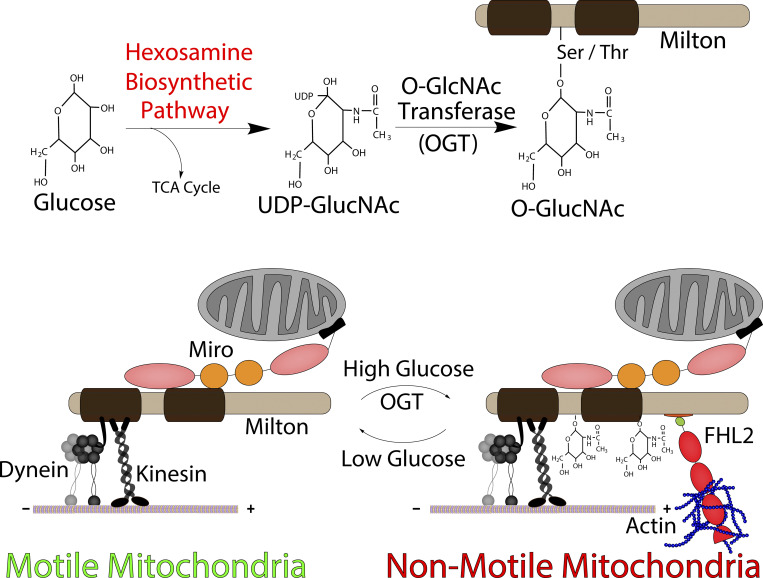
Basu et al. elucidate a mechanism for anchoring mitochondria to the actin cytoskeleton. The actin-associated protein FHL2 provides the necessary and sufficient anchor. They demonstrate that FHL2 is recruited to mitochondria by O-GlcNAcylated TRAK in response to glucose sensing. Anchored mitochondria are remobilized upon F-actin disruption or FHL2 knockdown.
Abstract
Mitochondrial movement and distribution are fundamental to their function. Here we report a mechanism that regulates mitochondrial movement by anchoring mitochondria to the F-actin cytoskeleton. This mechanism is activated by an increase in glucose influx and the consequent O-GlcNAcylation of TRAK (Milton), a component of the mitochondrial motor-adaptor complex. The protein four and a half LIM domains protein 2 (FHL2) serves as the anchor. FHL2 associates with O-GlcNAcylated TRAK and is both necessary and sufficient to drive the accumulation of F-actin around mitochondria and to arrest mitochondrial movement by anchoring to F-actin. Disruption of F-actin restores mitochondrial movement that had been arrested by either TRAK O-GlcNAcylation or forced direction of FHL2 to mitochondria. This pathway for mitochondrial immobilization is present in both neurons and non-neuronal cells and can thereby adapt mitochondrial dynamics to changes in glucose availability.
Graphical Abstract
Introduction
Mitochondria are highly dynamic organelles (Schwarz, 2013). They serve as hubs for calcium buffering, ATP production, nutrient sensing, and innate immune signaling (Beck et al., 2012; Friedman and Nunnari, 2014; Latorre-Muro et al., 2021; Sliter et al., 2018; Trigo et al., 2019; Williamson and Cleveland, 1999; Yu et al., 2020). They also participate in reactive oxygen species signaling, cell division, and cell migration (Chung et al., 2016; Cunniff et al., 2016; Debattisti et al., 2017; Katajisto et al., 2015; Schuler et al., 2017).
In most metazoans, mitochondria form distinct pools of stationary and motile organelles. The motile pool of mitochondria can move up to several microns every minute, pause repeatedly, and undergo fission and fusion. Signaling pathways, by regulating this behavior, serve to distribute healthy mitochondria to appropriate intracellular locations according to changing metabolic needs. Regulated mitochondrial motility and distribution are particularly important in neurons (Misgeld and Schwarz, 2017). Neuronal axons contain widely spaced energetically demanding synapses that require local mitochondrial activity (Economo et al., 2016). The mechanisms that regulate mitochondrial movement and positioning are as yet incompletely understood. They involve an interplay of movement driven by microtubule-bound molecular motors and proteins that inhibit this movement.
In mammalian cells, mitochondria move on microtubule tracks powered by microtubule-based molecular motors. Kinesin-1 heavy chain (KHC, KIF5A-C) and the cytoplasmic dynein-1–dynactin complex serve as the anterograde and retrograde motors, respectively. The molecular motors are recruited to the mitochondria by the adaptor protein Milton (whose two isoforms are known as TRAK1 and TRAK2), which in turn is bound to Miro (whose two isoforms are RHOT1 and RHOT2), a mitochondrial outer-membrane protein with a transmembrane domain (Brickley et al., 2005; Glater et al., 2006; Górska-Andrzejak et al., 2003; Hurd and Saxton, 1996; MacAskill et al., 2009a; Pilling et al., 2006; Stowers et al., 2002; Tanaka et al., 1998). This motor–adaptor complex serves as a molecular hub that is modified by many cellular pathways to modulate mitochondrial motility. Modifications to the motor–adaptor complex can switch the motors on or off (MacAskill et al., 2009b; Wang and Schwarz, 2009) or trigger the recruitment of anchoring proteins, such as syntaphilin, that can oppose motor-driven movements (Chen and Sheng, 2013; Gutnick et al., 2019; Kang et al., 2008).
Here we report a mechanism wherein a modification of the mitochondrial motor–adaptor machinery results in the anchoring of mitochondria to the F-actin cytoskeleton in response to increases in extracellular glucose. Glucose sensing is a prominent physiological signaling pathway that influences mitochondrial motility (Pekkurnaz et al., 2014). Normally, ∼1–3% of intracellular glucose enters the hexosamine biosynthetic pathway (Bond and Hanover, 2015; Chaveroux et al., 2016; Marshall et al., 1991). The hexosamine biosynthetic pathway results in the production of UDP-N-acetylglucosamine (UDP-GlcNAc). Increased glucose influx leads to increases in the flux of glucose through the hexosamine biosynthetic pathway, resulting in the up-regulation of UDP-GlcNAc concentrations. The enzyme O-GlcNAc transferase (OGT) transfers the GlcNAc to serine and threonine residues of target proteins (O-GlcNAcylations). This modification can be reversed by O-GlcNAcase (OGA; Ma and Hart, 2014). Milton (TRAK) is one such protein that is O-GlcNAcylated in various tissues, especially in the brain (Trinidad et al., 2012). In vivo, the GlcNAcylation state is enhanced by feeding, and in vitro TRAK becomes heavily O-GlcNAcylated following an increase in extracellular glucose (Pekkurnaz et al., 2014). The O-GlcNAc modification of TRAK suppresses mitochondrial motility (Pekkurnaz et al., 2014).
While investigating the consequences of the O-GlcNAcylation of TRAK to understand how it arrests mitochondrial motility, we uncovered a mechanism by which mitochondria bearing the O-GlcNAcylated TRAK become anchored to F-actin. We found that four and a half LIM domains protein 2 (FHL2), a member of the LIM domain family of proteins, associates with O-GlcNAcylated TRAK. We show that FHL2 is both necessary and sufficient to recruit F-actin to mitochondria and effectively anchor them. The motor–adaptor machinery in this anchored state remains functional, with the consequence that the mitochondria regain their motility when the F-actin network is disrupted. FHL2 recruitment thus provides a reversible mechanism for immobilizing mitochondria in response to changes in glucose availability.
Results
Mitochondrial motility arrest upon O-GlcNAcylation of Milton is mediated by F-actin
In rat hippocampal neurons, mitochondrial motility is suppressed following an acute increase in glucose supply (Pekkurnaz et al., 2014). We elected to study this phenomenon under conditions closely approximating the neuronal environment in vivo. To do this, we established neuronal cultures under low-glucose conditions. The neurons were initially dissected in typical neurobasal media containing 25 mM glucose, after which we washed, plated, and fed the neurons until day in vitro (DIV) 6 with media containing 5 mM glucose. At DIV 6, we switched the established cultures to a preconditioning 1 mM glucose medium for ∼72 h (3 d).
On DIV 9, we shifted the neurons (which were now conditioned to grow in 1 mM glucose) into a medium containing 5 mM glucose for 2 h and then monitored the consequences of the shift for mitochondrial motility (Fig. 1 A). We monitored mitochondria by expressing Mito-DsRed, which contains the mitochondrial matrix targeting sequence of COX8 fused to DsRed2. The shift in extracellular glucose concentrations from 1 mM to 5 mM causes an increase in the intracellular glucose concentration in neurons (Hou et al., 2011; Pekkurnaz et al., 2014) and represents the range of physiological variations reported for the extracellular space in mammalian brain (Silver and Erecińska, 1994).
Figure 1.
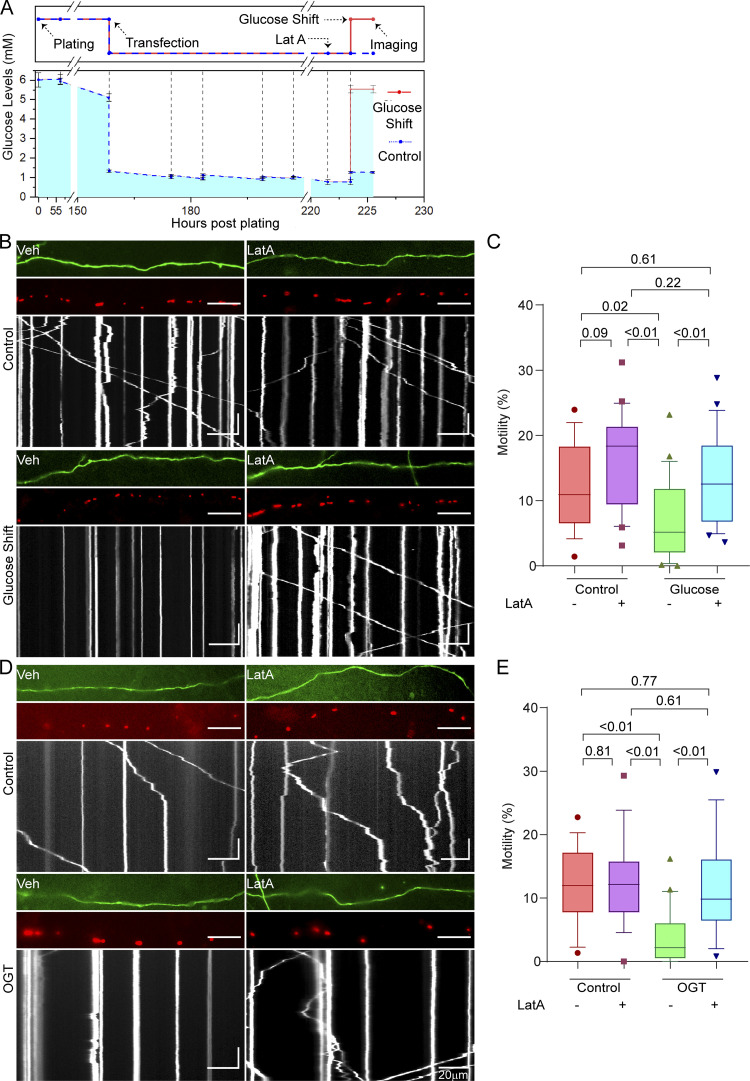
Glucose- and OGT-induced arrest of neuronal mitochondrial motility requires F-actin. (A) Experimental timeline and measured glucose levels for neuronal glucose shift experiments. Experimental outline (top) is aligned with measured glucose levels (bottom). Primary neurons were dissected in standard neurobasal media (containing 25 mM glucose). Neurons were then washed and plated in low-glucose media (containing 5 mM glucose). At the time of plating, the glucose levels are slightly higher than 5 mM due to incomplete washing. These cultures were then maintained by feeding with 5 mM glucose media until DIV 6. The neuronal cultures were then transfected with Mito-DsRed and GFP on DIV 6, at which time glucose was reduced to 1 mM. On DIV 9, the neurons were treated with 5 µM LatA or DMSO (vehicle control [Veh]) 2 h before being subjected to a glucose shift. Mitochondrial motility was quantified 2 h after the glucose shift. Glucose levels in the media were measured before every feeding or transfection and are marked with dotted vertical lines. (B) Mitochondrial motility in response to a glucose shift in neurons pretreated with LatA or vehicle control, as schematized in A. For each condition, representative images of axons (top), mitochondria (middle), and kymographs of mitochondrial motility (bottom) are shown and correspond to Video 1. Scale bars represent 20 µm (horizontal) and 30 s (vertical). Here and throughout, kymographs and axon segments are oriented such that rightward movement is toward the axon terminal. (C) Quantification of mitochondrial motility (each data point represents the average percentage of time spent in motion by all mitochondria in an axon segment) from kymographs such as those in B; n = 15–20 axons per condition from 3 independent animals. (D) Mitochondrial motility in neurons expressing GFP-2A-OGT with or without 4-h exposure to 5 µM LatA or DMSO (vehicle control) before imaging. Representative images as in B. Horizontal scale bars represent 20 µm, and vertical scale bars represent 30 s. (E) Quantification of mitochondrial motility from kymographs as in D; n = 15–20 axons per condition from 3 independent animals. Glucose measurements in A are represented as the mean of three measurements, with whiskers indicating SEM. All motility quantifications (C and E) are represented as box-and-whisker plots. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and were included in all statistical calculations. All P values were calculated from two-tailed unpaired t tests with Welch’s correction.
The shift to 5 mM glucose sharply decreased mitochondrial motility (Fig. 1, B and C). To determine whether F-actin is involved in this response, we treated the neurons with 5 µM Latrunculin A (LatA; a fungal toxin that depletes F-actin by blocking polymerization) or a vehicle control (DMSO). Disrupting the actin cytoskeleton with LatA rendered mitochondrial motility resistant to the shift in glucose (Fig. 1, B and C; and Video 1), but it had little effect on motility when the neurons were held in 1 mM glucose.
Video 1.
F-actin is necessary for glucose-induced arrest of neuronal mitochondrial motility (related to Fig. 1). The axon segments expressing Mito-DsRed and shown in Fig. 1 B were subjected to a glucose shift in the presence of LatA or its vehicle control (DMSO). The segments are oriented such that rightward movement is toward axon terminals. Scale bar represents 20 µm. Video is played at 30× real-time speed and rendered at 20 frames per second.
Overexpression of OGT increases O-GlcNAcylation of Milton, thereby mimicking the mitochondrial response to increased glucose (Pekkurnaz et al., 2014). To test the effect of F-actin depletion on mitochondrial motility arrest brought about by OGT, we treated neurons expressing GFP-2A-OGT (Fig. S1) or GFP (control) with 5 µM LatA. As expected, OGT significantly reduced mitochondrial motility. F-actin disruption had little effect in the absence of OGT expression. By contrast, LatA treatment in OGT-expressing neurons completely reversed the mitochondrial arrest brought about by OGT (Fig. 1, D and E). These results show that O-GlcNAcylation–mediated arrest of mitochondria is dependent on the F-actin cytoskeleton. Moreover, the ability of mitochondria to resume motility upon the disruption of F-actin implied that OGT overexpression or glucose shift does not inactivate the mitochondrial motors or release them from mitochondria but instead triggers an anchoring mechanism that overrides the motors.
Figure S1.
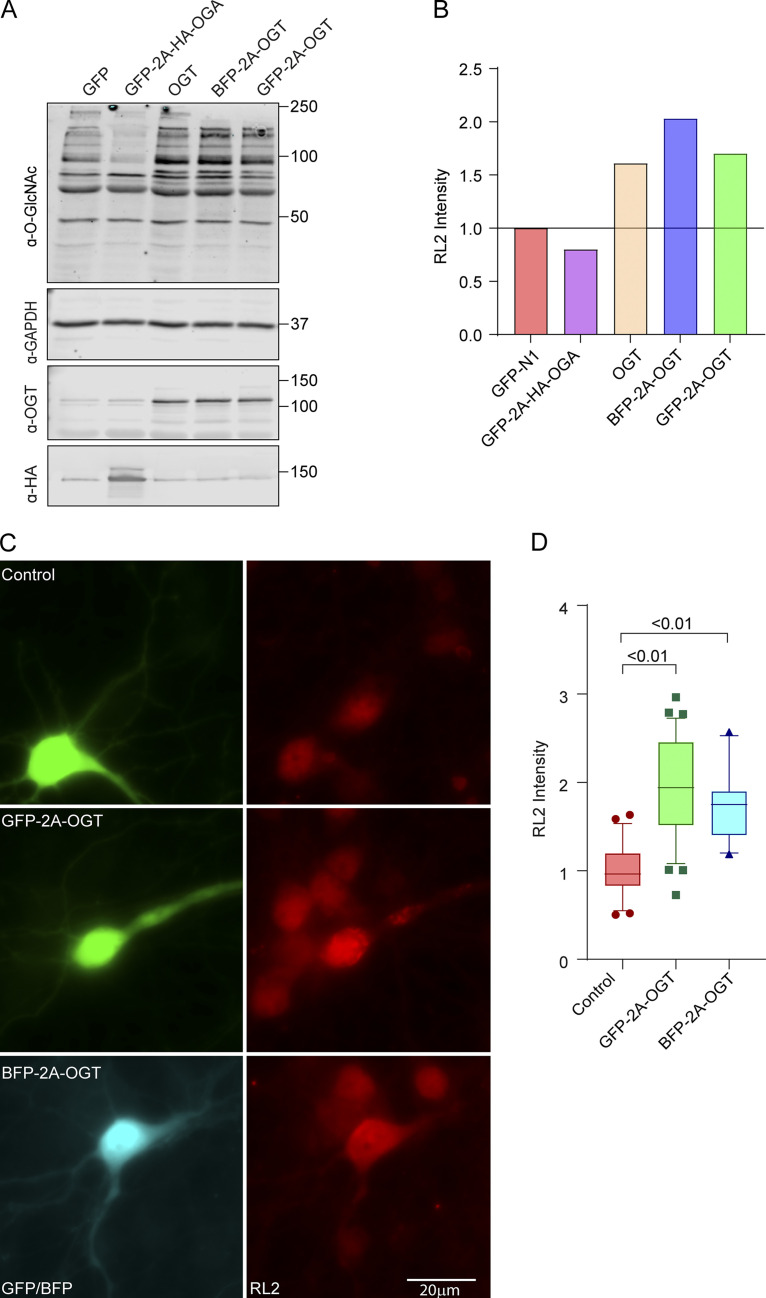
Verification of OGT and OGA constructs in COS-7 cells (related to Fig. 1). (A and B) Construct verification in COS-7 cells. COS-7 cells were transfected with OGT or OGA constructs and lysed after 2 d. (A) Representative Western blot of lysates probed for OGT, OGA (HA), GAPDH, and O-GlcNAcylation (with antibody RL2). Molecular weights (in kD) are indicated on the right. (B) O-GlcNAcylation was quantified by normalizing the intensity of RL2 staining (full lane) to that of the GAPDH band. Values were then expressed relative to those cells expressing only GFP. Each bicistronic construct was almost fully cleaved to release OGA or OGT. Expression of OGT increased, and OGA decreased, O-GlcNAcylation levels relative to the GFP (control)-expressing cells. (C and D) Construct verification in neurons. GFP-2A-OGT, BFP-2A-OGT, or GFP constructs were expressed in rat hippocampal neurons for 3 d. (C) Representative images of neurons stained with the O-GlcNAc antibody RL2. (D) To quantify the RL2 intensity, the BFP or GFP signal was used to mask the cell bodies, and total RL2 intensity inside the mask was measured and normalized to the control. n = 25–30 cells per condition from 3 independent animals.
