The molecule adopts a Z-shaped conformation with the carboxyl group nearly coplanar with the dihydroquinoline unit. In the crystal, two sets of C—H⋯O hydrogen bonds form chains along the b-axis direction, which are connected into corrugated layers parallel to (103) by additional C—H⋯O hydrogen bonds. The layers are connected by C—H⋯π(ring) interactions.
Keywords: crystal structure, alkyne, dihydroquinoline, hydrogen bond, Hirshfeld surface analysis
Abstract
The title molecule, C20H15NO3, adopts a Z-shaped conformation with the carboxyl group nearly coplanar with the dihydroquinoline unit. In the crystal, corrugated layers are formed by C—H⋯O hydrogen bonds and are stacked by C—H⋯π(ring) interactions. Hirshfeld surface analysis indicates that the most important contributions to the crystal packing are from H⋯H (43.3%), H⋯C/C⋯H (26.6%) and H⋯O/O⋯H (16.3%) interactions. The optimized structure calculated using density functional theory at the B3LYP/ 6–311 G(d,p) level is compared with the experimentally determined structure in the solid state. The calculated HOMO–LUMO energy gap is 4.0319 eV.
Chemical context
Nitrogen-based structures have attracted increased attention in recent years because of their interesting properties in structural and inorganic chemistry (Chkirate et al., 2019 ▸, 2020a
▸,b
▸, 2021 ▸). The family of quinolines, particularly those containing the 2-oxoquinoline moiety, is important in medicinal chemistry because of their wide range of pharmacological applications including as potential anticancer agents (Fang et al., 2021 ▸), anti-proliferative agents (Banu et al., 2017 ▸) and as potent modulators of ABCB1-related drug resistance of mouse T-lymphoma cells (Filali Baba et al., 2020 ▸). In particular, 2-oxoquinoline-4-carboxylate derivatives are active antioxidants (Filali Baba et al., 2019 ▸). Given the wide range of therapeutic applications for such compounds, and in a continuation of the work already carried out on the synthesis of compounds resulting from quinolin-2-one (Bouzian et al., 2020 ▸), a similar approach gave the title compound, benzyl 2-oxo-1-(prop-2-yn-1-yl)-1,2-dihydroquinoline-4-carboxylate, (I). Besides the synthesis, we also report the molecular and crystalline structures along with a Hirshfeld surface analysis and a density functional theory computational calculation carried out at the B3LYP/6– 311 G(d,p) level.
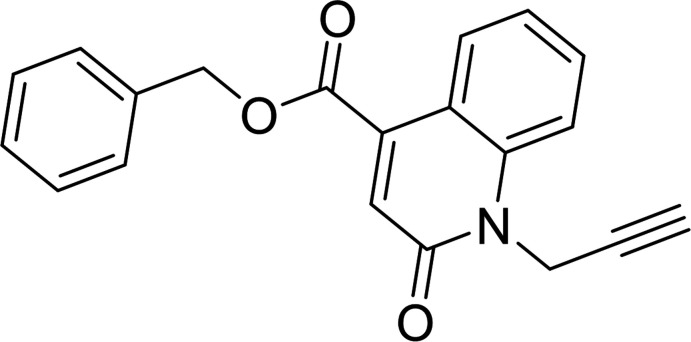
Structural commentary
The molecule adopts a Z-shaped conformation with the propynyl and benzyl substituents projecting from opposite sides of the mean plane of the dihydroquinoline moiety. This moiety is planar to within 0.0340 (6) Å (r.m.s. deviation = 0.0164) with N1 and C9 being, respectively, 0.0340 (6) and −0.0279 (7) Å from the mean plane, resulting in a slight twist at this location. The carboxyl group is nearly coplanar with the dihydroquinoline as seen from the 1.04 (5)° dihedral angle between the plane defined by C7/C13/O2/O3 and that of the dihydroquinoline (C1–C9/N1/O1). This is likely due, in part, to the intramolecular C5—H5⋯O2 interaction (Table 1 ▸ and Fig. 1 ▸). The propynyl substituent is rotated out of the mean plane of the dihydroquinoline moiety by 80.88 (3)°. The plane of the C15–C20 ring is inclined to that of the dihydroquinoline by 68.47 (2)°.
Table 1. Hydrogen-bond geometry (Å, °).
Cg2 and Cg3 are the centroids of the C1–C6 and C15–C20 benzene rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2⋯Cg3i | 0.95 | 2.94 | 3.8206 (10) | 154 |
| C4—H4⋯O2ii | 0.95 | 2.57 | 3.4846 (11) | 162 |
| C5—H5⋯O2 | 0.95 | 2.23 | 2.8917 (11) | 126 |
| C12—H12⋯O1iii | 0.95 | 2.25 | 3.1463 (14) | 157 |
| C14—H14A⋯Cg2iv | 0.99 | 2.65 | 3.4652 (9) | 140 |
| C16—H16⋯O1v | 0.95 | 2.50 | 3.3443 (12) | 148 |
Symmetry codes: (i) -x+1, -y+1, -z+1; (ii) -x, -y+1, -z+1; (iii) -x+{\script{3\over 2}}, y+{\script{1\over 2}}, -z+{\script{1\over 2}}; (iv) -x+{\script{1\over 2}}, y-{\script{1\over 2}}, -z+{\script{1\over 2}}; (v) -x+{\script{3\over 2}}, y-{\script{1\over 2}}, -z+{\script{1\over 2}}.
