Abstract
We present the first deep learning method to segment Multiple Sclerosis lesions and brain structures from MRI scans of any (possibly multimodal) contrast and resolution. Our method only requires segmentations to be trained (no images), as it leverages the generative model of Bayesian segmentation to generate synthetic scans with simulated lesions, which are then used to train a CNN. Our method can be retrained to segment at any resolution by adjusting the amount of synthesised partial volume. By construction, the synthetic scans are perfectly aligned with their labels, which enables training with noisy labels obtained with automatic methods. The training data are generated on the fly, and aggressive augmentation (including artefacts) is applied for improved generalisation. We demonstrate our method on two public datasets, comparing it with a state-of-the-art Bayesian approach implemented in FreeSurfer, and dataset specific CNNs trained on real data. The code is available at https://github.com/BBillot/SynthSeg.
Keywords: MS lesion, segmentation, contrast-agnostic
1. INTRODUCTION
Multiple Sclerosis (MS) is a presumed autoimmune disorder affecting one in ten thousand people in western countries [1]. MS attacks the central nervous system via a demyelinating process, which causes physical and mental disabilities. Additionally, MS correlates with abnormally fast cerebral atrophy, especially for the cortex and deep grey matter structures [2]. Because of its remarkable ability to image soft tissue and reveal white matter lesions, magnetic resonance imaging (MRI) is the technique of choice to diagnose MS and monitor its progression. Moreover, MRI is arguably the best technique to evaluate brain atrophy, and volumetric measurements obtained from MRI scans can be used to assess the efficacy of treatments [3]. For these reasons, joint segmentation of lesions and brain structures from MRI scans is of high importance in tracking MS progression. Although manual delineation is the gold standard, it remains time-consuming and suffers from inter- and intra-rater variability issues [4]. Therefore, there is great value in automated tools for accurate, fast, and reproducible segmentation of lesions and brain anatomy.
State-of-the-art methods for MS lesion segmentation mostly build on recent advances in convolutional neural networks (CNNs) [5, 6]. However, CNNs generalise poorly to test scans of unseen resolution or contrast, even within the same modality. Even if augmentation strategies can improve their robustness within modality [7, 8], supervised methods still need to be retrained for every new combination of contrast and resolution, often requiring new labelled data. This is especially problematic when analysing MS data, which are often acquired in clinical settings and thus may vary considerably in terms of resolution, pulse sequence, hardware, etc.
In comparison, Bayesian segmentation methods are robust to changes in contrast, and their unsupervised variants can segment lesions in any modality [9]. State-of-the-art results have been achieved with an atlas-based approach [10], where a variational autoencoder (VAE) is used as prior to model the spatial distribution of the lesions. However, these methods are slow compared to CNN-based approaches, and may be fragile when used on clinical scans with high slice thickness and thus subject to partial volume (PV) effects.
Here we present the first CNN to segment MS lesions and brain regions from (possibly multimodal) MRI of any contrast and resolution, without requiring any new data. Building on our recent work [11, 12], we train contrast-agnostic CNNs on synthetic scans with simulated lesions, to segment at any target resolution, by adjusting the amount of synthesised PV. Since the synthetic scans are built from training label maps, and thus perfectly aligned with them, we can use noisy segmentations obtained with automated methods. This enables us to train with public datasets, which have labels for MS lesions, but not for the brain anatomy. The results show that our method is more robust than CNNs trained with real scans, and segments lesions as accurately as the state-of-the-art in Bayesian contrast-adaptive methods, while being more accurate for brain regions and running 2 orders of magnitude faster.
2. METHODS
Segmenting MS lesions is challenging due to their highly varying appearance and location. We build on our recently proposed segmentation framework, which achieves great flexibility by training CNNs with synthetic scans [11, 12]. These scans are sampled on the fly with a generative model inspired from Bayesian segmentation, which uses a Gaussian mixture model (GMM) conditioned on label maps. Here, we adapt this method to MS lesions by modelling their appearance with a separate Gaussian component, while learning their spatial distribution from manual lesion annotations available from public datasets [4, 13]. Since the synthetic scans are perfectly aligned with their labels, we complement the training segmentations with automated rather than manual labels.
2.1. Generative model
In training, we assume the availability of a pool of 3D segmentations of J voxels at high resolution rh, with labels for MS lesions and brain structures (Fig. 1a). Image-segmentation pairs are generated at every minibatch as follows. First, we randomly select a segmentation S from the pool, and spatially deform it into L with a diffeomorphic transform ϕ, parametrised by θϕ: L = S ∘ ϕ(θϕ) (Fig 1b). The transform ϕ is the composition of three rotations, translations, shears, scalings, and a nonlinear transform obtained by integrating a random, smooth stationary velocity field [14].