O-GlcNAcylation–mediated arrest of mitochondrial motility occurs in non-neuronal cell types
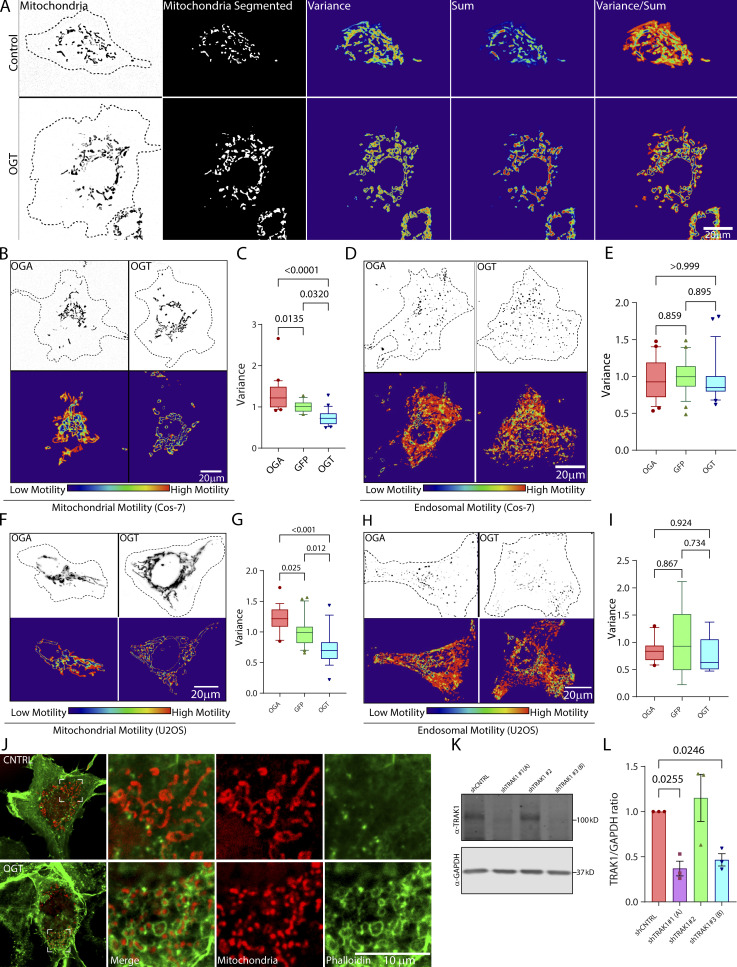
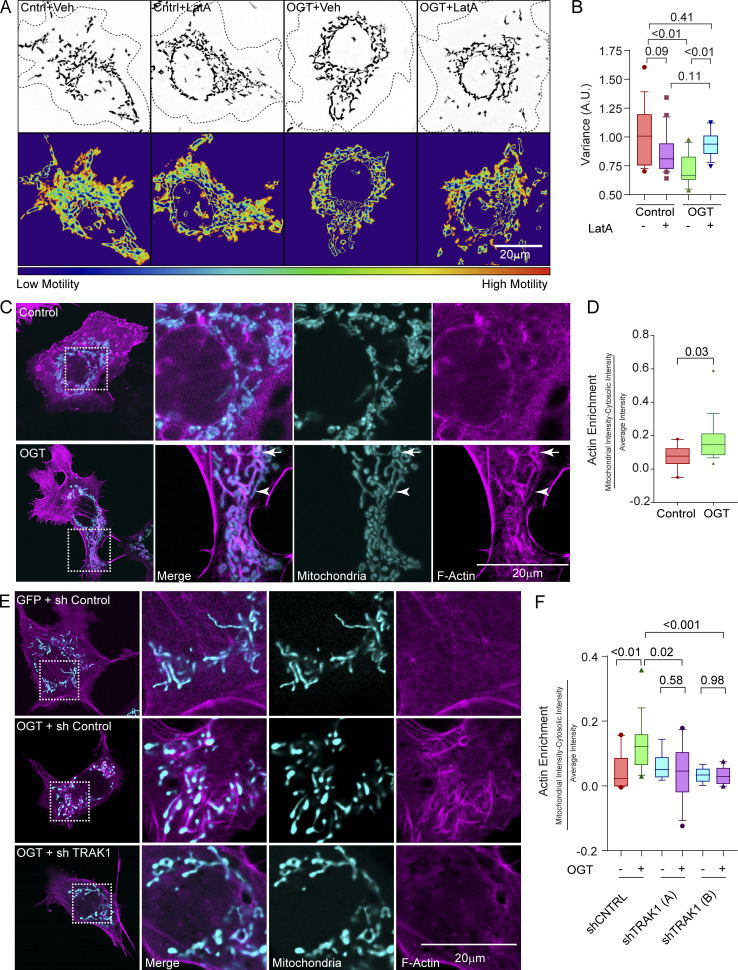
We also examined mitochondrial motility in COS-7 and U2OS cell lines. Mitochondria in these cells are extensively interconnected and make only short linear runs that are poorly analyzed by kymography. We therefore developed an algorithm (Basu and Schwarz, 2020) that reflects the extent to which they move by analyzing the variance in pixel occupancy of mitochondrial fluorescence over time (Fig. S2 A). The mitochondrial motility is depicted as a 2D heatmap, from which a motility score is derived for quantification. The variance could serve as the basis of the algorithm, because any pixel occupied by a stationary mitochondrion will have a constant value and low variance, whereas pixels that change between mitochondrion-occupied and -unoccupied states will have a high variance. By normalizing the variance in pixel occupancy to the total occupancy of each pixel over time, this algorithm also puts more weight on fast processive movements than on back-and-forth movements (see Materials and methods for more details).
Figure S2.
OGT suppresses mitochondrial motility in COS-7 and U2OS cells (related to Fig. 2). (A) Stages in the quantification of mitochondrial motility in non-neuronal cells, exemplified by representative COS-7 cells with high motility (control) and low motility (OGT expressing; for corresponding movie, see Video 2). The mitochondria are initially defined by a threshold that is based on the contrast of the fluorescence signal over the local background. This thresholding makes the measurement independent of the differences in the real fluorescence of the mitochondria. To eliminate noise from individual pixels, a lower size limit for a mitochondrion is also defined. After this segmentation step, the variance and the sum of fluorescence over time for each pixel are then calculated. The final motility heatmap is constructed as the ratio of variance over sum. Parts of the image that have high and processive movement display high variance and low sum and thereby appear as hot spots in the heatmap. To derive a motility value from such a heatmap, the sum of the values for all the pixels is normalized to the area occupied by the mitochondria. See Materials and methods for details. (B and C) Upon overexpression, OGT increases O-GlcNAcylation and decreases mitochondrial motility, whereas OGA decreases O-GlcNAcylation and increases mitochondrial motility. To monitor mitochondrial motility, COS-7 cells were transfected with Mito-DsRed along with GFP-2A-OGT or GFP-2A-HA-OGA or control (GFP) constructs. (B) Representative images of mitochondria in each outlined cell (top) and a heatmap for each cell of the variance in mitochondrial fluorescence over time (bottom; corresponds to Video 3). (C) Quantification of mitochondrial motility for cells such as those in B, as illustrated in A. n = 15–20 cells per condition from 3 independent transfections. (D and E) OGT and OGA constructs do not influence endosomal motility. COS-7 cells were cotransfected with Rab5-mCherry to label endosomes, along with GFP-2A-OGT or GFP-2A-HA-OGA or GFP and analyzed as in C. Representative images (D) and quantification of motility (E; similar cells expressing OGA and OGT are shown in Video 5). n = 15–20 cells per condition from 3 independent transfections. (F and G) In U2OS cells, OGT and OGA alter mitochondrial motility as they do in COS-7 cells. (F) Representative images of U2OS cells expressing Mito-DsRed and OGT or OGA are shown along with their variance heatmap. (G) Quantification of mitochondrial movement in the U2OS cells was done in the same manner as described for COS-7 cells. n = 10–15 cells per condition from 3 independent transfections. (H and I) OGT and OGA constructs do not influence endosomal motility in U2OS cells. U2OS cells were cotransfected with Rab5-mCherry to label endosomes, along with GFP-2A-OGT or GFP-2A-HA-OGA or GFP (control) and analyzed as before. (H) Representative images of endosomes and their heatmaps are shown. (I) Quantification of endosomal motility was done by measuring the variance of pixel occupancy as described in A. Neither OGA nor OGT changed the endosomal motility significantly from that observed for the cells expressing GFP (control; I). n = 10–15 cells per condition from 3 independent transfections. (J) Enrichment of phalloidin-labeled F-actin around mitochondria upon expression of OGT. Cells expressing OGT or GFP (control) were fixed and stained with phalloidin and an antibody against ATP5A. As seen with other F-actin markers (LifeAct-RFPt and F-Tractin), phalloidin staining revealed dense F-actin networks around mitochondria in the cells expressing OGT. (K and L) Validation of shRNA against TRAK1 in COS-7 cells. COS-7 cells were transfected with three different shRNA sequences against TRAK1 (shTRAK1 1–3) and a nontargeting control shRNA. Cell lysates were probed for TRAK1 to determine the efficiency of knockdown. (K) Representative Western blot. Molecular weights (in kD) are indicated on the right. (L) Quantification of TRAK1 knockdown from Western blots as in K. The intensity of the TRAK1 band was normalized to the intensity of the GAPDH band. Two shRNAs were effective at reducing TRAK1 levels and were chosen as shTRAK1 (A) and shTRAK1 (B). n = 3 independent transfections per condition. Quantifications in C, E, G, and I are represented as box-and-whisker plots. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. P values were determined with a two-tailed unpaired t test with Welch’s correction. In L, bars indicate mean ± SEM, and a ratio paired t test was employed to calculate P values.
As in neurons, overexpressing OGT caused a significant decrease in mitochondrial motility (Fig. 2, A and B; Fig. S2, B and C; Fig. S2, F and G; and Videos 2 and 3). This OGT-induced reduction was relieved by disrupting F-actin (LatA treatment; Fig. 2, A and B; and Video 4). Movement of Rab5-mCherry–labeled endosomes, however, was not altered by OGT expression (Fig. S2, D and E; Fig. S2, H and I; and Video 5). Thus O-GlcNAcylation–mediated arrest of mitochondria is a feature of multiple cell types, consistently dependent on the actin cytoskeleton, and selective for mitochondria.
Figure 2.
OGT expression results in F-actin accumulation around mitochondria. (A) Mitochondrial motility in COS-7 cells expressing OGT or a control plasmid (GFP; Cntrl) after treatment with 0.05 µM LatA or its vehicle (DMSO; Veh) for 90 min. Upper panels show representative images of mitochondria in outlined cells. Lower panels show heatmaps of the variance in pixel occupancy (of Mito-DsRed) over time. (B) Quantification of mitochondrial motility from cells as in A, as reflected in the variance heatmaps. n = 15–20 cells per condition from 3 independent transfections. (C and D) Enrichment of F-actin on mitochondria in COS-7 cells expressing GFP-2A-OGT, the F-actin marker LifeAct-RFPt (magenta), and the mitochondrial marker Mito-BFP (cyan). In representative images (C), some mitochondria appear encapsulated by actin (arrow), whereas others align with actin bundles (arrowhead). For quantification (D), F-actin intensity was calculated within the area of a mask formed from the mitochondrial signal and then dilated by 2 pixels (to include the F-actin–enriched area around the mitochondria). This intensity was then normalized to the total amount of F-actin within the cell. n = 10–15 cells per condition from 3 independent transfections. (E and F) shRNA to TRAK1 prevents the enrichment of F-actin on mitochondria in COS-7 cells expressing GFP-2A-OGT. (E) Representative images of cells expressing the F-actin marker F-Tractin-RFP (magenta) and the mitochondrial marker Mito-BFP (cyan). GFP-2A-OGT increases F-actin accumulation around mitochondria in the presence of a control shRNA but not in the presence of two unique shRNAs to TRAK1. Quantification in F is as in D. n = 10–15 cells per condition from 3 independent transfections. All quantifications are represented as box-and-whisker plots. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. The indicated P values are from the two-tailed unpaired t test in which Welch’s correction was used.
Video 2.
OGT suppresses mitochondrial motility in COS-7 cells expressing Mito-DsRed and OGT or GFP (control; related to Fig. S2). Corresponding images and motility heatmaps for these cells are shown in Fig. S2 A. Scale bar represents 20 µm. Video is played at 60× real-time speed and rendered at 60 frames per second.
Video 3.
Comparison of mitochondrial motility in COS-7 cells expressing OGT and OGA (related to Fig. S2). Mito-DsRed was expressed with either GFP-2A-OGT or GFP-2A-OGA. Corresponding images and motility heatmaps for these cells are shown in Fig. S2 B. Scale bar represents 20 µm. Video is played at 30× real-time speed and rendered at 60 frames per second.
Video 4.
F-actin disruption reverses OGT-mediated mitochondrial arrest in COS-7 cells (related to Fig. 2). A COS-7 cell coexpressing Mito-DsRed and GFP-2A-OGT is shown before and after its treatment with 0.05 µM LatA for 90 min. Scale bar represents 20 µm. Video is played at 60× real-time speed and rendered at 10 frames per second.
Video 5.
Hyper-O-GlcNAcylation by OGT or removal of O-GlcNAc by OGA expression does not influence endosomal motility (related to Fig. S2). COS-7 cells expressed Rab5-mCherry and either GFP-2A-OGT or GFP-2A-OGA. Scale bar represents 20 µm. Video is played at 30× real-time speed and rendered at 20 frames per second.
F-actin is enriched on mitochondria following O-GlcNAcylation–mediated motility arrest
To visualize any OGT-induced changes in the relationship of the actin cytoskeleton and mitochondria, we imaged F-actin in COS-7 cells by transfection with LifeAct-RFPt and labeled mitochondria with Mito-BFP. Consistent with an anchoring mechanism, OGT expression caused mitochondria to become associated with dense F-actin structures (Fig. 2, C and D). Some mitochondria appeared to be aligned with adjacent actin filaments (arrowhead), while others appeared to be surrounded by a ring of actin (arrow). The F-actin enrichment was also observed with phalloidin (Fig. S2 J) as well as F-Tractin (Fig. 2 E).
OGT-induced mitochondrial arrest depends on TRAK O-GlcNAcylation (Pekkurnaz et al., 2014). If the OGT-induced F-actin accumulation is part of the same arrest pathway, we predicted that it would require the presence of TRAK. We therefore knocked down TRAK1 in COS-7 cells with two independent shRNAs against TRAK1 (Fig. S2, K and L) and imaged F-actin with and without OGT overexpression. OGT expression did not increase F-actin density around mitochondria after knockdown of TRAK1 (Fig. 2, E and F). Thus, the OGT-induced F-actin rearrangement around the mitochondria following motility arrest is indeed mediated by TRAK1.
Mislocalizing the motor–adaptor complex to peroxisomes is sufficient to subject peroxisomes to regulation by OGT
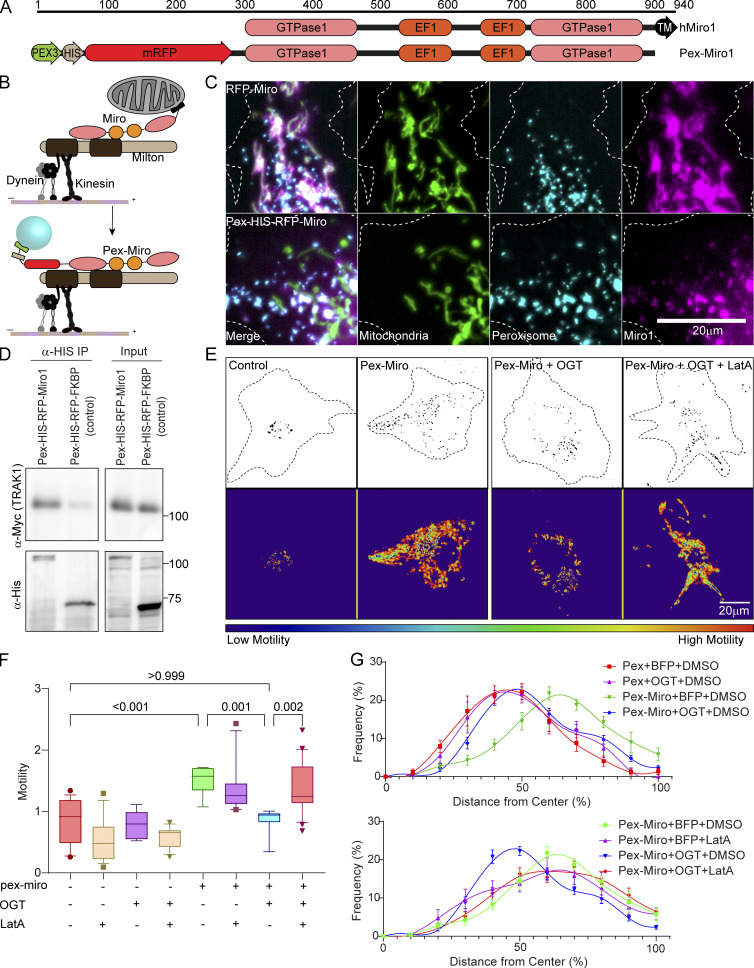
We hypothesized that misdirecting the TRAK1–Miro complex to peroxisomes would confer OGT-dependent regulation on that organelle. To test this hypothesis, we targeted the complex to the surface of peroxisomes by removing the transmembrane domain from the C-terminus of Miro, which normally attaches it to the mitochondrial outer membrane, and by adding the transmembrane domain of a peroxisomal membrane protein, peroxisomal biogenesis factor 3 (PEX3), to the N-terminus of Miro (Fig. 3, A and B). Peroxisomes were labeled by directing a fluorophore to their lumen by addition of a targeting sequence (serine-arginine/lysine-leucine [SRL, SKL]): mTurquoise-SRL or mCitrine-SKL. Upon expression in COS-7 cells, the peroxisome-targeted Miro (Pex-Miro) localized to peroxisomes (Fig. 3 C) and coprecipitated with TRAK1, indicating assembly of the complex on this surrogate organelle (Fig. 3 D). Consequently, the peroxisomes in cells expressing Pex-Miro were more mobile than those in cells expressing a control construct in which Miro was replaced by the inert, unrelated protein FK506-binding protein (Pex-FKBP; Fig. 3, E and F; and Video 6).
Figure 3.
Mislocalizing the mitochondrial motor–adaptor complex to peroxisomes is sufficient to make peroxisomal motility subject to regulation by OGT and F-actin. (A and B) Schematic of redirecting the TRAK1–Miro complex onto peroxisomes. (A) The transmembrane domain (N-terminal 44-aa residues) of a peroxisomal membrane protein (Pex3) was attached to the N-terminus of Miro1 (first 592 aa residues) lacking its own transmembrane domain. (B) In consequence, the complex assembles on the peroxisomal surface. (C) Representative images of cells demonstrating the localization of Pex-Miro to peroxisomes. The cells have been transfected with the mitochondrial marker Mito-BFP and the peroxisomal marker mCitrine-SKL in addition to RFP-tagged full-length Miro or Pex-Miro. Full-length WT Miro1 localized to mitochondria (upper panels), whereas Pex-Miro localized to peroxisomes (lower panels). (D) Myc-TRAK1 association with Pex-Miro. Pex-Miro (mol wt 105 kD) or Pex-FKBP (control; mol wt 60 kD) were immunoprecipitated by their 6XHis tags from HEK293T cells also expressing Myc-TRAK1. Immunoprecipitates were probed for His and Myc immunoreactivity to detect the Pex constructs and TRAK1. Molecular weights (in kD) are indicated on the right. (E–G) Expression of Pex-Miro conveys OGT regulation on the motility and dispersion of peroxisomes. COS-7 cells were transfected with BFP-2A-OGT or BFP (control) in addition to Pex-Miro or Pex-FKBP. The cells were also treated with 0.05 µM LatA or DMSO (control). Time-lapse images from these cells were quantified by variance analysis using the same software used for quantifying mitochondrial motility in COS-7 cells. (E) Representative images of the peroxisomes (upper panels) and a heatmap of the variance (lower panels) are shown and correspond to Video 6. (F) Quantification of peroxisomal motility from variance images as shown in E. Higher motility of peroxisomes yielded a higher variance signal in the cells expressing Pex-Miro. This was significantly reduced when BFP-2A-OGT was expressed in these cells. On treatment with LatA, the OGT-mediated arrest of Pex-Miro peroxisomes was reversed, thereby demonstrating the role of F-actin to bring about the arrest. (G) Quantification of peroxisomal distribution in COS-7 cells expressing constructs as in E. The distribution quantification was done by custom software that analyzes their distribution as a function of distance from the cell center, normalized for cell shape. Peroxisomes in control cells are close to the cell center but are broadly distributed in cells expressing Pex-Miro. Expression of OGT reduces peroxisomal dispersion, but this reduction is reversed by LatA. n = 15–21 cells per condition from 3 independent transfections. The data from the conditions Pex-Miro + BFP + DMSO and Pex-Miro + OGT + DMSO have been replicated in both panels for ease of comparison with the other datasets. For calculating peroxisomal distributions, each cell was divided into 10 shells, each representing 10% growth from the cell center. Peroxisomal frequencies are represented as the relative percentage of peroxisomes at each concentric shell. Each point indicates the means of all cells, and the whiskers indicate SEM. The motility quantifications in F are represented as box-and-whisker plots. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. A two-tailed unpaired t test with Welch’s correction was used to generate the indicated P values.