Figure 1.
The title molecule with labeling scheme and 50% probability ellipsoids. The intramolecular hydrogen bond is depicted by a dashed line.
Supramolecular features
In the crystal, C12—H12⋯O1 and C16—H16⋯O1 hydrogen bonds (Table 1 ▸) link the molecules into zigzag chains extending along the b-axis direction, which are connected by inversion-related pairs of C4—H4⋯O2 hydrogen bonds (Table 1 ▸) into corrugated layers parallel to the (103) plane (Fig. 2 ▸). The layers are stacked along the normal to (103) with C2—H2⋯Cg3 and C14—H14A⋯Cg2 interactions (Table 1 ▸ and Fig. 3 ▸).
Figure 2.
A portion of one layer viewed along the c axis with C—H⋯O hydrogen bonds depicted by dashed lines.
Figure 3.
Packing viewed parallel to (103) with the b axis horizontal and running from left to right. C—H⋯O hydrogen bonds and C—H⋯π(ring) interactions are depicted, respectively, by black and green dashed lines.
Hirshfeld surface analysis
The CrystalExplorer program (Turner et al., 2017 ▸) was used to investigate and visualize further the intermolecular interactions of (I). The Hirshfeld surface plotted over d norm in the range −0.3677 to 1.3896 a.u. is shown in Fig. 4 ▸ a. The electrostatic potential using the STO-3G basis set at the Hartree–Fock level of theory and mapped on the Hirshfeld surface over the range of ±0.05 a.u. clearly shows the positions of close intermolecular contacts in the compound (Fig. 4 ▸ b). The positive electrostatic potential (blue region) over the surface indicates hydrogen-donor potential, whereas the hydrogen-bond acceptors are represented by negative electrostatic potential (red region).
Figure 4.
(a) View of the three-dimensional Hirshfeld surface of the title compound, plotted over d norm in the range of −0.3677 to 1.3896 a.u. (b) View of the three-dimensional Hirshfeld surface of the title compound plotted over electrostatic potential energy in the range −0.0500 to 0.0500 a.u. using the STO-3 G basis set at the Hartree–Fock level of theory.
The overall two-dimensional fingerprint plot (McKinnon et al., 2007 ▸) is shown in Fig. 5 ▸ a, while those delineated into H⋯H, H⋯C/C⋯H, H⋯O/O⋯H, C⋯C, O⋯C/C⋯O, H⋯N/N⋯H, N⋯C/C⋯N and N⋯O/O⋯N contacts are illustrated in Fig. 5 ▸ b–i, respectively, together with their relative contributions to the Hirshfeld surface (HS). The most important interaction is H⋯H, contributing 43.3% to the overall crystal packing, which is reflected in Fig. 5 ▸ b as widely scattered points of high density due to the large hydrogen content of the molecule, with its tip at d e = d i = 1.19 Å. In the presence of C—H interactions, the pair of characteristic wings in the fingerprint plot delineated into H⋯C/C⋯H contacts (26.6% contribution to the HS, Fig. 5 ▸ c) has tips at d e + d i = 3.07 Å. The pair of scattered points of spikes in the fingerprint plot delineated into H⋯O/O⋯H contacts (Fig. 5 ▸ d, 16.3%) have tips at d e + d i = 2.08 Å. The C⋯C contacts (Fig. 5 ▸ e, 10.4%) have tips at d e + d i = 3.34 Å. The O⋯C/C⋯O contacts, Fig. 5 ▸ f, contribute 1.5% to the HS and appear as a pair of scattered points of spikes with tips at d e + d i = 3.55 Å. The H⋯N/N⋯H contacts (Fig. 5 ▸ g, 1.3%) have tips at d e + d i = 3.28 Å. Finally, the C⋯N/N⋯C and O⋯N/N⋯ O contacts, Fig. 5 ▸ h–i, contribute only 0.5% and 0.1% respectively to the HS and have a low-density distribution of points.
Figure 5.
The full two-dimensional fingerprint plots for the title compound, showing (a) all interactions, and delineated into (b) H⋯H, (c) H⋯C/C⋯H, (d) H⋯O/O⋯H, (e) C⋯C, (f) O⋯C/C⋯O, (g) H⋯N/N⋯H, (h) N⋯C/C⋯N and (i) N⋯O/O⋯N interactions. di and de values are the closest internal and external distances (in Å) from given points on the Hirshfeld surface.
