Abstract
Pregnant individuals infected with SARS-CoV-2 have higher rates of intensive care unit admission, oxygen requirement, need for mechanical ventilation, and death than nonpregnant individuals. Increased COVID-19 disease severity may be associated with an increased risk of viremia and placental infection. Maternal SARS-CoV-2 infection is also associated with pregnancy complications such as preeclampsia and preterm birth, which can be either placentally mediated or reflected in the placenta. Maternal viremia followed by placental infection may lead to maternal-fetal transmission (vertical), which affects 1% to 3% of exposed newborns. However, there is no agreed-upon or standard definition of placental infection. The National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development convened a group of experts to propose a working definition of placental infection to inform ongoing studies of SARS-CoV-2 during pregnancy. Experts recommended that placental infection be defined using techniques that allow virus detection and localization in placental tissue by one or more of the following methods: in situ hybridization with antisense probe (detects replication) or a sense probe (detects viral messenger RNA) or immunohistochemistry to detect viral nucleocapsid or spike proteins. If the abovementioned methods are not possible, reverse transcription polymerase chain reaction detection or quantification of viral RNA in placental homogenates, or electron microscopy are alternative approaches. A graded classification for the likelihood of placental infection as definitive, probable, possible, and unlikely was proposed. Manuscripts reporting placental infection should describe the sampling method (location and number of samples collected), method of preservation of tissue, and detection technique. Recommendations were made for the handling of the placenta, examination, and sampling and the use of validated reagents and sample protocols (included as appendices).
Key words: COVID-19, fetal death, immunohistochemistry, in situ hybridization, placental infection, placentitis, preeclampsia, preterm birth, SARS-CoV-2, stillbirth, syncytiotrophoblast, vertical transmission
Introduction
Pregnant individuals infected with SARS-CoV-2 have higher rates of admission to the intensive care unit, requirement for mechanical ventilation, extracorporeal membrane oxygenation, and death than nonpregnant individuals1, 2, 3 but the risk of placental, fetal, and pregnancy complications have been less well described. Studies show that maternal SARS-CoV-2 infection is associated with an increased risk of preterm birth and preeclampsia,4, 5, 6 both of which may be placentally mediated or placentally-reflected complications. Moreover, published reports7, 8, 9 show that first- and second-trimester infections with SARS-CoV-2 are possible and can result in pregnancy loss. Recent reports from Ireland found second-trimester miscarriage and stillbirth associated with presumed placental SARS-CoV-2 infection, placentitis, and specifically the B.1.1.7 variant.10 , 11 These reports suggest that placental infection does carry substantial perinatal morbidity and that clinical surveillance of exposed pregnancies and placental evaluation for infection are warranted. In addition, placental infection does not equate with vertical transmission but may cause placental damage, which leads to perinatal morbidity without infection. Many have noted the triad of histiocytic intervillositis, increased perivillous fibrin deposition, and trophoblast necrosis associated with placental infection.10 , 12, 13, 14, 15, 16, 17, 18, 19 Although these histologic findings are not specific for infection, they are characteristic enough to indicate a study to evaluate whether the placenta was infected.
Although early reports were focused on SARS-CoV-2 placental infection and in utero transmission,19, 20, 21, 22, 23, 24, 25, 26 larger studies and meta-analyses indicate that placental infection and vertical transmission were rare,4 , 9 , 27, 28, 29, 30, 31, 32 particularly in comparison with other maternal viral infections in pregnancy (such as cytomegalovirus, Zika, rubella, and untreated HIV33, 34, 35, 36). Diagnostic criteria for confirming placental infection have been inconsistent. Early reports conflated placental infection with vertical transmission22, highlighting the need for consensus definitions and nomenclature in this regard. One report found SARS-CoV-2 infection in 100% of placentas from mothers with COVID-19 based on positive immunohistochemistry studies,37 whereas others have rarely reported placental infection, if at all.17 , 29 , 38 , 39 Investigators have employed a variety of techniques to diagnose placental infection, including reverse transcription polymerase chain reaction (RT-PCR) on placental homogenates,9 , 40 , 41 immunohistochemistry,9 , 19 , 37 RNA in situ hybridization (RNA-ISH),9 , 21 , 29 , 39 and electron microscopy,21 , 42, 43, 44 making it difficult to compare across studies and establish definitive evidence regarding placental infection risks. The relative sensitivity and specificity of these techniques in the placenta have not been rigorously studied, but we have included what is known in the appendices. Given the patchy nature of infection of the placenta by SARS-CoV-2,45 legitimate concerns are raised about false positive and false negative results by various detection methods. Misclassification might lead to erroneous diagnoses with potential clinical and research implications. Indeed, knowing the true prevalence of placental infection depends on accurate diagnoses, and thus, these techniques are critically important to perform with validation and proper controls. Despite these caveats, a systematic review and meta-analysis of these disparate case reports and case series reported a placental infection rate of 7% (2 of 26 cases).46 Vertical transmission rates of 1.1% to 3% have been noted in cross-sectional studies ranging in size from 101 to 2399 mothers.4 , 27 , 30 , 32 , 47 There is clearly a need for standardization of methods to ensure robust and reproducible results and to facilitate cross-study comparisons.
