Abstract
Background
Plant height is an important architecture trait which is a fundamental yield-determining trait in crops. Variety with dwarf or semi-dwarf phenotype is a major objective in the breeding because dwarfing architecture can help to increase harvest index, increase planting density, enhance lodging resistance, and thus be suitable for mechanization harvest. Although some germplasm or genes associated with dwarfing plant type have been carried out. The molecular mechanisms underlying dwarfism in oilseed rape (Brassica napus L.) are poorly understood, restricting the progress of breeding dwarf varieties in this species. Here, we report a new dwarf mutant Bndwarf2 from our B. napus germplasm. We studied its inheritance and mapped the dwarf locus BnDWARF2.
Results
The inheritance analysis showed that the dwarfism phenotype was controlled by one semi-dominant gene, which was mapped in an interval of 787.88 kb on the C04 chromosome of B. napus by Illumina Brassica 60 K Bead Chip Array. To fine-map BnDWARF2, 318 simple sequence repeat (SSR) primers were designed to uniformly cover the mapping interval. Among them, 15 polymorphic primers that narrowed down the BnDWARF2 locus to 34.62 kb were detected using a F2:3 family population with 889 individuals. Protein sequence analysis showed that only BnaC04.BIL1 (BnaC04g41660D) had two amino acid residues substitutions (Thr187Ser and Gln399His) between ZS11 and Bndwarf2, which encoding a GLYCOGEN SYNTHASE KINASE 3 (GSK3-like). The quantitative real-time PCR (qRT-PCR) analysis showed that the BnaC04.BIL1 gene expressed in all tissues of oilseed rape. Subcellular localization experiment showed that BnaC04.BIL1 was localized in the nucleus in tobacco leaf cells. Genetic transformation experiments confirmed that the BnaC04.BIL1 is responsible for the plant dwarf phenotype in the Bndwarf2 mutants. Overexpression of BnaC04.BIL1 reduced plant height, but also resulted in compact plant architecture.
Conclusions
A dominant dwarfing gene, BnaC04.BIL1, encodes an GSK3-like that negatively regulates plant height, was mapped and isolated. Our identification of a distinct gene locus may help to improve lodging resistance in oilseed rape.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-03137-9.
Keyword: Brassica napus; semi-dominant; dwarf; single nucleotide polymorphism; gene mapping
Background
Oilseed rape (Brassica napus L.) is one of the most important oil crops worldwide, and provides high-quality vegetable oil for human diets, protein-rich feed for animals, and raw materials for industrial processes. Variety with dwarf or semi-dwarf phenotype is a major objective in the breeding because dwarfing architecture can help to increase harvest index, increase planting density, enhance lodging resistance, and thus be suitable for mechanization harvest [1]. To find available germplasm or genes associated with dwarfing plant type for B. napus breeding, some efforts have been carried out. For example, the dwarfness-associated genes in B. napus, including DS-1 [2], ndf-1 [3], DS-3 [4], DS-4 [5], G7 [6], BnaDwf.C9 [7], have been positioned or identified. Additionally, the Bndwf1 was fine-mapped on the A9 chromosome to a 152-kb interval [8]. However, the molecular mechanism(s) underlying the development of the dwarf phenotype in B. napus remain elusive. The lack of innovation on B. napus ideal type breeding is mainly due to absence of successfully applied cultivar in vast oilseed rape production region.
Dwarfism is usually related to plant hormone biosynthesis and signal transduction, such as auxin [9, 10], gibberellin (GA) [1, 11], and brassinosteroid (BR) [12, 13]. Auxin affects plant height by regulating cell division, elongation, and differentiation [14]. GA mainly affects the elongation of stem and internode to regulate plant height [1]. Relationship between plant dwarf stature and genes in auxin and GA biosynthesis and signal transduction pathways has been well-documented [4, 15–19]. Defects in BR biosynthesis and signaling pathways can lead to dwarfing phenotypes. During BR biosynthesis, many synthases belong to the cytochrome P450 monooxygenase (CYP) gene. Defects of these synthases can lead to dwarfing phenotypes. For instance, CPD (constitutive photomorphogenesis and dwarfism) encodes a steroid 23α-hydroxylase enzyme, a member of CYP90A family, which acts in the conversion of cathasterone to teasterone in the BR biosynthetic pathway [20]. The loss-of-function mutations of AtCPD gene leads to the dwarfing phenotype, when over-expression of CPD gene can restore the plant height and plant type [20]. BRD1 (BR-deficient dwarf 1) gene encodes the final catalytic enzyme (BR-C6 oxidase) in BR biosynthesis, mutation of which cause dwarfing phenotype. Rice brd1 was the first report to describe the phenotypic characterization of a BR-deficient mutant in monocot plants, and showed the phenotype of leaf sheath small, leaves wrinkled, internodes short, fewer tillers. The exogenous application of BL can restore the plant type of the brd1 mutants [21]. The dwf4 (CYP90B1) in Arabidopsis, dwarf2 (CYP90D) and dwarf11 (CYP724B1) in rice encode P450 monooxygenase to involve in BR biosynthesis, loss-of-function mutations of which reduce the endogenous BR levels and consequently confer reduced plant height [13, 22, 23]. The Arabidopsis DET2 (de-etiolated 2) is a key gene in BR biosynthesis and allow an assignment for this steroid’s role in plant development [24, 25]. The Arabidopsis det2 mutant [24] and the maize na1 (nana plant1) mutant [26] were the loss-of-function of DET2 gene lead to the dwarfing phenotype, dark green leaves, and have reduced fertility.