Video 6.
Mislocalizing the mitochondrial motor–adaptor complex to peroxisomes increases peroxisomal motility and is sufficient to make peroxisomal motility subject to regulation by OGT (related to Fig. 3). COS-7 cells expressing Pex-HIS-RFP-Miro or Pex-HIS-RFP-FKBP (control) along with OGT or GFP (control) and treated with LatA or DMSO. Corresponding images and motility heatmaps for these cells are shown in Fig. 3 E. Scale bar represents 20 µm. Video is played at 30× real-time speed and rendered at 20 frames per second.
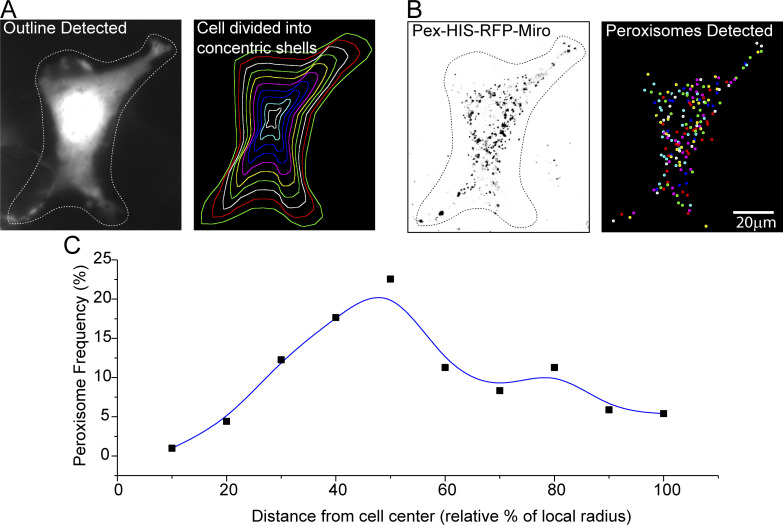
We also analyzed the distribution of these peroxisomes by developing software that calculates the frequency of occurrence of peroxisomes within a series of concentric shells that expand from the cell’s center to the cell membrane in parallel to the outline of the cell (Fig. S3). The peroxisomes with Pex-Miro were more broadly dispersed throughout the cytoplasm than those in control cells whose peroxisomes were typically concentrated closer to the nucleus (Fig. 3 G); this redistribution was a likely consequence of their increased motility.
Figure S3.
Quantification of peroxisomal distribution in COS-7 cells (related to Fig. 3). (A) Division of the cell into concentric shells. The cell outline is automatically detected from fluorescence intensity above a defined global threshold. The outline can then be manually refined. A user-defined cell center is then selected; for these studies, it was placed in the center of the nucleus. The distance of each point on the cell outline to the cell center is calculated using a path that does not go outside the cell outline. These values are used to make equally spaced concentric shells that are parallel to the cell outline. As illustrated here, 10 shells were used for each cell in this study. (B) The peroxisomes are detected based on their intensity over their local background and with upper and lower size limits to eliminate pixel noise. (C) Representative graph showing the frequency of peroxisomal occurrence in each shell of the cell illustrated in A and B. The average of multiple such frequency histograms from different cells was used to quantify each condition depicted in Fig. 3 G.
Consistent with our hypothesis, the motile peroxisomes (in cells expressing Pex-Miro) were arrested by OGT expression, and their distribution became more perinuclear (Fig. 3, E–G). To determine whether F-actin was required for the effect of OGT, we treated the cells with LatA. As predicted, the Pex-Miro peroxisomes regained their motility and broad distribution despite the expression of OGT (Fig. 3, E–G). F-actin disruption in control cells had minimal effect on the peroxisomal motility. Thus, relocating the TRAK1–Miro complex to peroxisomes is sufficient to confer properties of mitochondrial motility on peroxisomes, including the actin-dependent regulation of motility by OGT.
O-GlcNAcylated Milton associates with FHL2
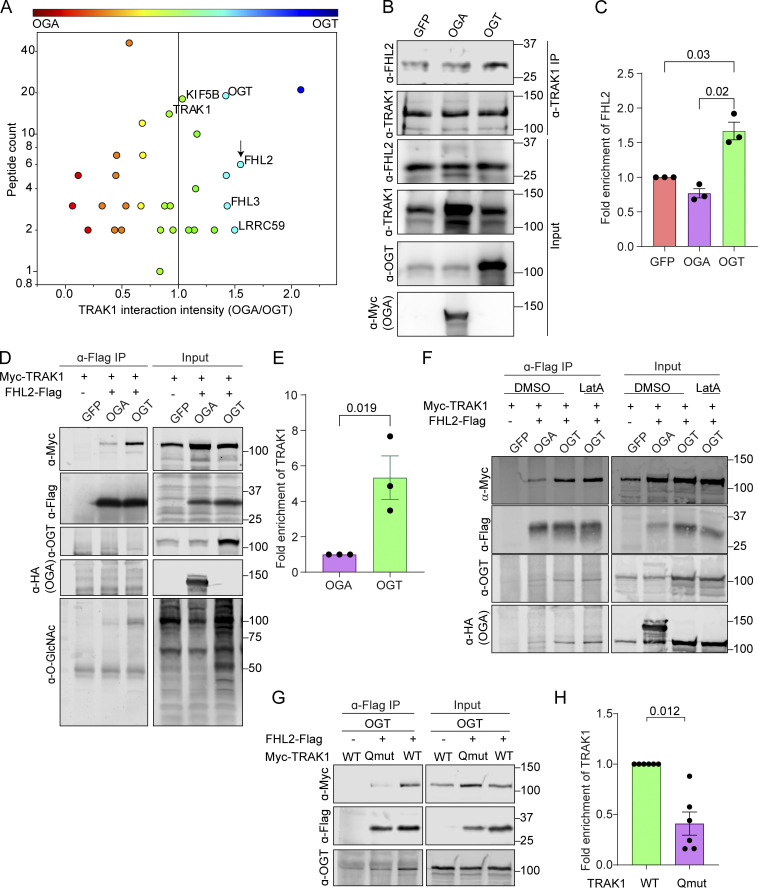
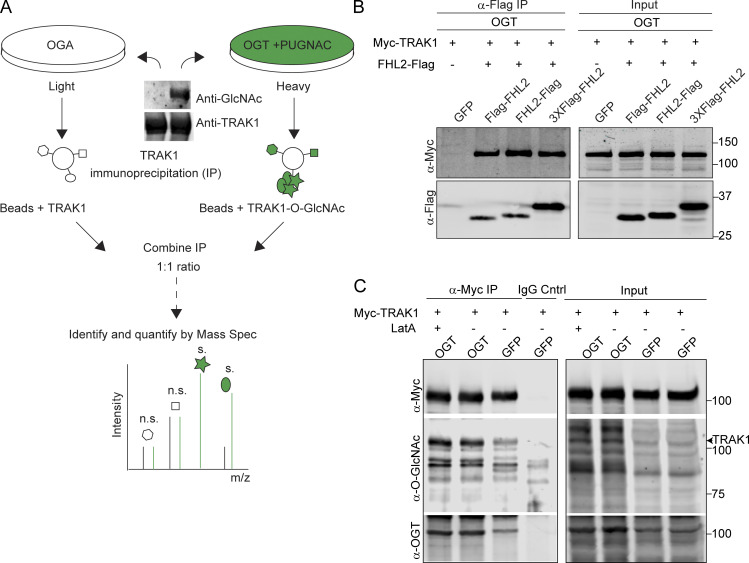
The finding that OGT-stimulated mitochondrial arrest was mediated by the TRAK1–Miro complex and F-actin suggested that an additional protein might be recruited to the O-GlcNAcylated complex to bring about the actin recruitment. We therefore employed stable isotope labeling by amino acids in cell culture mass spectrometry (SILAC-MS) to compare endogenous TRAK1 immunoprecipitates from HEK293T/17 cells expressing either OGT or OGA, representing conditions in which TRAK1 would be maximally or minimally O-GlcNAcylated (Fig. 4 A and Fig. S4 A; and Table S1). Two related proteins from the family of the four and a half LIM domains proteins, FHL2 and FHL3, were enriched in the TRAK1 immunoprecipitate from the OGT-expressing cells. FHL2 and FHL3 can directly or indirectly bind to F-actin and can influence F-actin polymerization and bundling (Boateng et al., 2016; Coghill et al., 2003; Johannessen et al., 2006; Li et al., 2001; Ng et al., 2014; Olson and Nordheim, 2010; Tran et al., 2016). Because FHL3 is not detectably expressed in neurons (Zhang et al., 2014), we focused on FHL2. We confirmed on the basis of Western blots that 60% more FHL2 coprecipitated with endogenous TRAK1 from HEK293T/17 cells expressing OGT than from those expressing OGA (Fig. 4, B and C). The OGT-dependent association was also observed upon coexpression of Flag-tagged FHL2 and Myc-tagged TRAK1 (Fig. 4, D and E; and Fig. S4 B). In anti-Flag immunoprecipitates, there was fivefold greater Myc-TRAK1 in the hyper-O-GlcNAcylated condition of OGT overexpression than with OGA overexpression. Treatment of these cells with LatA before the immunoprecipitation (IP) neither prevented the association of FHL2-Flag with Myc-TRAK1 (Fig. 4 F) nor altered the degree of TRAK1 O-GlcNAcylation (Fig. S4 C). Therefore, the FHL2–TRAK1 interaction is not a secondary consequence of actin recruitment to the mitochondria, but rather might serve as the anchor connecting mitochondria to F-actin when the OGT pathway is activated.
Figure 4.
GlcNAcylated Milton coprecipitates with FHL2. (A) TRAK1-interacting proteins identified by SILAC-MS after IP of endogenous TRAK1 from HEK293T cells expressing either OGT or OGA. The OGT-expressing cells were also treated with 100 µM PUGNAc (an inhibitor of OGA) to maximize O-GlcNAcylation. Protein enrichment is plotted against peptide counts. Commonly detected contaminants were removed from the plot. (See Table S1 for the full list of proteins.) The constituents of the motor–adaptor complex and key hits (enriched in immunoprecipitates from cells expressing OGT) are labeled. FHL2 is marked by an arrow. (B and C) Endogenous TRAK1 was immunoprecipitated from HEK293T cells expressing OGT, OGA, or GFP (control). (B) Representative Western blots demonstrating the expression and co-IP efficiency of FHL2 with TRAK1 are shown for OGT-, OGA-, and GFP-expressing cells. (C) Quantification of co-IP efficiency of FHL2 with TRAK1 as the ratio of the intensity of the FHL2 band to that of TRAK1, normalized to the GFP condition. Immunoreactivity for endogenous FHL2 was enriched by ∼60% in the immunoprecipitates from cells expressing OGT. n = 3 independent transfections per condition. (D and E) OGT expression enhanced interaction of Myc-TRAK1 and FHL2-Flag coexpressed in HEK293T cells. HEK293T cells were cotransfected with Myc-TRAK1, FHL2-Flag, and OGT or OGA. After IP of FHL2-Flag, immunoblots (D) were probed for Myc-TRAK1, FHL2-Flag, OGT, HA-tagged OGA, and O-GlcNAc moieties (using the antibody RL2). Cells expressing GFP instead of FHL2-Flag were used as a control for nonspecific binding of Myc-TRAK1 to the Sepharose beads. The IP (E) was quantified as the ratio of the intensity of the Myc-TRAK1 band to that of the FHL2-Flag, normalized to the OGA condition. Myc-TRAK1 coprecipitated with FHL2-Flag strongly in the presence of OGT but weakly in the presence of OGA. n = 3 independent transfections per condition. (F) Interaction of Myc-TRAK1 and FHL2-Flag is independent of the actin cytoskeleton. Co-IP as in D, but cells were treated with DMSO or 0.05 µM LatA for 90 min before lysis. During IP, DMSO or 5 µM LatA was added to the IP buffer to prevent actin repolymerization in the lysate. Disruption of the actin cytoskeleton via LatA did not prevent the OGT-enhanced association of FHL2-Flag with Myc-TRAK1. (G and H) Interaction of Myc-TRAK1 and FHL2-Flag is dependent on the O-GlcNAcylation state of Myc-TRAK1. FHL2-Flag and OGT were coexpressed in HEK293T cells with either Myc-TRAK1WT or Myc-TRAK1QMut. (G) Representative Western blot of the FHL2-Flag immunoprecipitates. (H) Quantification of co-IP efficiency of Myc- TRAK1WT or Myc-TRAK1QMut with FHL2-Flag. Immunoprecipitates were quantified by measuring the ratio of the intensity of the Myc-TRAK1 band to that of the FHL2-Flag, normalized to the Myc-TRAK1WT condition. Although Myc-TRAK1WT coprecipitated with FHL2-Flag strongly in the presence of OGT, Myc-TRAK1QMut showed a 60% decrease in the FHL2-Flag interaction even in the presence of OGT. n = 6 from 3 independent transfections per condition. For all quantifications, P values are from ratio paired t tests. All data points are shown. Bars indicate mean ± SEM. For panels showing Western blots, molecular weights (in kD) are indicated on the right.
Figure S4.
Identification of FHL2 as an interactor of O-GlcNAcylated TRAK1 (related to Fig. 4). (A) Schematic for TRAK1 interactome analysis by SILAC-based quantitative proteomics. HEK293T cells expressing either OGA or OGT were cultured through five doublings in media containing L-arginine and L-lysine made of heavy and light C and N isotopes. OGT-expressing cells were cultured in the presence of the OGA inhibitor PUGNAc (100 µM). Cells were then immunoprecipitated with 2 μg of anti-TRAK1 antibody. A Western blot of the precipitate probed for TRAK1 and O-GlcNAc is shown and illustrates the differences in the extent of TRAK1 O-GlcNAcylation in the two conditions. The immunoprecipitates were then combined in a 1:1 ratio to ensure equal amounts of immunoprecipitated TRAK1 and interactors were quantified using mass spectrometry (Mass Spec). m/z, mass-to-charge ratio. (B) TRAK1 binds to Flag-tagged FHL2 in OGT-expressing cells. HEK293T cells were cotransfected with OGT, Myc-TRAK1, and one of three Flag-tagged FHL2 constructs (FHL2-Flag, Flag-FHL2, 3XFlag-FHL2) or GFP as a control. A representative Western blot is shown for the IP with anti-Flag antibody and subsequent probing with anti-Flag and anti-Myc. FHL2-Flag was most effective in immunoprecipitating TRAK1 and was chosen for further experiments. (C) LatA treatment does not change O-GlcNAcylation levels on TRAK1. HEK293T cells expressing OGT or GFP (control) along with Myc-TRAK1 were treated with 0.05 μM LatA or DMSO (control). Myc-TRAK1 was then immunoprecipitated from these cells and probed for levels of O-GlcNAcylation on a Western blot. Myc-TRAK1 immunoprecipitated from cells expressing OGT show higher levels of O-GlcNAcylation than control cells. This difference persisted in the presence of LatA. The TRAK1 band is indicated with an arrowhead. For panels showing Western blots, molecular weights (in kD) are indicated on the right.
We had previously shown that both glucose-mediated and OGT-mediated arrest of mitochondria depend on O-GlcNAcylation of TRAK1 at four identified serine and threonine residues. The OGT-induced mitochondrial arrest can be prevented by mutating these four sites to alanine (Pekkurnaz et al., 2014). We used this quadruple-mutant TRAK1 (TRAK1QMut) to determine whether the recruitment of FHL2 to TRAK1 is consistent with the mechanism that regulates mitochondrial motility. We immunoprecipitated FHL2-Flag from HEK293T/17 cells also expressing OGT and either Myc-TRAK1QMut or Myc-TRAK1WT. The association of FHL2 with Myc-TRAK1QMut was 60% less than with Myc-TRAK1WT (Fig. 4, G and H). Thus, the O-GlcNAcylation state of TRAK1 regulates the association of FHL2 with the TRAK1–Miro complex. The residual association of FHL2 with the complex, even when Myc-TRAK1QMut or OGA is expressed, may represent FHL2–TRAK interactions arising from other signaling events unrelated to the OGT pathway.
OGT recruits FHL2 to mitochondria to mediate mitochondrial association with actin
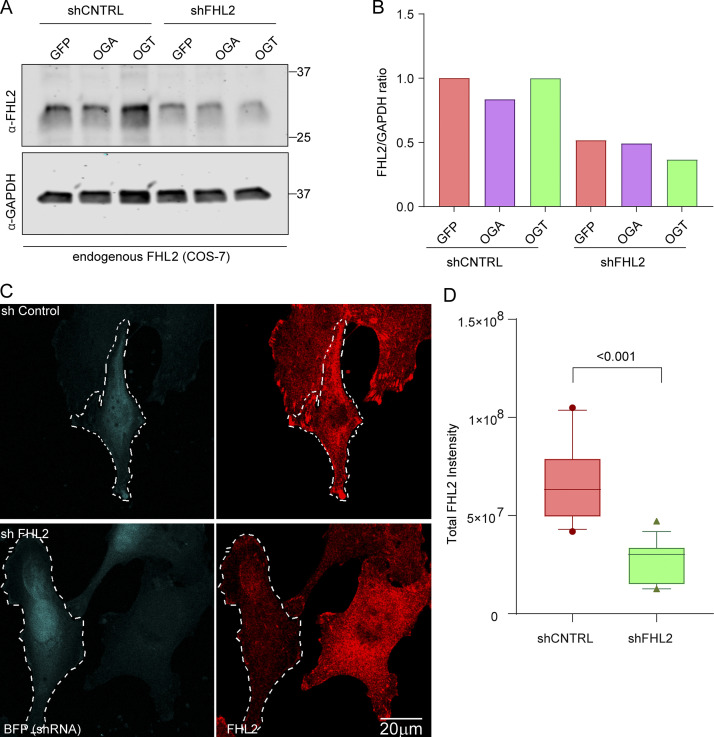
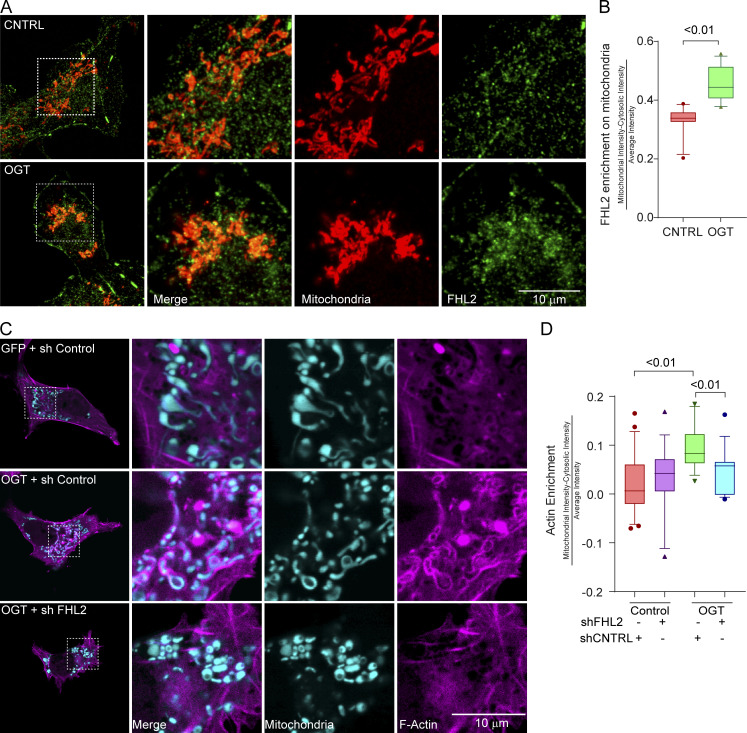
The OGT-driven association of TRAK1 and FHL2 predicted that FHL2 will be enriched on mitochondria when OGT is expressed. We therefore stained COS-7 cells expressing either OGT or a control plasmid with antibodies to endogenous FHL2 (for details of antibody validation, see Fig. S5, A–D) and the mitochondrial marker ATP5A. FHL2, normally diffuse in the cytosol and concentrated at focal adhesions (Boateng et al., 2016; Nakazawa et al., 2016), was significantly enriched on mitochondria in OGT-expressing cells (Fig. 5, A and B). We then proceeded to validate shRNAs against FHL2 (Fig. S5) and knockdown FHL2 in COS-7 cells to determine if the TRAK1–FHL2 interaction was necessary for the OGT-evoked association of actin with mitochondria. Knockdown of FHL2 prevented the mitochondrial accumulation of F-actin due to OGT expression (Fig. 5, C and D).