Although a definition of transplacental infection has been proposed (World Health Organization reference number: WHO/2019-nCoV/mother-to-child transmission/2021.148), the criteria for placental infection have not. Although not all viruses infect the placenta before infecting the fetus,49 , 50 placental infection seems to be a necessary intermediate step for in utero transmission of SARS-CoV-2 to the fetus.15 Objective, robust, consensus-driven criteria for the documentation of placental infection are needed to provide guidelines for research and clinical care. Herein, we present consensus definitions, tiered by rigor, formulated by a team of experts on the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)–assembled task force, for what constitutes placental infection by SARS-CoV-2 for clinical and research applications. We include, in appendices, protocols for implementation with specifics and references.
Methods
A group of active investigators with expertise in placental pathology, virology, obstetrics, infectious diseases, immunology, and molecular biology convened virtually to discuss and critique methods used to diagnose placental infection with SARS-CoV-2. In this multidisciplinary virtual workshop, experts in each field presented the following methods for documenting placental infection by SARS-CoV-2 (highlighting the strengths and limitations of each method): RNA RT-PCR or quantitative RT-PCR from placental homogenates, immunohistochemistry, RNA-ISH, electron microscopy, and histopathology. A round-table discussion followed the presentations and dialogue continued over several days. A 3-person team (D.J.R., A.G.E., R.J.R.) then devised a ranked template of specific diagnostic techniques and procedures in descending order of rigor (Table 1 ). This was then distributed to the larger committee for further discussion until consensus was achieved on the recommendations that follow.
Table 1.
Definition categories of placental infection with SARS-CoV-2
| Definite: evidence of active replicating virus with location in the placental tissues |
| Probable: evidence of viral RNA or protein located in placental tissues |
| Possible: evidence of viral RNA in placental homogenates or viral-like particles by electron microscopy in placental tissues |
| Unlikely: no evidence of any of the above |
| No testing: testing not done |
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
Recommendations for Definition of Placental Infection
Recognizing that there may be a difference between what represents the most stringent criteria for defining placental infection and what can be done in the greatest number of real-world settings, we seek to offer guidance on definitions of placental SARS-CoV-2 infection in clinical settings and in biomedical investigation.
As such, we offer 5 levels of rigor for detecting placental infection with SARS-CoV-2 (both acute and current or persistent) (Table 1), followed by a pragmatic approach for defining placental infection. For each individual investigation, we suggest choosing 1 (or more) of the 5 categories based on the level of rigor desired by the investigator or clinician and what tools are available for implementation. Our recommended diagnostic criteria are provided in Table 2 . In addition, we provide instruction for handling, processing, and examination of the placenta (with additional sample collection, handling, and storage information in NICHD, National Institutes of Health, and United States Department of Health and Human Services (2021). “Promoting Data Harmonization to Accelerate COVID-19 Pregnancy Research,” Biospecimens Common Data Elements section. Retrieved from: https://www.niehs.nih.gov/research/programs/disaster/database/nihpromotin_dataharmonizationacceleratecovid19pregnancyresearchbiomedicalpsychosocialbiospecimens_vf.pdf.) The recommendations for reporting in scientific manuscripts are presented in Table 3 .
Table 2.
Definition specifics of placental infection with SARS-CoV-2 in order of rigor
| Definite: documentation of viral presence, location in the placenta tissues, and replication, by: |
|
| or |
|
| Probable: documentation of viral proteins or RNA within placental tissues, without evidence of active replication via: |
|
| or |
|
| Possible: less specific detection of virus. These approaches could be detecting viral particles engulfed by macrophages rather than actively replicating virus. RT-PCR of placental homogenates theoretically may have a positive result owing to maternal viremia (although this is a rare entity), rather than placental involvement. |
|
| Note that an alternative approach is a 2-step approach, in which RT-PCR is used as a screen and then followed up with one of the methods recommended to confirm “definite” or “probable” infection. This hybrid/2-step approach would be more rigorous than RT-PCR alone and potentially more sensitive than the “definite” and “probable” approaches. |
|
| Unlikely: Negative results from any of the above tests |
| No testing: placenta not tested |
PBS, phosphate-buffered saline; RNA-ISH, RNA in situ hybridization; RT-PCR, reverse transcription polymerase chain reaction.
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
Table 3.