BR signal transduction is a signaling cascade from the BR receptor to the expression of BR target genes, which plays an important role in various developmental and growth processes in plants [27]. Researches during the past several decades have accumulated extensive knowledge of BR signaling pathways in model plants [28–30], such as Arabidopsis and rice. It is well documented that BRs are perceived extracellularly by the BR-INSENSITIVE1/BRI1-ASSOCIATED KINASE1 (BRI1-BAK1) [31–33] complex. Afterwards, the binding between BRs and BRI1-BAK1 complex could initiate signal transduction to BRASSINAZOLERESISTANT1/BRI1-EMS-SUPPRESSOR1 (BZR1/BES1) [34, 35] through CONSTITUTIVE DIFFERENTIAL GROWTH1 (CDG1) [36] and BR SIGNALING KINASE1 (BSK1) [37], then BRI-SUPPRESSOR1 (BSU1) [38], BRASSINOSTEROID INSENSITIVE2 (BIN2) [39, 40], as well somehow PROTEIN PHOSPHATASE 2A (PP2A) [41]. The transcriptional factor BES1/BZR1 affects plant growth and development in various aspects through the regulating expression of thousands of BR responsive genes. Among these genes, glycogen synthase kinase-3 (GSK3)-like kinase BIN2 is a key suppressor that regulates plant growth and development by determining the phosphorylation status of BES1 and BZR1 [33, 34, 39, 40]. GSK3-like kinases are a highly conserved Ser/Thr kinases that are implicated in a wide range of cellular and developmental processes [42]. In Arabidopsis, the GSK3/SHAGGY-like family has 10 gene members that can be classified into four subgroups [43]. In this family, the Arabidopsis GSK3-like kinase (AT4G18710, BIN2/UCU1/DWF12/AtSK21) which belongs to the group II, has activity to negatively regulate the BR signal transduction by phosphorylating BZR1/BES1 [39, 40, 44]. The gain-of-function bin2 mutant was discovered to be insensitive to BRs in Arabidopsis and has the shaggy phenotypic characteristic of dwarfing architecture. It also confers curved leaves, and an impaired cell elongation [45]. The coding sequence of the BIN2 gene, substitutes consecutive glutamate residues in the highly conserved TREE domain, which results in the negatively regulating growth by phosphorylating the BES1 and BZR1 proteins, that result in the degradation of BZR1 to reduce its activity [40]. Based on sequence similarity of BIN2 with its two closest group II Arabidopsis homologs, BIN2-Like1 (BIL1) and BIN2-Like2 (BIL2), which belong to the AtSKs group [40]. It was further suggested that BIL1 and BIL2 may also be involved in BR signaling. Overexpression of BIL1 or BIL2 gene driven by their native promoters in wild-type Arabidopsis plants exhibits the dwarf phenotype [46]. However, the evidence of BIL1 and BIL2 genes involved in BR signal transduction is still insufficient, and the mechanism of plant dwarf phenotype caused by overexpression of BIL1 and BIL2 genes remains to be elucidated. Therefore, it is urgent to further explore their participation and even related mechanism.
In this study, a pure dwarf mutant, Bndwarf2, was found in advanced selfing generation in a nearly pure line CB1501-1 in B. napus. To expedite this study, the dwarf gene BnaC04.BIL1 was isolated using map-based cloning. The BnaC04.BIL1 gene encoding a GSK3-like kinase, belongs to GSK II subfamily. Genetic transformation experiments confirmed that the BnaC04.BIL1 was responsible for the plant dwarf phenotype in the Bndwarf2 mutants. Our study clarifies the role of BnaC04.BIL1 in the regulation of plant height, which may help to improve lodging resistance in oilseed rape.
Results
Characterization of the Bndwarf2 Mutant
A pure dwarf mutant, Bndwarf2 was obtained in advanced selfing generation in a nearly pure line CB1501-1 in B. napus. The Bndwarf2 mutant showed obvious dwarf phenotype after 6 d dark germination compared to Zhongshuang 11 (ZS11, a conventional B. napus cultivar), which was used as a parent to map-based clone the gene responsible for the dwarfism (Fig. 1a). At seedling stage, the Bndwarf2 mutant plants had shorter hypocotyls and shorter petioles (Fig. 1b, c). The leaves of Bndwarf2 mutants showed darker green, thickened, and wrinkled leaves, and had significant higher Chl a, Chl b, and Chl contents than those of ZS11 (Table S1). At flowering stage, the Bndwarf2 mutant showed significant difference in plant height from ZS11 (Figure S1). While at maturity stage, the Bndwarf2 mutant showed dwarf stature (33.62 ± 1.12 cm) with no apical dominance, that was significantly lower than that for ZS11 (193.54 ± 4.80 cm) (Fig. 1d). The siliques of Bndwarf2 mutants were significantly shorter compared to that of ZS11 (Fig. 1e). In addition, the Bndwarf2 mutants had lower 1000-seeds weight and compact plant architecture (Fig. 1f; Table S2). The F1 plants (105.30 ± 5.16 cm) generated by cross of ZS11 with Bndwarf2 were in-between that of ZS11 and Bndwarf2.
Fig. 1.
Phenotypic comparison among ZS11, F1, and Bndwarf2 mutant. a The root length and hypocotyl length of ZS11 (left), F1 (middle), and Bndwarf2 (right) in the dark for 6 d. b The performance of ZS11 (left), F1 (middle), and Bndwarf2 (right) at seedling stage. c The petioles comparison at seedling stage. d The plant height comparison at maturity stage. e The siliques comparison of at maturity stage. f The seeds comparison of ZS11 (up) and Bndwarf2 mutant (down). Bars = 2 cm
Inheritance of the dwarf trait
To investigate the genetic regulation mechanism for Bndwarf2, the F1 (ZS11 × Bndwarf2) and RF1 (Bndwarf2 × ZS11) plants were obtained by crossing Bndwarf2 with ZS11, all had the dwarf trait, indicating that dwarf trait was controlled by dominant genes. The phenotypic segregation ratio of dwarf plants to tall plants in the F2 population was in a Mendelian model of 1:2:1 (69 homozygous dwarf plants vs. 140 hybrid dwarf plants vs. 78 tall plants, χ2 < χ20.05) (Figure S2). Among 289 BC1 individuals, 139 as dwarf types and 150 as tall types, also approximately fitted an expected Mendelian inheritance ratio of 1:1 (dwarf plants vs. tall plants). In subsequent segregating F2:3 populations, the genetic regulation was confirmed (Table 1). These results indicated that the dwarf trait was controlled by a semi-dominant nuclear gene, which was named as BnDWARF2 in the subsequent study.
Table 1.