Figure S5.
Validation in COS-7 cells of shRNA and antibody against FHL2 (related to Fig. 5). (A and B) COS-7 cells were transfected with OGT, OGA, and GFP along with an shRNA to FHL2. The cells were then lysed, and the lysates were probed for FHL2 and GAPDH to confirm FHL2 knockdown (A). In all cases, the shRNA against FHL2 was effective in reducing the levels of FHL2 (as compared with GAPDH, loading control) by >50% (B). (C and D) Validation of antibody against FHL2 used for immunocytochemistry in COS-7 cells. The FHL2 shRNA plasmid was modified to coexpress BFP to mark the transfected cells. Cultures expressing the FHL2 shRNA or a control shRNA were fixed and stained with the antibody against FHL2 (C). Left panels indicate BFP-tagged cells; right panels show the FHL2 staining as a sum projection of confocal slices. (D) The total intensity of FHL2 staining was quantified in the transfected cells. n = 10–15 cells from 3 independent transfections. Quantifications are represented as box-and whisker plots. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. P value is from two-tailed unpaired t test with Welch’s correction.
Figure 5.
FHL2 accumulation on mitochondria is necessary for OGT-induced mitochondrial recruitment of F-actin. (A) FHL2 enrichment on mitochondria in COS-7 cells expressing OGT. COS-7 cells expressing GFP-2A-OGT or a control (CNTRL) GFP plasmid were fixed and probed with antibodies against FHL2 (green) and mitochondria (ATP5A, red). Full cells (left) and enlargements (right) of the indicated regions are shown. (B) Quantification of FHL2 enrichment on mitochondria in cells as in A. The enrichment of FHL2 on mitochondria was quantified as the intensity of the FHL2 in the mitochondrial mask normalized to the average intensity of FHL2 in the cytosol. n = 10–15 cells per condition from 3 independent transfections. (C and D) Mitochondrial F-actin enrichment in COS-7 cells expressing GFP-2A-OGT is dependent on FHL2. (C) Representative images, full cells (left panels), and enlarged regions of cells expressing the F-actin marker F-Tractin-RFP (magenta) and the mitochondrial marker Mito-BFP (cyan) along with GFP-2A-OGT or GFP (control). (D) Quantifications of F-actin enrichment around mitochondria were done as described for Fig. 2. When FHL2 is knocked down (shFHL2), the OGT-mediated increase of F-actin around mitochondria is prevented. n = 10–15 cells per condition from 3 independent transfections. Data are represented as box-and-whisker plots. The line indicates the median, the box indicates interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. For significance testing, a two-tailed unpaired t test with Welch’s correction was used, and P values are indicated.
FHL2 is required for mitochondrial arrest induced by OGT expression or by glucose shift
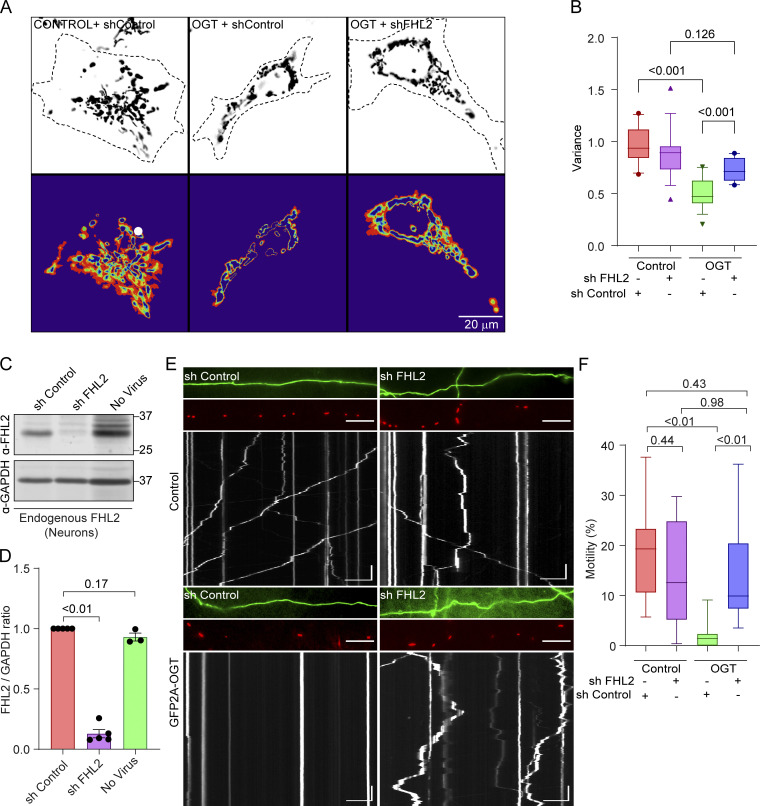
To ask whether FHL2 is required for the mitochondrial arrest driven by TRAK1 O-GlcNAcylation, we monitored mitochondrial motility in COS-7 cells when FHL2 was knocked down (Fig. 6, A and B). Consistent with the FHL2-dependent mitochondrial arrest model, OGT expression failed to suppress mitochondrial movement in COS-7 cells expressing the shRNA to FHL2.
Figure 6.
Knockdown of FHL2 prevents OGT-mediated mitochondrial arrest. (A and B) Mitochondrial motility in COS-7 cells expressing OGT or a control plasmid (GFP) along with shRNA to FHL2. (A) Representative images of mitochondria (upper panels) and heatmaps of the variance in Mito-DsRed over time (lower panels). (B) Quantification of mitochondrial motility as determined from the variance. The expression of OGT decreased mitochondrial motility in control cells, but not in those expressing shRNA to FHL2. n = 15–20 cells per condition from 3 independent transfections. (C and D) Knockdown of endogenous FHL2 in rat hippocampal neurons by lentiviral expression of shRNA. (C) Representative neuronal cell lysates on Western blots probed for FHL2 and GAPDH (as a loading control) are shown. Molecular weights (in kD) are indicated on the right. (D) Knockdown efficiency was quantified by measuring the ratio of the intensity of the FHL2 band to that of GAPDH, normalized to the control shRNA condition. n = 3–5 independent transductions per condition. P values were determined by ratio paired t tests. All data points are shown. Bars indicate mean ± SEM. (E and F) Effect of FHL2 knockdown in neurons on OGT-mediated mitochondrial arrest. Neurons were cotransfected with Mito-DsRed, GFP-2A-OGT or GFP, and shRNA against FHL2 or a control nontargeting shRNA. (E) Representative images of axons with GFP (top, green) and Mito-DsRed (middle, red) and kymographs (bottom) of mitochondrial motility. (F) Quantification of mitochondrial motility (percentage of time spent in motion for all mitochondria in the imaged regions) from kymographs. n = 10–12 axons per condition from 3 independent animals. Horizontal scale bars represent 20 µm, and vertical scale bars represent 30 s. All data except for bar graphs in D are represented as box-and-whisker plots. The line indicates the median, the box indicates the interquartile range, whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. Significance testing was done using a two-tailed unpaired t test with Welch’s correction, and P values are indicated.
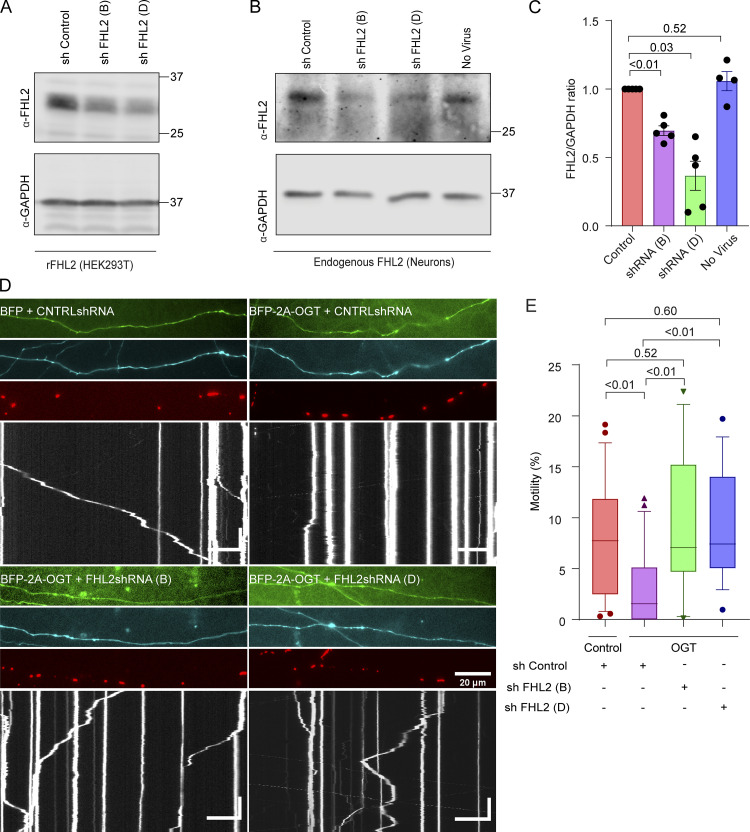
To test whether this mechanism also pertained to neurons, we identified shRNA sequences effective against endogenous rat FHL2 upon lentiviral transduction. The most effective shRNA construct or a control construct (Fig. 6, C and D) was transfected into rat hippocampal neurons along with a mitochondrial marker, Mito-DsRed, and OGT (Fig. 6, E and F). As expected, neurons overexpressing OGT showed significantly suppressed mitochondrial motility compared with neurons expressing GFP alone (control). However, in the presence of the FHL2 shRNA construct, mitochondrial motility was resistant to OGT. To rule out sequence-specific and backbone-specific off-target effects, we tested two additional shRNA sequences built on different vector backbones (Fig. S6) and verified their efficacy in knocking down both overexpressed and endogenous FHL2 (Fig. S6, A–C). These additional shRNA constructs also effectively suppressed the OGT-mediated mitochondrial arrest in rat hippocampal neurons (Fig. S6, D and E). Thus, FHL2 is required for O-GlcNAcylation–induced mitochondrial movement arrest in neurons and non-neuronal cells.
Figure S6.
Knockdown of FHL2 prevents OGT-mediated mitochondrial arrest (related to Fig. 6). (A) Validation of shRNA sequences against rat (r)FHL2 in HEK293T cells transfected with a rat FHL2 overexpression vector and the indicated shRNA constructs. On Western blots probed for FHL2 and GAPDH (as a loading control), shRNA B and shRNA D (out of a set of four shRNA constructs tested) potently reduced FHL2 levels. (B and C) Validation of knockdown of endogenous neuronal FHL2 by shRNA. At DIV 1, neurons were transduced with lentiviruses encoding shRNA constructs B and D or a control construct. (B) Representative Western blot of neuronal lysates harvested on DIV 6 and probed for FHL2 and GAPDH. (C) Knockdown efficiency was quantified as the ratio of the intensity of FHL2 bands to that of GAPDH and normalized to the control shRNA. n = 5 independent transductions per condition. P values are from ratio paired t tests, and all data points are shown. Bars indicate mean ± SEM. (D and E) Mitochondria in OGT-expressing neurons remain motile if FHL2 shRNA is also expressed using constructs distinct from those in Fig. 6. Hippocampal neurons were transfected with Mito-DsRed, BFP-2A-OGT or BFP (control), and shRNA against FHL2 or a control nontargeting shRNA. A GFP expression cassette in the shRNA vector backbone reported expression levels of shRNA. (D) Representative axonal images of shRNA expression (green), OGT expression (cyan), mitochondria (red), and kymographs constructed from the mitochondrial channel (bottom). (E) Quantification of mitochondrial motility from kymographs as in D. n = 15–20 axons per condition from 3 independent animals. Horizontal scale bars represent 20 µm, and vertical scale bars represent 30 s. OGT expression suppresses mitochondrial motility in the presence of the control shRNA. However, in the presence of either FHL2 shRNA construct, mitochondrial motility was unimpaired by OGT. Data are represented as box-and-whisker plot. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. P values are from two-tailed unpaired t tests with Welch’s correction. For panels showing Western blots, molecular weights (in kD) are indicated on the right.
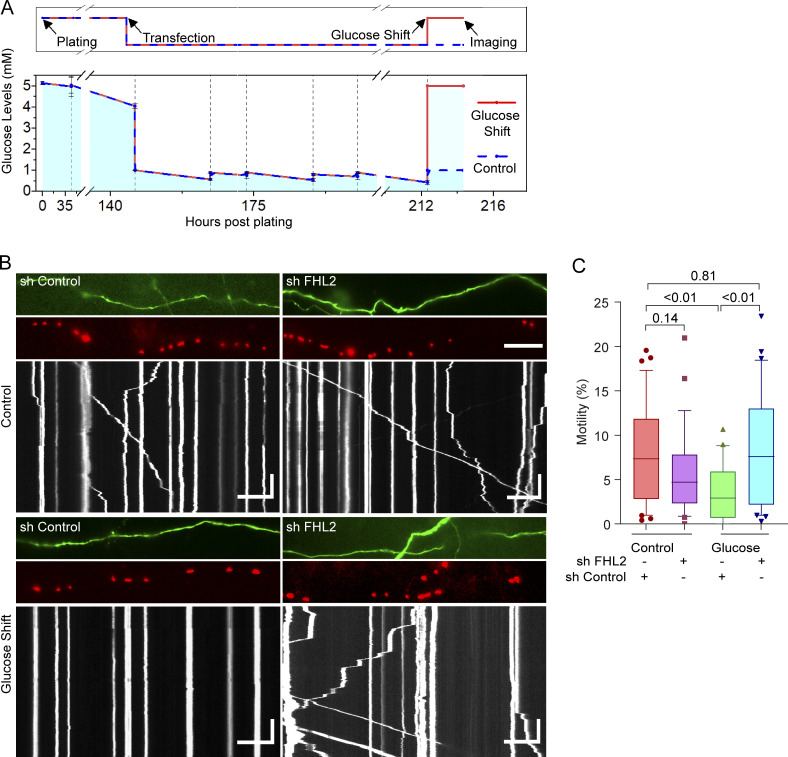
The requirement for FHL2 in OGT-mediated mitochondrial arrest predicted that FHL2 would also be required for the decrease in motility observed in response to increases in extracellular glucose. We therefore transfected neurons with Mito-DsRed, GFP, and either shRNA against FHL2 or a nontargeting control shRNA. As in Fig. 1, neurons were grown in 1 mM glucose before DIV 9, at which time the extracellular glucose was increased to 5 mM. Mitochondrial motility was measured 2 h after the glucose shift (Fig. 7 A). As we observed before, mitochondrial motility decreased in response to the glucose shift in neurons expressing the control shRNA. The expression of FHL2 shRNA had minimal effect on mitochondrial motility in neurons that were not subjected to the glucose shift (with a slight trend toward a decrease in motility). In contrast, neurons expressing the FHL2 shRNA and subjected to the glucose shift were resistant to decreases in mitochondrial motility (Fig. 7, B and C; and Video 7).
Figure 7.
Glucose-mediated mitochondrial arrest requires FHL2. (A) Schematic of experimental timeline (top) and measured glucose levels (bottom). Neurons were maintained at 5 mM glucose until DIV6, transfected, and then grown in 1 mM glucose. After 72 h, the glucose levels were either shifted back to 5 mM (glucose shift) or maintained in 1 mM (control) and imaged 2 h later. Glucose levels were measured before and after every feeding. n = 3 biological replicates, and mean and SEM are shown. (B and C) Neurons, cultured as in A, were transfected with shRNA against FHL2 or a nontargeting control shRNA, in addition to Mito-DsRed and GFP. (B) Images and kymographs of mitochondrial motility in neurons transfected as indicated (corresponding to Video 7). Horizontal scale bars represent 20 µm, and vertical scale bars represent 30 s. (C) Quantification of the percentage motility from kymographs as in B. The glucose shift did not decrease motility in neurons expressing FHL2 shRNA. n = 27–30 axons per condition from 4 independent animals. All quantifications are represented as box-and-whisker plots. The line indicates the median, the box indicates interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. The indicated P values are from two-tailed unpaired t tests with Welch’s correction.
Video 7.
FHL2 is necessary for glucose-mediated arrest of neuronal mitochondria (related to Fig. 7). Axon segments expressing Mito-DsRed and either an shRNA toward FHL2 or a nontargeting control shRNA were subjected to a glucose shift. Corresponding kymographs for these axons are shown in Fig. 7 B. The segments are oriented such that rightward movement is toward axon terminals. Scale bar represents 20 µm. Video is played at 30× real-time speed and rendered at 20 frames per second.
Localizing FHL2 to mitochondria causes actin-dependent mitochondrial arrest independent of O-GlcNAcylation
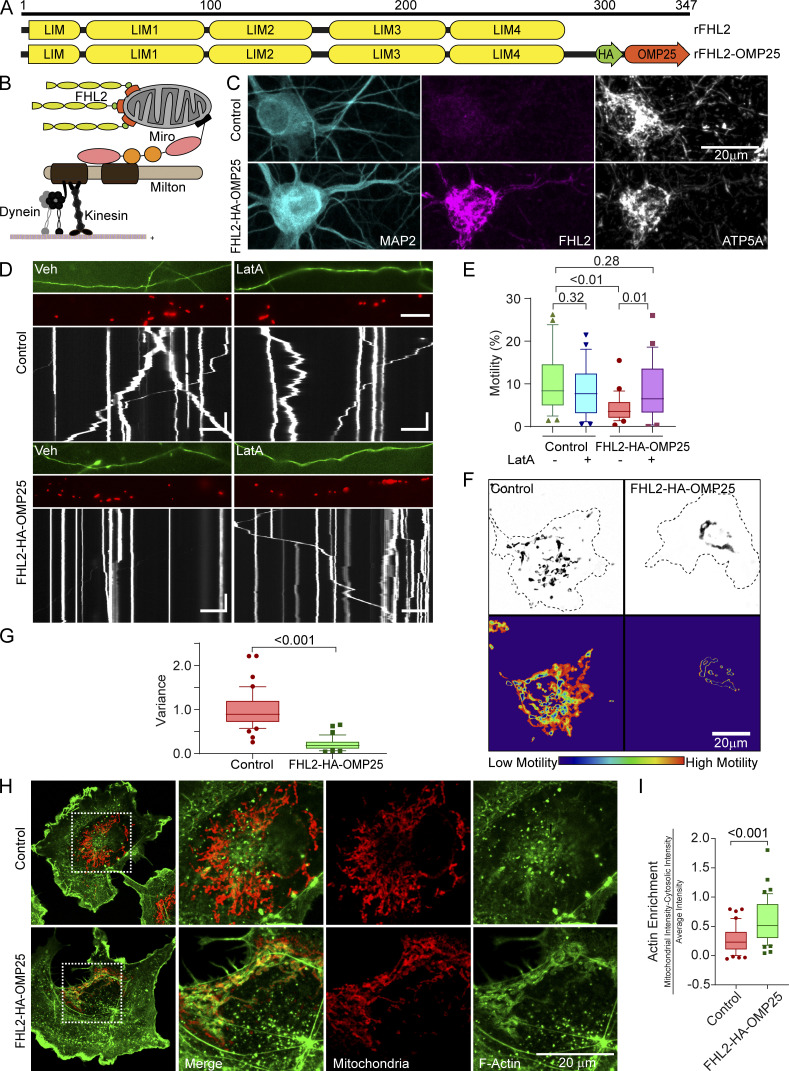
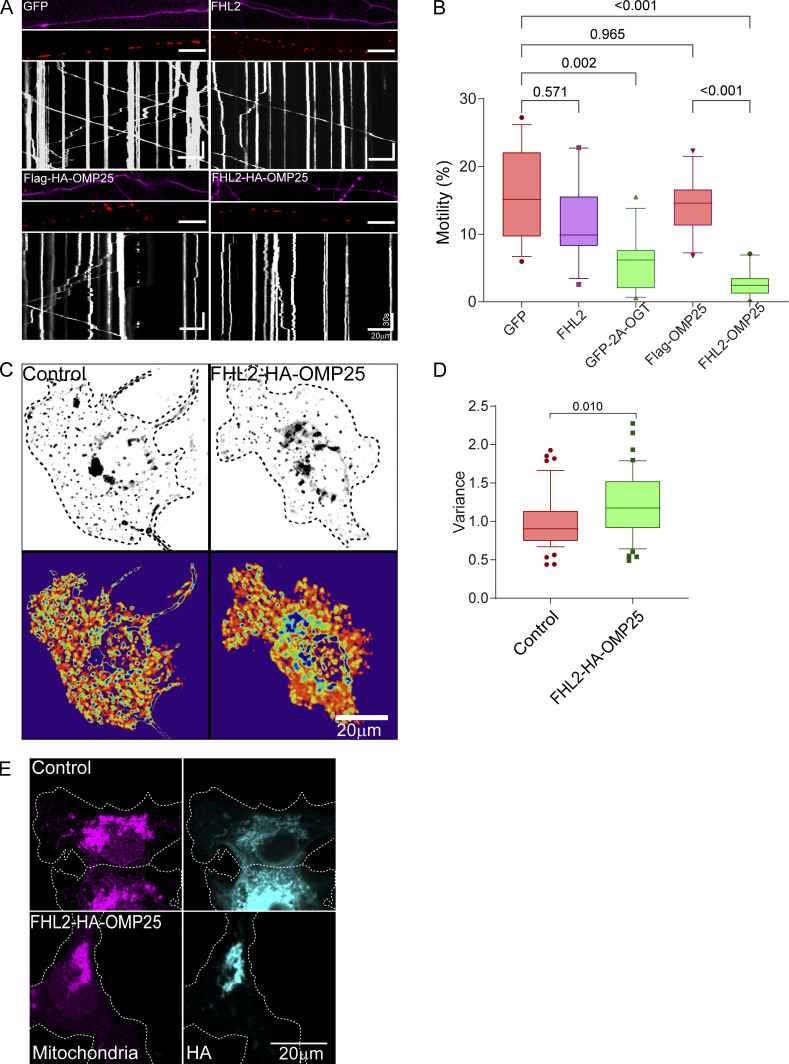
Having demonstrated that FHL2 was required for the glucose/OGT pathway to arrest mitochondria, we asked whether the presence of FHL2 on mitochondria was sufficient to recruit actin and arrest mitochondria. We artificially directed FHL2 to the mitochondrial surface by fusing FHL2 to the transmembrane domain of OMP25, a mitochondrial outer membrane protein (FHL2-HA-OMP25; Fig. 8, A and B), and confirmed by immunostaining that FHL2-HA-OMP25 localizes to mitochondria (Fig. 8 C). We transfected FHL2-HA-OMP25 or a control construct (SNAP-OMP25) into neurons cotransfected with Mito-DsRed and GFP (Fig. 8, D and E). Mitochondrial motility was greatly reduced in axons that expressed FHL2-HA-OMP25 compared with those expressing a control construct or cytosolic FHL2 (Fig. 8 E; and Fig. S7, A and B). Thus, mitochondrial FHL2 was sufficient to arrest motility even in the absence of induced O-GlcNAcylation. When we depleted F-actin in these neurons with LatA, mitochondrial motility was indistinguishable from axons without FHL2-HA-OMP25 (Fig. 8, D and E). Thus, the direct localization of FHL2 to the mitochondrial surface, like that mediated by O-GlcNAcylated TRAK1, immobilizes mitochondria by an F-actin–dependent mechanism.