Fig. 1.
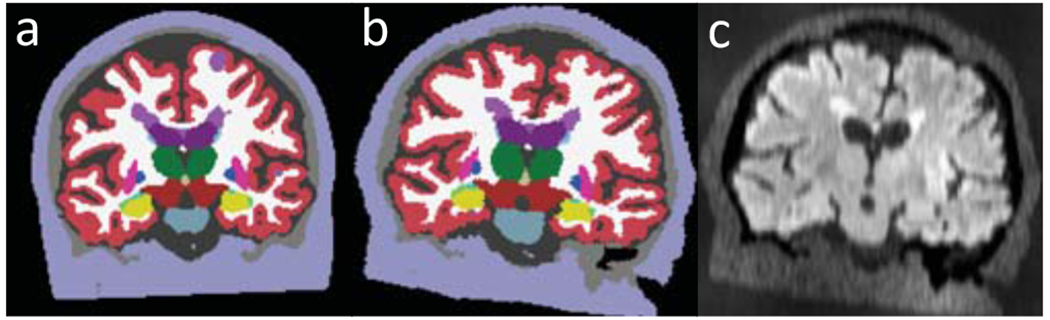
Generation of a FLAIR scan: (a) Training label map with MS lesions (bright purple); the segmentation of the brain regions are automated and thus imperfect. (b) Spatial augmentation. (c) GMM sampling, artefact and PV modelling.
We then generate an intensity image G (possibly multimodal), by sampling a GMM of K classes conditioned on L, which we corrupt with a random, smooth bias field B defined in logarithmic domain and parametrised by θB:
| (1) |
where j indexes voxels and θG = {μk, ∑k}1≤k≤K groups all the parameters of the GMM. Next, we normalise G between 0 and 1, and raise its voxel intensities by a random power γ (centred around 1) to yield an image Ih at high resolution rh.
We then model resolution and PV by forming a low resolution image Il defined on a coarser grid of J′ < J voxels. We first simulate voxel thickness by independently blurring each channel n of Ih with a Gaussian kernel of standard deviation σn that divides the power by 10 at the cut-off frequency:
| (2) |
where is the target low resolution of the n-th channel, and α is a random factor (centred around 1) that increases the variability of σn, thus making the method robust to deviations from the nominal target resolution. Il is obtained by down-sampling each blurred channel to to simulate slice spacing. Finally, we upsample Il back to rh (, Fig. 1c). This upsampling step, also applied at test time, enables to train the CNN with (, L) pairs, so it learns to segment at high resolution.
2.2. Parameter sampling and lesion modelling
The parameters of the generative model are all independently sampled at each minibatch for enhanced augmentation. In practice, θϕ, θB, γ, α are drawn from uniform distributions of wide range to expose the segmentation CNN to highly varied samples, thus improving its generalisation ability.
For the GMM parameters θG, modelling the contrasts of lesions and anatomy, we consider three sampling strategies:
SynthSeg:
means and variances are drawn from uniform priors (25, 255) and (5, 25). This approach yields scans of random contrast, which we use to train modality-agnostic networks [11] to segment anatomy and lesions of any contrast.
SynthSeg-rule:
θG is sampled from modality-specific normal prior distributions with hyperparameters coarsely estimated from a few scans of the target contrast and resolution. Specifically, we (i) segment them with a publicly available tool [10], (ii) use these labels to robustly estimate means and variances for each label class, (iii) correct the variances to account for the resolution gap between rh and , and (iv) artificially increase the variances by a factor of 5 for improved generalisation. SynthSeg-rule aims to simulate realistic contrasts for lesions and anatomical regions in a given target modality.
SynthSeg-mix:
we randomly select between the two previous strategies at each minibatch with equal probability. This method combines the tailored contrast modelling of SynthSeg-rule with the higher generalisation of SynthSeg.
2.3. Learning
We train a different network for each new target resolution (and modality, depending on the sampling of θG), using the same architecture as in our previous works [11, 12]. Specifically, we use a 3D UNet [15] with 5 levels, each consisting in 2 convolutional layers (3×3×3 kernels, ELU activations) and batch normalisation. The first layer counts 24 kernels, and this number is doubled after each max-pooling and halved after each upsampling. Probabilistic predictions are obtained by appending a softmax activation layer. The loss function is the average soft Dice computed over all predicted labels. Both the generative model and the network are coded in Tensorflow.
3. EXPERIMENTS AND RESULTS
3.1. Experimental setup
We use two publicly available datasets in our experiments:
MSSeg:
15 unpreprocessed T1 and FLAIR scans of varying resolution, resampled to 1 mm isotropic, with consensus lesion tracings obtained from seven human raters [4]. We use these to apply a lesion filling method to the T1 scans [16], which are then processed with FreeSurfer to obtain whole-brain segmentations [17]. These are combined with the consensus tracings into “silver standard” labels for evaluation.