Recommended reporting guidelines for scientific manuscripts
| Scientific manuscripts that report on placental detection of SARS-CoV-2 should report on: |
|
|
|
|
| Other issues of importance to report: |
|
|
|
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
Recommended placental handling, processing, and examination
For RT-PCR testing, we recommend at least 2 fresh placental samples (0.5 cm3) taken within 20 minutes of placental delivery (and not >1 hour after placental delivery to avoid RNA degradation), best obtained in the delivery room by appropriately protected and trained individuals, from the fetal side of the placenta (Figures 1 and 2 ) rinsed in sterile normal saline or phosphate-buffered saline, and then either snap-frozen (in liquid nitrogen or on dry ice until placed in −80°C) or placed in 5 to 10 volumes of RNAlater (Thermo Fisher Scientific, Waltham, MA) for subsequent freezing and storage at −80°C. Biopsies should be taken from a midpoint between the chorionic and basal plates and midway between the cord insertion and the placental edge (at least 3 cm from the cord insertion and 3 cm from the placental edge) to ensure viable villous parenchyma is collected (Figures 1 and 2). The fetal membranes should be dissected off the fetal surface and not included in the biopsy. Notably, 2 samples are recommended for protection against failure of RNA extraction and for biological sampling diversity of different placental regions, given that patchy infection is possible.45 Snap-frozen biopsies may permit single-cell RNA-Seq/nuc-Seq approaches, other RNA analyses, and protein analyses, whereas biopsies preserved in RNAlater can be used for RNA/DNA analyses and protein analyses, but not for scRNA-Seq/nuc-Seq. RNA quality is likely better for longer periods of time with the use of RNAlater. For RNA-ISH and immunohistochemistry studies, formalin-fixed paraffin-embedded full-thickness placental parenchyma cut at 5 μm onto glass slides should be used. All studied placentas should be examined and reported promptly by pathology following the Amsterdam Criteria for sampling and histologic diagnoses.52
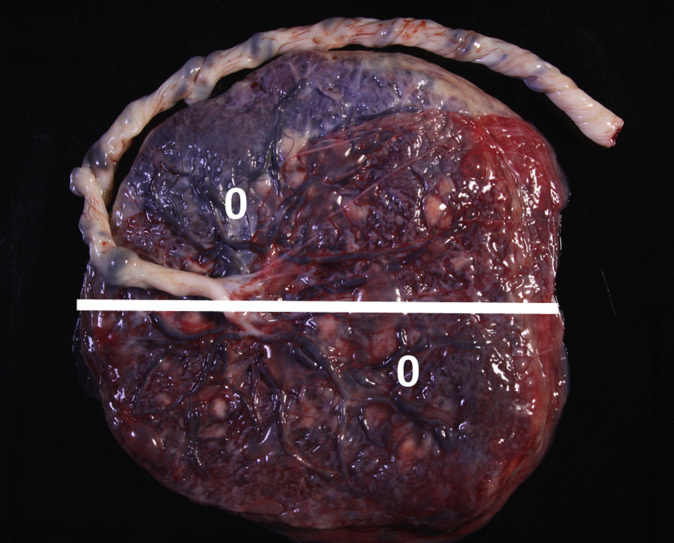
Figure 1.
Gross photograph of the chorionic plate (fetal side) of a normal (uninfected) placenta
The white line is cut site for Figure 2 slab section. The white zeroes are possible biopsy sites.
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
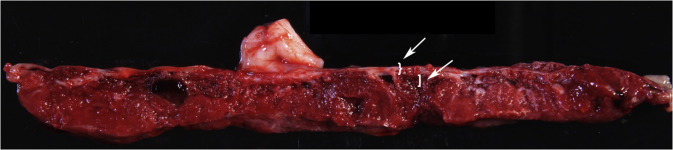
Figure 2.
Gross photograph of the full-thickness slab section through the white line in Figure 1
The white brace highlights chorionic plate—fetal membranes should be excluded from the biopsy. The white bracket indicates fetal parenchyma as target for sampling; approximately 0.5 cm depth is recommended. The white arrows highlight the brace and the bracket.
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
Conclusion
We submit this set of recommendations to standardize the definition of SARS-CoV-2 infection of the placenta. These recommendations are made by a consensus panel of experts in the fields of obstetrics, virology, placental pathology, infectious disease, immunology, and molecular biology. The definitions are tiered by rigor of the diagnostic technique. Investigators and clinical care providers are encouraged to use the most rigorous method available for documenting placental infection in studies and in clinical diagnosis. Comparisons across studies are more valuable when the studies use the same standardized methods. We understand that not all, or perhaps not any, of these methods will be available at all institutions, but we predict that most institutions will have at least one recommended method at their disposal, and collaboration or consultation is advised for those that do not have any recommended method available. We anticipate that utilization of these recommended definitions of placental infection by SARS-COV-2 will facilitate comparisons of results among studies and that improved interpretation will follow, allowing for maximum impact of research in this area and optimization of patient care.
Glossary.
Immunohistochemistry (IHC): antibody-mediated localization of proteins in the tissue.
Negative-sense strand: provides evidence of viral replication in a positive-sense RNA virus (eg, SARS-CoV-2).
N-protein: the nucleocapsid RNA binding protein of the SARS-CoV-2 virus.
RNA in situ hybridization (RNA-ISH): molecular localization of the RNA transcript in the tissue.
RT-PCR: reverse transcription polymerase chain reaction for amplification of RNA.
Positive-sense strand: provides evidence of viral presence but not necessarily viral replication in a positive-sense RNA virus as in SARS-CoV-2.
RT-qPCR: quantitative reverse transcription polymerase chain reaction for amplification of RNA.
S-protein: the spike surface glycoprotein of the SARS-CoV-2 virus.
Viral replication: active/infectious viral reproduction.
Acknowledgments
The authors thank the task force at the NICHD and Dr Diana Bianchi for the foresight to organize this workshop. We are indebted to Drs Nahida Chakhtoura, Jessica Gleason, and Caroline Signore who contributed greatly to the organization and this workshop.
