity of 33 nM for this first index case which is extremely high relative to random peptides which typically bind with an affinity of 20,000–40,000 nM. Importantly, as this affinity is dependent on both fusion peptide sequence and HLA types, HLA types with a high affinity for the neoantigen fusion of index case 1 are present in one-third of the advanced cancer population in a sampling of the overall cancer database. This may suggest that for BRD4-NUT NC cases that possess this neoantigenic fusion peptide, one third may have the HLA type that permits high pMHC affinity and possibly correlate to response to immune checkpoint inhibitors. Given the overall poor prognosis of NC cases, further investigation is still warranted to explore and validate this therapeutic hypothesis for even a subset of NC cases. Notably however, as in Yang et al., this therapeutic hypothesis of a high pMHC affinity fusion peptide may predict response to immune checkpoint inhibitors and thus could be broadly applicable to any cancer patient who harbors a fusion with high pMHC affinity. A cardinal test case could be alveolar soft parts sarcoma, given the recent findings of preliminary anti-tumor activity in a trial of axitinib+pembrolizumab in this disease also characterized by low TMB and PD-L1 expression.
A limitation of the current study was the inability to assess of the peptide:MHC affinity of the second index case as the specific fusion sequence could not be resolved with the NGS platform used in this study. The most updated version of the platform used for this study baits both NUT as well BRD4 with DNA probes and may provide superior resolution of fusion peptides for NC cases, and a follow-up study. Other studies have reported low TMB but response to immune checkpoint inhibitors in NC and should be similarly assessed for pMHC affinity [23,24]. Another limitation of this study is the modest duration of response in these index cases (4 and 5 months), which is nonetheless clinically significant given the projected short median survival of NC carcinoma patients (7 months).
Conclusions
NC is a likely underdiagnosed but highly aggressive carcinoma that is de facto defined by NUT fusions, most frequently BRD4-NUT. CGP can aid with diagnosis of this disease, as shown by possible reclassification of 80% of cases in this series as NC from their initial diagnoses as based on identification of BRD4-NUT. The index cases suggests that identification of BRD4-NUT may be relevant to benefit from checkpoint blockade if in silico pMHC affinity is assessed high. Moreover, the mechanism of sensitivity of BRD4-NUT-positive checkpoint blockade may lie in the intersection of the fusion sequence and HLA type to yield high-affinity pMHC binding. Future studies in this rare malignancy should replicate and expand on these findings to help alleviate the otherwise very poor prognosis of NC patients [18].
Competing interests
SMA, RM, ABS, AMH, SET, JHC, BA, CE, GMF, LAA, DXJ, ESS, JPG, VAM, SRamakissoon, CE, JSR are employees/former employees of Foundation Medicine, a wholly owned subsidiary of Roche Pharmaceuticals in which they have equity interest. JPG is a consultant for Foundation medicine. VAM is a board member for Revolution Medicines and employee of EQRx. SMA is an employee of EQRx and has consulted for Takeda, ArcherDx and is a SAB member for In8bio, Elevation Oncology, and Pillar Biosciences. JSR is a board observer for Celsius Therapeutics. SJK has received consulting/advisory fees from Eli Lilly, Astellas, Boston Biomedical, Bristol Myers Squibb, Foundation Medicine, Inc., and holds stock/equity in Turning Point Therapeutics. JHC is an employee of GSK. SRahman holds patents relating to BRD4 and NSD3 and is an employee of EQRx. NP is an advisor for and received honorarium from AstraZeneca, AID Genomics, Boehringer-Ingelheim, BMS, E.Lilly, MSD, Novartis, Pfizer, Roche, NovellusDx, FMI, Guardant360. JWR has received honoraria from Celgene, Spectrum, Loxo Oncology, Boehringer Ingelheim, Heron Pharmaceuticals and has received research funding (to institution) from Merck, Boehringer Ingelheim, Spectrum, AstraZeneca, Novartis, Revolution Medicine). SSK and WK have nothing to declare. LCR received honoraria from Roche, AZ, Pfizer, Abbvie and is advisor to Allscripts.
Abbreviations used
NC: NUT carcinoma, formerly known as NUT midline carcinoma,
Funding
No funding was used for this case report.
Availability of data and methods
Please contact the author for data requests.