Figure 8.
FHL2 localization to mitochondria is sufficient for actin-dependent mitochondrial immobilization. (A and B) Schematic of the rat (r)FHL2-HA-OMP25 construct (A) and strategy (B) for localizing FHL2 on the mitochondrial outer membrane. Full-length FHL2 was conjugated to an HA tag and the last 40 aa of OMP25. (C) Representative images showing that hippocampal mitochondria become highly immunoreactive for FHL2 when FHL2-HA-OMP25, but not SNAP-OMP25 (as a control), is expressed. The neurons were fixed and stained with antibodies toward FHL2, MAP2, and ATP5A. (D and E) Effect of FHL2-HA-OMP25 on mitochondrial motility and its dependence on F-actin. Neurons were transfected with FHL2-HA-OMP25 or a control vector (SNAP-OMP25) in addition to GFP and Mito-DsRed. FHL2-HA-OMP25 expression inhibited motility, which could be reversed by 5 µM LatA. (D) Representative images of axons (GFP, top), Mito-DsRed (middle), and kymographs (bottom) of mitochondrial motility in hippocampal axons. (E) Quantification of mitochondrial motility (average percentage of time spent in motion by all mitochondria in an axon segment) was calculated from kymographs as in D. n = 20–25 axons per condition from 3 independent animals. Horizontal scale bars represent 20 µm and vertical scale bars represent 30 s. (F and G) Mitochondrial motility in COS-7 cells expressing FHL2-HA-OMP25 or FLAG-HA-OMP25 (control). (F) Representative images of Mito-GFP (top) and heatmap of the temporal variance in the mitochondrial fluorescence (bottom). (G) Quantification of motility from analyzing the temporal variance of mitochondrial fluorescence from images as in F. n = 40–45 cells per condition from 3 independent transfections. (H and I) Actin enrichment on mitochondria bearing FHL2-HA-OMP25. COS-7 cells expressing FHL2-HA-OMP25 or Flag-HA-OMP25 (control) were fixed and stained with antibodies to HA and ATP5A (red) and with phalloidin conjugated to Alexa Fluor 488 (green). (H) Representative images of full cells (left) and enlarged insets of the indicated area (right). (I) Quantification of mitochondrial F-actin enrichment as in Fig. 2. F-actin around the mitochondria was significantly enriched in cells expressing FHL2-HA-OMP25 compared with control cells. n = 40–45 cells per condition from 3 independent transfections. All quantifications are represented as box-and-whisker plots. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. P values are from a two-tailed unpaired t test with Welch’s correction.
Figure S7.
FHL2-HA-OMP25 expression in neurons and COS-7 cells (related to Fig. 8). (A and B) Mito-FHL2 (FHL2-OMP25) inhibits mitochondrial motility in neurons, whereas cytosolic FHL2 does not. Hippocampal neurons were transfected with FHL2, FHL2-HA-OMP25, GFP-2A-OGT, GFP (control), or Flag-HA-OMP25 (control) along with Mito-DsRed. (A) Representative axonal images of FHL2 or OGT expression (magenta), mitochondria (red), and kymographs constructed from the mitochondrial channel (bottom). (B) Quantification of mitochondrial motility (average percentage of time spent in motion by all mitochondria in an axon segment) from kymographs shown in A. Although FHL2-HA-OMP25 and OGT significantly reduce mitochondrial motility, the expression of cytosolic FHL2 does not. n = 15–20 cells per condition from 3 independent animals. Horizontal scale bars represent 20 µm, and vertical scale bars represent 30 s. (C and D) Endosomal motility in COS-7 cells was not inhibited by expression of FHL2-HA-OMP25. FHL2-HA-OMP25 or FLAG-HA-OMP25 (control) were expressed together with Rab5-mEmerald. (C) Representative images of endosomes (top) and heatmap of variance representing endosomal motility (bottom). (D) Motility was quantified from the variance and normalized to the control cells. n = 40–45 cells per condition from 3 independent transfections. The quantifications in B and D are represented as box-and-whisker plots. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. P values are from a two-tailed unpaired t test with Welch’s correction. (E) Mitochondrial localization of FHL2-HA-OMP25 and Flag-HA-OMP25. Transfected COS-7 cells were fixed and stained with antibodies to HA and to the mitochondrial protein ATP5A.
To ask whether FHL2 is sufficient to increase the amount of actin associated with mitochondria (similar to that caused by the overexpression of OGT; Fig. 2), we expressed the FHL2-HA-OMP25 construct in COS-7 cells. As seen in neurons, FHL2-HA-OMP25 strongly suppressed mitochondrial motility in COS-7 cells (Fig. 8, F and G). This reduction in motility was specific to mitochondria; it was not observed for Rab5-labeled endosomes (Fig. S7, C and D). Staining with phalloidin-488 revealed that the F-actin intensity associated with the mitochondria was increased by expression of FHL2-HA-OMP25 (Fig. 8, H and I; and Fig. S7 E). Thus, mitochondrially bound FHL2 suffices to recruit F-actin and thereby immobilize mitochondria.
Discussion
Mitochondrial dynamics can bolster mitochondrial efficiency. By regulating their movement, mitochondria position themselves at areas of high metabolic demand, such as neuronal synapses (Gutnick et al., 2019; Shlevkov et al., 2019) or the leading edge of migrating cells (Cunniff et al., 2016). In addition, as proteins from older mitochondria are turned over, newer mitochondria can arrive to replace them or fuse with them to maintain a steady distribution of healthy mitochondria throughout the cell (Misgeld and Schwarz, 2017; Schwarz, 2013). To achieve a proper distribution, cells have distinct stationary and motile pools of mitochondria. Under certain conditions, mitochondria become stationary by losing their molecular motors (Chung et al., 2016; Wang et al., 2011). Many mitochondria, however, are stationary despite retaining their motors (Wang and Schwarz, 2009). This may involve inactivation of their motors (Wang and Schwarz, 2009) or anchoring to immovable cytoskeletal elements. One such anchoring protein that has been studied extensively is syntaphilin (Chen and Sheng, 2013; Kang et al., 2008; Lin et al., 2017; Park et al., 2016; Seo et al., 2018). It is predominantly expressed in neuronal axons and anchors the mitochondria to microtubules in response to metabolic signaling.
We have now elucidated an additional anchoring mechanism, one that tethers the mitochondria to F-actin in many cell types. We have studied this process in the context of the mitochondrial arrest resulting from increases in extracellular glucose concentrations. An increase in extracellular glucose levels leads to O-GlcNAcylation of Milton (TRAK1; Pekkurnaz et al., 2014; Su and Schwarz, 2017), a protein on the outer mitochondrial membrane. We have found the resulting arrest to be mediated by actin via FHL2. We demonstrate the following. (1) Polymerized actin is responsible for immobilizing O-GlcNAcylated mitochondria, which can be remobilized upon actin depolymerization (Fig. 1). (2) F-actin accumulation around O-GlcNAcylated mitochondria is dependent on TRAK1. (3) FHL2 coimmunoprecipitates with TRAK1 and is detectably recruited to mitochondria under conditions that promote TRAK1 O-GlcNAcylation (Fig. 4). (4) Mutation of the O-GlcNAcylation sites on TRAK1 that are required for arrest also reduce its association with FHL2 (Fig. 4). (5) Knockdown of FHL2 prevents F-actin accumulation around O-GlcNAcylated mitochondria and prevents mitochondrial arrest by OGT expression or glucose elevation (Figs. 5, 6, and 7). (6) Artificially tethering FHL2 to mitochondria is sufficient to drive their association with actin and to stop their movement (Fig. 8).
A growing number of studies have addressed the interactions of mitochondria with actin (Chada and Hollenbeck, 2004; Kruppa et al., 2018; Moore et al., 2016; Pathak et al., 2010). Some of the reported actin–mitochondria interactions assist in fission (Manor et al., 2015) and short-range movements powered by Myo19 (López-Doménech et al., 2018; Shneyer et al., 2016). Others likely entail anchoring and contribute to the stationary pool of mitochondria. In neurons, F-actin anchoring is known to hold mitochondria at specific axonal and dendritic sites (Gutnick et al., 2019; Pathak et al., 2010). In non-neuronal cells, circulating pools of F-actin dynamically associate with mitochondria (Moore et al., 2016) and help position them at areas requiring high ATP, such as the leading edge (Cunniff et al., 2016). F-actin binding proteins, such as myosin V and myosin VI, have previously been implicated as the mitochondria-bound links to actin (Kruppa et al., 2018; López-Doménech et al., 2018; Pathak et al., 2010; Shneyer et al., 2016). However, with the exception of Myo19 (Bocanegra et al., 2020; López-Doménech et al., 2018; Oeding et al., 2018; Shneyer et al., 2016), it is largely unclear how myosins associate with mitochondria. In other situations, the proteins bridging the gap between actin and mitochondria are not known (Cunniff et al., 2016; Gutnick et al., 2019).
We took an unbiased approach to discover proteins that are enriched on highly O-GlcNAcylated mitochondria and might therefore mediate their interaction with actin. By immunoprecipitating TRAK1 and quantifying its binding partners with SILAC-MS, we discovered FHL2 to be the primary actin-associated protein enriched on O-GlcNAcylated TRAK1 (Fig. 4). This FHL2–TRAK1 association provides a mechanism by which mitochondria anchor to F-actin in response to increased nutrient availability. Although FHL2 is also known to have transcriptional activities (Boateng et al., 2016; Johannessen et al., 2006; Ng et al., 2014) that influence cell metabolism and actin dynamics (Olson and Nordheim, 2010), we find that the transcription-related functions of FHL2 are not essential for mitochondrial arrest. Our artificial construct, FHL2-OMP25, that forces FHL2 specifically on the mitochondrial outer membrane is sufficient to stop the mitochondria and recruit F-actin (Fig. 8). Additionally, the FHL2-OMP25 construct demonstrates that O-GlcNAcylation–induced changes of actin and cofilin, which may occur in response to an up-regulation of glucose flux (Hédou et al., 2009; Hedou et al., 2007; Huang et al., 2013) are not necessary for mitochondrial arrest. Apart from influencing F-actin structure and dynamics, OGT (and O-GlcNAcylation) influences several other proteins on mitochondria, thereby impacting mitochondrial structure, tethering to the ER, import, export, and turnover (Latorre-Muro et al., 2021; Murakami et al., 2021; Sacoman et al., 2017). The FHL2-OMP25 construct, however, demonstrates that these additional OGT actions are not required for mitochondrial anchoring to F-actin.
Our data using the Pex-Miro construct demonstrates that the FHL2- and TRAK1-mediated tethering to F-actin can occur even on surrogate organelles such as peroxisomes. Although studies have localized small amounts of Miro1 to peroxisomes (with overexpression; Castro et al., 2018; Covill-Cooke et al., 2020), the Miro1 that is normally present on peroxisomes appears to mediate not peroxisomal motility but instead peroxisomal fission (Covill-Cooke et al., 2020) and contacts with ER. Indeed, ER contacts can be greatly increased by directing more Miro to peroxisomes (Guillén-Samander et al., 2021). In agreement with these reports, we show that peroxisomes normally have low motility in COS-7 cells. Upon misdirection of excess Miro1 and consequently TRAK1 to peroxisomes, their motility increases and becomes subject to regulation by OGT and F-actin. Thus, O-GlcNAcylation–triggered anchoring to F-actin requires proteins associated with the mitochondrial motor protein–adaptor complex but is independent of other mitochondrial proteins.
The expression of OGT caused a substantial accumulation of F-actin around mitochondria in non-neuronal cells (Fig. 2). A gross change in the organization of F-actin, however, is not a priori necessary for mitochondrial arrest. In axons, which are inherently actin rich, no significant OGT-induced enrichment of F-actin on mitochondria was detectable (Fig. S8), although the OGT-induced arrest required F-actin (Fig. 1). Small accumulations of F-actin may have escaped detection, but it is likely that axonal mitochondria can be acutely anchored to preexisting F-actin filaments, without any large structural changes to the cytoskeleton. This feature of the FHL2 mechanism would allow mitochondrial motility to be highly responsive; transient changes in glucose availability could thus modulate mitochondrial movement without necessitating large cytoskeletal rearrangement.
Figure S8.
OGT-mediated arrest in neurons does not require large redistribution of axonal F-actin (related to the Discussion text). (A and B) Neurons were transfected with LifeAct-RFPt (actin marker), OGT or GFP (control), and Mito-BFP. Top: Representative images of LifeAct-RFPt (magenta) and mitochondria (cyan). The F-actin intensity was measured along each axon and normalized to the average intensity in that axon. Bottom: Normalized F-actin intensity (magenta) along the corresponding axons and the locations of mitochondria (cyan). (C) Quantification of F-actin colocalization with mitochondria. To calculate F-actin enrichment, the ratio of F-actin intensity on mitochondria to the mean F-actin intensity (along the axon) was considered. Although there is a trend of increased F-actin on mitochondria in axons expressing OGT, the increase is slight. The quantification is represented as a box-and-whisker plot. The line indicates the median, the box indicates the interquartile range, and whiskers indicate the 10th and 90th percentiles. Outliers are represented as individual dots and are included in all statistical calculations. P values are from a two-tailed unpaired t test with Welch’s correction.
We found small amounts of FHL2 to be associated with TRAK1 in cells where OGA is expressed and O-GlcNAcylation is low (Fig. 4). Some O-GlcNAcylation sites are resistant to the expression of OGA and have extremely slow turnover rates (Qin et al., 2017). These may partially explain the residual FHL2. The residual association of FHL2 on TRAK1 with OGA overexpression or on TRAK1QMut (a version of TRAK1 that is partially resistant to O-GlcNAcylation) may also be a reflection of FHL2 and TRAK1 interactions arising from other cellular events unrelated to glucose and OGT. For example, mitochondrial capture and enrichment at developing presynapses (Lewis et al., 2016) involves actin (Gutnick et al., 2019) and may involve FHL2. Mitochondrial motility responds to many other signals, including reactive oxygen species, ATP/ADP levels, and growth factors (Beck et al., 2012; Debattisti et al., 2017; Schuler et al., 2017; Su et al., 2014; Trigo et al., 2019), for which the mechanisms are not fully established. Any of these modes of regulation might result in the direct or indirect binding of FHL2 to TRAK1 independent of its O-GlcNAcylation. TRAK also undergoes multiple phosphorylations that alter its binding partners (Chung et al., 2016; Hornbeck et al., 2012). It will be interesting to learn whether these events influence O-GlcNAcylation and FHL2 binding.
This mechanism of anchoring mitochondria in response to glucose supply likely operates in vivo. Several observations support this model: (1) The F-actin–based anchoring mechanism is activated when glucose levels are varied to mimic normal blood and brain glucose variations (Silver and Erecińska, 1994; Tirosh et al., 2005; Figs. 1 and 7); (2) previous work has shown that the GlcNAcylation state of TRAK1 in mammalian brains can be altered by a fasting-refeeding protocol; and (3) Drosophila lines overexpressing OGT show decreased mitochondrial movement (Pekkurnaz et al., 2014). Such a mechanism that tunes mitochondrial distribution to glucose levels is vital to efficient ATP generation by oxidative phosphorylation of glucose. In vivo, the misregulation of this pathway would therefore be expected to disrupt mitochondrial efficacy and potentially impair the function of cells with high energy demands, such as neurons (Su and Schwarz, 2017) and muscle cells (Shi et al., 2018). The misregulation of mitochondrial motility may therefore be a factor in the neurological phenotypes reported in OGT mutant mice. These phenotypes include altered electrophysiology and function in the brain regions that govern appetite and body weight (Lagerlöf et al., 2017; Lagerlöf et al., 2016; Ruan et al., 2014) and poor development and survival of peripheral neurons (Su and Schwarz, 2017). Indeed, diabetic patients having abnormal O-GlcNA as a result of poor glucose regulation often suffer from peripheral and central neuropathies and myopathies (Banerjee et al., 2015; Ma et al., 2016; Ma and Hart, 2013; Semba et al., 2014; Vincent et al., 2011). Understanding the influence of GlcNAcylation (Sheng, 2014) on mitochondrial behavior may therefore reveal therapeutic avenues for conditions in which the GlcNAcome is misregulated.
The actin-anchoring mechanism described here adds to an expanding catalog of mechanisms that regulate mitochondrial motility through changes to the motor–adaptor complex. The FHL2–TRAK1 association is driven by a reversible protein modification and is thus well suited for responding to transient changes in glucose availability. In this regard, it is similar to the reversible change in kinesin–Miro interaction that underlies Ca2+-mediated arrest of mitochondria or the transient shedding of motor proteins that occurs during mitosis (Chung et al., 2016; Wang and Schwarz, 2009) or in response to inhibitory substrates for axon growth (Kalinski et al., 2019). The FHL2-dependent mechanism stands in contrast to the irreversible dissolution of the motor–adaptor complex that occurs when the PINK1/Parkin pathway causes the degradation of Miro on damaged mitochondria (Course et al., 2018; Newman and Shadel, 2018; Pickrell and Youle, 2015; Wang et al., 2011). Elucidating the different ways by which the motor–adaptor complex, and thereby mitochondrial motility, is fine-tuned is essential to explain the normal distribution of mitochondria and to understand the pathological states that arise from its misregulation (Devine and Kittler, 2018; Mattson et al., 2008; Misgeld and Schwarz, 2017; Pickrell and Youle, 2015; Schwarz, 2013; Vanhauwaert et al., 2019).
