ABSTRACT
We report the complete genome sequence and base modification analysis of the Shewanella algae type strain CECT 5071 (= OK-1 = ATCC 51192 = DSM 9167 = IAM 14159). The genome is composed of a single chromosome of 4,924,764 bp, with a GC content of 53.10%.
ANNOUNCEMENT
The gammaproteobacterium Shewanella algae was first described as a tetrodotoxin-producing epiphyte of the red alga Jania sp. (1), and the type strain was designated OK-1 following the original nomenclature by Kotaki et al. (2). S. algae is the most relevant human pathogen within the genus Shewanella, causing bacteremia, otitis, skin and soft tissue infections, and a variety of other diseases, with the emergence of multidrug-resistant isolates being a concern (3, 4). S. algae is also known for its ability to produce secondary metabolites like iron-scavenging siderophores (5). Its versatile physiology is considered a hallmark of the genus Shewanella. In the past 3 years, genomes of several clinical and environmental isolates of S. algae have been sequenced (6–9), but no complete genome sequence of a reference strain has been available. We have investigated S. algae from different angles (10–14). Here, we report the complete genome sequence of the S. algae type strain OK-1 (1), which was obtained from the Spanish Type Culture Collection (strain CECT 5071T).
The strain was grown in LB medium at 37°C to exponential phase, and the DNA was isolated with the Genomic-tip 500/G kit (Qiagen). Multiplexed sequencing libraries with a target insert size of 8 kb were prepared using the SMRTbell Express template preparation kit v2.0 and barcoded overhang adapter kit 8A (Pacific Biosciences [PacBio], Menlo Park, CA, USA). After removal of adapter dimers, the libraries were sequenced on a Sequel single-molecule real-time (SMRT) cell following the manufacturer’s recommendations (PacBio). Barcode splitting was done using SMRT Link v8.0 (PacBio) and resulted in 622,115 reads totaling 3,045,548,325 bases, with an N50 value of 7,709 bases. A total of 693,216,159 bases from unique reads were used in the microbial assembly pipeline included in SMRT Link v8.0 (PacBio) with default parameters.
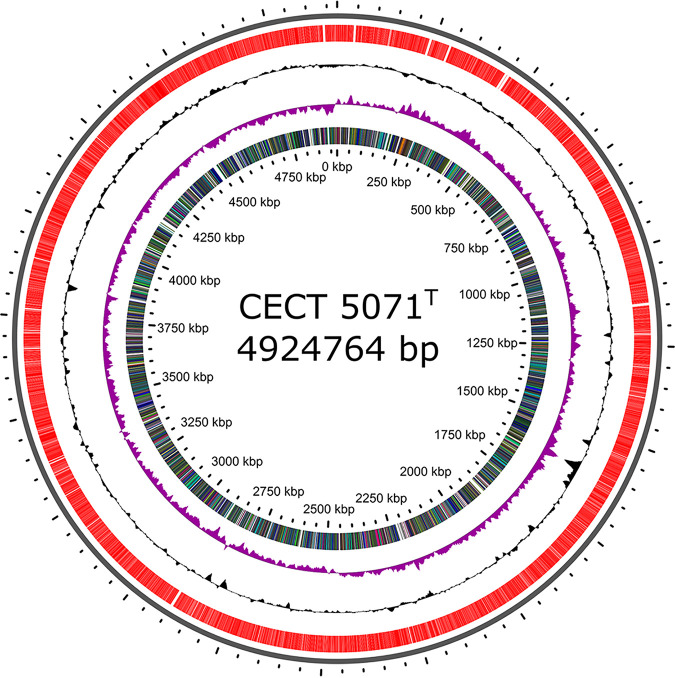
The genome of the S. algae type strain CECT 5071 is composed of a single chromosome of 4,924,764 bp, with a GC content of 53.10% (Fig. 1). No extrachromosomal elements were detected. Genome annotation by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v5.0 (15) predicted 4,400 genes, of which 4,225 are protein-coding genes. A total of 136 RNAs were predicted, including 107 tRNAs, 25 rRNAs (5S, 9 copies; 16S, 8 copies; 23S, 8 copies), and 4 noncoding RNAs. SMRT sequencing offers the possibility of assessing genome-wide DNA methylation patterns. Detected methylation motifs are summarized in Table 1, and the methylome is available in the REBASE database (16).