Methods
Approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20,152,817). Next-generation-sequencing (NGS) based comprehensive genomic profiling (CGP) was prospectively performed on 181,838 cancer cases during routine clinical care. CGP was conducted from formalin-fixed, paraffin-embedded tissues using a hybrid capture-based next generation sequencing platform (FoundationOneTM) at a CLIA-certified, New York State and CAP-accredited laboratory (Foundation Medicine, Cambridge, MA) on Illumina HiSeq2500 instruments. 50 ng of DNA was extracted and adaptor-ligated, and capture was performed for all coding exons of 186, 236 or 315 cancer-related genes and selected introns of 14, 19, or 28 genes frequently rearranged in cancer. Captured libraries were sequenced to a median exon coverage depth of >650x, and resultant sequences were analyzed for base substitutions, short insertions, deletions, gene copy number alterations (focal amplifications and homozygous deletions) and gene fusions, as previously described [25,26]. For TMB (mutations per megabase), the number of somatic mutations detected on comprehensive genomic profiling (interrogating up to 1.1 Mb of the genome) was quantified, and that value was extrapolated to the whole exome using a validated algorithm [27]. Alterations likely or known to be oncogenic drivers and germline polymorphisms were excluded.
Peptide MHC affinity was assessed as in a previous study [8,9]. The patient-specific 4-digit MHC-I HLA types were determined with OptiType, and binding affinity predictions were performed on the fusion protein sequence with NetMHCPan 4.0 [10].
Binding predictions for the VVSPP|ASAL neoantigen (index case #1) were performed for all HLA-A/B/C types available in the NetMHCPan 4.0 database (2915). 3% (105/2915) of HLA types were predicted to strongly bind the fusion neoantigen (predicted affinity <50 nM); these strong binders clustered into only three 2-digit HLA groups: HLA-C*03, HLA-C*12, HLA-C*16 [28].
Authors contributions
SMA, JSR, VAM, AMH, SRahman, NP, SET, JWR generated the study concept and designed the study. AMH, RM, SRamakissoon, JPG, JWR, NP, SET, ESS, DXJ, WK acquired the data. RM, WK, ESS, ABS, DXJ, performed quality control of the data and assessed algorithms. AH, RM, SRahman, ABS, SMA, SRamakissoon, JSR, VAM, NP, WK, SSK, LCR ES, JWR analyzed and interpreted data. SMA, ESS, SRahman, NP, WK, SSK, LCR prepared the manuscript. SMA, JHC, RM, ABS, CE, ESS, SRahman, NP, WK, SSK, LCR, BA, JSR, VAM, JWR edited the manuscript. All authors reviewed the manuscript.
Declarations
Approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817). For the two deceased patients highlighted in this study, for one, permission was granted by institutional IRBs to retrospectively include patient in a deidentified fashion in this report, and for the other the patient provided consent to an institutional IRB approved biorepository.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101184.
Appendix. Supplementary materials
Supplemental Table 1: Genomic profiles of BRD4-NUT patients. Diagnosis, age, gender, TMB score, Microsatellite (MS) status, and genomic alterations beyond BRD4-NUT for each case in this series. No other point mutations, insertions/deletions, copy number alterations or genomic rearrangements were observed other than those listed here.
References
- 1.Giridhar P., Mallick S., Kashyap L., Rath G.K. Patterns of care and impact of prognostic factors in the outcome of NUT midline carcinoma: a systematic review and individual patient data analysis of 119 cases. Eur. Arch. Oto-Rhino-Laryngol. Off. J. Eur. Fed Oto-Rhino-Laryngol. Soc. EUFOS Affil. Ger. Soc. Oto-Rhino-Laryngol. - Head Neck Surg. 2018;275:815–821. doi: 10.1007/s00405-018-4882-y. [DOI] [PubMed] [Google Scholar]
- 2.French C.A. The importance of diagnosing NUT midline carcinoma. Head Neck Pathol. 2013;7:11–16. doi: 10.1007/s12105-013-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.French C.A., Kutok J.L., Faquin W.C., Toretsky J.A., Antonescu C.R., Griffin C.A. Midline carcinoma of children and young adults with NUT rearrangement. J Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2004;22:4135–4139. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 4.French C.A., Rahman S., Walsh E.M., Kühnle S., Grayson A.R., Lemieux M.E. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov. 2014;4:928–941. doi: 10.1158/2159-8290.CD-14-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haack H., Johnson L.A., Fry C.J., Crosby K., Polakiewicz R.D., Stelow E.B. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am. J. Surg. Pathol. 2009;33:984–991. doi: 10.1097/PAS.0b013e318198d666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer D.E., Mitchell C.M., Strait K.M., Lathan C.S., Stelow E.B., Lüer S.C. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012;18:5773–5779. doi: 10.1158/1078-0432.CCR-12-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stathis A., Zucca E., Bekradda M., Gomez-Roca C., Delord J.