Materials and methods
Primers and shRNA sequences are presented in Table S2.
Plasmid constructs
Previously published constructs
The following previously published DNA constructs were used in this study: pEGFP-N1 (Clontech), Mito-DsRed, eBFP2-N1, LifeAct-RFPt, mCitrine-Peroxisomes-2 (a gift from Michael Davidson, Florida State University, Tallahassee, FL; Addgene plasmids 55838, 54595, 54586, and 54672, respectively), Mito-BFP (a gift from Gia Voeltz, University of Colorado Boulder, Boulder, CO; Addgene plasmid 49151; Friedman et al., 2011), pmTurquoise2-Perox (a gift from Dorus Gadella, Swammerdam Institute for Life Sciences, Amsterdam, Netherlands; Addgene plasmid 36203; Goedhart et al., 2012), FHL2-Flag, Flag-FHL2 (a gift from Beat Schäfer, University of Zurich, Zurich, Switzerland; Scholl et al., 2000), 3XFlag-FHL2 (a gift from Patrick Mehlen, Centre Léon Bérard, Lyon, France; Mille et al., 2009), Myc-TRAK1, Myc-TRAK1QMut, OGT, GFP-OGT, Myc-OGA (Pekkurnaz et al., 2014), Mito-GFP (Chung et al., 2016), and SNAP-OMP25 (a gift from David Sabatini, Whitehead Institute, Cambridge, MA; Addgene plasmid 69599; Katajisto et al., 2015).
Purchased constructs
The lentiviral GFP vectors containing shRNA against FHL2 and the corresponding control were purchased from OriGene Technologies, Inc. (TL711478). The untagged lentiviral vectors containing the shRNA construct against FHL2, TRAK1, and the corresponding control shRNA were purchased from the Sigma-Aldrich MISSION shRNA library.
Constructs cloned in this study
We used six constructs in this study: (1) GFP-2A-OGT: The GFP-2A-OGT construct was made by standard Gibson assembly (Gibson et al., 2009)–based cloning strategies from the OGT constructs described in Pekkurnaz et al. (2014). To build the GFP-2A-OGT construct, the GFP expression cassette was amplified from the GFP-OGT vector (Pekkurnaz et al., 2014) using the primers GFP_ForPrimer and GFP_RevPrimerP2AOverhang. The untagged OGT vector (Pekkurnaz et al., 2014) was linearized by PCR using the primers OGT_ForPrimerP2AOverhang and OGT_RevPrimerGFPOverhang and was recircularized with the GFP and 2A sequences fused to the N-terminus of OGT by Gibson assembly. The following P2A sequence was incorporated in the primers to situate it between the two coding regions (i.e., between GFP and OGT): 5′-GGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGCGACGTGGAGGAGAACCCTGGACCT-3′. (2) BFP-2A-OGT: The GFP-2A-OGT construct was used as a template to make BFP-2A-OGT. To do this, the GFP was excised from the GFP-2A-OGT vector using XmaI and HindIII digestion, and the BFP expression cassette was amplified from the Mito-BFP construct described above, using the primers BFP_ForPrimerOGTOverhang and BFP_RevPrimerP2AOverhang that contained appropriate overhangs to the backbone of the OGT vector. The BFP was then fused in place of the GFP using Gibson assembly. (3) GFP-2A-HA-OGA: To make the GFP-2A-HA-OGA construct, the Myc-OGA construct described in Pekkurnaz et al. (2014) was linearized by PCR with the primers OGA_ForPrimerHAOverhang and OGA_RevPrimerGFPOverhang. These primers were designed to omit the Myc tag in the backbone and instead code for an HA tag. The GFP-2A cassette was amplified from the GFP-2A-OGT vector using primers GFP_ForPrimer and P2A_RevPrimerHAOverhang that had appropriate overhangs to the OGA vector. The linearized OGA vector containing the HA tag was recircularized after incorporating the GFP-2A cassette at the N-terminus of HA-OGA by Gibson assembly. This construct was used to overexpress OGA in all IP experiments except for the IP of endogenous TRAK1 (Fig. 4, A and B) where Myc-OGA was overexpressed. The GlcNAcylation activities of the OGT and OGA vectors and the cleavage and liberation of GFP from 2A-OGT or 2A-OGA vectors were confirmed by Western blotting and immunocytochemistry (Fig. S1). (4) FHL2-HA-OMP25: To generate the FHL2-HA-OMP25 construct, the rat FHL2 ORF was initially cloned from neuronal cDNA into a pcDNA3.1 expression vector. The primers FHL2_Rev_pcDNA3.1Overhang and FHL2_For_pcDNA3.1Overhang were used to amplify the FHL2 coding sequence with an overhang to the pcDNA3.1 backbone. The vector backbone was linearized by digestion with Xho1 and BamHI and then recircularized by inserting the FHL2 coding sequence by Gibson assembly. The pcDNA 3.1 vector having rat FHL2 was then linearized by PCR using primers rFHL2-OMP25_ForPrimer1 and rFHL2-OMP25_revPrimer1 followed by recircularization by inserting the OMP25 mitochondrial targeting domain (aa 110–145, amplified from the SNAP-OMP25 with the primers rFHL2-OMP25_forPrimer2 and rFHL2-OMP25_revPrimer2) along with the HA tag at the C-terminus of FHL2 by Gibson assembly. (5) Flag-HA-OMP25: To make the Flag-HA-OMP25 construct, primers encoding the Flag tag were used to amplify the FHL2-OMP25 vector backbone (GSLinker-HA-OMP25_ForPrimer and Flag-GSLinker-OMP25_RevPrimer), excluding the FHL2 sequence. The ends of this amplicon containing the polylinker, HA tag, and OMP25 sequence were then ligated to each other using the KLD (kinase-ligase-Dpn1) kit (E0554S; New England Biolabs). All these constructs were validated by sequencing and expressing in HEK293T/17 cells followed by Western blotting with antibodies to FHL2, HA, and Flag. Our rat FHL2, cloned from three independent hippocampal cultures, had a silent single-base difference at the R59 amino acid residue (from CGC to CGG) as compared with the Rattus norvegicus mRNA annotation in the GenBank database (accession no. 63839). (6) Pex3-HIS-mRFP-Miro1: The pex3-Miro1 construct was made by amplifying the first 592 amino acid residues from isoform 3 of human Miro1 as cloned in Fransson et al. (2003) with the primers pex-miro1_ForPrimer and pex-miro1_RevPrimer. The pex3 peroxisomal targeting domain (aa 1–42) along with mRFP (i.e., Pex3-mRFP) was obtained by excising the FKBP from the pex3-mRFP-FKBP construct (a gift from Casper Hoogenraad, Utrecht University, Utrecht, Netherlands; Kapitein et al., 2010) using EcoR1 and AscI. This was followed by fusing the Miro1 PCR product into the Pex3-mRFP backbone by Gibson assembly.
Sequences and plasmid constructs will be made available for redistribution through Addgene (https://www.addgene.org/Thomas_Schwarz/) and directly upon request.
Antibodies used for Western blotting
The following primary antibodies were used at the stated dilutions: anti-human TRAK1 at 1:2,000 (HPA005853; Sigma-Aldrich), anti-Myc at 1:5,000 (05-724; EMD Millipore), anti-OGT at 1:2,000 (DM-17; Sigma-Aldrich), anti-HA at 1:5,000 (ab9110; Abcam), anti-FLAG 1:5,000 (F7425; Sigma-Aldrich), anti-FHL2 1:1,000 (HPA006028; Sigma-Aldrich), anti-GAPDH 1:5,000 (6C5; EMD Millipore), and anti–6X-HIS at 1:1,000 (MA1-21315; Thermo Fisher Scientific). For chemiluminescence detection of proteins on Western blots, HRP-conjugated secondary antibodies to mouse, rabbit, and rat (Jackson ImmunoResearch Laboratories, Inc.) were used at 1:5,000 along with Pierce SuperSignal West Dura (Thermo Fisher Scientific). For fluorescence detection (used for all quantitative blots), 800CW donkey anti-rabbit, 680RD donkey anti-rabbit, 680RD donkey anti-mouse, 800CW donkey anti-mouse, and 800CW goat anti-rat were used at 1:5,000 (LI-COR Biosciences), and all blots were scanned by the Odyssey CLx Imaging System.
Antibodies used for immunocytochemistry
The primary antibodies used for immunocytochemistry and their working dilutions are as follows: anti–microtubule-associated protein 2 (anti-MAP2) at 1:500 (NB300-213; Novus Biologicals), anti-HA at 1:500 (ab9110; Abcam), anti-ATP5a at 1:500 (ab14748; Abcam), and anti-FHL2 at 1:100 (HPA006028, Sigma-Aldrich; and ab202584, Abcam). For fluorescence detection of epitopes, Alexa Fluor 405–, Alexa Fluor 568–, and Alexa Fluor 647–conjugated secondary antibodies (Thermo Fisher Scientific) were used at a dilution of 1:500.
GlcNAcylation assays
From cell lysates by Western blotting
To assess the degree of O-GlcNAcylation upon OGT and OGA expression in HEK293T/17 cells and COS-7 cells, the constructs were expressed in these cell lines for 2 d (Fig. S1, A and B). The cells were lysed, and proteins were separated by SDS-PAGE and transferred onto a nitrocellulose membrane for Western blotting with an antibody against O-GlcNAcylation [RL2] (ab2739; Abcam). O-GlcNAcylation was quantified by the ratio of RL2 immunoreactivity in the entire lane to an anti-GAPDH band and then normalized to lysates from cells expressing GFP alone.
In fixed cells by immunocytochemistry
To determine whether the OGT constructs increase O-GlcNAcylation in rat hippocampal neurons, 3 d after transfection, neurons were fixed and stained with the antibody against O-GlcNAcylation (RL2) and anti-OGT (Fig. S1, C and D). Neurons positive for the OGT expression also had high levels of RL2 staining, indicating increased cellular GlcNAcylation levels.
Cell cultures and transfections
Cell lines
HEK293T/17 and COS-7 cells were cultured in DMEM supplemented with L-glutamine, penicillin-streptomycin (Life Technologies), and 10% FBS (Atlanta Premium). Plasmid DNA transfections in HEK293T/17 cells were performed with calcium phosphate (Kingston et al., 2003) and in other cell lines with TransIT-LT1 reagent (MIR 2300; Mirus Bio) using the manufacturer’s protocol. These cell lines were generally transfected 16–18 h after plating and assayed 2–3 d later.
Hippocampal neurons
Hippocampal neurons were dissected and dissociated from E18 rat (Charles River) embryos as previously described (Nie and Sahin, 2012) and plated at a density of 5–7 × 104 cells/cm2 on glass-bottomed dishes (D35-20-1.5-N; Cellvis) coated with 20 µg/ml poly-L-lysine (Sigma-Aldrich) and 4 µg/ml laminin (Life Technologies). The neurons were maintained in neurobasal medium supplemented with B27 (Life Technologies), L-glutamine, and penicillin-streptomycin, unless specified otherwise. The hippocampal neurons were transfected 5 or 6 d after plating (DIV 5 or DIV 6) using Lipofectamine 2000 (11668-019; Life Technologies) and imaged 2–3 d later. The coexpression of multiple constructs transfected into neurons was validated by retrospective immunostaining after live-cell imaging.
IP
For all IPs, HEK293T/17 cells were plated at 5.5 × 105 cells/well density in a six-well plate and transfected with the indicated plasmid constructs the next day. HEK293T/17 cells expressing OGT were also treated with 100 µm Thiamet-G (110165CBC; EMD Biosciences). 2 d after transfection, cells were washed once with ice-cold PBS and lysed in 600 µl buffer containing 1% NP-40 (Calbiochem), 15 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 40 mM GlcNAc, 1 µM Thiamet-G, 1 mM DTT (GBiosciences), 0.1 mg/ml PMSF, and protease inhibitor cocktail set III (539134-1SET; EMD Millipore) at 1:500 dilution. Lysates were centrifuged for 10 min at 13,000 ×g at 4°C, and the clarified supernatants were collected. For IPs of endogenous TRAK1, FHL2-Flag, Pex-His-Miro, or Myc-TRAK1, 2.0 µg anti-human TRAK1 antibody (HPA005853; Sigma-Aldrich), anti-Flag antibody (F7425; Sigma-Aldrich), anti-6X-HIS antibody, or anti-Myc antibody (NB600-302; Novus Biologicals) was incubated with 500 µl of the clarified supernatants for 2 h at 4°C with constant tumbling. Following the antibody incubation, the lysates were mixed with protein A Sepharose beads and incubated for another 1 h at 4°C with constant tumbling. The beads were then washed three times with lysis buffer and resuspended in 1× Laemmli buffer. 80–90% of this resuspension was then separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were then incubated overnight at 4°C with primary antibodies in the blocking buffer (3% BSA [wt/vol] in TBS with 0.1% Tween-20). The blot was then washed three times with TBS with 0.1% Tween-20 and blotted with secondary antibodies for 1 h at RT before ECL imaging or scanning with the Odyssey CLx Imaging System.
Milton interactome analysis by SILAC-based quantitative proteomics
We applied SILAC-MS to profile O-GlcNAcylation–dependent protein–protein interactions for TRAK1, as summarized in Fig. S4.
SILAC IP
Lyophilized preparation of heavy amino acids L-lysine-2HCl (13C6, 15N2, 88209; Thermo Fisher Scientific) and L-arginine-HCl (13C6, 15N4, 89990; Thermo Fisher Scientific) and light amino acids L-lysine-2HCl (89987; Thermo Fisher Scientific) and L-arginine-HCl (89989; Thermo Fisher Scientific) were dissolved in SILAC-DMEM (88364; Thermo Fisher Scientific) according to the protocol provided by the manufacturer to prepare the “heavy” and “light” SILAC media. HEK293T/17 cells were cultured in the heavy and light SILAC media (supplemented with L-glutamine, penicillin-streptomycin [Life Technologies], and 10% FBS [Atlanta Premium]). To achieve >95% heavy L-lysine and L-arginine incorporation efficiency, cells are passaged for at least five cell doublings using enzyme-free cell dissociation buffer (13151014; Gibco). IPs of endogenous TRAK1 from heavy or light labeled HEK293T/17 cells, expressing either OGT or OGA, were performed as described above with 2 μg anti-hTRAK1 antibody (HPA005853 Lot A61497; Sigma-Aldrich) for each well. The OGT-expressing cells were also treated overnight with 100 µm O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino-Z-N-phenylcarbamate (PUGNAc; 3384; Tocris Bioscience), an inhibitor of OGA, to further increase the levels of O-GlcNAcylation (Pekkurnaz et al., 2014). For mass spectrometry analysis, four wells from a six-well plate for each condition were combined. Upon validation of similar IP efficiencies by Western blot analysis, the heavy and light TRAK1 samples were combined at a ratio of 1:1. 80–90% of the combined immunoprecipitates were separated by SDS-PAGE and stained with SimplyBlue SafeStain (LC6060; Thermo Fisher Scientific). Protein bands were then excised, excluding IgGs, and in gel digested using trypsin according to Shevchenko et al. (2006).
Mass spectrometry analysis
The resultant peptides were analyzed by online C18 nanoflow reversed-phase HPLC (NanoLC 2D; Eksigent) linked to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific). Samples were loaded onto an in-house packed 100-µm inner diameter × 15-cm C18 column (Magic C18, 5 µm, 200 Å; Michrom Bioresource) and separated at 200 nl/min with 60-min linear gradients from 5% to 35% acetonitrile in 0.4% formic acid. Survey spectra were acquired in the Orbitrap with the resolution set to a value of 30,000. Up to six of the most intense ions per cycle were fragmented and analyzed in the linear trap. Raw files were processed using version 1.1.1.21 of Cox et al. (2009). Cysteine carbamidomethylation was used as a fixed modification, and oxidation (M), deamidation (N), and N-acetyl (protein N-term) were set as variable modifications. Two missed tryptic cleavages were allowed, and the minimal length required for a peptide was six amino acids. The initial precursor mass tolerance was set to 10–20 parts per million, and the fragment mass tolerance was set to 0.5 D. The peptide false discovery rate was set to 0.05, the protein false discovery rate was 0.01, and all other parameters were default settings. The datasets were searched against the International Protein Index human database.
Image acquisition parameters for motility analysis in live cells
Neurons
For live-cell imaging of axonal mitochondria, hippocampal neuronal cultures were grown on glass-bottomed dishes (D35-20-1.5-N; Cellvis) at a density of 75,000 cells/cm2 and imaged on a Nikon Ti-Eclipse microscope equipped with an environmental chamber that was supplied with humidified 5% CO2 and maintained at 37°C. The neurons were illuminated by a Lumencore Sola Light Engine light-emitting diode and imaged with an Andor Zyla scientific complementary metal–oxide–semiconductor camera with a 63×/NA 1.4 plan apochromat (Plan Apo) objective. The light-emitting diode intensity was always kept at minimum (<5%). For each image, a 250–300-µm axon segment was selected that was ∼300 µm away from the growth cone and ∼1 mm away from the cell body. Images were captured every 0.5 s. Kymographs were generated from 3- to 5-min time-lapse movies and analyzed with Kymolyzer (Basu et al., 2020), a custom-written ImageJ macro for the percentage of time spent in motion, velocity, total distance traveled, mitochondrial density, and length. For final quantifications, the average measurement of all mitochondria in each axon was considered as a single data point. The percentage motility quantifications represent the average percentage of time spent in motion by all mitochondria in each axon segment.
Cell lines
The imaging and environmental parameters for live-cell imaging of mitochondria, peroxisomes, and endosomes in COS-7, HEK293T/17, and U2OS cells were the same as those for the neuronal cultures.
Quantifications of organelle motility in cell lines
The time-lapse images of organellar movement in cell lines were analyzed with software we developed for this purpose, QuoVadoPro (Basu and Schwarz, 2020). Briefly, the images are initially segmented into binary masks. The masking allows the algorithm to examine pixel occupancy instead of raw fluorescence levels. The motility can be inferred from the variation in pixel occupancy over time by the following formula:
where variancetime represents the occupancy variance of a single pixel over time, sumtime represents the total time each pixel is occupied over the entire time lapse, and nPixelssegmentedfirstframe represents the number of pixels that were segmented in the first frame (indicating the total amount of area occupied by the organelles). Pixels having sumtime = 0 were not considered in the calculations.
Cells with fluorescently tagged intracellular objects with high motility show a high pixel variance over time. This is because multiple pixels within the cell area have a high probability of being transiently illuminated by a moving object. Conversely, a cell having fluorescently tagged objects that are nonmotile or less motile have pixels that that are constantly occupied or constantly empty. The pixels in such cases will have low occupancy variance. Based on this principle, the pixel occupancy variance over time provides a proxy readout for the motility of objects within the cells. It is important to note that QuoVadoPro also normalizes the variance in pixel occupancy to the total occupancy during a time lapse. This normalization technique allows QuoVadoPro to distinguish between processive movements and back-and-forth movements. During processive movements, a series of pixels get transiently illuminated and thereby show high occupancy variance but low total occupancy. However, when an object is moving back and forth, it occupies the same set of pixels repeatedly by turns. These pixels will therefore have a high total occupancy. Thus, normalizing for the total occupancy allows QuoVadoPro to give more weight to processive movements than to back-and-forth jitters (Basu and Schwarz, 2020).
The approach taken by QuoVadoPro can be applied to multiple intracellular organelles whose movements cannot be measured by conventional approaches such as kymography or particle tracking. QuoVadoPro is available to download as an installable ImageJ macro package with detailed use instructions. It is important to note that although it is a useful tool for analyzing mitochondrial movement in COS-7 and U2OS cells, QuoVadoPro provides not absolute measures of processive movement but a proxy readout.
Quantifications of organelle distribution in cell lines
For quantification of the peroxisomal distribution, we developed another custom software, DoveSonoPro. In this software package, the cell outline and cell center are selected by the user. Each point on the cell outline is then projected to the cell center by a curved line that never leaves the confines of the cell outline. Each of these connections is then broken down into 10 segments of equal length. The segments are then connected to define concentric zones parallel to the cell membrane (i.e., that follow the cell shape). The peroxisomal frequency within each ring is then calculated (see Fig. S3).
Kymolyzer, QuoVadoPro, and DoveSonoPro are available to download from GitHub (https://github.com/ThomasSchwarzLab).