Inheritance of the plants height trait in populations derived from the two parents in B. napus
| Population | Homozygous dwarf plants | Hybrid dwarf plants | Tall plants | Expectation | χ2 | P value |
|---|---|---|---|---|---|---|
| F1 | 0 | 30 | 0 | |||
| RF1 | 0 | 30 | 0 | |||
| F2 | 69 | 140 | 78 | 1:2:1 | 0.74 | 0.69 |
| BC1 | 0 | 139 | 150 | 1:1 | 0.35 | 0.56 |
| F2:3 | 206 | 451 | 232 | 1:2:1 | 1.71 | 0.43 |
Map-based cloning
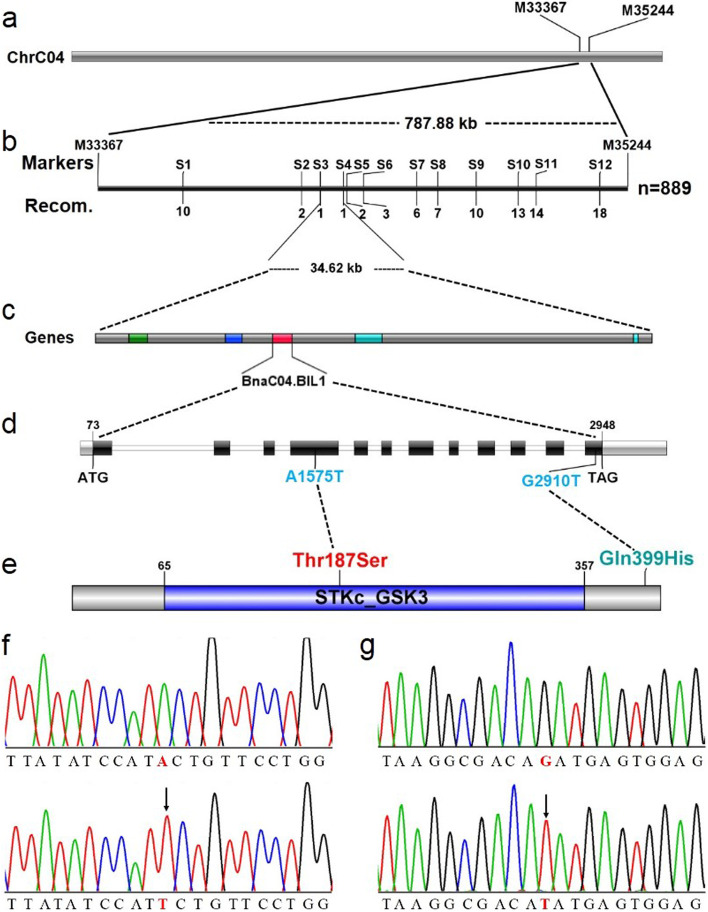
To map BnDWARF2, 94 plants (70 dwarf plants and 24 tall plants) from the F2 population were used for single nucleotide polymorphism (SNP) marker genotyping. Although the chip (Illumina, Inc) has 52,157 SNP markers, only 7457 polymorphic markers were used to construct the SNP genetic linkage map after removing the invalid or non-polymorphism markers. The BnDWARF2 locus was primarily mapped within the 787.88-kb on C04 chromosome between the SNP marker M33367 and M35244 (Fig. 2a). To fine map the BnDWARF2 locus, 318 primer pairs of simple sequence repeat (SSR) markers were designed to uniformly cover the preliminary mapping interval. A further 889 individuals from the F2:3 populations, finally narrowed down the BnDWARF2 locus to a 34.62-kb region between SSR markers S3 and S4 (Fig. 2b). No other markers to further narrow the mapping interval were found for this mapping population and its parents. A total of 5 putative genes (BnaC04g41640D, BnaC04g41650D, BnaC04g41660D, BnaC04g41670D, and BnaC04g41680D) were localized in the 34.62-kb region according to the gene annotation of the B. napus reference genome (Fig. 2c; Table 2). Gene cloning was performed for the mapping interval, and the results showed that only BnaC04g41660D (BnaC04.BIL1) gene had 10 SNPs differences between ZS11 and Bndwarf2. The BnaC04.BIL1 had two amino acid residues substitutions at aa-187 (Thr-to-Ser mutation, named Thr187Ser) and aa-399 (Gln-to-His mutation, named Gln399His) (Fig. 2e).
Fig. 2.
Map-based cloning of BnDWARF2. a The BnDWARF2 locus was mapped primarily on C04 chromosome between the SNP markers M33367 and M35244. b The BnDWARF2 locus was fine-mapped in the 34.62 kb region between SSR markers S3 and S4. The numerals indicate the number of recombinants. c The genes in the mapping interval. d The gene structure and the mutation sites in BnaC04.BIL1. e The protein structure and the mutation sites of the BnaC04.BIL1 protein, and the STKc_GSK3 superfamily domain was predicted. Solid lines show the position of the amino acid transition. f The chromatogram of BnaC04.BIL1 at 1565–1585 bp in ZS11 and Bndwarf2 mutant, respectively. g The chromatogram of BnaC04.BIL1 at 2900–2920 bp in ZS11 and Bndwarf2 mutant, respectively. The black arrows denote the A1575T and G2910T substitutions, respectively
Table 2.
Information of 5 putative genes in the mapping interval
| Gene in B. napus | Homologue in A. thaliana | Gene function |
|---|---|---|
| BnaC04g41640D | AT2G29770.1 | Galactose oxidase/kelch repeat superfamily protein |
| BnaC04g41650D | unknown protein | |
| BnaC04g41660D | AT2G30980 | Encodes a GSK3-like protein kinase |
| BnaC04g41670D | AT2G30990.1 | Arginine N-methyltransferase, putative (DUF688) |
| BnaC04g41680D | unknown protein |
BnaC04.BIL1 contains a 1233-bp open reading frame (ORF) with 11 introns in B. napus (Fig. 2d; Figure S3). BnaC04.BIL1 is a homologous gene of the Arabidopsis AT2G30980 gene, which encodes a GSK3-like [47]. The conservative domain analysis showed that the amino acid sequence 65–357 was the conserved domain of STKc_GSK3, and Thr187Ser is in the conserved domain (Fig. 2e). The amino acid multiple sequence analysis showed that BnaC04.BIL1 had a series of amino acid residues conserved in GSK3 kinase, such as GSK3 domain signature SYICSR and plant-specific TREE motif (Figure S3a). It was perfectly aligned with the genes for GSK3/Shaggy kinases with regarding to a series of amino acid residues such as the GSK3 signature SYICSR within domain VIII that was absent from MAP kinase sequences [48]. The E-K mutation in the highly conserved TREE motif is thought to preventing the BR-mediated BIN2 inhibition [49], thus resulting in the increased BIN2 stability [50, 51]. The phylogenetic tree clustering and construction were analyzed by MEGA 7.0 selection Neighbor-joining method. The results showed that BnaC04.BIL1 and Arabidopsis BIN2 were homologous, belonging to GSK3 II subfamily (Figure S3b). These suggested that the BnaC04.BIL1 gene may be responsible for the dwarf trait of Bndwarf2.
Expression patterns of BnaC04.BIL1 and the subcellular localization
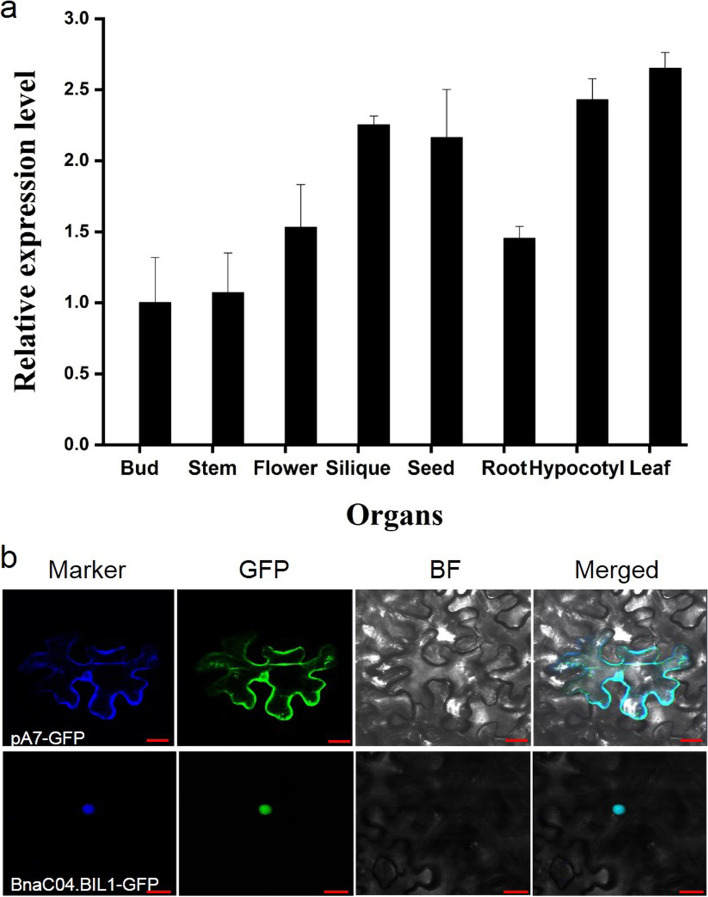
To explore the possible function of BnaC04.BIL1 gene from Bndwarf2 mutant in different tissues, the transcription levels of BnaC04.BIL1 in leaves, roots, hypocotyls, stems, buds, flowers, siliques, and seeds were analyzed. The qRT-PCR analysis showed that the BnaC04.BIL1 gene was expressed in all tissues, which indicated that BnaC04.BIL1 expressed constitutively (Fig. 3a). The expression level of BnaC04.BIL1 was higher in leaves, hypocotyls, siliques, and seeds, while its level in buds and stems were lower.