-.P., de La Motte Rouge T. Clinical Response of Carcinomas Harboring the BRD4-NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer Discov. 2016;6:492–500. doi: 10.1158/2159-8290.CD-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmaier R.J., Charo J., Fabrizio D., Goldberg M.E., Albacker L.A., Pao W. Genomic analysis of 63,220 tumors reveals insights into tumor uniqueness and targeted cancer immunotherapy strategies. Genome Med. 2017;9:16. doi: 10.1186/s13073-017-0408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenzer S., Peters B., Bulik S., Schoor O., Lemmel C., Schatz M.M. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol. Life Sci. CMLS. 2005;62:1025–1037. doi: 10.1007/s00018-005-4528-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szolek A., Schubert B., Mohr C., Sturm M., Feldhahn M., Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinform. Oxf. Engl. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chau N.G., Hurwitz S., Mitchell C.M., Aserlind A., Grunfeld N., Kaplan L. Intensive treatment and survival outcomes in NUT midline carcinoma of the head and neck. Cancer. 2016;122:3632–3640. doi: 10.1002/cncr.30242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko L.N., Weng Q.Y., Song J.S., Asel M., Granter S.R., Mostaghimi A. A 48-year-old male with cutaneous metastases of nut midline carcinoma misdiagnosed as herpes zoster. Case Rep. Oncol. Karger Publ. 2017;10:987–991. doi: 10.1159/000481429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewin J., Soria J.-.C., Stathis A., Delord J.-.P., Peters S., Awada A. Phase Ib trial with Birabresib, a small-molecule inhibitor of bromodomain and extraterminal proteins, in patients with selected advanced solid tumors. J. Clin. Oncol. Am. Soc. Clin. Oncol. 2018;36:3007–3014. doi: 10.1200/JCO.2018.78.2292. [DOI] [PubMed] [Google Scholar]
- 14.Piha-Paul S.A., Hann C.L., French C.A., Cousin S., Braña I., Cassier P.A. Phase 1 study of molibresib (GSK525762), a bromodomain and extra-terminal domain protein inhibitor, in NUT carcinoma and other solid tumors. JNCI Cancer Spectr. [Internet]. Oxf. Acad. 2020:4. doi: 10.1093/jncics/pkz093. https://academic.oup.com/jncics/article/4/2/pkz093/5613897 [cited 2020 Oct 18]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilton J., Cristea M.C., Voskoboynik M., Postel-Vinay S., Edenfield W., Gavai A. Initial results from a phase I/IIa trial evaluating BMS-986158, an inhibitor of the bromodomain and extra-terminal (BET) proteins, in patients (pts) with advanced cancer. Ann. Oncol. 2018;29:viii134. Elsevier. [Google Scholar]
- 16.Mahoney K.M., Freeman G.J., McDermott D.F. The next immune-checkpoint inhibitors: PD-1/PD-L1 blockade in melanoma. Clin. Ther. 2015;37:764–782. doi: 10.1016/j.clinthera.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brogden K.A., Parashar D., Hallier A.R., Braun T., Qian F., Rizvi N.A. Genomics of NSCLC patients both affirm PD-L1 expression and predict their clinical responses to anti-PD-1 immunotherapy. BMC Cancer. 2018;18:225. doi: 10.1186/s12885-018-4134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marabelle A., Fakih M., Lopez J., Shah M., Shapira-Frommer R., Nakagawa K. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–1365. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 20.McGranahan N., Furness A.J.S., Rosenthal R., Ramskov S., Lyngaa R., Saini S.K. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samstein R.M., Lee C.-.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W., Lee K.-.W., Srivastava R.M., Kuo F., Krishna C., Chowell D. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat. Med. 2019;25:767. doi: 10.1038/s41591-019-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie X.-.H., Wang L.-.Q., Qin Y.-.Y., Lin X.-.Q., Xie Z.-.H., Liu M. Clinical features, treatment, and survival outcome of primary pulmonary NUT midline carcinoma. Orphanet. J. Rare. Dis. 2020;15:183. doi: 10.1186/s13023-020-01449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He M., Chernock R., Zhou S., Gondim M., Dehner L.P., Pfeifer J.D. Tumor mutation burden and checkpoint immunotherapy markers in NUT midline carcinoma. Appl. Immunohistochem. Mol. Morphol. AIMM. 2020;28:495–500. doi: 10.1097/PAI.0000000000000781. [DOI] [PubMed] [Google Scholar]
- 25.Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J., Abdel-Wahab O., Nahas M.K., Wang K., Rampal R.K., Intlekofer A.M. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127:3004–3014. doi: 10.1182/blood-2015-08-664649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen M., Lundegaard C., Blicher T., Lamberth K., Harndahl M., Justesen S. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLOS ONE. Public Lib. Sci. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Genomic profiles of BRD4-NUT patients. Diagnosis, age, gender, TMB score, Microsatellite (MS) status, and genomic alterations beyond BRD4-NUT for each case in this series. No other point mutations, insertions/deletions, copy number alterations or genomic rearrangements were observed other than those listed here.