Density Functional Theory calculations
The structure in the gas phase of the title compound was optimized by means of density functional theory. The density functional theory calculation was performed by the hybrid B3LYP method and the 6–311 G(d,p) basis-set, which is based on Becke’s model (Becke, 1993 ▸) and considers a mixture of the exact (Hartree–Fock) and density functional theory exchange utilizing the B3 functional, together with the LYP correlation functional (Lee et al., 1988 ▸). After obtaining the converged geometry, the harmonic vibrational frequencies were calculated at the same theoretical level to confirm that the number of imaginary frequencies is zero for the stationary point. Both the geometry optimization and harmonic vibrational frequency analysis of the title compound were performed with the Gaussian 09 program (Frisch et al., 2009 ▸). Theoretical and experimental results related to bond lengths and angles are in good agreement, and are summarized in Table 2 ▸. Calculated numerical values for the title compound including electronegativity (χ), hardness (η), ionization potential (I), dipole moment (μ), electron affinity (A), electrophilicity (ω) and softness (σ) are collated in Table 3 ▸. The electron transition from the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO) energy level is shown in Fig. 6 ▸. The HOMO and LUMO are localized in the plane extending over the whole benzyl 2-oxo-1-(prop-2-yn-1-yl)-1,2-dihydroquinoline-4-carboxylate system. The energy band gap (ΔE = E LUMO − E HOMO) of the molecule is 4.0319 eV, and the frontier molecular orbital energies, E HOMO and E LUMO, are −6.3166 and −2.2847 eV, respectively.
Table 2. Comparison (X-ray and DFT) of selected bond lengths and angles (Å, °).
| X-ray | B3LYP/6–311G(d,p) | |
|---|---|---|
| O1—C9 | 1.2355 (10) | 1.223 |
| O3—C13 | 1.3375 (10) | 1.3447 |
| N1—C9 | 1.3788 (10) | 1.4042 |
| N1—C10 | 1.4730 (10) | 1.4725 |
| O2—C13 | 1.2058 (10) | 1.2092 |
| O3—C14 | 1.4588 (10) | 1.4611 |
| N1—C1 | 1.3999 (10) | 1.3953 |
| C13—O3—C14 | 116.87 (7) | 117.1258 |
| C9—N1—C10 | 115.85 (6) | 115.6313 |
| N1—C1—C2 | 119.87 (7) | 120.5532 |
| O1—C9—N1 | 121.42 (7) | 121.7499 |
| N1—C9—C8 | 116.04 (7) | 115.2168 |
| O2—C13—C7 | 125.74 (7) | 125.0357 |
| O3—C14—C15 | 112.63 (7) | 111.678 |
| C9—N1—C1 | 123.16 (6) | 123.4431 |
| C1—N1—C10 | 120.93 (6) | 120.911 |
| N1—C1—C6 | 120.08 (6) | 120.1155 |
| O1—C9—C8 | 122.54 (7) | 123.0317 |
| C11—C10—N1 | 111.46 (7) | 113.9875 |
| O2—C13—O3 | 123.21 (7) | 123.6586 |
| O3—C13—C7 | 111.05 (6) | 111.3015 |
Table 3. Calculated energies.
| Molecular energy | Compound (I) |
|---|---|
| Total energy TE (eV) | −28621.0571 |
| E HOMO (eV) | −6.3166 |
| E LUMO (eV) | −2.2847 |
| Gap, ΔE (eV) | 4.0319 |
| Dipole moment, μ (Debye) | 1.9469 |
| Ionization potential, I (eV) | 6.3166 |
| Electron affinity, A | 2.2847 |
| Electronegativity, χ | 4.3007 |
| Hardness, η | 2.0160 |
| Electrophilicity index, ω | 4.5873 |
| Softness, σ | 0.4960 |
| Fraction of electron transferred, ΔN | 0.6695 |
Figure 6.
The energy band gap of benzyl 2-oxo-1-(prop-2-yn-1-yl)-1,2-dihydroquinoline-4-carboxylate.
Database survey
A search of the Cambridge Structural Database (CSD version 5.42, updated May 2021; Groom et al., 2016 ▸) with the 2-oxo-1-(prop-2-yn-1-yl)-1,2-dihydroquinoline-4-carboxylate fragment yielded multiple matches. Of these, two had an alkyl substituent on O3 comparable to (I). The first compound (refcode OKIGAT; Hayani et al., 2021 ▸) carries an ethyl group on O3, while the second one (refcode OKIGOH; Hayani et al., 2021 ▸) carries a cyclohexyl group. The ethyl carboxylate in OKIGAT forms a dihedral angle of −8.3 (7)° with the dihydroquinoline unit. In OKIGOH, the dihedral angle between the mean planes of the cyclohexyl carboxylate and dihydroquinoline rings is 37.3 (8)°. As previously mentioned, the carboxyl group in (I) is nearly coplanar with the dihydroquinoline [dihedral angle of 1.04 (5)°], which is approximately the same as in OKIGAT, but less tilted than in OKIGOH.