Fig. 3.
Expression pattern of BnaC04.BIL1 and subcellular localization of its encoding protein. a Expression pattern of BnaC04.BIL1 detected by qRT-PCR in bud, stem, flower, silique, seed, root, hypocotyl, and leaf from Bndwarf2. The BnActin gene was used as a reference gene and the expression level of bud was set to 1. The bud, stem, and flower samples are from flowering stage. The silique samples are from podding stage. The seed samples are from maturity stage. The root and hypocotyl samples are from 7-day-old seedlings grown on medium, and the leaf samples are from seedling stage. b Subcellular localization of BnaC04.BIL1 protein in tobacco leaf cells. Plasmids pA7-GFP and BnaC04.BIL1-GFP were introduced into tobacco leaf cells by particle bombardment, respectively. Bars = 20 μm
Previous research showed that Arabidopsis BIN2 [52] was localized in the nucleus. To define the subcellular location of expression, pA7-GFP and BnaC04.BIL1-GFP constructs were then introduced into the tobacco leaf cells by the particle bombardment method. The merged image of BnaC04.BIL1-GFP and nuclear localization signal (NLS)-mCherry signals showed that BnaC04.BIL1 was localized to the nucleus (Fig. 3b). The result showed the BnaC04.BIL1 gene functions in the nucleus.
Overexpression of BnaC04.BIL1 leads to plant dwarf
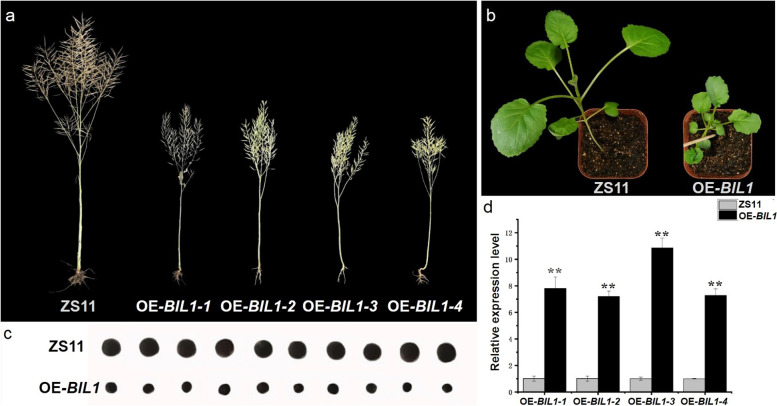
To investigate BnaC04.BIL1 functioning in plant height, a construct was generated by inserting a 1233 bp BnaC04.BIL1 ORF fragment from Bndwarf2 into the vector pBI-121 under the control of the CaMV35S promotor. The construct was introduced into ZS11 plants by Agrobacterium-mediated transformation. The plant height trait was compared between the ZS11 and OE-BnaC04.BIL1 (OE-BIL1) transgenic plants by overexpressing the BnaC04.BIL1 gene. Notably, plant height in the OE-BIL1 transgenic plants was similar to the expected Bndwarf2 phenotype with obvious dwarf stature; meanwhile, the transgenic plants also displayed dramatically smaller seeds than the ZS11 plants (Fig. 4a-c; Table S3). At the seedling stage, the OE-BIL1 transgenic lines displayed darker green and wrinkled leaves compared to those of ZS11 (Fig. 4b). These results suggest that the BnaC04.BIL1 gene not only controls the plant height, but also regulates the seed size. It follows that, the yield of per OE-BIL1 transgenic plants showed a significantly reduction compared to that of ZS11 (Table S3). The T2 progeny plants were examined from six independent T1 transgenic lines in growth chamber, which showed the expected Mendelian inheritance ratio of 3:1 in T2 progeny (dwarf vs. tall plants, χ2 < χ20.05, 1 = 3.84; P > 0.05; Table S4). The T2 progeny plants displayed perfect co-segregation between the transgene and the dwarf phenotype. Consistently, the expressions of BnaC04.BIL1 gene in homozygous T3 lines (OE-BnaC04.BIL1 transgenic genes) were significantly higher than those of ZS11 plants (Fig. 4d). These results confirmed that the BnaC04.BIL1 is the causal mutation for the dwarfism and controls smaller seeds, which were also observed in Bndwarf2.
Fig. 4.
Phenotype comparison and qRT-PCR analysis between the ZS11 and OE-BIL1 transgenic plants. a The phenotype of ZS11 (left) and OE-BIL1 (right) transgenic plants at maturity stage. b The phenotype of ZS11 (left) and OE-BIL1 (right) transgenic plants at seedling stage. c The seeds of ZS11 (up) and OE-BIL1 (down) plants. d qRT-PCR analysis of BnaC04.BIL in ZS11 and OE-BIL1 transgenic lines
Discussion
Plant height is an important growth habit that is a fundamental yield determining trait in crops. In the 1960s and 1970s, the dwarf trait genes (Rht1 and sd1) were introduced into wheat and rice that were crucial to the first “Green Revolution” [1, 53]. The semi-dwarf architecture can help to increase harvest index, increase planting density, enhance lodging resistance, and thus be suitable for mechanization harvest [54]. However, there are few studies with respect to dwarf oilseed rape. Because of the lower mechanization level of oilseed rape production and few varieties suitable for mechanization harvest, oilseed rape production faces severe challenge.