Participants: the authors and Andrew Bremer, MD, PhD; Samantha Calabrese, MPH; Nahida Chakhtoura, MD; Gaya Dowling, PhD; Susan Fisher, PhD; Michelle Freund, PhD; Jessica Gleason, PhD; Katherine Grantz, MD; Corinne Hausmann, PhD; Rohan Hazra, MD; John Ilekis, PhD; George Miller, PhD; David Pepin, PhD; Richard K. Miller, PhD; Yoel Sadovsky, MD; Caroline Signore, MD, MPH; Fasil Tekola-Ayele, PhD; and David Weinberg, PhD.
Footnotes
D.J.R., A.G.E., and R.J.R. share first authorship.
Meeting date: March 9, 2021.
D.J.R. received royalties for authorship on perinatal pathology topics for UpToDate and Cambridge University Press, neither of which should impact this manuscript. J.L.H. is a consultant for Aadi Bioscience and TRACON Pharmaceuticals, unrelated to the topic of this manuscript. D.T.T. is a consultant for ROME Therapeutics, NanoString Technologies, Pfizer, Merrimack Pharmaceuticals, Ventana Roche, Foundation Medicine, Inc, and EMD Millipore Sigma, which are not related to this work. D.T.T. is a founder and has equity in ROME Therapeutics, PanTher Therapeutics, and TellBio, Inc, which is not related to this work. D.T.T. receives sponsored research support from ACD Bio-Techne for RNA in situ hybridization, which is related to this manuscript. D.T.T.’s interests were reviewed and are managed by Mass General Brigham in accordance with their conflict of interest policies. T.D.M. is a site principal investigator (PI) and medical consultant for a Pfizer trial of SARS-CoV-2 vaccination in pregnancy. T.D.M. receives UpToDate royalties and was the site PI for a Novavax and GestVision trial, which are unrelated to this work. The remaining authors report no conflict of interest.
This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy ShriverNational Institute of Child Health and Human Development, National Institutes of Health, United States Department of Health and Human Services (NICHD/NIH/DHHS), and, in part, with federal funds from NICHD/NIH/DHHS under contract number HHSN275201300006C. R.J.R. has contributed to this work as part of his official duties as an employee of the United States Federal Government.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Supplementary Data
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
Appendix A
Recommended guide for RNA in situ hybridization
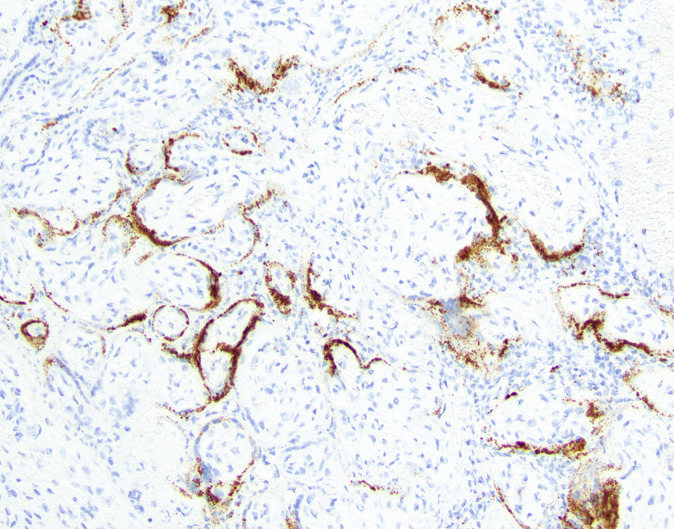
RNA in situ hybridization (RNA-ISH) is performed on 5 μm thick formalin-fixed paraffin-embedded (FFPE) sections. SARS-CoV-2 RNA-ISH is performed using the RNAscope 2.5 LS Probe-V-nCoV2019-S catalog number 848568 and RNAscope 2.5 LS Reagent Kit-RED catalog number 322150 (Advanced Cell Diagnostics, Bio-Techne, Minneapolis, MN). RNA probes include RNAscope, hybridization chain reactions (HCRs) designed toward N1/N2/N3 cDNA of SARS-CoV-2 and human RNase P gene loci on the BOND RX automated staining platform (Leica Biosystems, Wetzlar, Germany). For dsRNA probes example reagent: Jena Bioscience (Jena, Germany) Anti-dsRNA monoclonal antibody J2, catalog number RNT-SCI-10010200. Protocol to be provided in appendix. RNA-ISH that detects the replication strand is performed with the RNAscope 2.5 LS Probe- V-nCoV2019-S- Sense Probe, catalog number 845701 (Advanced Cell Diagnostics). RNA-ISH is performed per the standard protocol from ACD Bio-Techne on the Leica BOND RX automated staining platform. The protocol begins with a 1-hour incubation at 60°C followed by pretreatment using the BOND Epitope Retrieval Solution 2 for 15 minutes at 95°C. Then RNA is unmasked using protease for 15 minutes and probe hybridization for 2 hours. Signal is amplified by a series of branched DNA amplification steps followed by color development in red using the BOND Polymer Refine Red Detection kit (Leica Biosystems). Hematoxylin is then performed to counterstain the nuclei on the BOND RX machine. Such RNA probes include RNAscope, HCRs designed toward N1/N2/N3 cDNA of SARS-CoV-2 and human RNase P gene loci.
Note that the gold standard of SARS-CoV-2 titer demonstration via plaque assay or TCID50 require a bio-safety level 3 laboratory setting, which is not achievable in most real-world settings.