TRAK1 shRNA validation
Five TRAK1 shRNA constructs (SHCLND-NM_014965; Sigma-Aldrich Mission shRNA library) were tested in both HEK293T/17 cells and COS-7 cells. These constructs were transfected into HEK293T/17 cells using the calcium phosphate method or into COS-7 cells using TransIT-LT1 reagent. After 3 d of expression, the two constructs, TRCN0000036275 and TRCN0000036277, were found effective in knocking down TRAK1 in both cell lines. These constructs were then chosen for further experiments.
FHL2 shRNA validation
Five FHL2 shRNA constructs were obtained from the Sigma-Aldrich Mission shRNA library (SHCLNG-NM_001450) and were initially tested in HEK293T/17 cells for efficient knockdown. They were transfected in HEK293T/17 cells using the calcium phosphate method. 3 d later, the cells were lysed and analyzed by SDS-PAGE. The shRNA construct, TRCN0000005773, was found to be the most effective of the constructs from Sigma-Aldrich; it reduced endogenous FHL2 by >50%. This shRNA was capable of knocking down FHL2 in COS-7 cells and in rat hippocampal neurons and was used in Figs. 5, S5, 6, and 7.
Four other shRNA constructs against FHL2 were tested from OriGene Technologies, Inc. (TL711478). These constructs specifically targeted rat FHL2. To initially select the most efficacious sequences, these shRNAs were transfected in HEK293T/17 cells along with the rat FHL2 expression vector (as described above). These cells were lysed, and cell lysates were analyzed by SDS-PAGE followed by Western blotting. Two sequences, TL711478B (referred to as shRNA B) and TL711478D (referred to as shRNA D), were capable of significantly knocking down rat FHL2 (Fig. S6 A) and were chosen for further experiments in hippocampal neurons.
TRCN0000005773, TL711478B, and TL711478D were also tested for efficacy of knockdown of FHL2 in hippocampal neurons. This was done by transducing the neurons with viral particles containing the shRNA vectors. All the shRNA constructs were in lentiviral vector backbones. To generate lentiviral particles, we cotransfected the shRNA plasmids with viral packaging plasmids (ENV, GAG-POL, RSV-REV, and TAT) into HEK293T/17 cells in 15-cm dishes. 2 d later, 100 ml of culture supernatant containing the viral particles was collected and filtered through a 0.45-μm filter. The viral particles were then pelleted by centrifugation at 82,000 ×g for 90 min at 4°C and resuspended in 50 μl of 0.001% Pluronic F-68 in PBS solution. Aliquots were snap frozen in liquid nitrogen and stored at −80°C. To test the shRNA efficacy, we infected neuronal cultures on DIV 1. On DIV 6, these cultures were washed once with ice-cold PBS and lysed and analyzed by SDS-PAGE. All three shRNAs tested showed efficient knockdown of FHL2 in neurons compared with their respective control sequences (Fig. 6 and Fig. S6).
Low-glucose neuronal cultures
To establish low-glucose neuronal cultures, hippocampal neurons were dissected in media containing the typical 25 mM glucose. Following dissection, the neurons were washed once with media containing 5 mM glucose and plated. At the time of plating, the final glucose amount in the media was, in some cases, as high as 6.3 mM due to incomplete washing. These neuronal cultures were then maintained by feeding at the time points shown in Figs. 1 and 7 with media containing either 1 mM or 5 mM glucose. During feeding, 75% of the old media was replaced with fresh media. The feedings ensured that the neurons were maintained at the intended 1 mM or 5 mM glucose level.
To make neuronal media with differing glucose concentrations, glucose-free neurobasal A medium (A2477501; Thermo Fisher Scientific) supplemented with penicillin-streptomycin, 1.22 M pyruvate (P8574; Sigma-Aldrich), 1 M lactate (71718; Sigma-Aldrich), and 1 mM or 5 mM D-glucose was used.
For the glucose shift, the initial glucose levels in the low-glucose neuronal cultures were measured. Fresh D-glucose was then added to these cultures to a final concentration of 5 mM or 1 mM (control). The control cultures also received equivalent molar concentrations of D-sorbitol to prevent differences in osmolarity.
FHL2 immunocytochemistry
The protocol for immunostaining neurons or COS-7 cells was adapted from that described in Whelan and Bell (2015) and Xu et al. (2013). Cells cultured in 20-mm glass-bottomed dishes were fixed with 4% PFA in PEM buffer (80 mM Pipes at pH 6.8, 5 mM EGTA, and 2 mM MgCl2 in water) prewarmed to 37°C. The fixed neurons or COS-7 cells were then permeabilized and blocked by treatment with 0.5% (vol/vol) Triton X-100 in PBS for 30 min at RT followed by 5% (wt/vol) BSA in PBS for 1 h at RT. To detect neuronal cell bodies, mitochondria, and FHL2, primary antibodies to MAP2, ATP5a, and FHL2 (described above) diluted in antibody dilution buffer (3% [wt/vol] BSA, 0.2% Triton X-100 in PBS) were applied to the blocked samples overnight at 4°C. The samples were then washed three times in PBS and incubated with Alexa Fluor 405–, Alexa Fluor 568–, and Alexa Fluor 647–conjugated secondary antibodies diluted in antibody dilution buffer for 2 h at RT. The cells were then further washed three times in PBS and mounted with Fluoromount-G (SouthernBiotech).
Actin staining for fixed cells
The protocol for visualizing mitochondrially associated actin in fixed COS-7 cells was adapted from Xu et al. (2013). The COS-7 cells cultured in 20-mm glass-bottomed dishes (D35-20-1.5-N; Cellvis) were fixed for 90–120 s with 0.4% (vol/vol) glutaraldehyde and 0.25% Triton X-100 (wt/vol) in cytoskeleton buffer (150 mM NaCl, 10 mM MES, pH 6.1, 15 mM MgCl2, 5 mM EGTA, and 5 mM sucrose in water) and then postfixed for 15 min with 3% (vol/vol) glutaraldehyde in cytoskeleton buffer. Fixed samples were treated with freshly prepared 0.5% (wt/vol) sodium borohydride in PBS to minimize background fluorescence. Samples were then blocked and permeabilized in blocking buffer (3% [wt/vol] BSA and 0.2% [vol/vol] Triton X-100 in PBS) for 1 h at RT. To detect mitochondria (ATP5a) and HA epitopes, primary antibodies (described above) diluted in blocking buffer were applied for 48 h at 4°C. The cells were then washed three times with blocking buffer (5 min/wash) and incubated with Alexa Fluor 568– and Alexa Fluor 647–conjugated secondary antibodies overnight at 4°C. Actin filaments were simultaneously stained overnight at 4°C with Alexa Fluor 488–conjugated phalloidin (sc-363791; Santa Cruz Biotechnology). The samples were then washed three times with blocking buffer (5 min/wash) and mounted with Fluoromount-G.
Image acquisition parameters for F-actin quantification in live cells
Live COS-7 cells expressing LifeAct-RFPt or F-Tractin-RFP were imaged on a Leica SP8 laser-scanning confocal microscope. The cells also expressed Mito-BFP to visualize the mitochondria and cytosolic GFP. This combination of fluorophores was chosen to minimize bleed-through from the mitochondrial channel into the F-actin channel and vice versa. For all experiments, bleed-through controls, such as cells expressing Mito-BFP alone, cells expressing LifeAct-RFPt, or cells expressing F-Tractin-RFP alone and cells expressing GFP alone were used to set up imaging parameters.
To visualize F-actin, mitochondria, and the cytosol, the cells were illuminated by a 405-nm diode laser, an argon laser tuned to 488 nm, and a white light laser tuned to 560 nm. Fluorescence emissions were collected using hybrid and photomultiplier detectors. A Plan Apo 63×/1.40 NA oil objective was used. During image acquisition, the live cells were kept within a humidified chamber maintained at 37°C and 5% CO2 (from Okolab).
Image acquisition parameters for imaging F-actin staining and FHL2 immunocytochemistry in fixed cells
Fixed COS-7 cells stained for mitochondria (ATP5a) and F-actin (phalloidin) or FHL2 were also imaged on a Leica SP8 laser-scanning confocal microscope. Similar to the imaging of live cells, the fixed cells were also imaged using a Plan Apo 63×/1.40 NA oil objective. The cells were illuminated by a 405-nm diode laser, an argon laser tuned to 488 nm, and a white light laser tuned to 560 nm and 647 nm. Fluorescence was detected using hybrid and photomultiplier detectors.
Actin quantification
Whole COS-7 cells were reconstructed from serial Z-slices. All actin quantifications were performed with a custom-written MATLAB script. In this analytical method, cell masks were drawn in the actin channel for each Z-slice by tracing the perimeter of the cell cytoplasm and excluding the peripheral cortical actin. The mitochondria were then masked by applying a global threshold on the mitochondrially localized marker, Mito-BFP or the HA signal. The mitochondrial mask was then expanded by 2 or 3 pixels to account for the actin enrichment around the mitochondria while the area of the mitochondrial matrix was excluded from the mask. Slices containing no discernible mitochondrial signal were discarded from the analysis. Cytoplasmic masks were determined by subtracting the mitochondrial matrix from the cell mask of each slice. The actin enrichment on mitochondria in each cell was then determined as:
where Actinmitochondria represents the density of F-actin in the dilated mitochondrial mask across all slices, Actincytosol represents the density of F-actin in the cytoplasmic mask across all slices, and Actintotal represents the density of F-actin in the cell mask (i.e., summation of the mitochondrial and cytoplasmic mask).
Statistical analysis
Quantification of organelle motility
Data regarding quantifications of organelle motility in neurons are expressed as median ± interquartile range. The error bars represent the 10th and 90th percentiles of the data. Outliers are shown as individual dots and are included in all calculations. Each data point represents a separate neuron. The percentage of time spent in motion by all mitochondria in a particular axon segment is averaged to generate one such data point. The distribution of these data points was assumed to be normal (as demonstrated to be the case in WT neurons; Basu et al., 2020). Thus, a two-tailed unpaired t test with Welch’s correction was used to determine the significance of differences between populations.
Western blot quantification
Data regarding Western blot quantifications are represented as mean ± SEM. All data points (each arising from a separate biological repeat) are shown. For finding significant differences, pairwise t tests were used.
Online supplemental material
Fig. S1 (related to Fig. 1) shows the efficacy of OGT and OGA constructs in increasing or decreasing O-GlcNAcylation. Fig. S2 (related to Fig. 2) illustrates the steps used for quantifying mitochondrial motility in non-neuronal cells, demonstrates the specificity of OGT in suppressing mitochondrial motility in COS-7 cells and U2OS cells, illustrates phalloidin staining in COS-7 cells in the presence of OGT or a control plasmid, and shows the testing of various shRNA constructs used to knock down TRAK1. Fig. S3 (related to Fig. 3) illustrates the steps to quantify peroxisomal distribution in COS-7 cells. Fig. S4 (related to Fig. 4) depicts a schematic of the workflow used to analyze the changes in the TRAK1 interactome as a result of O-GlcNAcylation, demonstrates the association of O-GlcNAcylated TRAK1 to different Flag-tagged versions of FHL2, and demonstrates that O-GlcNAcylation of TRAK1 is independent of F-actin depolymerization. Fig. S5 (related to Fig. 5) shows the validation of FHL2 shRNA and the validation of the FHL2 antibody for staining endogenous FHL2 in COS-7 cells. Fig. S6 (related to Fig. 6) shows that the knockdown of FHL2 prevents OGT-mediated mitochondrial arrest using a set of shRNA sequences and vector backbones different from that used in Fig. 6. Fig. S7 (related to Fig. 8) demonstrates that cytosolic FHL2 does not inhibit mitochondrial motility, unlike FHL2-HA-OMP25 (mitochondrial FHL2); that expression of FHL2-HA-OMP25 in COS-7 cells does not inhibit endosomal motility; and the localization of FHL2-HA-OMP25 to mitochondria. Fig. S8 (related to the Discussion section of text) demonstrates that OGT-mediated mitochondrial arrest in neurons does not involve a detectable rearrangement of F-actin around the mitochondria. Table S1 (related to Fig. 4 A) lists high-confidence interactors of TRAK1 as detected by SILAC-MS. Table S2 (related to the Materials and methods section of text) lists primer sequences and shRNA sequences used in this study. Video 1 (related to Fig. 1) shows that F-actin is necessary for glucose-induced arrest of neuronal mitochondrial motility. Video 2 (related to Fig. S2) shows that OGT suppresses mitochondrial motility in COS-7 cells. Video 3 (related to Fig. S2) shows the comparison of mitochondrial motility in COS-7 cells expressing OGT and OGA. Video 4 (related to Fig. 2) demonstrates that F-actin disruption reverses OGT-mediated mitochondrial arrest in COS-7 cells. Video 5 (related to Fig. S2) shows that O-GlcNAcylation does not influence endosomal motility. Video 6 (related to Fig. 3) demonstrates that mislocalizing the mitochondrial motor–adaptor complex to peroxisomes increases peroxisomal motility and makes it subject to regulation by OGT. Video 7 (related to Fig. 7) demonstrates the necessity of FHL2 for glucose-induced mitochondrial arrest.
Supplementary Material
lists high-confidence interactors of TRAK1 as detected by SILAC-MS (related to Fig. 4 A).
lists primer sequences and shRNA sequences used in this study (related to Materials and methods text).
Acknowledgments
We are grateful to Beat Schäfer for sharing the plasmids encoding Flag-tagged FHL2 and to Patrick Mehlen for sharing the 3X-Flag-FHL2 construct. We thank L. Mkhitaryan and Pedro Augusto for technical support, Kayla Davis (Boston Children's Hospital, Boston, MA) for the full-length RFP-Miro construct, and the members of the T.L. Schwarz laboratory for fruitful discussions and critical reading of the manuscript. We also thank S. Vasquez for help with hippocampal dissections from embryonic rats.
We are grateful to the Harvard NeuroDiscovery Center’s Enhanced Neuroimaging Core (National Institute of Neurological Disorders and Stroke P30 Core Center grant NS072030) and the Cellular Imaging Core Intellectual and Developmental Disabilities Research Center at Boston Children’s Hospital (National Institutes of Health grant U54 HD090255) for support with imaging. This research was generously supported by National Institutes of Health grant R01 GM11200 (to J. Steen) and National Institute of General Medical Sciences grant R01 GM069808 (to T.L. Schwarz).
The authors declare no competing financial interests.
Author contributions: G. Pekkurnaz, T.L. Schwarz, and H. Basu conceived the project. G. Pekkurnaz and J. Steen conducted the SILAC experiment, M. Chin and J. Falk conducted and analyzed the actin recruitment resulting from FHL2-OMP25 expression, W. Wei constructed the GFP-2A-OGT vector and helped optimize the FHL2 shRNA expression and antibody staining, and H. Basu performed all other experiments. T.L. Schwarz and H. Basu wrote the manuscript with feedback from all authors.
