Abstract
Antibiotic resistance in bacteria limits the effect of corresponding antibiotics, and the classification of antibiotic resistance genes (ARGs) is important for the treatment of bacterial infections and for understanding the dynamics of microbial communities. Although several methods have been developed to classify ARGs, none of them work well when the ARGs diverge from those in the reference ARG databases. We develop a novel method, ARG-SHINE, for ARG classification. ARG-SHINE utilizes state-of-the-art learning to rank machine learning approach to ensemble three component methods with different features, including sequence homology, protein domain/family/motif and raw amino acid sequences for the deep convolutional neural network. Compared with other methods, ARG-SHINE achieves better performance on two benchmark datasets in terms of accuracy, macro-average f1-score and weighted-average f1-score. ARG-SHINE is used to classify newly discovered ARGs through functional screening and achieves high prediction accuracy. ARG-SHINE is freely available at https://github.com/ziyewang/ARG_SHINE.
INTRODUCTION
With the wide spread use and misuse of antibiotics in clinical and agricultural practices, antibiotic resistance (AR), the resistance of bacterial pathogens to antimicrobials, has become an urgent public health problem (1,2). According to US Centers for Disease Control (CDC), over 2.8 million people are infected by AR pathogens and over 35 000 people die from antimicrobial resistance each year in US alone. An estimated annual cost of $20–35 billion is spent on antibiotic-resistant pathogens (https://www.cdc.gov/drugresistance/index.html). Therefore, it is essential to find the antibiotic-resistant genes (ARGs) from the clinical and environmental samples and to identify ARGs’ type to develop targeted treatment or control measures (3–6). Moreover, the rapid identification of ARGs in the pathogens can help optimize the antibacterial treatment (3).
Culture-based antimicrobial susceptibility testing (AST) can provide phenotypic resistance results of the microbes, but it may take weeks, and it is less informative than sequencing-based methods in terms of resistance gene epidemiology (7). Culture-based methods are not applicable to the unculturable bacteria (8). Functional metagenomics approaches select antibiotic resistance DNA sequences in a metagenomic library by transforming the candidate fragments into the recombinant expressed host and exposing the host to antimicrobials (7,9). However, the original host and the recombinant expression host with the same gene may have different phenotypes for the antimicrobials, limiting the use of functional genomics approaches. Finally, the selected fragments need to be annotated by alignment-based methods or machine learning methods.
With the development of next-generation sequencing (NGS) technologies, sequencing-based methods for antimicrobial resistance identification spring up as a complement to culture-based and functional metagenomics methods (7). According to the input, sequencing-based ARG identification methods can be divided into two categories: assembly-based methods and read-based methods. In assembly-based methods, the reads are first assembled into contiguous regions (contigs) using assembly programs and then these contigs are aligned to known reference ARG databases or hidden Markov models (HMM) for ARGs. The read-based methods, on the other hand, directly map the reads to sequences in reference ARG databases. Boolchandani et al. (7) presented an excellent review on experimental and computational methods for ARG identification in NGS data.
Although these alignment-based and map-based methods can successfully identify known ARGs in the reference databases, they cannot identify or classify ARGs that are highly different from those in the reference databases resulting in high rate of false negatives. To overcome this issue, two machine learning based methods, DeepARG (10) and TRAC (11), have been developed to classify ARGs into different classes. DeepARG aligns a query sequence to the reference ARG database to obtain the similarity distribution of a query sequence to known ARGs and uses the similarity distribution as features for a deep learning model. DeepARG was shown to be able to identify and classify ARGs that do not have high similarity with sequences in the reference ARG database. To further increase the classification accuracy for ARGs, Hamid et al. (11) first built an antibiotic resistance gene database, COALA, by integrating 15 available antibiotic resistance gene databases. They then developed a transfer learning based deep neural network model, TRAC (TRansfer learning for Antibiotic resistance gene Classification), for ARG classification and showed that TRAC achieved much better performance than other alignment-based methods and their self-attention based recurrent neural network model (11). Despite the successes of DeepARG and TRAC on the classification of ARG sequences, they possess several limitations that can be further improved to increase ARG classification accuracy.