Although the test characteristics for SARS-CoV-2 RNA-ISH in placental samples are not well defined given the limited data in these samples, the D.T.T. laboratory has performed a paired analysis of the Centers for Disease Control and Prevention (CDC) approved quantitative reverse transcription polymerase chain reaction (RT-qPCR) assay and the RNA-ISH in a variety of human autopsy materials from patients who died of COVID-19.53 Using 90 human autopsy samples, the D.T.T. laboratory performed the RT-qPCR assay using a cycle threshold (Ct) cutoff of 42.5 for both N1 and N2 primer sets as the gold standard. RNA-ISH in these paired samples had a specificity of 96.7% and a sensitivity of 76.7%. A paired analysis of placenta will be needed to determine the work test characteristics of RNA-ISH in this setting.
Appropriate positive control should be a FFPE specimen that is from an individual confirmed to be positive for SARS-CoV-2 by RT-PCR or RNA-Seq and show positive signal by RNA-ISH in the appropriate tissues. Appropriate negative control should be a similar FFPE specimen that is confirmed to be negative for SARS-CoV-2 by RT-qPCR. Appropriate positive controls include infected lung tissues from patients who died of COVID-19 (autopsy cases) and known infected placentas appropriately preserved; appropriate negative controls include tissues with similar pathologic processes (eg, diffuse alveolar damage in the lung and placental tissues of the same gestational age with similar pathologic features).
Controls should be performed on each lot of RNA-ISH reagents and on each batch run of cases.
An example image of RNA-ISH performed using the above technique is provided in Figure 3.
Appendix B
Recommended protocol for immunohistochemistry
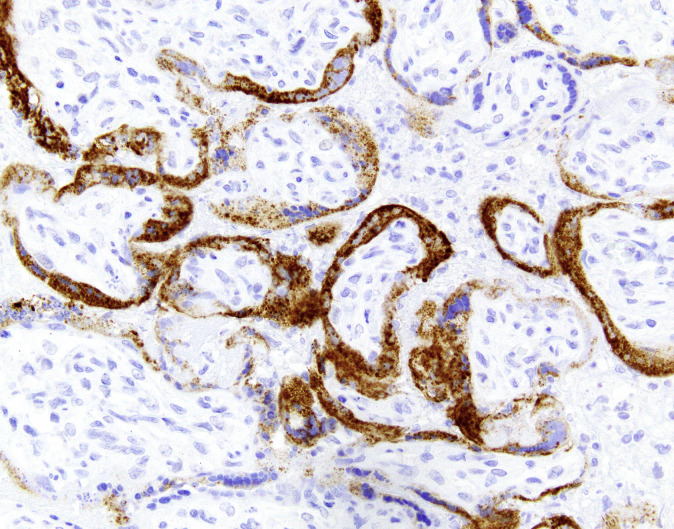
The following antibodies have been validated for immunohistochemistry (IHC) in formalin-fixed paraffin-embedded tissue sections: (1) a rabbit polyclonal antibody raised against the nucleocapsid protein (Novus Biologicals, Littleton, CO), (2) a rabbit monoclonal antibody raised against the nucleocapsid protein (clone #001; Sino Biological, Beijing, China), and (3) a mouse monoclonal antibody raised against the S2 subunit of the spike protein (clone 1A9; GeneTex, Irvine, CA) (1-3). Sensitivity for the spike protein antibody was validated using immunofluorescence and western blotting (available at: https://www.genetex.com/Product/Detail/SARS-CoV-SARS-CoV-2-COVID-19-spike-antibody-1A9/GTX632604) but such studies on tissue sections using brightfield IHC are lacking. IHC using these antibodies requires careful validation for optimal detection and to avoid nonspecific and background staining. For analytical validation of IHC laboratory-developed tests, the College of American Pathologists recommends testing a minimum of 10 positive and 10 negative tissues; when the laboratory medical director determines that fewer than 20 cases are sufficient for a particular marker (eg, rare antigens), the rationale for that decision should be documented (4). The IHC expert in this consensus group (J.L.H.) used infected lung tissues from patients who died of COVID-19 (autopsy cases) as positive controls; negative control cases should include tissues with similar pathologic processes (eg, diffuse alveolar damage in the lung). The appropriate placental negative control in this context would be one with chronic histiocytic intervillositis or massive perivillous fibrin deposition at a similar gestational age in a placenta from prepandemic times. In J.L.H.’s laboratory at the Brigham and Women’s Hospital, Boston, Massachusetts, IHC was performed after pressure cooker antigen retrieval (pH 6.1 citrate buffer; Target Retrieval Solution, Dako, Agilent Technologies, Santa Clara, CA) using the antispike protein antibody (1A9; 1:1000 dilution) and the EnVision+ detection system (Dako).
An example image of placental Spike IHC performed using the above technique is provided in Figure 4.