ISBI15:
15 skull-stripped T1 (0.82 × 0.82 × 1.17 mm) and FLAIR scans (0.82 × 0.82 × 2.2 mm), resampled to 1 mm isotropic [13]. Using distance maps, we combine manual labels of the lesions from two raters into consensus tracings. These are merged with FreeSurfer segmentations (computed as above) to obtain silver standard labels for evaluation.
We evaluate the three variants of our method, namely SynthSeg, SynthSeg-rule and SynthSeg-mix, with two experiments. First, we conduct a cross-validation study on the MSSeg dataset, by dividing it in 3 folds of 5 subjects each. We separately segment the T1, FLAIR, and multimodal T1-FLAIR scans of each fold after having trained our method on the labels of the two remaining folds. Importantly, we train two SynthSeg models for each fold (one for the unimodal scans, and one for the T1-FLAIR pairs), whereas we train SynthSeg-rule and SynthSeg-mix three times each (once for every modality) with intensity distributions estimated on the scans of the corresponding training folds. We assess performance by computing the Dice scores for the MS lesions and 12 representative brain regions of interest (ROI).
In the second experiment we evaluate the robustness of our method by retraining every model on all MSSeg scans, and testing them on the held-out ISBI15 dataset. As before, we compute Dice scores obtained for 1 mm resolution segmentations of the T1, FLAIR and T1-FLAIR scans. However, we now train different SynthSeg models for the T1 and FLAIR scans, as they do not have the same native resolutions.
We compare the proposed approach against two competing methods. First, we train supervised CNNs on real annotated images of the target contrast and resolution. Importantly, we use the same 3D UNet architecture and apply the same augmentation as for our method. We emphasise that this approach requires supervised training data, which is seldom available for a given combination of contrast and resolution. We also compare our method against the state-of-the-art Bayesian tool for MS lesion and anatomy segmentation “SAMSEG-lesion” [10], which does not require any retraining. Even if its lesion model is trained on a private dataset of 212 scans (more than 10 times larger than any of our training sets) [10], SAMSEG-lesion is a natural competitor to the SynthSeg variants as it is fully contrast adaptive, only requires label maps to be trained, and segments most brain regions.
3.2. Results
The box plots of Figure 2a and 2b show that SynthSeg and its variants accurately segment MS lesions and brain ROIs, despite having only been trained on synthetic data generated from automated segmentations of brain anatomy. These scores also approach the inter-rater precision median Dice score of 0.68 (computed on the seven manual delineations). Interestingly, training with random or realistic contrasts yields very similar results (no statistical difference for two-sided non-parametric Wilcoxon signed-rank test), while mixing both strategies leads to significant improvements (p < 0.01 for all scores). Compared to the state-of-the-art tool SAMSEG-lesion, our method obtains similar results for MS lesions (no statistical difference, except with SynthSeg-mix for FLAIR), and is more accurate for brain ROIs while running two orders of magnitude faster. Being trained on the exact intensities, the supervised CNNs considerably outperform the other approaches (except SynthSeg-mix for the lesions, where no statistical difference can be inferred), but are only an option when supervised training data are available.
Fig. 2.
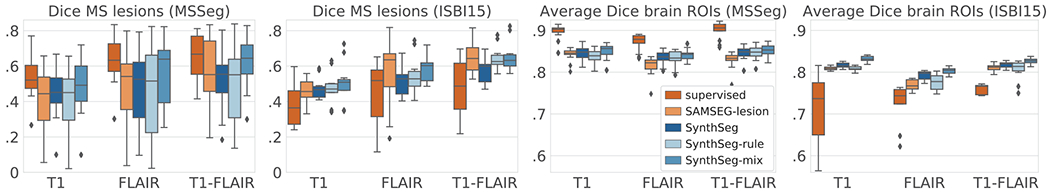
(a) Box plots of the cross-validation Dice scores for the MS lesions and (c) for the average over 12 brain ROIs: cerebral cortex and white matter, lateral ventricle, cerebellar cortex and white matter, thalamus, caudate, putamen, pallidum, brainstem, hippocampus, and amygdala. (b) and (d) show the results obtained when training on MSSeg and testing on ISBI15.