Most of our knowledge about BIN2 functions came mostly from gain-of-function results. For example, genetic screening in Arabidopsis for BR-insensitive dwarf mutants resulted in the isolation of eight gain-of-function bin2 alleles [39, 40, 45]. Based on sequence similarity of BIN2 with its two closest group II Arabidopsis homologs, BIN2-Like1 (BIL1) and BIN2-Like2 (BIL2), which belong to the AtSKs group [46]. Overexpression of BIL1 or BIL2 gene driven by their native promoters in wild-type Arabidopsis plants exhibits the dwarf phenotype [46]. In our study, a gain-of-function mutation for BIL1 in oilseed rape has been discovered, and most importantly, it exhibits the BR-insensitive dwarf phenotype. For example, the Bndwarf2 mutant displayed the BR signaling phenotypes: shorter hypocotyls, shorter petioles, wrinkled leaves, and obvious dwarf compared with the ZS11 (Fig. 1; Figure S1; Table S2). These characteristics were similar to the phenotypes of BR-insensitive mutants such as bri1 [27], dwf12 [39], and ucu1 [45]. Through map-based cloning, the BnaC04.BIL1 was identified to be a BIN2-Like1 (BIL1), showing a Thr187Ser amino acid substitution residing in the conserved region (Fig. 2; Figure S3a). Genetic transformation experiments confirmed that the BnaC04.BIL1 was responsible for the plant dwarf phenotype in the Bndwarf2 mutants. Overexpression of BnaC04.BIL1 under the background of ZS11 reduced plant height compared with ZS11 (Fig. 4; Table S3). This result was consistent with previous reports, showing that overexpressing BIL1 gene confers the dwarf phenotype in Arabidopsis [46]. The genetic evidence clarifies the BnaC04.BIL1 can sharply change plant architecture in natural plant accessions in allotetraploid.
Further study has identified Bndwarf2, a dwarf and compact mutant in B. napus, and the dwarf trait is controlled by a semi-dominant nuclear gene (Table 1). The plant height of F1 derived from the cross of Bndwarf2 with the tall parent, decreased by about 50% compared to that of tall plant (Table S2). Particularly, the Bndwarf2 displayed an extreme reduction in height at maturity, which is different from the previously reported dwarf mutants in B. napus [4, 8, 55–57]. For example, the dwarf locus of bnC.dwf mutant was controlled by a recessive gene [56]. And, the dwarf trait of Bndwf1 mutant was controlled by a semi-dominant gene [8]. The F1 plants have compact properties such as shortened branch, shortened gap between siliques, shortened gap between branches and dwarfing plant height by BnDWARF2 gene (Fig. 1; Figure S1; Table S2). This finding implicates that the plant architecture of homozygous or heterozygous individuals derived Bndwarf2 mutant is compact (Fig. 1). This kind of compact architecture can be undoubtedly helpful to increase planting density, enhance lodging resistance and increase planting density, therefore the compact plant architecture is ideal for machinery production of oilseed rape.
The germplasm Bndwarf2 has compact plant type, and lacks strong growth vigor. However, the compact plant architecture can be used in hybrid cultivar development in which the compact type and hybrid vigor can be combined well. This is helpful to breeding of variety breeding with the objectives such as high-yield, good quality and suitable for machinery. On the other hand, the growth vigor in pure line or cultivar may be improved in some genetic background. Some reports have demonstrated that the genes in BIN2 regulation network can also interact with BIN2, leading to improvement of the growth inhibition caused by BIN2 gene overexpression caused by natural biological accession state or by transgenic [58–61]. We speculate that some gene may interact with BnaC04.BIL1 to attenuate its role in limit growth vigor as that the Arabidopsis homolog BIN2 crosstalk experiments have shown. Furthermore, expressions of some regulator genes may probably alter the expression level of BnaC04.BIL1 that is constitutively expressed in the various organs, and reduced expression level may improve the growth vigor. The subcellular localization analysis demonstrated that BnaC04.BIL1 exists in the nucleus (Fig. 3). Consistently, the Arabidopsis BIN2 functioned in nucleus to negatively regulate BR signaling [52]. In fact, previous results revealed that many genes regulated by BZR1 and/or BES1, and some proteins interacting with BZR1/BES1, were closely associated with the BR signaling [29, 62]. The BR signal transduction pathways was impairment to lead to the dwarfing phenotype.
Methods
Plant materials and growth conditions
A pure dwarf mutant, Bndwarf2 was found in advanced selfing generation in a nearly pure line CB1501-1 in B. napus from our germplasm bank of our lab in Nanjing Agricultural University. The populations for mapping the BnDWARF2 locus, were generated from the crosses between Bndwarf2 and the canola variety Zhongshuang 11 (ZS11). All oilseed rape materials were grown in growth chamber and the fields of the Jiangpu Agricultural Experimental Station at Nanjing Agricultural University.
Tobacco was grown in growth chamber. The illumination period was 14 h with temperature at 26 °C and 10 h with temperature at 20 °C. When tobacco leaves at 5-leaf stage were used for the subcellular localization.
Map-based cloning
SNP and SSR markers were used to map the dwarf gene. 70 dwarf plants, 24 tall plants and parents from F2 population were genotyped using a Brassica 60 K SNP Bead Chip Array (Illumina, Inc), which have a total of 52,157 SNP markers. The SNP genetic map was constructed by JoinMap 4.1 mapping software [63], then the BnDWARF2 locus was primarily mapped onto physical and genetic map. The mapping interval sequence was downloaded from the Brassica napus Genome Browser (http://www.genoscope.cns.fr/brassicanapus/cgi-bin/gbrowse/colza/). Using this genomic sequence, SSR marker primers were designed by aid of SSR Hunter 1.3 [64], and Primer Premier 5.0 [65]. A total of 318 polymorphic SSR markers were obtained. These SSR markers helped to fine-map the BnDWARF2 locus using a size-enlarged population comprised of F2:3 plants.
To identify genes associated to the dwarf trait, sequence of the fine mapping interval was obtained from the Brassica napus Genome Browser for reference to next-step experiments. Then, all of the genes in the fine mapping interval were cloned from Bndwarf2 and parent ZS11. And, the resulting sequences were aligned using ClustalX 1.83 and GeneDoc software. The specific primers of the genes are listed in Table S5.
Sequence analysis
The B. napus BIL1 genes were obtained by screening the B. napus Genome Browser (http://www.genoscope.cns.fr/brassicanapus/) with known A. thaliana BIL1 gene as a query. The Conserved Domain Database was used to search the protein functional in the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov). Predicted A. thaliana BIL1 amino acid sequences were obtained from the TAIR website (http://www.arabidopsis.org/Blast). Moreover, the protein sequences of other species were obtained from the NCBI using the A. thaliana BIL1 protein sequence as a query. All obtained protein sequences were aligned using ClustalX 1.83 [66]. Additionally, a phylogenetic tree was constructed using MEGA 7.0 [67] with maximum likelihood method, and the bootstrap values were estimated with 1000 replicates.