Synthesis and crystallization
A mixture of 2-oxo-1-(prop-2-yn-1-yl)-1,2-dihydroquinoline-4-carboxylic acid (0.7 g, 3 mmol), K2CO3 (0.51 g, 3.6 mmol), benzyl chloride (0.76 ml, 6 mmol) and tetra n-butylammonium bromide as a catalyst in DMF (30 mL) was stirred at room temperature for 48 h. After removal of the salts by filtration, the solvent was evaporated under reduced pressure and the residue obtained was dissolved in dichloromethane. The organic phase was dried over Na2SO4 and concentrated under vacuum. The crude product obtained was purified by chromatography on a column of silica gel (eluent: hexane/ ethyl acetate: 9/1). 1H NMR (300 MHz, DMSO-d 6) δ ppm: 3.08 (t, 1H, CH≡); 4.37 (d, 2H, CH2—N); 5.12 (s, 2H, CH2—O); 7.08–8.74 (m, 10H, CHarom); 13C NMR (75 MHz, DMSO-d 6) δ ppm: 34.3 (CH3—N); 66.2 (CH2—O); 72.1 (–C≡); 73.2 (CH≡); 115.6-148.7 (CHarom and Cquat arom); 162. 5 (C=Oquinol); 168.2 (C=Ocarboxyl). MS (ESI): m/z = 318 (M + H)+.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. H atoms attached to carbon were placed in calculated positions (C—H = 0.95–1.00 Å), and were included as riding contributions with isotropic displacement parameters 1.2 or 1.5 times those of the attached atoms. Two reflections affected by the beamstop were omitted from the final refinement.
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C20H15NO3 |
| M r | 317.33 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 150 |
| a, b, c (Å) | 8.2284 (3), 13.7693 (4), 13.9230 (4) |
| β (°) | 96.155 (1) |
| V (Å3) | 1568.37 (9) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.09 |
| Crystal size (mm) | 0.44 × 0.35 × 0.32 |
| Data collection | |
| Diffractometer | Bruker D8 QUEST PHOTON 3 diffractometer |
| Absorption correction | Numerical (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.93, 0.97 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 80207, 6020, 5304 |
| R int | 0.025 |
| (sin θ/λ)max (Å−1) | 0.774 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.046, 0.133, 1.03 |
| No. of reflections | 6020 |
| No. of parameters | 217 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.46, −0.21 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989021007416/tx2040sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021007416/tx2040Isup3.hkl
Supporting information file. DOI: 10.1107/S2056989021007416/tx2040Isup3.cml
CCDC reference: 2097267
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
JTM thanks Tulane University for support of the Tulane Crystallography Laboratory. Authors’ contributions are as follows. Conceptualization, YB; methodology, YB and NHA; investigation, KC; theoretical calculations, KC; writing (original draft), KC; writing (review and editing of the manuscript), FHAO; supervision, EME; crystal-structure determination and validation, JTM.
supplementary crystallographic information
Crystal data
| C20H15NO3 | F(000) = 664 |
| Mr = 317.33 | Dx = 1.344 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.2284 (3) Å | Cell parameters from 9939 reflections |
| b = 13.7693 (4) Å | θ = 2.5–33.3° |
| c = 13.9230 (4) Å | µ = 0.09 mm−1 |
| β = 96.155 (1)° | T = 150 K |
| V = 1568.37 (9) Å3 | Block, colourless |
| Z = 4 | 0.44 × 0.35 × 0.32 mm |
Data collection
| Bruker D8 QUEST PHOTON 3 diffractometer | 6020 independent reflections |
| Radiation source: fine-focus sealed tube | 5304 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.025 |