Appendix C
Sample protocol for quantitative reverse transcription polymerase chain reaction
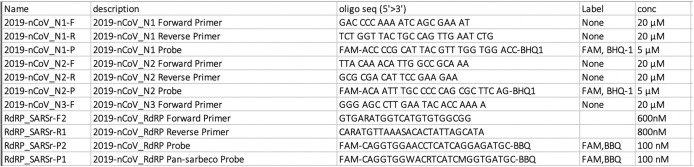
Assessment of SARS-CoV-2 RNA was performed using SuperScript III One-Step RT-PCR System with Platinum Taq Polymerase (Invitrogen, Thermo Fisher Scientific, Waltham, MA; containing 0.4 mM of each deoxyribonucleotide triphosphates and 3.2 mM magnesium sulfate), 1 μL of reverse transcriptase/Taq mixture from the kit, 0.4 μL of a 50 mM magnesium sulfate solution (Invitrogen—not provided with the kit), and 1 μg of nonacetylated bovine serum albumin (Roche, Basel, Switzerland). Specific detection of 2019-nCoV was performed using N1 and N2 assays per the CDC guidelines (https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html). For further validation of the specimens, RNA-dependent RNA polymerase (RdRp) assay was performed. RT-qPCR primers for both RdRp and N are recommended to confirm SARS-CoV-2 viral infection, rather than the presence of remnant or residual N genes persisting after viral clearance.54 A housekeeping gene, such as RNaseP, should be used to validate the extraction of RNA from the specimen.
A specimen was considered positive if the gene target ha a cycle threshold based on true positive and negative controls in each laboratory generating a receiver operating characteristic curve. We recommend a Ct cutoff of <40 for the limit of detection of 50 copies/mL. The use of semiquantitative methods such as inferring viral burden from Ct values should be confirmed by assessing infectious virions from the specimen (Vero E6 culture). The most rigorous approach is quantitative RT-qPCR, where a standard curve of RNA is generated for quantifying viral load using a CDC positive control. For quantitative RT-qPCR, SARS-CoV-2 viral loads below 40 RNA copies/mL should be categorized as undetectable and set at 1.0 log10 RNA copies/mL. Positive controls are commercially available for several SARS-CoV-2 target genes in a number of formats (eg, New England Biolabs, Ipswich, MA; N2117S, Enzo ENZ-GEN218-0004). Of note, although respiratory and serum or plasma viral loads have been demonstrated to correlate with disease severity and death in a cohort of nonpregnant older adults,55 available data suggest that viremia is rare in pregnancy,29 and there is limited data regarding placental viral load and an increased risk of adverse obstetrical or neonatal outcomes, including but not limited to vertical transmission. Regardless, quantitative methods are recommended over semiquantitative or binary (positive or negative) methods, both because of their increased rigor and because quantification of placental viral loads could facilitate future research correlating viral burden with obstetrical or neonatal outcomes.
Supplemental Figure.
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
References
- 1.Zambrano L.D., Ellington S., Strid P., et al. Update: characteristics of symptomatic women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status - United States, January 22-October 3, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1641–1647. doi: 10.15585/mmwr.mm6944e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellington S., Strid P., Tong V.T., et al. Characteristics of women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status - United States, January 22-June 7, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:769–775. doi: 10.15585/mmwr.mm6925a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delahoy M.J., Whitaker M., O'Halloran A., et al. Characteristics and maternal and birth outcomes of hospitalized pregnant women with laboratory-confirmed COVID-19 - COVID-NET, 13 states, March 1-August 22, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1347–1354. doi: 10.15585/mmwr.mm6938e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodworth K.R., Olsen E.O., Neelam V., et al. Birth and infant outcomes following laboratory-confirmed SARS-CoV-2 infection in pregnancy—SET-NET, 16 jurisdictions, March 29-October 14, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:1635–1640. doi: 10.15585/mmwr.mm6944e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villar J., Ariff S., Gunier R.B., et al. Maternal and neonatal morbidity and mortality among pregnant women with and without COVID-19 infection: the INTERCOVID multinational cohort study. JAMA Pediatr. 2021;175:817–826. doi: 10.1001/jamapediatrics.2021.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roseboom J.I., Raghuraman N., Carter E.B., Kelly J.C. Coronavirus disease 2019 and hypertensive disorders of pregnancy. Am J Obstet Gynecol. 2021;224:623–624. doi: 10.1016/j.ajog.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosma S., Carosso A.R., Cusato J., et al. Coronavirus disease 2019 and first-trimester spontaneous abortion: a case-control study of 225 pregnant patients. Am J Obstet Gynecol. 2021;224:391.e1–391.e7. doi: 10.1016/j.ajog.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shende P., Gaikwad P., Gandhewar M., et al. Persistence of SARS-CoV-2 in the first trimester placenta leading to transplacental transmission and fetal demise from an asymptomatic mother. Hum Reprod. 2021;36:899–906. doi: 10.1093/humrep/deaa367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colson A., Depoix C.L., Dessilly G., et al. Coronavirus disease 2019 during pregnancy: clinical and in vitro evidence against placenta infection at term by severe acute respiratory syndrome coronavirus 2. Am J Pathol. 2021 doi: 10.1016/j.ajpath.2021.05.009. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linehan L., O’Donoghue K., Dineen S., White J., Higgins J.R., Fitzgerald B. SARS-CoV-2 placentitis: an uncommon complication of maternal COVID-19. Placenta. 2021;104:261–266. doi: 10.1016/j.placenta.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Royal College of Physicians of Ireland. FoPatIoOaG. Covid placentitis: statement from the RCPI Faculty of Pathology and the Institute of Obstetricians and Gynaecologists Online; 2021.