RNA extraction and qRT-PCR
Total RNA was extracted from various samples using TRIzol reagent (Sigma; http://www.sigmaaldrich.com/). First-strand cDNA synthesis was carried out using a Reverse Transcription System (Takara, Tokyo, Japan). The cDNA was used as the template for qRT-PCR analysis with specific primers (Table S5). The qRT-PCRs were carried out with SYBR Green Real-time PCR Master mix using a CFX96-2 PCR machine (BIO-RAD, USA). Relative expression levels were calculated using the 2−ΔΔCt method with Actin as an internal control.
Plant transformation
The 1223-bp BnaC04.BIL1 open reading frame was amplified from Bndwarf2 using the primers BnaC04.BIL1-F/R (Table S5) and cloned into the Xba I-BamH I sites of the overexpression pBI121 vector with CaMV35S promotor to construct the 35S::BnaC04.BIL1-pBI121 plasmid. The 35S::BnaC04.BIL1-pBI121 plasmid was introduced into Agrobacterium tumefaciens strain EHA105 by a heat shock method. The positive A. tumefaciens were transformed into ZS11 with a modified floral dip method. Briefly, agrobacteria cultures carrying a target construct were collected by centrifugation and then resuspended in a solution containing 1/2 MS salts containing 3% Suc, 0.1% Silwet L-77, 2 ng/L 6-benzyladenine, and 8 mg/L acetosyringone. The ZS11 plants at the flowering stage were used for the transformation. The head of a flowering plant was bent downward and dipped into a beaker containing the agrobacterial culture liquid for 3 min, and the treated plant head was loosely wrapped with a vegetable parchment paper. The plant for transformation was treated every week 1 to 2 times and then continued to grow until maturation. Seeds that experienced transgenic treatment were harvested. The transformant leaf were collected for PCR detection [68]. The 35S::BnaC04.BIL1-pBI121 sequence was detected by PCR in transgenic plants, the transgenic plants were named OE-BnaC04.BIL1 (OE-BIL1).
Supplementary Information
Additional file 1: Figure S1. The plant height of ZS11, F1, and Bndwarf2 at flowering stage. Figure S2. The distribution of plant height in the F2 population. Figure S3. Sequence analysis of BnaC04.BIL1 protein. a Multiple sequence alignment of amino acid sequences of BnC04.BIL1 protein. The conserved TREE and SYICSR motif were boxed in red. At, Cs, Cr, Aa, Es, Bo, Br, Bn, Cs, Cc, Qs, Pp, Hu, Cp, Ap, Gm, Vv, Ac, and Mn denote Arabidopsis thaliana, Camelina sativa, Capsella rubella, Arabis alpine, Raphanus sativus, Eutrema salsugineum, Brassica oleracea, Brassica rapa, Brassica napus,Citrus sinensis, Quercus suber, Prunus persica,Herrania umbratica, Carica papaya, Abrus precatorius, Glycine max, Vitis vinifera, Actinidia chinensis, Morus notabilis, respectively. b Phylogenetic tree analysis of BnaC04.BIL1 using Neighbor-joining method in MEGA 7.0 program. Bootstrap values from 1000 replicates were indicated at each node. The GSK3 group I is marked with green, and the GSK3 group II is marked with red, and the GSK3 group III is marked with yellow, and the GSK3 group IV is marked with blue. The BnaC04.BIL1, BnaC04.BIL1-Mut, and BnaC04.BIL1-WT are labeled. Table S1. Leaf chlorophyll contents in the leaves of ZS11 and Bndwarf2 mutant. Table S2. Agronomic trait comparisons in ZS11, F1, and Bndwarf2. Table S3. Agronomic traits of OE-BIL1 transgenic lines. Table S4. Genetic analysis of T2 progeny derived from six independent T1transgenic plants.
Additional file 2: Table S5. Primers used in this study.
Acknowledgements
We would like to thank Dr Evan Evans (University of Tasmania; tassiebeerdr@gmail.com) for the English editing of this paper.
Abbreviations
- B. napus
Brassica napus
- GSK3
GLYCOGEN SYNTHASE KINASE 3
- BIN2
BRASSINOSTEROID INSENSITIVE2
- BIL1
BIN2-LIKE1
- qRT-PCR
quantitative real-time PCR
- ZS11
Zhongshuang 11
- SNP
Single nucleotide polymorphism
- SSR
Simple sequence repeat
- Thr187Ser
Thr-to-Ser mutation at 187
- Gln399His
Gln-to-His mutation at 399
- GA
gibberellin
- BR
brassinosteroid
- CYP
cytochrome P450 monooxygenase
- CPD
CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM
- BRD1
BR-DEFICIENT DWARF 1
- DET2
DE-ETIOLATED 2
- BRI1-BAK1
BR-INSENSITIVE1/BRI1-ASSOCIATED KINASE 1
- BZR1/BES1
BRASSINAZOLERESISTANT1/BRI1-EMS-SUPPRESSOR1
- CDG1
CONSTITUTIVE -DIFFERENTIAL GROWTH 1
- BSK1
BR SIGNALING KINASE1
- BSU1
BRI-SUPPRESSOR1
- PP2A
PROTEIN PHOSPHATASE 2A
- ORF
Open reading frame
- OE-BIL1
OE-BnaC04.BIL1
- NCBI
National Center for Biotechnology Information
Authors’ contributions
MY and RG designed the research. MY, SW, WL, WC, YW, XJ, PC, and PC performed the experiments. JH analyzed the SNP and sequencing data. RG advised on the experiments. MY wrote the manuscript. WS and RG revised the manuscript. All authors read and approved of the final manuscript.
Funding
This research was supported financially by the National Science and Technology Major Project of the Ministry of Science and Technology of China (2018ZX08020001-010), the National Key Research and Development Plan (2016YFD0101306, 2016YFD0101300), the National Natural Science Foundation of China (31771832), the Fundamental Research Funds for the Central Universities (Y02021001), and the Cyrus Tang Seed Innovation Center, Nanjing Agricultural University. The funders provided the financial support to the research, but had no role in the design of the study, analysis, interpretations of data and in writing the manuscript.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Figshare repository (https://doi.org/10.6084/m9.figshare.14679021).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mao Yang, Email: 2017201033@njau.edu.cn.
Jianbo He, Email: hjbxyz@gmail.com.
Shubei Wan, Email: 2016201079@njau.edu.cn.
Weiyan Li, Email: 2018101062@njau.edu.cn.
Wenjing Chen, Email: 2016201058@njau.edu.cn.
Yangming Wang, Email: 2015201029@njau.edu.cn.
Xiaomei Jiang, Email: 2019101077@njau.edu.cn.
Pengfei Cheng, Email: 2020216037@njau.edu.cn.
Pu Chu, Email: chupu@njau.edu.cn.
Wenbiao Shen, Email: wbshenh@njau.edu.cn.
Rongzhan Guan, Email: guanrzh@njau.edu.cn.