| Detector resolution: 7.3910 pixels mm-1 | θmax = 33.4°, θmin = 2.9° |
| φ and ω scans | h = −12→12 |
| Absorption correction: numerical (SADABS; Krause et al., 2015) | k = −21→21 |
| Tmin = 0.93, Tmax = 0.97 | l = −21→21 |
| 80207 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.133 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0739P)2 + 0.3685P] where P = (Fo2 + 2Fc2)/3 |
| 6020 reflections | (Δ/σ)max = 0.001 |
| 217 parameters | Δρmax = 0.46 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Experimental. The diffraction data were obtained from 9 sets of frames, each of width 0.5° in ω or φ, collected with scan parameters determined by the "strategy" routine in APEX3. The scan time was 7 sec/frame. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. H-atoms attached to carbon were placed in calculated positions (C—H = 0.95 - 1.00 Å). All were included as riding contributions with isotropic displacement parameters 1.2 - 1.5 times those of the attached atoms. Two reflections affected by the beamstop were omitted from the final refinement. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.79130 (8) | 0.56932 (5) | 0.29079 (6) | 0.03386 (17) | |
| O2 | 0.15346 (8) | 0.39853 (5) | 0.36012 (6) | 0.03159 (16) | |
| O3 | 0.34027 (8) | 0.35057 (5) | 0.26450 (5) | 0.02547 (14) | |
| N1 | 0.63622 (8) | 0.63640 (5) | 0.39987 (5) | 0.01811 (12) | |
| C1 | 0.49088 (9) | 0.64053 (5) | 0.44357 (5) | 0.01656 (13) | |
| C2 | 0.46483 (10) | 0.71713 (6) | 0.50694 (6) | 0.02098 (14) | |
| H2 | 0.544815 | 0.766658 | 0.518906 | 0.025* | |
| C3 | 0.32280 (11) | 0.72050 (6) | 0.55192 (6) | 0.02469 (16) | |
| H3 | 0.306866 | 0.771800 | 0.595567 | 0.030* | |
| C4 | 0.20287 (10) | 0.64925 (7) | 0.53375 (6) | 0.02508 (16) | |
| H4 | 0.106080 | 0.651845 | 0.565229 | 0.030* | |
| C5 | 0.22538 (10) | 0.57466 (6) | 0.46961 (6) | 0.02108 (14) | |
| H5 | 0.142234 | 0.527154 | 0.456509 | 0.025* | |
| C6 | 0.36952 (9) | 0.56798 (5) | 0.42340 (5) | 0.01629 (13) | |
| C7 | 0.40167 (8) | 0.49162 (5) | 0.35579 (5) | 0.01659 (13) | |
| C8 | 0.54184 (9) | 0.49270 (6) | 0.31341 (6) | 0.02043 (14) | |
| H8 | 0.560136 | 0.442273 | 0.269253 | 0.025* | |
| C9 | 0.66584 (10) | 0.56733 (6) | 0.33219 (6) | 0.02148 (15) | |
| C10 | 0.76472 (10) | 0.71018 (6) | 0.42091 (6) | 0.02241 (15) | |
| H10A | 0.775440 | 0.725750 | 0.490720 | 0.027* | |
| H10B | 0.870536 | 0.683646 | 0.405222 | 0.027* | |
| C11 | 0.72727 (11) | 0.79935 (6) | 0.36498 (7) | 0.02598 (17) | |
| C12 | 0.69298 (13) | 0.87027 (8) | 0.31880 (9) | 0.0356 (2) | |
| H12 | 0.665529 | 0.927061 | 0.281827 | 0.043* | |
| C13 | 0.28317 (9) | 0.41024 (5) | 0.32886 (6) | 0.01886 (14) | |
| C14 | 0.23208 (11) | 0.27253 (7) | 0.22635 (6) | 0.02652 (17) | |
| H14A | 0.266692 | 0.250123 | 0.164117 | 0.032* | |
| H14B | 0.119369 | 0.298095 | 0.213628 | 0.032* | |
| C15 | 0.23223 (10) | 0.18782 (6) | 0.29404 (6) | 0.02316 (15) | |
| C16 | 0.36596 (12) | 0.12502 (8) | 0.30625 (7) | 0.0318 (2) | |
| H16 | 0.457268 | 0.135594 | 0.271197 | 0.038* | |
| C17 | 0.36579 (17) | 0.04679 (9) | 0.36979 (9) | 0.0430 (3) | |
| H17 | 0.457907 | 0.004905 | 0.378888 | 0.052* | |
| C18 | 0.2320 (2) | 0.02998 (9) | 0.41959 (9) | 0.0502 (3) | |
| H18 | 0.232480 | −0.023368 | 0.462895 | 0.060* | |
| C19 | 0.09713 (18) | 0.09064 (9) | 0.40660 (9) | 0.0442 (3) | |
| H19 | 0.004674 | 0.078535 | 0.440243 | 0.053* | |