Currently available ARG classification methods have several limitations. First, the protein functional information is not used for ARG classification. Protein domains/motifs are the fundamental units of the proteins and they contain information about the ARG classes. However, neither DeepARG nor TRAC uses protein domain/family/motif information for ARG classification. Second, currently available ARG classification methods use only one source of information, either alignment to known ARGs or amino acid composition, but do not integrate the predictions from different approaches. Although DeepARG (10) uses low identity cutoff and a deep learning method, it does not learn representation over raw sequences, limiting its performance. TRAC (11) learns representation over raw sequences using deep Recurrent Neural Network (RNN). However, the authors did not evaluate the performance of the methods for the proteins with high sequence identity scores against the database. Furthermore, the machine learning methods may not work as well as the alignment-based methods on the sequences with close homolog against the genes in the ARG database (see Supplementary Table S1 and Table 4 for details). We hypothesize that ARG classification accuracy can be improved by integrating multiple data sources.
Table 4.
The prediction accuracy of the different methods on the sequences of the COALA90 test data stratified by identity scores against the training database
| Identity score | None | ≤50% | >50% |
|---|---|---|---|
| Number of sequences | 142 | 338 | 1223 |
| BLAST best hit | 0.0000 | 0.6243 | 0.9542 |
| DIAMOND best hit | 0.0000 | 0.5740 | 0.9534 |
| DeepARG | 0.0000 | 0.5266 | 0.9419 |
| HMMER | 0.0563 | 0.2751 | 0.6051 |
| TRAC | 0.3521 | 0.6124 | 0.9199 |
| ARG-CNN | 0.4577 | 0.6538 | 0.9452 |
| ARG-InterPro | 0.4085 | 0.6509 | 0.9141 |
| ARG-KNN | 0.0000 | 0.6361 | 0.9542 |
| ARG-SHINE | 0.4648 | 0.6864 | 0.9558 |
The best results among all the methods and best results among the stand-alone methods are in bold. The lowest identity score among the test data is 21.32%. ‘None’ means that the sequences do not have any alignment against the training database with the e-value no more than 1e-3.
To overcome the limitations of the available ARG classification methods mentioned above, we developed a novel ensemble method, ARG-SHINE, for antibiotic resistance class prediction. ARG classification is a multi-class prediction problem. Learning to Rank (LTR) (12) is widely used to solve the multi-label prediction problems and has been successfully used for protein function prediction integrating multiple information sources (13,14). A multi-class problem can be regarded as the simplification of the multi-label problem by ranking the correct label before the incorrect labels. Therefore, ARG-SHINE utilizes LTR to integrate three component methods, ARG-CNN, ARG-InterPro and ARG-KNN, using raw sequences, protein functional information, and sequence homology information for ARG classification, respectively.
We developed ARG-CNN, ARG-InterPro and ARG-KNN for ARG classification, as the component methods for the ensemble model. ARG-CNN applies a deep convolutional neural network (CNN) over raw protein sequences. Deep convolutional neural networks have achieved good performance on multiple classical machine learning tasks such as image classification (15,16), object detection (17) and sentence classification (18). Convolutional neural networks can extract local features (19). They are suitable for ARG classification because the phenotype is related to several specific antibiotic resistance mechanisms, such as antibiotic efflux and antibiotic modification. ARG-InterPro applies InterProScan (20) to find the domain, family and motif information from sequences and uses the obtained functional signatures for logistic regression. The domain/family/motif information represents biological domain knowledge. The method has been proven useful in protein function prediction (13). Antibiotic resistance is related to some protein functions, such as antibiotic efflux. Our ARG-InterPro model can be regarded as the version of the corresponding method used in protein function prediction (13) trained on the ARG database. ARG-KNN aligns the sequences against the database generated from the training data with BLAST (21), and the k-nearest neighbor (KNN) method is used for achieving the final classification. The method can utilize homology-based information.