References
- 1.Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19(1):5–9. doi: 10.1016/S0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 2.Liu C, Wang JL, Huang TD, Wang F, Yuan F, Cheng XM, Zhang Y, Shi SW, Wu JS, Liu KD. A missense mutation in the VHYNP motif of a DELLA protein causes a semi-dwarf mutant phenotype in Brassica napus. Theor Appl Genet. 2010;121(2):249–258. doi: 10.1007/s00122-010-1306-9. [DOI] [PubMed] [Google Scholar]
- 3.Li HP, Wang Y, Li XC, Gao Y, Wang ZJ, Zhao Y, et al. A GA-insensitive dwarf mutant of Brassica napus L. correlated with mutation in pyrimidine box in the promoter of GID1. Mol Biol Rep. 2011;38(1):191–7. [DOI] [PubMed]
- 4.Zhao B, Li H, Li J, Wang B, Dai C, Wang J, Liu KD. Brassica napus DS-3, encoding a DELLA protein, negatively regulates stem elongation through gibberellin signaling pathway. Theor Appl Genet. 2017;130(4):727–741. doi: 10.1007/s00122-016-2846-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhao B, Wang B, Li Z, Guo T, Zhao J, Guan Z, Liu KD. Identification and characterization of a new dwarf locus DS-4 encoding an Aux/IAA7 protein in Brassica napus. Theor Appl Genet. 2019;132(5):1435–1449. doi: 10.1007/s00122-019-03290-8. [DOI] [PubMed] [Google Scholar]
- 6.Cheng H, Jin F, Zaman QU, Ding B, Hao M, Wang Y, Huang Y, Wells R, Dong Y, Hu Q. Identification of Bna.IAA7.C05 as allelic gene for dwarf mutant generated from tissue culture in oilseed rape. BMC Plant Biol. 2019;19(1):500. [DOI] [PMC free article] [PubMed]
- 7.Wang X, Zheng M, Liu H, Zhang L, Hua W. Fine-mapping and transcriptome analysis of a candidate gene controlling plant height in Brassica napus L. Biotechnol Biofuels. 2020;13:42. [DOI] [PMC free article] [PubMed]
- 8.Wang YK, Chen WJ, Chu P, Wan SB, Yang M, Wang MM, Guan RZ. Mapping a major QTL responsible for dwarf architecture in Brassica napus using a single-nucleotide polymorphism marker approach. BMC Plant Biol. 2016;16(1):178. doi: 10.1186/s12870-016-0865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timpte C, Wilson AK, Estelle M. The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics. 1994;138(4):1239–1249. doi: 10.1093/genetics/138.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133(1):177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416(6882):701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 12.Yamamuro C, Ihara Y, Xiong W, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Matsuoka KM. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12(9):1591–1605. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Iwasaki Y. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell. 2005;17(3):776–790. doi: 10.1105/tpc.104.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y. Auxin biosynthesis and its role in plant development. Ann Rev Plant Biol. 2010;61(1):49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Y, Dai XH, Zhao Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006;20(13):1790–1799. doi: 10.1101/gad.1415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cassani E, Bertolini E, Badone FC, Landoni M, Gavina D, Sirizzotti A, Pilu R. Characterization of the first dominant dwarf maize mutant carrying a single amino acid insertion in the VHYNP domain of the dwarf8 gene. Mol Breed. 2009;24(4):375–385. doi: 10.1007/s11032-009-9298-3. [DOI] [Google Scholar]
- 17.Chen Y, Hou M, Liu L, Wu S, Shen Y, Ishiyama K, Kobayashi M, Mccarty DR, Tan BC. The maize DWARF1 encodes a gibberellin 3-Oxidase and is dual localized to the nucleus and cytosol. Plant Physiol. 2014;166(4):2028. doi: 10.1104/pp.114.247486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing A, Gao Y, Ye L, Zhang W, Cai L, Ada C, Victor L, Blaine J, Liu L, Yang X. A rare SNP mutation in Brachytic2 moderately reduces plant height and increases yield potential in maize. J Exp Botany. 2015;66(13):3791–3802. doi: 10.1093/jxb/erv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z, Ding T, Liu K, Miao C, Zhuo X, Li Y, Tan X, Sun M, Luo Q, Cheng Z. Characterization of a new semi-dominant dwarf allele of SLR1 and its potential application in hybrid rice breeding. J Exp Botany. 2018;69:(20):4703–13. [DOI] [PMC free article] [PubMed]
- 20.Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85(2):171–182. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 21.Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2010;32(4):495–508. doi: 10.1046/j.1365-313X.2002.01438.x. [DOI] [PubMed] [Google Scholar]
- 22.Choe S, Dikes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-Hydroxylation steps in Brassinosteroid biosynthesis. Plant Cell. 1998;10(2):231–244. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H. Matsuoka: The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17(8):2243–2254. doi: 10.1105/tpc.105.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.2307/3869351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JM, Nagpal P, Vitart V, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272(5260):398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 26.Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DP, et al. Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci U S A. 2011;2011(108):19814–19819. doi: 10.1073/pnas.1108359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clouse SD, Mcmorris LTC. A Brassinosteroid-Insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111(3):671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belkhadir Y, Jaillais Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015;206(2):522–540. doi: 10.1111/nph.13269. [DOI] [PubMed] [Google Scholar]
- 29.Guo H, Lei L, Aluru M, Aluru S, Yin Y. Mechanisms and networks for brassinosteroid regulated gene expression. Curr Opin Plant Biol. 2013;16(5):545–553. doi: 10.1016/j.pbi.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Wang WF, Bai MY. ZY: The brassinosteroid signaling network - a paradigm of signal integration. Curr Opin Plant Biol. 2014;21:147–153. doi: 10.1016/j.pbi.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90(5):929–938. doi: 10.1016/S0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Wen J, Lease KA, Doke JT, Walker JC. BAK1, an Arabidopsis LRR Receptor-like Protein Kinase, Interacts with BRI1 and Modulates Brassinosteroid Signaling. Cell. 2002;110(2):213–222. doi: 10.1016/S0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 33.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110(2):203–212. doi: 10.1016/S0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhi YW, Nakano T, Gendron J, He J, Chory J. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2(4):505–513. doi: 10.1016/S1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 35.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109(2):181–191. doi: 10.1016/S0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 36.Kim TW, Guan S, Burlingame AL, Wang ZY. The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Mol Cell. 2011;43(4):561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang W, Kim TW, Oses-Prieto JA, Yu S, Wang ZY. BSKs Mediate Signal Transduction from the Receptor Kinase BRI1 in Arabidopsis. Science. 2008;321(5888):557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mora-Garcia S, Vert G, Yin YH, Delgado A, Cheong H, Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18(4):448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choe S, Schmita RJ, Fujioka S, Takatsuto S, Lee M, Yoshida S, Feldmann K, Tax FE. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol. 2002;130(3):1506–1515. doi: 10.1104/pp.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Nam KH. Regulation of Brassinosteroid Signaling by a GSK3/SHAGGY-Like Kinase. Science. 2002;295(5558):1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 41.Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13(2):124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woodgett JR. Judging a Protein by More Than Its Name: GSK-3. Stke. 2001;100(100):re12. [DOI] [PubMed]