- 12.Schwartz D.A., Dhaliwal A. Coronavirus diseases in pregnant women, the placenta, fetus, and neonate. Adv Exp Med Biol. 2021;1318:223–241. doi: 10.1007/978-3-030-63761-3_14. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz D.A., Baldewijns M., Benachi A., et al. Chronic histiocytic intervillositis with trophoblast necrosis is a risk factor associated with placental infection from coronavirus disease 2019 (COVID-19) and intrauterine maternal-fetal severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission in live-born and stillborn infants. Arch Pathol Lab Med. 2021;145:517–528. doi: 10.5858/arpa.2020-0771-SA. [DOI] [PubMed] [Google Scholar]
- 14.Argueta LB, Lacko LA, Bram Y, et al. SARS-CoV-2 infects syncytiotrophoblast and activates inflammatory responses in the placenta. bioRxiv. Preprint posted online June 17, 2021. https://doi.org/10.1101/2021.06.01.446676
- 15.Schwartz D.A., Morotti D. Placental pathology of COVID-19 with and without fetal and neonatal infection: trophoblast necrosis and chronic histiocytic intervillositis as risk factors for transplacental transmission of SARS-CoV-2. Viruses. 2020;12:1308. doi: 10.3390/v12111308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pulinx B., Kieffer D., Michiels I., et al. Vertical transmission of SARS-CoV-2 infection and preterm birth. Eur J Clin Microbiol Infect Dis. 2020;39:2441–2445. doi: 10.1007/s10096-020-03964-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debelenko L., Katsyv I., Chong A.M., Peruyero L., Szabolcs M., Uhlemann A.C. Trophoblast damage with acute and chronic intervillositis: disruption of the placental barrier by severe acute respiratory syndrome coronavirus 2. Hum Pathol. 2021;109:69–79. doi: 10.1016/j.humpath.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mongula J.E., Frenken M.W.E., van Lijnschoten G., et al. COVID-19 during pregnancy: non-reassuring fetal heart rate, placental pathology and coagulopathy. Ultrasound Obstet Gynecol. 2020;56:773–776. doi: 10.1002/uog.22189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vivanti A.J., Vauloup-Fellous C., Prevot S., et al. Transplacental transmission of SARS-CoV-2 infection. Nat Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong L., Tian J., He S., et al. Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. JAMA. 2020;323:1846–1848. doi: 10.1001/jama.2020.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosier H., Farhadian S.F., Morotti R.A., et al. SARS-CoV-2 infection of the placenta. J Clin Invest. 2020;130:4947–4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patanè L., Morotti D., Giunta M.R., et al. Vertical transmission of coronavirus disease 2019: severe acute respiratory syndrome coronavirus 2 RNA on the fetal side of the placenta in pregnancies with coronavirus disease 2019-positive mothers and neonates at birth. Am J Obstet Gynecol MFM. 2020;2:100145. doi: 10.1016/j.ajogmf.2020.100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penfield C.A., Lighter J., Roman A.S. Intrauterine vertical transmissibility of SARS-CoV-2: the evidence is evolving. Am J Obstet Gynecol MFM. 2020;2:100227. doi: 10.1016/j.ajogmf.2020.100227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamouroux A., Attie-Bitach T., Martinovic J., Leruez-Ville M., Ville Y. Evidence for and against vertical transmission for severe acute respiratory syndrome coronavirus 2. Am J Obstet Gynecol. 2020;223:91.e1–91.e4. doi: 10.1016/j.ajog.2020.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alzamora M.C., Paredes T., Caceres D., Webb C.M., Valdez L.M., La Rosa M. Severe COVID-19 during pregnancy and possible vertical transmission. Am J Perinatol. 2020;37:861–865. doi: 10.1055/s-0040-1710050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng H., Xu C., Fan J., et al. Antibodies in infants born to mothers with COVID-19 pneumonia. JAMA. 2020;323:1848–1849. doi: 10.1001/jama.2020.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adhikari E.H., Moreno W., Zofkie A.C., et al. Pregnancy outcomes among women with and without severe acute respiratory syndrome coronavirus 2 infection. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.29256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dumitriu D., Emeruwa U.N., Hanft E., et al. Outcomes of neonates born to mothers with severe acute respiratory syndrome coronavirus 2 infection at a large medical center in New York City. JAMA Pediatr. 2021;175:157–167. doi: 10.1001/jamapediatrics.2020.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edlow A.G., Li J.Z., Collier A.Y., et al. Assessment of maternal and neonatal SARS-CoV-2 viral load, transplacental antibody transfer, and placental pathology in pregnancies during the COVID-19 pandemic. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.30455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flaherman V.J., Afshar Y., Boscardin J., et al. Infant outcomes following maternal infection with SARS-CoV-2: first report from the PRIORITY study. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1411. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotlyar A.M., Tal O., Tal R. Vertical transmission of coronavirus disease 2019, a response. Am J Obstet Gynecol. 2021;224:329–330. doi: 10.1016/j.ajog.