We compared ARG-SHINE and our component methods with several available ARG classification methods including BLAST (21), DIAMOND (22), DeepARG (10), RGI (23), HMMER (24) and TRAC (11). ARG-CNN and ARG-InterPro can achieve better performance compared with other available methods when the query sequence is not highly similar to the known ARG sequences. Our results show that ARG-KNN and BLAST best hit achieve better performance compared with DIAMOND best hit and DeepARG for sequences with high similarity with known ARG sequences. Compared with other methods, ARG-SHINE achieves the best performance on the benchmark datasets in terms of accuracy, macro-average f1-score, and weighted-average f1-score in general. Compared with BLAST best hit, our final model can achieve much better performance on the sequences with low identity scores against the database, and slightly better performance on the sequences with high identity scores against the database.
MATERIALS AND METHODS
In this section, we present (i) the descriptions of the benchmark datasets; (ii) the overview of ARG-SHINE; (iii) the implementation of three proposed component methods used for integration and the ensemble model; (iv) the methods we choose for comparison and (v) the metrics to evaluate the performance.
Datasets
Hamid et al. (11) created the COALA (COllection of ALl Antibiotic resistance gene databases) dataset curated from 15 available antimicrobial gene databases (see Supplementary Material for the details) to provide benchmark datasets for improving antibiotic resistance class prediction. They performed CD-HIT (25) clustering of ARG sequences with different identity thresholds, 40% and 70%, respectively, on the original COALA dataset to remove similar sequences and generated two datasets.
We built the COALA90 dataset to include the sequences with higher identity scores than those used in (11) by performing CD-HIT with 90% identity threshold based on the COALA dataset. A fasta file of the representative sequences for the clusters that CD-HIT generated was kept. After removing the ARG classes containing less than 15 sequences, 17 023 proteins from 16 antibiotic resistance classes remain. Each pair of sequences in the COALA90 dataset has at most 90% identity (using CD-HIT).
In addition, we used CD-HIT with 100% identify threshold to build a complete dataset, COALA100, by just removing the duplicate sequences. We removed ARG classes with less than 45 sequences resulting in 41 851 proteins from 17 ARG classes. We used different thresholds for the numbers of sequences in ARG classes for the COALA90 and COALA100 datasets since the sequences in COALA100 are more similar than that in COALA90. These thresholds yield similar numbers of ARG classes of interest.
To train and evaluate the component methods and the ensemble model, we randomly divided each dataset into four parts: (i) training data for the component methods (70%). We used the training data to build the training database for the BLAST or DIAMOND alignment-based methods; (ii) validation data (10%); (iii) test data (10%) and (iv) training data for the LTR model (10%). Table 1 and Supplementary Table S2 present the names of the ARG classes and the corresponding number of sequences for each ARG class in the COALA90 and COALA100 datasets, respectively.
Table 1.
The numbers of sequences for each class in the COALA90 dataset
| ARG Class | Whole dataset | Training data for component methods | Training data for LTR | Validation data | Test data |
|---|---|---|---|---|---|
| MULTIDRUG | 382 | 263 | 34 | 37 | 48 |
| AMINOGLYCOSIDE | 1189 | 844 | 110 | 122 | 113 |
| MACROLIDE | 756 | 563 | 64 | 66 | 63 |
| BETA-LACTAM | 5845 | 4051 | 606 | 586 | 602 |
| GLYCOPEPTIDE | 2304 | 1638 | 193 | 243 | 230 |
| TRIMETHOPRIM | 666 | 424 | 91 | 71 | 80 |
| FOLATE-SYNTHESIS- | 2448 | 1730 | 249 | 249 | 220 |
| INHIBITOR (FSI) | |||||
| TETRACYCLINE | 2056 | 1448 | 205 | 185 | 218 |
| SULFONAMIDE | 315 | 217 | 32 | 36 | 30 |
| FOSFOMYCIN | 138 | 102 | 15 | 10 | 11 |
| PHENICOL | 460 | 318 | 50 | 46 | 46 |
| QUINOLONE | 229 | 154 | 27 | 23 | 25 |
| STREPTOGRAMIN | 19 | 11 | 2 | 3 | 3 |
| BACITRACIN | 127 | 90 | 16 | 11 | 10 |
| RIFAMYCIN | 23 | 15 | 3 | 3 | 2 |
| MACROLIDE/LINCOSAMIDE/ | 66 | 48 | 5 | 11 | 2 |
| STREPTOGRAMIN (MLS) |
Overview of ARG-SHINE
We developed a novel method, ARG-SHINE, for antibiotic resistance class prediction. Figure 1 shows the framework of ARG-SHINE, which consists of two modules: (i) ‘Component module’ for obtaining the prediction scores of three different component methods: ARG-CNN (deep learning), ARG-InterPro (domain/family/motif), and ARG-KNN (homology) and (ii) ‘Ensemble module’ for integrating the predictions generated from the ‘Component module’ by the learning to rank framework to improve the overall performance. More descriptions of ARG-SHINE are as follows.