- 43.Jonak C, Hirt H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Ence. 2002;7(10):457–461. doi: 10.1016/S1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- 44.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci U S A. 2002;99(15):10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Perez JM, Ponce MR, Micol JL. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol. 2002;242(2):161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- 46.Yan Z, Zhao J, Peng P, Chihara RK, Li J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009;150(2):710–721. doi: 10.1104/pp.109.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charrier B, Champion A, Henry Y, Kreis M. Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 2002;130(2):577–590. doi: 10.1104/pp.009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dornelas MC, Wittich P, Recklinghausen IV, Lammeren AV, Kreis M. Characterization of three novel members of the Arabidopsis SHAGGY-related protein kinase (ASK) multigene family. Plant Mol Biol. 1999;39(1):137–147. doi: 10.1023/A:1006102812280. [DOI] [PubMed] [Google Scholar]
- 49.Peng P, Li J. Brassinosteroid Signal Transduction: A Mix of Conservation and Novelty. J Plant Growth Regul. 2003;22(4):298–312. doi: 10.1007/s00344-003-0059-y. [DOI] [PubMed] [Google Scholar]
- 50.Peng P, Yan ZY, Zhu YY, Li JM. Regulation of the Arabidopsis GSK3-like Kinase BRASSINOSTEROID-INSENSITIVE 2 through Proteasome-Mediated Protein Degradation. Mol Plant. 2008;1(2):338–346. doi: 10.1093/mp/ssn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441(7089):96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 52.Ryu H, Kim K, Cho H, Hwang I. Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol Cells. 2010;29(3):291. doi: 10.1007/s10059-010-0034-y. [DOI] [PubMed] [Google Scholar]
- 53.Khush GS. Green revolution: the way forward. Nat Rev Genet. 2001;2(10):815. [DOI] [PubMed]
- 54.Islam N, Evans EJ. Influence of lodging and nitrogen rate on the yield and yield attributes of oilseed rape (Brassica napus L.). Theor Appl Genet. 1994;88(5):530–4. [DOI] [PubMed]
- 55.Foisset N, Delourme R, Barret P, Renard M. Molecular tagging of the dwarf BREIZH (Bzh) gene in Brassica napus. Theor Appl Genet. 1995;91(5):756–761. doi: 10.1007/BF00220955. [DOI] [PubMed] [Google Scholar]
- 56.Zeng X, Zhu L, Chen Y, Qi L, Fu T. Identification, fine mapping and characterisation of a dwarf mutant (bnaC.dwf) in Brassica napus. Theor Appl Genet. 2010;122(2):421–8. [DOI] [PubMed]
- 57.Wang YK, He JB, Yang L, Wang Y, Chen WJ, Wan SB, Chu P, Guan RZ. Fine mapping of a major locus controlling plant height using a high-density single-nucleotide polymorphism map in Brassica napus. Theor Appl Genet. 2016;129(8):1479–1491. doi: 10.1007/s00122-016-2718-y. [DOI] [PubMed] [Google Scholar]
- 58.Ling JJ, Li J, Zhu D, Deng XW. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc Natl Acad Sci U S A. 2017;114(13):3539–3544. doi: 10.1073/pnas.1700850114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun S, Wang T, Wang L, Li X, Jia Y, Liu C, Huang X, Xie W, Wang X. Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling. Nat Commun. 2018;9(1):2523. doi: 10.1038/s41467-018-04952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He G, Liu J, Dong H, Sun J. The Blue Light Receptor CRY1 Interacts with BZR1 and BIN2 to Modulate the Phosphorylation and Nuclear Function of BZR1 in Repressing BR Signaling in Arabidopsis. Mol Plant. 2019;12(5):689–703. doi: 10.1016/j.molp.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 61.He YQ, Hong GJ, Zhang HH, Tan XX, Li LL, Kong Y, Sang T, Xie K, Sun Z. The OsGSK2 Kinase Integrates Brassinosteroid and Jasmonic Acid Signaling by Interacting with OsJAZ4. Plant Cell. 2020;32(9):2806–2822. doi: 10.1105/tpc.19.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J. Regulation of the nuclear activities of brassinosteroid signaling. Curr Opin Plant Biol. 2010;13(5):540–547. doi: 10.1016/j.pbi.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ooijen J, Van JW. JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma, BV; 2006. https://www.kyazma.nl/index.php/JoinMap/.
- 64.Li Q. SSRHUNTER: Development of a local searching software for SSR sites. Hereditas(Beijing). 2005;27:808–10. [PubMed]
- 65.Singh VK, Mangalam AK, Dwivedi S, Naik S. Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques. 1998;24(2):318–319. doi: 10.2144/98242pf02. [DOI] [PubMed] [Google Scholar]
- 66.Crooks G. E: WebLogo: A Sequence Logo Generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sudhir K, Glen S, Koichiro T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. [DOI] [PMC free article] [PubMed]
- 68.Tan H, Yang X, Zhang F, Zheng X, Qu C, Mu J, Fu F, Li J, Guan R, Zhang H. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011;156(3):1577–1588. doi: 10.1104/pp.111.175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The plant height of ZS11, F1, and Bndwarf2 at flowering stage. Figure S2. The distribution of plant height in the F2 population. Figure S3. Sequence analysis of BnaC04.BIL1 protein. a Multiple sequence alignment of amino acid sequences of BnC04.BIL1 protein. The conserved TREE and SYICSR motif were boxed in red. At, Cs, Cr, Aa, Es, Bo, Br, Bn, Cs, Cc, Qs, Pp, Hu, Cp, Ap, Gm, Vv, Ac, and Mn denote Arabidopsis thaliana, Camelina sativa, Capsella rubella, Arabis alpine, Raphanus sativus, Eutrema salsugineum, Brassica oleracea, Brassica rapa, Brassica napus,Citrus sinensis, Quercus suber, Prunus persica,Herrania umbratica, Carica papaya, Abrus precatorius, Glycine max, Vitis vinifera, Actinidia chinensis, Morus notabilis, respectively. b Phylogenetic tree analysis of BnaC04.BIL1 using Neighbor-joining method in MEGA 7.0 program. Bootstrap values from 1000 replicates were indicated at each node. The GSK3 group I is marked with green, and the GSK3 group II is marked with red, and the GSK3 group III is marked with yellow, and the GSK3 group IV is marked with blue. The BnaC04.BIL1, BnaC04.BIL1-Mut, and BnaC04.BIL1-WT are labeled. Table S1. Leaf chlorophyll contents in the leaves of ZS11 and Bndwarf2 mutant. Table S2. Agronomic trait comparisons in ZS11, F1, and Bndwarf2. Table S3. Agronomic traits of OE-BIL1 transgenic lines. Table S4. Genetic analysis of T2 progeny derived from six independent T1transgenic plants.
Additional file 2: Table S5. Primers used in this study.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Figshare repository (https://doi.org/10.6084/m9.figshare.14679021).