FIG 1.
Circular map of the Shewanella algae CECT 5071T chromosome. Circles from outside to inside represent the coding sequences (red), GC content (black), GC skew (purple), and color-coded Clusters of Orthologous Groups (COG) assignments of protein-coding genes. The map was generated with CGView (20).
TABLE 1.
Summary of methylation motifs detected in the complete genome sequence of S. algae CECT 5071T and associated methyltransferases
| Motif | Center position | Modification type | No. detected | No. in genome | Type | Methyltransferase locus tag |
|---|---|---|---|---|---|---|
| GATC | 2 | m6A | 41,451 | 41,456 | II | —a |
| TGANNNNNNNTTCC | 3 | m6A | 827 | 827 | Iγ | E1N14_007370 |
| TGGCCA | 4 | m4C | 4,218 | 8,820 | IIα | E1N14_009720 |
—, GATC could not be matched unambiguously by REBASE because there is more than one candidate.
Of note, the rpoS gene is truncated in the type strain of S. algae (10), consistent with evidence from the draft genomes of equivalent strains, namely, ATCC 51192 (GenBank accession no. GCA_012396675.1), NBRC 103173 (GenBank accession no. GCA_001598875.1), and JCM 21037 (GenBank accession no. GCA_000615045.1), suggesting that this truncation was already present in the original OK-1 isolate. In Escherichia coli, the RpoS protein is the stress sigma factor of the RNA polymerase required for stationary-phase transcription (17). However, natural E. coli rpoS mutants exist (18, 19). The ecological and physiological significance of the rpoS truncation in the S. algae type strain remains to be determined.
Data availability.
The complete genome sequence of S. algae CECT 5071T was deposited in DDBJ/ENA/GenBank under the accession no. CP068230. Sequencing raw data are available at the SRA under the accession no. SRR14739658. The methylome of S. algae CECT 5071T is available at the REBASE database under the organism accession no. 46337.
ACKNOWLEDGMENTS
This work was supported by grants from Stiftelsen Lars Hiertas Minne (grant FO2019‐0293), Stiftelsen Längmanska Kulturfonden (grant BA20‐0736), the Karolinska Institutet Research Foundation (grant 2020‐01556), Stiftelsen Anna och Gunnar Vidfelts Fond för Biologisk Forskning (grant 2019-051-Vidfelts fond/SOJOH), and Hans Dahlbergs Stiftelsen för Miljö och Hälsa to A.J.M.-R. M.Y.G. was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health. We acknowledge the National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX for providing assistance in massive parallel sequencing and computational infrastructure. Work performed at NGI/Uppsala Genome Center has been funded by the Council for Research Infrastructure/Swedish Research Council and Science for Life Laboratory, Sweden.
Contributor Information
Alberto J. Martín-Rodríguez, Email: jonatan.martin.rodriguez@ki.se.
David A. Baltrus, University of Arizona
REFERENCES
- 1.Simidu U, Kita-Tsukamoto K, Yasumoto T, Yotsu M. 1990. Taxonomy of four marine bacterial strains that produce tetrodotoxin. Int J Syst Bacteriol 40:331–336. doi: 10.1099/00207713-40-4-331. [DOI] [PubMed] [Google Scholar]
- 2.Kotaki Y, Oshima Y, Yasumoto T. 1985. Bacterial transformation of paralytic shellfish toxins in coral reef crabs and a marine snail. Bull Jpn Soc Sci Fish 51:1009–1013. doi: 10.2331/suisan.51.1009. [DOI] [Google Scholar]
- 3.Martín-Rodríguez AJ, Martín-Pujol O, Artiles-Campelo F, Bolaños-Rivero M, Römling U. 2017. Shewanella spp. infections in Gran Canaria, Spain: retrospective analysis of 31 cases and a literature review. JMM Case Rep 4:e005131. doi: 10.1099/jmmcr.0.005131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janda JM, Abbott SL. 2014. The genus Shewanella: from the briny depths below to human pathogen. Crit Rev Microbiol 40:293–312. doi: 10.3109/1040841X.2012.726209. [DOI] [PubMed] [Google Scholar]