References
- Banerjee, P.S., Ma J., and Hart G.W.. 2015. Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. USA. 112:6050–6055. 10.1073/pnas.1424017112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, H., and Schwarz T.L.. 2020. QuoVadoPro, an autonomous tool for measuring intracellular dynamics using temporal variance. Curr. Protoc. Cell Biol. 87:e108. 10.1002/cpcb.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, H., Ding L., Pekkurnaz G., Cronin M., and Schwarz T.L.. 2020. Kymolyzer, a semi-autonomous kymography tool to analyze intracellular motility. Curr. Protoc. Cell Biol. 87:e107. 10.1002/cpcb.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, H., Flynn K., Lindenberg K.S., Schwarz H., Bradke F., Di Giovanni S., and Knöll B.. 2012. Serum response factor (SRF)-cofilin-actin signaling axis modulates mitochondrial dynamics. Proc. Natl. Acad. Sci. USA. 109:E2523–E2532. 10.1073/pnas.1208141109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng, L.R., Bennin D., De Oliveira S., and Huttenlocher A.. 2016. Mammalian actin-binding protein-1/Hip-55 interacts with FHL2 and negatively regulates cell invasion. J. Biol. Chem. 291:13987–13998. 10.1074/jbc.M116.725739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocanegra, J.L., Fujita B.M., Melton N.R., Cowan J.M., Schinski E.L., Tamir T.Y., Major M.B., and Quintero O.A.. 2020. The MyMOMA domain of MYO19 encodes for distinct Miro-dependent and Miro-independent mechanisms of interaction with mitochondrial membranes. Cytoskeleton (Hoboken). 77:149–166. 10.1002/cm.21560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond, M.R., and Hanover J.A.. 2015. A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208:869–880. 10.1083/jcb.201501101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickley, K., Smith M.J., Beck M., and Stephenson F.A.. 2005. GRIF-1 and OIP106, members of a novel gene family of coiled-coil domain proteins: association in vivo and in vitro with kinesin. J. Biol. Chem. 280:14723–14732. 10.1074/jbc.M409095200 [DOI] [PubMed] [Google Scholar]
- Castro, I.G., Richards D.M., Metz J., Costello J.L., Passmore J.B., Schrader T.A., Gouveia A., Ribeiro D., and Schrader M.. 2018. A role for Mitochondrial Rho GTPase 1 (MIRO1) in motility and membrane dynamics of peroxisomes. Traffic. 19:229–242. 10.1111/tra.12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chada, S.R., and Hollenbeck P.J.. 2004. Nerve growth factor signaling regulates motility and docking of axonal mitochondria. Curr. Biol. 14:1272–1276. 10.1016/j.cub.2004.07.027 [DOI] [PubMed] [Google Scholar]
- Chaveroux, C., Sarcinelli C., Barbet V., Belfeki S., Barthelaix A., Ferraro-Peyret C., Lebecque S., Renno T., Bruhat A., Fafournoux P., et al. 2016. Nutrient shortage triggers the hexosamine biosynthetic pathway via the GCN2-ATF4 signalling pathway. Sci. Rep. 6:27278. 10.1038/srep27278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., and Sheng Z.H.. 2013. Kinesin-1-syntaphilin coupling mediates activity-dependent regulation of axonal mitochondrial transport. J. Cell Biol. 202:351–364. 10.1083/jcb.201302040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J.Y., Steen J.A., and Schwarz T.L.. 2016. Phosphorylation-induced motor shedding is required at mitosis for proper distribution and passive inheritance of mitochondria. Cell Rep. 16:2142–2155. 10.1016/j.celrep.2016.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghill, I.D., Brown S., Cottle D.L., McGrath M.J., Robinson P.A., Nandurkar H.H., Dyson J.M., and Mitchell C.A.. 2003. FHL3 is an actin-binding protein that regulates α-actinin-mediated actin bundling: FHL3 localizes to actin stress fibers and enhances cell spreading and stress fiber disassembly. J. Biol. Chem. 278:24139–24152. 10.1074/jbc.M213259200 [DOI] [PubMed] [Google Scholar]
- Course, M.M., Scott A.I., Schoor C., Hsieh C.H., Papakyrikos A.M., Winter D., Cowan T.M., and Wang X.. 2018. Phosphorylation of MCAD selectively rescues PINK1 deficiencies in behavior and metabolism. Mol. Biol. Cell. 29:1219–1227. 10.1091/mbc.E18-03-0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covill-Cooke, C., Toncheva V.S., Drew J., Birsa N., López-Doménech G., and Kittler J.T.. 2020. Peroxisomal fission is modulated by the mitochondrial Rho-GTPases, Miro1 and Miro2. EMBO Rep. 21:e49865. 10.15252/embr.201949865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J.V., and Mann M.. 2009. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4:698–705. 10.1038/nprot.2009.36 [DOI] [PubMed] [Google Scholar]
- Cunniff, B., McKenzie A.J., Heintz N.H., and Howe A.K.. 2016. AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Mol. Biol. Cell. 27:2662–2674. 10.1091/mbc.e16-05-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debattisti, V., Gerencser A.A., Saotome M., Das S., and Hajnóczky G.. 2017. ROS control mitochondrial motility through p38 and the motor adaptor Miro/Trak. Cell Rep. 21:1667–1680. 10.1016/j.celrep.2017.10.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine, M.J., and Kittler J.T.. 2018. Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci. 19:63–80. 10.1038/nrn.2017.170 [DOI] [PubMed] [Google Scholar]
- Economo, M.N., Clack N.G., Lavis L.D., Gerfen C.R., Svoboda K., Myers E.W., and Chandrashekar J.. 2016. A platform for brain-wide imaging and reconstruction of individual neurons. eLife. 5:e10566. 10.7554/eLife.10566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson, A., Ruusala A., and Aspenström P.. 2003. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 278:6495–6502. 10.1074/jbc.M208609200 [DOI] [PubMed] [Google Scholar]
- Friedman, J.R., and Nunnari J.. 2014. Mitochondrial form and function. Nature. 505:335–343. 10.1038/nature12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Glater, E.E., Megeath L.J., Stowers R.S., and Schwarz T.L.. 2006. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173:545–557. 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart, J., von Stetten D., Noirclerc-Savoye M., Lelimousin M., Joosen L., Hink M.A., van Weeren L., Gadella T.W. Jr., and Royant A.. 2012. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 3:751. 10.1038/ncomms1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Górska-Andrzejak, J., Stowers R.S., Borycz J., Kostyleva R., Schwarz T.L., and Meinertzhagen I.A.. 2003. Mitochondria are redistributed in Drosophila photoreceptors lacking Milton, a kinesin-associated protein. J. Comp. Neurol. 463:372–388. 10.1002/cne.10750 [DOI] [PubMed] [Google Scholar]
- Guillén-Samander, A., Leonzino M., Hanna M.G., Tang N., Shen H., and De Camilli P.. 2021. VPS13D bridges the ER to mitochondria and peroxisomes via Miro. J. Cell Biol. 220:e202010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutnick, A., Banghart M.R., West E.R., and Schwarz T.L.. 2019. The light-sensitive dimerizer zapalog reveals distinct modes of immobilization for axonal mitochondria. Nat. Cell Biol. 21:768–777. 10.1038/s41556-019-0317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedou, J., Cieniewski-Bernard C., Leroy Y., Michalski J.C., Mounier Y., and Bastide B.. 2007. O-linked N-acetylglucosaminylation is involved in the Ca2+ activation properties of rat skeletal muscle. J. Biol. Chem. 282:10360–10369. 10.1074/jbc.M606787200 [DOI] [PubMed] [Google Scholar]
- Hédou, J., Bastide B., Page A., Michalski J.C., and Morelle W.. 2009. Mapping of O-linked β-N-acetylglucosamine modification sites in key contractile proteins of rat skeletal muscle. Proteomics. 9:2139–2148. 10.1002/pmic.200800617 [DOI] [PubMed] [Google Scholar]
- Hornbeck, P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M.. 2012. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40:D261–D270. 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, B.H., Takanaga H., Grossmann G., Chen L.Q., Qu X.Q., Jones A.M., Lalonde S., Schweissgut O., Wiechert W., and Frommer W.B.. 2011. Optical sensors for monitoring dynamic changes of intracellular metabolite levels in mammalian cells. Nat. Protoc. 6:1818–1833. 10.1038/nprot.2011.392 [DOI] [PubMed] [Google Scholar]
- Huang, X., Pan Q., Sun D., Chen W., Shen A., Huang M., Ding J., and Geng M.. 2013. O-GlcNAcylation of cofilin promotes breast cancer cell invasion. J. Biol. Chem. 288:36418–36425. 10.1074/jbc.M113.495713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd, D.D., and Saxton W.M.. 1996. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 144:1075–1085. 10.1093/genetics/144.3.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen, M., Møller S., Hansen T., Moens U., and Van Ghelue M.. 2006. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell. Mol. Life Sci. 63:268–284. 10.1007/s00018-005-5438-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski, A.L., Kar A.N., Craver J., Tosolini A.P., Sleigh J.N., Lee S.J., Hawthorne A., Brito-Vargas P., Miller-Randolph S., Passino R., et al. 2019. Deacetylation of Miro1 by HDAC6 blocks mitochondrial transport and mediates axon growth inhibition. J. Cell Biol. 218:1871–1890. 10.1083/jcb.201702187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.S., Tian J.H., Pan P.Y., Zald P., Li C., Deng C., and Sheng Z.H.. 2008. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 132:137–148. 10.1016/j.cell.2007.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein, L.C., Schlager M.A., van der Zwan W.A., Wulf P.S., Keijzer N., and Hoogenraad C.C.. 2010. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys. J. 99:2143–2152. 10.1016/j.bpj.2010.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajisto, P., Döhla J., Chaffer C.L., Pentinmikko N., Marjanovic N., Iqbal S., Zoncu R., Chen W., Weinberg R.A., and Sabatini D.M.. 2015. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 348:340–343. 10.1126/science.1260384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston, R.E., Chen C.A., and Rose J.K.. 2003. Calcium phosphate transfection. Curr. Protoc. Mol. Biol. Chapter 9:Unit 9.1. [DOI] [PubMed] [Google Scholar]
- Kruppa, A.J., Kishi-Itakura C., Masters T.A., Rorbach J.E., Grice G.L., Kendrick-Jones J., Nathan J.A., Minczuk M., and Buss F.. 2018. Myosin VI-dependent actin cages encapsulate Parkin-positive damaged mitochondria. Dev. Cell. 44:484–499.e6. 10.1016/j.devcel.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerlöf, O., Slocomb J.E., Hong I., Aponte Y., Blackshaw S., Hart G.W., and Huganir R.L.. 2016. The nutrient sensor OGT in PVN neurons regulates feeding. Science. 351:1293–1296. 10.1126/science.aad5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerlöf, O., Hart G.W., and Huganir R.L.. 2017. O-GlcNAc transferase regulates excitatory synapse maturity. Proc. Natl. Acad. Sci. USA. 114:1684–1689. 10.1073/pnas.1621367114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Muro, P., O’Malley K.E., Bennett C.F., Perry E.A., Balsa E., Tavares C.D.J., Jedrychowski M., Gygi S.P., and Puigserver P.. 2021. A cold-stress-inducible PERK/OGT axis controls TOM70-assisted mitochondrial protein import and cristae formation. Cell Metab. 33:598–614.e7. 10.1016/j.cmet.2021.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, T.L. Jr., Turi G.F., Kwon S.K., Losonczy A., and Polleux F.. 2016. Progressive decrease of mitochondrial motility during maturation of cortical axons in vitro and in vivo. Curr. Biol. 26:2602–2608. 10.1016/j.cub.2016.07.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.Y., Ng E.K., Lee S.M., Kotaka M., Tsui S.K., Lee C.Y., Fung K.P., and Waye M.M.. 2001. Protein-protein interaction of FHL3 with FHL2 and visualization of their interaction by green fluorescent proteins (GFP) two-fusion fluorescence resonance energy transfer (FRET). J. Cell. Biochem. 80:293–303. [DOI] [PubMed] [Google Scholar]
- Lin, M.Y., Cheng X.T., Tammineni P., Xie Y., Zhou B., Cai Q., and Sheng Z.H.. 2017. Releasing syntaphilin removes stressed mitochondria from axons independent of mitophagy under pathophysiological conditions. Neuron. 94:595–610.e6. 10.1016/j.neuron.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Doménech, G., Covill-Cooke C., Ivankovic D., Halff E.F., Sheehan D.F., Norkett R., Birsa N., and Kittler J.T.. 2018. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 37:321–336. 10.15252/embj.201696380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and Hart G.W.. 2013. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev. Proteomics. 10:365–380. 10.1586/14789450.2013.820536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., and Hart G.W.. 2014. O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics. 11:8. 10.1186/1559-0275-11-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., Banerjee P., Whelan S.A., Liu T., Wei A.C., Ramirez-Correa G., McComb M.E., Costello C.E., O’Rourke B., Murphy A., et al. 2016. Comparative proteomics reveals dysregulated mitochondrial O-GlcNAcylation in diabetic hearts. J. Proteome Res. 15:2254–2264. 10.1021/acs.jproteome.6b00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAskill, A.F., Brickley K., Stephenson F.A., and Kittler J.T.. 2009a. GTPase dependent recruitment of Grif-1 by Miro1 regulates mitochondrial trafficking in hippocampal neurons. Mol. Cell. Neurosci. 40:301–312. 10.1016/j.mcn.2008.10.016 [DOI] [PubMed] [Google Scholar]
- MacAskill, A.F., Rinholm J.E., Twelvetrees A.E., Arancibia-Carcamo I.L., Muir J., Fransson A., Aspenstrom P., Attwell D., and Kittler J.T.. 2009b. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 61:541–555. 10.1016/j.neuron.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor, U., Bartholomew S., Golani G., Christenson E., Kozlov M., Higgs H., Spudich J., and Lippincott-Schwartz J.. 2015. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife. 4:e08828. 10.7554/eLife.08828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, S., Bacote V., and Traxinger R.R.. 1991. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem. 266:4706–4712. 10.1016/S0021-9258(19)67706-9 [DOI] [PubMed] [Google Scholar]
- Mattson, M.P., Gleichmann M., and Cheng A.. 2008. Mitochondria in neuroplasticity and neurological disorders. Neuron. 60:748–766. 10.1016/j.neuron.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mille, F., Thibert C., Fombonne J., Rama N., Guix C., Hayashi H., Corset V., Reed J.C., and Mehlen P.. 2009. The Patched dependence receptor triggers apoptosis through a DRAL-caspase-9 complex. Nat. Cell Biol. 11:739–746. 10.1038/ncb1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misgeld, T., and Schwarz T.L.. 2017. Mitostasis in neurons: maintaining mitochondria in an extended cellular architecture. Neuron. 96:651–666. 10.1016/j.neuron.2017.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, A.S., Wong Y.C., Simpson C.L., and Holzbaur E.L.. 2016. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission–fusion balance within mitochondrial networks. Nat. Commun. 7:12886. 10.1038/ncomms12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, K., Kurotaki D., Kawase W., Soma S., Fukuchi Y., Kunimoto H., Yoshimi R., Koide S., Oshima M., Hishiki T., et al. 2021. OGT regulates hematopoietic stem cell maintenance via PINK1-dependent mitophagy. Cell Rep. 34:108579. 10.1016/j.celrep.2020.108579 [DOI] [PubMed] [Google Scholar]
- Nakazawa, N., Sathe A.R., Shivashankar G.V., and Sheetz M.P.. 2016. Matrix mechanics controls FHL2 movement to the nucleus to activate p21 expression. Proc. Natl. Acad. Sci. USA. 113:E6813–E6822. 10.1073/pnas.1608210113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, L.E., and Shadel G.S.. 2018. Pink1/Parkin link inflammation, mitochondrial stress, and neurodegeneration. J. Cell Biol. 217:3327–3329. 10.1083/jcb.201808118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, C.F., Xu J.Y., Li M.S., and Tsui S.K.. 2014. Identification of FHL2-regulated genes in liver by microarray and bioinformatics analysis. J. Cell. Biochem. 115:744–753. 10.1002/jcb.24714 [DOI] [PubMed] [Google Scholar]
- Nie, D., and Sahin M.. 2012. A genetic model to dissect the role of Tsc-mTORC1 in neuronal cultures. Methods Mol. Biol. 821:393–405. 10.1007/978-1-61779-430-8_25 [DOI] [PubMed] [Google Scholar]
- Oeding, S.J., Majstrowicz K., Hu X.P., Schwarz V., Freitag A., Honnert U., Nikolaus P., and Bähler M.. 2018. Identification of Miro1 and Miro2 as mitochondrial receptors for myosin XIX. J. Cell Sci. 131:jcs219469. 10.1242/jcs.219469 [DOI] [PubMed] [Google Scholar]
- Olson, E.N., and Nordheim A.. 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11:353–365. 10.1038/nrm2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C., Lee S.A., Hong J.H., Suh Y., Park S.J., Suh B.K., Woo Y., Choi J., Huh J.W., Kim Y.M., et al. 2016. Disrupted-in-schizophrenia 1 (DISC1) and Syntaphilin collaborate to modulate axonal mitochondrial anchoring. Mol. Brain. 9:69. 10.1186/s13041-016-0250-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak, D., Sepp K.J., and Hollenbeck P.J.. 2010. Evidence that myosin activity opposes microtubule-based axonal transport of mitochondria. J. Neurosci. 30:8984–8992. 10.1523/JNEUROSCI.1621-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkurnaz, G., Trinidad J.C., Wang X., Kong D., and Schwarz T.L.. 2014. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell. 158:54–68. 10.1016/j.cell.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell, A.M., and Youle R.J.. 2015. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 85:257–273. 10.1016/j.neuron.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling, A.D., Horiuchi D., Lively C.M., and Saxton W.M.. 2006. Kinesin-1 and dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 17:2057–2068. 10.1091/mbc.e05-06-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, W., Lv P., Fan X., Quan B., Zhu Y., Qin K., Chen Y., Wang C., and Chen X.. 2017. Quantitative time-resolved chemoproteomics reveals that stable O-GlcNAc regulates box C/D snoRNP biogenesis. Proc. Natl. Acad. Sci. USA. 114:E6749–E6758. 10.1073/pnas.1702688114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, H.B., Dietrich M.O., Liu Z.W., Zimmer M.R., Li M.D., Singh J.P., Zhang K., Yin R., Wu J., Horvath T.L., et al. 2014. O-GlcNAc transferase enables AgRP neurons to suppress browning of white fat. Cell. 159:306–317. 10.1016/j.cell.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacoman, J.L., Dagda R.Y., Burnham-Marusich A.R., Dagda R.K., and Berninsone P.M.. 2017. Mitochondrial O-GlcNAc transferase (mOGT) regulates mitochondrial structure, function, and survival in HeLa cells. J. Biol. Chem. 292:4499–4518. 10.1074/jbc.M116.726752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl, F.A., McLoughlin P., Ehler E., de Giovanni C., and Schäfer B.W.. 2000. DRAL is a p53-responsive gene whose four and a half LIM domain protein product induces apoptosis. J. Cell Biol. 151:495–506. 10.1083/jcb.151.3.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M.H., Lewandowska A., Caprio G.D., Skillern W., Upadhyayula S., Kirchhausen T., Shaw J.M., and Cunniff B.. 2017. Miro1-mediated mitochondrial positioning shapes intracellular energy gradients required for cell migration. Mol. Biol. Cell. 28:2159–2169. 10.1091/mbc.e16-10-0741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, T.L. 2013. Mitochondrial trafficking in neurons. Cold Spring Harb. Perspect. Biol. 5:a011304. 10.1101/cshperspect.a011304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semba, R.D., Huang H., Lutty G.A., Van Eyk J.E., and Hart G.W.. 2014. The role of O-GlcNAc signaling in the pathogenesis of diabetic retinopathy. Proteomics Clin. Appl. 8:218–231. 10.1002/prca.201300076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.H., Agarwal E., Bryant K.G., Caino M.C., Kim E.T., Kossenkov A.V., Tang H.Y., Languino L.R., Gabrilovich D.I., Cohen A.R., et al. 2018. Syntaphilin ubiquitination regulates mitochondrial dynamics and tumor cell movements. Cancer Res. 78:4215–4228. 10.1158/0008-5472.CAN-18-0595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, Z.H. 2014. Mitochondrial trafficking and anchoring in neurons: new insight and implications. J. Cell Biol. 204:1087–1098. 10.1083/jcb.201312123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko, A., Tomas H., Havlis J., Olsen J.V., and Mann M.. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Shi, H., Munk A., Nielsen T.S., Daughtry M.R., Larsson L., Li S., Høyer K.F., Geisler H.W., Sulek K., Kjøbsted R., et al. 2018. Skeletal muscle O-GlcNAc transferase is important for muscle energy homeostasis and whole-body insulin sensitivity. Mol. Metab. 11:160–177. 10.1016/j.molmet.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov, E., Basu H., Bray M.A., Sun Z., Wei W., Apaydin K., Karhohs K., Chen P.F., Smith J.L.M., Wiskow O., et al. 2019. A high-content screen identifies TPP1 and Aurora B as regulators of axonal mitochondrial transport. Cell Rep. 28:3224–3237.e5. 10.1016/j.celrep.2019.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneyer, B.I., Ušaj M., and Henn A.. 2016. Myo19 is an outer mitochondrial membrane motor and effector of starvation-induced filopodia. J. Cell Sci. 129:543–556. 10.1242/jcs.175349 [DOI] [PubMed] [Google Scholar]
- Silver, I.A., and Erecińska M.. 1994. Extracellular glucose concentration in mammalian brain: continuous monitoring of changes during increased neuronal activity and upon limitation in oxygen supply in normo-, hypo-, and hyperglycemic animals. J. Neurosci. 14:5068–5076. 10.1523/JNEUROSCI.14-08-05068.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter, D.A., Martinez J., Hao L., Chen X., Sun N., Fischer T.D., Burman J.L., Li Y., Zhang Z., Narendra D.P., et al. 2018. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 561:258–262. 10.1038/s41586-018-0448-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers, R.S., Megeath L.J., Górska-Andrzejak J., Meinertzhagen I.A., and Schwarz T.L.. 2002. Axonal transport of mitochondria to synapses depends on Milton, a novel Drosophila protein. Neuron. 36:1063–1077. 10.1016/S0896-6273(02)01094-2 [DOI] [PubMed] [Google Scholar]
- Su, C., and Schwarz T.L.. 2017. O-GlcNAc transferase is essential for sensory neuron survival and maintenance. J. Neurosci. 37:2125–2136. 10.1523/JNEUROSCI.3384-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, B., Ji Y.S., Sun X.L., Liu X.H., and Chen Z.Y.. 2014. Brain-derived neurotrophic factor (BDNF)-induced mitochondrial motility arrest and presynaptic docking contribute to BDNF-enhanced synaptic transmission. J. Biol. Chem. 289:1213–1226. 10.1074/jbc.M113.526129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Kanai Y., Okada Y., Nonaka S., Takeda S., Harada A., and Hirokawa N.. 1998. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 93:1147–1158. 10.1016/S0092-8674(00)81459-2 [DOI] [PubMed] [Google Scholar]
- Tirosh, A., Shai I., Tekes-Manova D., Israeli E., Pereg D., Shochat T., Kochba I., and Rudich A.. Israeli Diabetes Research Group . 2005. Normal fasting plasma glucose levels and type 2 diabetes in young men. N. Engl. J. Med. 353:1454–1462. 10.1056/NEJMoa050080 [DOI] [PubMed] [Google Scholar]
- Tran, M.K., Kurakula K., Koenis D.S., and de Vries C.J.. 2016. Protein-protein interactions of the LIM-only protein FHL2 and functional implication of the interactions relevant in cardiovascular disease. Biochim. Biophys. Acta. 1863:219–228. 10.1016/j.bbamcr.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Trigo, D., Goncalves M.B., and Corcoran J.P.T.. 2019. The regulation of mitochondrial dynamics in neurite outgrowth by retinoic acid receptor β signaling. FASEB J. 33:7225–7235. 10.1096/fj.201802097R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad, J.C., Barkan D.T., Gulledge B.F., Thalhammer A., Sali A., Schoepfer R., and Burlingame A.L.. 2012. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics. 11:215–229. 10.1074/mcp.O112.018366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhauwaert, R., Bharat V., and Wang X.. 2019. Surveillance and transportation of mitochondria in neurons. Curr. Opin. Neurobiol. 57:87–93. 10.1016/j.conb.2019.01.015 [DOI] [PubMed] [Google Scholar]
- Vincent, A.M., Callaghan B.C., Smith A.L., and Feldman E.L.. 2011. Diabetic neuropathy: cellular mechanisms as therapeutic targets. Nat. Rev. Neurol. 7:573–583. 10.1038/nrneurol.2011.137 [DOI] [PubMed] [Google Scholar]
- Wang, X., and Schwarz T.L.. 2009. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 136:163–174. 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Winter D., Ashrafi G., Schlehe J., Wong Y.L., Selkoe D., Rice S., Steen J., LaVoie M.J., and Schwarz T.L.. 2011. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 147:893–906. 10.1016/j.cell.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan, D.R., and Bell T.D.. 2015. Image artifacts in single molecule localization microscopy: why optimization of sample preparation protocols matters. Sci. Rep. 5:7924. 10.1038/srep07924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, T.L., and Cleveland D.W.. 1999. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat. Neurosci. 2:50–56. 10.1038/4553 [DOI] [PubMed] [Google Scholar]
- Xu, K., Zhong G., and Zhuang X.. 2013. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 339:452–456. 10.1126/science.1232251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C.H., Davidson S., Harapas C.R., Hilton J.B., Mlodzianoski M.J., Laohamonthonkul P., Louis C., Low R.R.J., Moecking J., De Nardo D., et al. 2020. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 183:636–649.e18. 10.1016/j.cell.2020.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists high-confidence interactors of TRAK1 as detected by SILAC-MS (related to Fig. 4 A).
lists primer sequences and shRNA sequences used in this study (related to Materials and methods text).