The robustness of the proposed method is demonstrated in the second experiment, where SynthSeg and its variants sustain their high performances when tested on the ISBI15 dataset (Fig. 2c,d and 3). In contrast, the scores of the supervised CNNs drastically degrade compared to the first experiment, thus showing poor generalisation abilities, even within the same modality (Fig. 3b). Although it was trained on a much larger dataset, SAMSEG-lesion achieves a level of performance similar to our method for MS lesions (no statistical difference), but is significantly outperformed for the brain ROIs. For example, SAMSEG-lesion segments the cerebral cortex poorly (red arrows in Figure 3c), which is remarkably well recovered by all the variants of SynthSeg.
Fig. 3.

Segmentation of an ISBI15 FLAIR scan: (a) ground truth, (b) supervised, (c) SAMSEG-lesion, (d) SynthSeg, (e) SynthSeg-rule, (f) SynthSeg-mix. MS lesions are in bright purple. Arrows indicate major segmentation errors (yellow for MS lesions, red for brain ROIs).
4. CONCLUSION
We have presented the first deep learning method for joint segmentation of MS lesions and whole-brain regions for scans of any contrast at any predefined resolution, without requiring any new supervised data. Despite being trained on partly automated labels, our method achieves state-of-the-art results in contrasts-agnostic segmentation, and remarkably generalises to unseen datasets. While mixing synthetic scans of realistic and random contrasts in training gives slightly more accurate results for a fixed contrast and resolution, we found that using totally random contrasts enables to readily segment scans of any contrast at a given resolution. Future work will focus on extending the proposed method to other types of white matter lesions (e.g., in stroke), and further enriching the model by replacing manual segmentations of lesions with randomly generated masks. Thanks to its flexibility, we believe that the presented approach has the potential to facilitate large studies on the progression of MS with clinical data.
5. ACKNOWLEDGEMENTS
Supported by the EU (ERC Starting Grant 677697, Marie Curie 765148), the EPSRC (EP-L016478/1), and the NIH (1RF1MH123195-01, 1R01AG0-64027-01A1, R01NS112161).
Footnotes
6. COMPLIANCE WITH ETHICAL STANDARDS
We used publicly available datasets of brain MRI and ethical approval was not required as per the corresponding licenses.
7. REFERENCES
- [1].Confavreux C et al. “Rate of Pregnancy-Related Relapse in MS,” J. of Med, vol. 339, pp. 285–291, 1998. [DOI] [PubMed] [Google Scholar]
- [2].Fisher E et al. “Gray matter atrophy in multiple sclerosis study,” Annals of Neur, vol. 64, pp. 255–265, 2008. [DOI] [PubMed] [Google Scholar]
- [3].Barkhof F et al. “Imaging outcomes for neuroprotection and repair in multiple sclerosis trials,” Nature Reviews. Neurology, 5, pp. 256–266, 2009. [DOI] [PubMed] [Google Scholar]
- [4].Commowick O et al. “Objective Evaluation of MS Lesion Segmentation using a Data Management and Processing Infrastructure,” Scientific Reports, vol. 8, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kamnitsas K et al. “Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation,” Medical Im. Analysis, vol. 36, pp. 61–78, 2017. [DOI] [PubMed] [Google Scholar]
- [6].McKinley R et al. “Automatic detection of lesion load change in MS using convolutional neural networks with segmentation confidence,” NeuroImage, vol. 25, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Valverde S et al. “One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks,” NeuroImage, vol. 21, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Salem M et al. “Multiple Sclerosis Lesion Synthesis in MRI Using an Encoder-Decoder U-NET,” IEEE Access, vol. 7, pp. 25171–25184, 2019. [Google Scholar]
- [9].Sudre C et al. “Bayesian model selection for pathological neuroimaging data applied to white matter lesion segmentation,” IEEE TMI, vol. 34, pp. 79–102, 2015. [DOI] [PubMed] [Google Scholar]
- [10].Cerri S et al. “A Contrast-Adaptive Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis,” NeuroImage, 2020, (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Billot B et al. “A Learning Strategy for Contrast-agnostic MRI Segmentation,” in MIDL, 2020. [Google Scholar]
- [12].Billot B et al. “PV segmentation of brain MRI scans of any resolution and contrast,” in MICCAI, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Carass A et al. “Longitudinal MS lesion segmentation data resource,” D. in Brief, vol. 12, pp. 346–350, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Arsigny V et al. “A Log-Euclidean Framework for Statistics on Diffeomorphisms,” in MICCAI, 2006. [DOI] [PubMed] [Google Scholar]
- [15].Ronneberger O et al. “U-Net: Convolutional Networks for Biomedical Image Segmentation,” in MICCAI, 2015. [Google Scholar]
- [16].Prados F et al. “A multi-time-point modality agnostic patch-based method for lesion filling in multiple sclerosis,” NeuroImage, vol. 139, pp. 376–384, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fischl B, “FreeSurfer,” NeuroImage, vol. 62, pp. 774–781, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]