| C20 | 0.09774 (12) | 0.16940 (7) | 0.34402 (7) | 0.02989 (19) | |
| H20 | 0.005332 | 0.211073 | 0.335298 | 0.036* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0257 (3) | 0.0298 (3) | 0.0496 (4) | −0.0088 (3) | 0.0203 (3) | −0.0130 (3) |
| O2 | 0.0222 (3) | 0.0270 (3) | 0.0474 (4) | −0.0083 (2) | 0.0127 (3) | −0.0108 (3) |
| O3 | 0.0263 (3) | 0.0234 (3) | 0.0276 (3) | −0.0089 (2) | 0.0067 (2) | −0.0090 (2) |
| N1 | 0.0171 (3) | 0.0150 (3) | 0.0224 (3) | −0.0026 (2) | 0.0026 (2) | −0.0004 (2) |
| C1 | 0.0176 (3) | 0.0148 (3) | 0.0171 (3) | 0.0000 (2) | 0.0009 (2) | 0.0012 (2) |
| C2 | 0.0242 (3) | 0.0177 (3) | 0.0208 (3) | −0.0006 (3) | 0.0015 (3) | −0.0023 (2) |
| C3 | 0.0270 (4) | 0.0233 (4) | 0.0240 (3) | 0.0028 (3) | 0.0041 (3) | −0.0051 (3) |
| C4 | 0.0219 (3) | 0.0277 (4) | 0.0265 (4) | 0.0025 (3) | 0.0070 (3) | −0.0035 (3) |
| C5 | 0.0182 (3) | 0.0223 (3) | 0.0232 (3) | 0.0000 (2) | 0.0040 (2) | −0.0010 (3) |
| C6 | 0.0160 (3) | 0.0154 (3) | 0.0174 (3) | 0.0005 (2) | 0.0011 (2) | 0.0012 (2) |
| C7 | 0.0165 (3) | 0.0145 (3) | 0.0186 (3) | −0.0011 (2) | 0.0013 (2) | 0.0007 (2) |
| C8 | 0.0196 (3) | 0.0164 (3) | 0.0260 (3) | −0.0028 (2) | 0.0059 (3) | −0.0032 (2) |
| C9 | 0.0191 (3) | 0.0178 (3) | 0.0284 (4) | −0.0025 (2) | 0.0068 (3) | −0.0027 (3) |
| C10 | 0.0197 (3) | 0.0192 (3) | 0.0281 (4) | −0.0051 (3) | 0.0013 (3) | −0.0009 (3) |
| C11 | 0.0237 (3) | 0.0219 (4) | 0.0332 (4) | −0.0062 (3) | 0.0072 (3) | 0.0002 (3) |
| C12 | 0.0313 (4) | 0.0291 (4) | 0.0484 (6) | −0.0021 (4) | 0.0138 (4) | 0.0106 (4) |
| C13 | 0.0183 (3) | 0.0167 (3) | 0.0213 (3) | −0.0018 (2) | 0.0008 (2) | −0.0001 (2) |
| C14 | 0.0295 (4) | 0.0249 (4) | 0.0247 (4) | −0.0092 (3) | 0.0008 (3) | −0.0067 (3) |
| C15 | 0.0230 (3) | 0.0217 (3) | 0.0250 (3) | −0.0049 (3) | 0.0034 (3) | −0.0082 (3) |
| C16 | 0.0268 (4) | 0.0337 (5) | 0.0345 (4) | 0.0020 (3) | 0.0016 (3) | −0.0126 (4) |
| C17 | 0.0522 (7) | 0.0316 (5) | 0.0426 (6) | 0.0117 (5) | −0.0067 (5) | −0.0085 (4) |
| C18 | 0.0824 (10) | 0.0299 (5) | 0.0385 (6) | −0.0013 (6) | 0.0066 (6) | 0.0026 (4) |
| C19 | 0.0619 (7) | 0.0333 (5) | 0.0409 (6) | −0.0112 (5) | 0.0214 (5) | −0.0035 (4) |
| C20 | 0.0309 (4) | 0.0253 (4) | 0.0355 (4) | −0.0054 (3) | 0.0123 (3) | −0.0084 (3) |
Geometric parameters (Å, º)
| O1—C9 | 1.2355 (10) | C8—H8 | 0.9500 |
| O2—C13 | 1.2058 (10) | C10—C11 | 1.4687 (12) |
| O3—C13 | 1.3375 (10) | C10—H10A | 0.9900 |
| O3—C14 | 1.4588 (10) | C10—H10B | 0.9900 |
| N1—C9 | 1.3788 (10) | C11—C12 | 1.1865 (14) |
| N1—C1 | 1.3999 (10) | C12—H12 | 0.9500 |
| N1—C10 | 1.4730 (10) | C14—C15 | 1.4995 (13) |
| C1—C2 | 1.4062 (10) | C14—H14A | 0.9900 |
| C1—C6 | 1.4192 (10) | C14—H14B | 0.9900 |
| C2—C3 | 1.3846 (11) | C15—C20 | 1.3922 (12) |
| C2—H2 | 0.9500 | C15—C16 | 1.3955 (13) |
| C3—C4 | 1.3953 (12) | C16—C17 | 1.3940 (17) |
| C3—H3 | 0.9500 | C16—H16 | 0.9500 |
| C4—C5 | 1.3864 (11) | C17—C18 | 1.382 (2) |
| C4—H4 | 0.9500 | C17—H17 | 0.9500 |
| C5—C6 | 1.4116 (10) | C18—C19 | 1.385 (2) |
| C5—H5 | 0.9500 | C18—H18 | 0.9500 |
| C6—C7 | 1.4543 (10) | C19—C20 | 1.3914 (16) |
| C7—C8 | 1.3507 (10) | C19—H19 | 0.9500 |
| C7—C13 | 1.5062 (10) | C20—H20 | 0.9500 |
| C8—C9 | 1.4520 (11) | ||
| C13—O3—C14 | 116.87 (7) | N1—C10—H10A | 109.3 |
| C9—N1—C1 | 123.16 (6) | C11—C10—H10B | 109.3 |
| C9—N1—C10 | 115.85 (6) | N1—C10—H10B | 109.3 |