- 5.Rütschlin S, Gunesch S, Böttcher T. 2017. One enzyme, three metabolites: Shewanella algae controls siderophore production via the cellular substrate pool. Cell Chem Biol 24:598–604.e10. doi: 10.1016/j.chembiol.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Chen YJ, Tung KC, Hong YK, Chen SY, Huang YT, Liu PY. 2019. Genome sequence of colistin-resistant bacteremic Shewanella algae carrying the beta-lactamase gene blaOXA-55. Can J Infect Dis Med Microbiol 2019:3840563. doi: 10.1155/2019/3840563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamez AM, McLaughlin RW, Li J, Wan XL, Zheng JS. 2021. Searching for putative virulence factors in the genomes of Shewanella indica and Shewanella algae. Arch Microbiol 203:683–692. doi: 10.1007/s00203-020-02060-1. [DOI] [PubMed] [Google Scholar]
- 8.de Andrade Alves VB, Carvalho E, Madureira PA, Marino ED, Vaz ACN, Vidal AMC, de Azevedo Ruiz VL. 2020. First isolation and whole-genome sequencing of a Shewanella algae strain from a swine farm in Brazil. BMC Microbiol 20:360. doi: 10.1186/s12866-020-02040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endimiani A, Bernasconi OJ, Büdel T, Campos-Madueno EI, Kuenzli E, Hatz C, Carattoli A. 2020. Whole-genome characterization of a Shewanella algae strain coharboring blaCTX-M-15 and armA genes on a novel IncC plasmid. Antimicrob Agents Chemother 64:e00267-20. doi: 10.1128/AAC.00267-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martín-Rodríguez AJ, Suárez-Mesa A, Artiles-Campelo F, Römling U, Hernández M. 2019. Multilocus sequence typing of Shewanella algae isolates identifies disease-causing Shewanella chilikensis strain 6I4. FEMS Microbiol Ecol 95:fiy210. doi: 10.1093/femsec/fiy210. [DOI] [PubMed] [Google Scholar]
- 11.Thorell K, Meier-Kolthoff JP, Sjöling Å, Martín-Rodríguez AJ. 2019. Whole-genome sequencing redefines Shewanella taxonomy. Front Microbiol 10:1861. doi: 10.3389/fmicb.2019.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martín-Rodríguez AJ, Villion K, Yilmaz-Turan S, Vilaplana F, Sjöling Å, Römling U. 2021. Regulation of colony morphology and biofilm formation in Shewanella algae. Microb Biotechnol 14:1183–1200. doi: 10.1111/1751-7915.13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín-Rodríguez AJ, Reyes-Darias JA, Martín-Mora D, González JM, Krell T, Römling U. 2021. Reduction of alternative electron acceptors drives biofilm formation in Shewanella algae. NPJ Biofilms Microbiomes 7:9. doi: 10.1038/s41522-020-00177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martín-Rodríguez AJ, González-Orive A, Hernández-Creus A, Morales A, Dorta-Guerra R, Norte M, Martín VS, Fernández JJ. 2014. On the influence of the culture conditions in bacterial antifouling bioassays and biofilm properties: Shewanella algae, a case study. BMC Microbiol 14:102. doi: 10.1186/1471-2180-14-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W, O’Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C, Marchler-Bauer A, Thibaud-Nissen F. 2021. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res 49:D1020–D1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts RJ, Vincze T, Posfai J, Macelis D. 2015. REBASE: a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter MQ, Louie JW, Huynh S, Parker CT. 2014. Natural rpoS mutations contribute to population heterogeneity in Escherichia coli O157:H7 strains linked to the 2006 US spinach-associated outbreak. Food Microbiol 44:108–118. doi: 10.1016/j.fm.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Uhlich GA, Chen CY, Cottrell BJ, Hofmann CS, Dudley EG, Strobaugh TP, Nguyen LH. 2013. Phage insertion in mlrA and variations in rpoS limit curli expression and biofilm formation in Escherichia coli serotype O157:H7. Microbiology (Reading) 159:1586–1596. doi: 10.1099/mic.0.066118-0. [DOI] [PubMed] [Google Scholar]
- 20.Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of S. algae CECT 5071T was deposited in DDBJ/ENA/GenBank under the accession no. CP068230. Sequencing raw data are available at the SRA under the accession no. SRR14739658. The methylome of S. algae CECT 5071T is available at the REBASE database under the organism accession no. 46337.