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullins E., Hudak M.L., Banerjee J., et al. Pregnancy and neonatal outcomes of COVID-19: coreporting of common outcomes from PAN-COVID and AAP-SONPM registries. Ultrasound Obstet Gynecol. 2021;57:573–581. doi: 10.1002/uog.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenneson A., Cannon M.J. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol. 2007;17:253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 34.Honein M.A., Dawson A.L., Petersen E.E., et al. Birth defects among fetuses and infants of US women with evidence of possible Zika virus infection during pregnancy. JAMA. 2017;317:59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- 35.Dontigny L., Arsenault M.Y., Martel M.J. Clinical Practice Obstetrics Committee. Rubella in pregnancy. J Obstet Gynaecol Can. 2008;30:152–158. doi: 10.1016/S1701-2163(16)32740-2. [DOI] [PubMed] [Google Scholar]
- 36.Connor E.M., Sperling R.S., Gelber R., et al. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. N Engl J Med. 1994;331:1173–1180. doi: 10.1056/NEJM199411033311801. [DOI] [PubMed] [Google Scholar]
- 37.Taglauer E., Benarroch Y., Rop K., et al. Consistent localization of SARS-CoV-2 spike glycoprotein and ACE2 over TMPRSS2 predominance in placental villi of 15 COVID-19 positive maternal-fetal dyads. Placenta. 2020;100:69–74. doi: 10.1016/j.placenta.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tolu L.B., Ezeh A., Feyissa G.T. Vertical transmission of severe acute respiratory syndrome coronavirus 2: a scoping review. PLoS One. 2021;16 doi: 10.1371/journal.pone.0250196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hecht J.L., Quade B., Deshpande V., et al. SARS-CoV-2 can infect the placenta and is not associated with specific placental histopathology: a series of 19 placentas from COVID-19-positive mothers. Mod Pathol. 2020;33:2092–2103. doi: 10.1038/s41379-020-0639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stonoga E.T.S., de Almeida Lanzoni L., Rebutini P.Z., et al. Intrauterine transmission of SARS-CoV-2. Emerg Infect Dis. 2021;27:638–641. doi: 10.3201/eid2702.203824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valk J.E., Chong A.M., Uhlemann A.C., Debelenko L. Detection of SARS-CoV-2 in placental but not fetal tissues in the second trimester. J Perinatol. 2021;41:1184–1186. doi: 10.1038/s41372-020-00877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Algarroba G.N., Rekawek P., Vahanian S.A., et al. Visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am J Obstet Gynecol. 2020;223:275–278. doi: 10.1016/j.ajog.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldsmith C.S., Miller S.E., Martines R.B., Bullock H.A., Zaki S.R. Electron microscopy of SARS-CoV-2: a challenging task. Lancet. 2020;395:e99. doi: 10.1016/S0140-6736(20)31188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Algarroba G.N., Rekawek P., Vahanian S.A., et al. Alternative interpretation to the findings reported in visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am J Obstet Gynecol. 2020;223:786–788. doi: 10.1016/j.ajog.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanna N., Lin X., Thomas K., et al. Underestimation of SARS-CoV-2 infections in placental samples. Am J Obstet Gynecol. 2021 doi: 10.1016/j.ajog.2021.07.010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotlyar A.M., Grechukhina O., Chen A., et al. Vertical transmission of coronavirus disease 2019: a systematic review and meta-analysis. Am J Obstet Gynecol. 2021;224:35–53.e3. doi: 10.1016/j.ajog.2020.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dumitriu D., Gyamfi-Bannerman C. Understanding risk for newborns born to SARS-CoV-2-positive mothers. JAMA. 2021;325:2051–2052. doi: 10.1001/jama.2021.6210. [DOI] [PubMed] [Google Scholar]
- 48.Sexual and reproductive health and research WH. Definition and categorization of the timing of mother-to-child transmission of SARS-C0V-2COVID-19 Scientific briefs. Newletter 2021.
- 49.Pereira L. Congenital viral infection: traversing the uterine-placental interface. Annu Rev Virol. 2018;5:273–299. doi: 10.1146/annurev-virology-092917-043236. [DOI] [PubMed] [Google Scholar]
- 50.Bhat P., Anderson D.A. Hepatitis B virus translocates across a trophoblastic barrier. J Virol. 2007;81:7200–7207. doi: 10.1128/JVI.02371-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.National Institutes of Health COVID-19 treatment guidelines. Clinical Spectrum of SARS-CoV-2 Infection. 2021. https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ Available at: [PubMed]
- 52.Khong T.Y., Mooney E.E., Ariel I., et al. Sampling and definitions of placental lesions: Amsterdam placental workshop group consensus statement. Arch Pathol Lab Med. 2016;140:698–713. doi: 10.5858/arpa.2015-0225-CC. [DOI] [PubMed] [Google Scholar]
- 53.Desai N., Neyaz A., Szabolcs A., et al. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun. 2020;11:6319. doi: 10.1038/s41467-020-20139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das Adhikari U., Eng G., Farcasanu M., et al. Fecal SARS-CoV-2 RNA is associated with decreased COVID-19 survival. Clin Infect Dis. 2021 doi: 10.1093/cid/ciab623. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fajnzylber J., Regan J., Coxen K., et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.
Roberts et al. Consensus definition of SARS-CoV-2 placental infection. Am J Obstet Gynecol 2021.