| C1—N1—C10 | 120.93 (6) | H10A—C10—H10B | 108.0 |
| N1—C1—C2 | 119.87 (7) | C12—C11—C10 | 178.18 (10) |
| N1—C1—C6 | 120.08 (6) | C11—C12—H12 | 180.0 |
| C2—C1—C6 | 120.05 (7) | O2—C13—O3 | 123.21 (7) |
| C3—C2—C1 | 120.12 (7) | O2—C13—C7 | 125.74 (7) |
| C3—C2—H2 | 119.9 | O3—C13—C7 | 111.05 (6) |
| C1—C2—H2 | 119.9 | O3—C14—C15 | 112.63 (7) |
| C2—C3—C4 | 120.62 (7) | O3—C14—H14A | 109.1 |
| C2—C3—H3 | 119.7 | C15—C14—H14A | 109.1 |
| C4—C3—H3 | 119.7 | O3—C14—H14B | 109.1 |
| C5—C4—C3 | 119.80 (8) | C15—C14—H14B | 109.1 |
| C5—C4—H4 | 120.1 | H14A—C14—H14B | 107.8 |
| C3—C4—H4 | 120.1 | C20—C15—C16 | 119.00 (9) |
| C4—C5—C6 | 121.24 (7) | C20—C15—C14 | 120.58 (8) |
| C4—C5—H5 | 119.4 | C16—C15—C14 | 120.41 (8) |
| C6—C5—H5 | 119.4 | C17—C16—C15 | 120.11 (10) |
| C5—C6—C1 | 118.15 (7) | C17—C16—H16 | 119.9 |
| C5—C6—C7 | 124.21 (7) | C15—C16—H16 | 119.9 |
| C1—C6—C7 | 117.65 (6) | C18—C17—C16 | 120.18 (11) |
| C8—C7—C6 | 119.87 (6) | C18—C17—H17 | 119.9 |
| C8—C7—C13 | 117.37 (7) | C16—C17—H17 | 119.9 |
| C6—C7—C13 | 122.76 (6) | C17—C18—C19 | 120.23 (11) |
| C7—C8—C9 | 123.06 (7) | C17—C18—H18 | 119.9 |
| C7—C8—H8 | 118.5 | C19—C18—H18 | 119.9 |
| C9—C8—H8 | 118.5 | C18—C19—C20 | 119.68 (11) |
| O1—C9—N1 | 121.42 (7) | C18—C19—H19 | 120.2 |
| O1—C9—C8 | 122.54 (7) | C20—C19—H19 | 120.2 |
| N1—C9—C8 | 116.04 (7) | C19—C20—C15 | 120.76 (10) |
| C11—C10—N1 | 111.46 (7) | C19—C20—H20 | 119.6 |
| C11—C10—H10A | 109.3 | C15—C20—H20 | 119.6 |
| C9—N1—C1—C2 | −175.56 (7) | C1—N1—C9—C8 | −4.77 (11) |
| C10—N1—C1—C2 | 1.37 (10) | C10—N1—C9—C8 | 178.16 (7) |
| C9—N1—C1—C6 | 3.97 (11) | C7—C8—C9—O1 | −177.70 (9) |
| C10—N1—C1—C6 | −179.10 (7) | C7—C8—C9—N1 | 2.64 (12) |
| N1—C1—C2—C3 | −178.65 (7) | C9—N1—C10—C11 | 96.66 (8) |
| C6—C1—C2—C3 | 1.82 (11) | C1—N1—C10—C11 | −80.48 (9) |
| C1—C2—C3—C4 | −1.16 (13) | C14—O3—C13—O2 | 4.71 (12) |
| C2—C3—C4—C5 | −0.41 (13) | C14—O3—C13—C7 | −175.41 (7) |
| C3—C4—C5—C6 | 1.33 (13) | C8—C7—C13—O2 | 179.78 (8) |
| C4—C5—C6—C1 | −0.66 (11) | C6—C7—C13—O2 | −1.07 (12) |
| C4—C5—C6—C7 | 179.70 (7) | C8—C7—C13—O3 | −0.10 (10) |
| N1—C1—C6—C5 | 179.56 (7) | C6—C7—C13—O3 | 179.05 (7) |
| C2—C1—C6—C5 | −0.91 (11) | C13—O3—C14—C15 | −80.18 (10) |
| N1—C1—C6—C7 | −0.77 (10) | O3—C14—C15—C20 | 107.82 (9) |
| C2—C1—C6—C7 | 178.76 (7) | O3—C14—C15—C16 | −73.57 (10) |
| C5—C6—C7—C8 | 178.41 (7) | C20—C15—C16—C17 | −1.82 (13) |
| C1—C6—C7—C8 | −1.23 (11) | C14—C15—C16—C17 | 179.56 (8) |
| C5—C6—C7—C13 | −0.72 (11) | C15—C16—C17—C18 | 1.23 (16) |
| C1—C6—C7—C13 | 179.64 (6) | C16—C17—C18—C19 | 0.10 (18) |
| C6—C7—C8—C9 | 0.28 (12) | C17—C18—C19—C20 | −0.80 (19) |
| C13—C7—C8—C9 | 179.45 (7) | C18—C19—C20—C15 | 0.19 (17) |
| C1—N1—C9—O1 | 175.57 (8) | C16—C15—C20—C19 | 1.12 (14) |
| C10—N1—C9—O1 | −1.50 (12) | C14—C15—C20—C19 | 179.74 (9) |
Hydrogen-bond geometry (Å, º)
Cg2 and Cg3 are the centroids of the C1–C6 and C15–C20 benzene rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···Cg3i | 0.95 | 2.94 | 3.8206 (10) | 154 |
| C4—H4···O2ii | 0.95 | 2.57 | 3.4846 (11) | 162 |
| C5—H5···O2 | 0.95 | 2.23 | 2.8917 (11) | 126 |
| C12—H12···O1iii | 0.95 | 2.25 | 3.1463 (14) | 157 |
| C14—H14A···Cg2iv | 0.99 | 2.65 | 3.4652 (9) | 140 |
| C16—H16···O1v | 0.95 | 2.50 | 3.3443 (12) | 148 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x, −y+1, −z+1; (iii) −x+3/2, y+1/2, −z+1/2; (iv) −x+1/2, y−1/2, −z+1/2; (v) −x+3/2, y−1/2, −z+1/2.
References
- Banu, S., Bollu, R., Bantu, R., Nagarapu, L., Polepalli, S., Jain, N., Vangala, R. & Manga, V. (2017). Eur. J. Med. Chem. 125, 400–410. [DOI] [PubMed]
- Becke, A. D. (1993). J. Chem. Phys. 98, 5648–5652.
- Bouzian, Y., Karrouchi, K., Sert, Y., Lai, C.-H., Mahi, L., Ahabchane, N. H., Talbaoui, A., Mague, J. T. & Essassi, E. M. (2020). J. Mol. Struct. 1209, 127940.
- Brandenburg, K. & Putz, H. (2012). DIAMOND, Crystal Impact GbR, Bonn, Germany.
- Bruker (2020). APEX3 and SAINT. Bruker AXS LLC, Madison, Wisconsin, USA.
- Chkirate, K., Azgaou, K., Elmsellem, H., El Ibrahimi, B., Sebbar, N. K., Anouar, E. H., Benmessaoud, M., El Hajjaji, S. & Essassi, E. M. (2021). J. Mol. Liq. 321, 114750.
- Chkirate, K., Fettach, S., El Hafi, M., Karrouchi, K., Elotmani, B., Mague, J. T., Radi, S., Faouzi, M. E. A., Adarsh, N. N., Essassi, E. M. & Garcia, Y. (2020a). J. Inorg. Biochem. 208, 21–28. [DOI] [PubMed]
- Chkirate, K., Fettach, S., Karrouchi, K., Sebbar, N. K., Essassi, E. M., Mague, J. T., Radi, S., Faouzi, M. E. A., Adarsh, N. N. & Garcia, Y. (2019). J. Inorg. Biochem. 191, 21–28. [DOI] [PubMed]
- Chkirate, K., Karrouchi, K., Dege, N., Sebbar, N. K., Ejjoummany, A., Radi, S., Adarsh, N. N., Talbaoui, A., Ferbinteanu, M., Essassi, E. M. & Garcia, Y. (2020b). New J. Chem. 44, 2210–2221.
- Fang, Y., Wu, Z., Xiao, M., Wei, L., Li, K., Tang, Y., Ye, J., Xiang, J. & Hu, A. (2021). Bioorg. Chem. 106, 104469. [DOI] [PubMed]
- Filali Baba, Y., Misbahi, H., Kandri Rodi, Y., Ouzidan, Y., Essassi, E. M., Vincze, K., Nové, M., Gajdács, M., Molnár, J., Spengler, G. & Mazzah, A. (2020). Chem. Data Collect. 29, 100501.
- Filali Baba, Y., Sert, Y., Kandri Rodi, Y., Hayani, S., Mague, J. T., Prim, D., Marrot, J., Ouazzani Chahdi, F., Sebbar, N. K. & Essassi, E. M. (2019). J. Mol. Struct. 1188, 255–268.
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloino, J., Zheng, G., Sonnenberg, J. L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery, J. A. Jr, Peralta, J. E., Ogliaro, F., Bearpark, M., Heyd, J. J., Brothers, E., Kudin, K. N., Staroverov, V. N., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A., Burant, J. C., Iyengar, S. S., Tomasi, J., Cossi, M., Rega, N., Millam, J. M., Klene, M., Knox, J. E., Cross, J. B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R. E., Yazyev, O., Austin, A. J., Cammi, R., Pomelli, C., Ochterski, J. W., Martin, R. L., Morokuma, K., Zakrzewski, V. G., Voth, G. A., Salvador, P., Dannenberg, J. J., Dapprich, S., Daniels, A. D., Farkas, O., Foresman, J. B., Ortiz, J. V., Cioslowski, J. & Fox, D. J. (2009). Gaussian 09. Revision A. 02. Gaussian Inc, Wallingford, CT, US.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hayani, S., Sert, Y., Filali Baba, Y., Benhiba, F., Ouazzani Chahdi, F., Laraqui, F.-Z., Mague, J. T., El Ibrahimi, B., Sebbar, N. K., Kandri Rodi, Y. & Essassi, E. M. (2021). J. Mol. Struct. 1227, 129520.
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Lee, C., Yang, W. & Parr, R. G. (1988). Phys. Rev. B, 37, 785–789. [DOI] [PubMed]
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. The University of Western Australia.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989021007416/tx2040sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989021007416/tx2040Isup3.hkl
Supporting information file. DOI: 10.1107/S2056989021007416/tx2040Isup3.cml
CCDC reference: 2097267
Additional supporting information: crystallographic information; 3D view; checkCIF report