Figure 1.
The framework of ARG-SHINE for ARG class prediction. In the first module, we develop three component methods: ARG-CNN, ARG-InterPro and ARG-KNN for the classification of the ARG sequences. In the second module, we use learning to rank (LTR) to integrate the three component prediction scores from the first module for ARG classification (‘None’ means that the query sequence does not have any alignment against the training database with an e-value no more than 1e-3).
The component module: three different component methods using different features—ARG-CNN, ARG-InterPro and ARG-KNN
ARG-CNN: deep Convolutional Neural Network (CNN) model for ARG classification
Supplementary Figure S1 shows the architecture of ARG-CNN. It consists of four main layers: (i) an embedding layer for representing each amino acid with a dense vector; (ii) a convolution layer for capturing local information of the sequences; (iii) a two-layer self-attention network for capturing the most relevant parts of the given protein sequence for ARG classification and (iv) a fully connected layer and softmax output. Cross-entropy loss is used as the loss function during the training process. More details about the training strategy of ARG-CNN is described in the Supplementary Material. The detailed explanations for each part of ARG-CNN are as follows.
Embedding Layer
We used a trainable dense vector to represent each amino acid in the embedding layer, which can capture rich semantic information of amino acids (26). For a given L-length protein p, the output of the embedding layer, 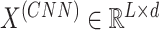 , is as follows:
, is as follows:
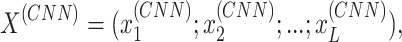 |
(1) |
where 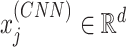 is the d-dimensional embedding of the j-th amino acid in the protein sequence.
is the d-dimensional embedding of the j-th amino acid in the protein sequence.
Convolutional Layer
We used a one-dimensional convolutional layer to extract the local information of the given protein sequence. The output 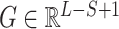 of the convolutional layer for each filter of size S is defined as:
of the convolutional layer for each filter of size S is defined as:
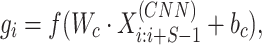 |
(2) |
 |
(3) |
where 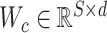 is the weight matrix of the filter, bc is the bias, ‘·’ indicates dot multiplication and f is the ReLU (27) activation function. Then we obtained the output of the convolutional layer
is the weight matrix of the filter, bc is the bias, ‘·’ indicates dot multiplication and f is the ReLU (27) activation function. Then we obtained the output of the convolutional layer 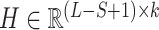 for k filters as follows:
for k filters as follows:
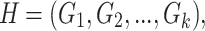 |
(4) |
Self-attention layer
We used a two-layer fully connected neural network to select key input information for processing, and it is similar to the self-attention network used in (11,28). For a given H, to extract r important parts from the sequences using the attention layer, we generated the weights 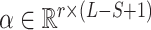 (attentions) for outputs as follows:
(attentions) for outputs as follows:
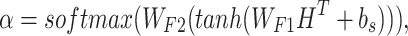 |
(5) |
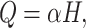 |
(6) |
where 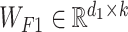 and
and 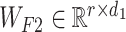 are the weight matrices of the two-layer feed-forward neural network, d1 denotes the number of the hidden units of the network, bs denotes the bias and tanh is the activation function. In Equation (5), softmax() is performed along the second dimension of its input. We flattened the output matrix Q into a T (=r × k) size vector, M.
are the weight matrices of the two-layer feed-forward neural network, d1 denotes the number of the hidden units of the network, bs denotes the bias and tanh is the activation function. In Equation (5), softmax() is performed along the second dimension of its input. We flattened the output matrix Q into a T (=r × k) size vector, M.
Fully connected layer and softmax
Let NC denote the number of antibiotic resistance classes. We used a fully connected layer and softmax to obtain the final prediction scores 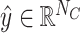 for all classes as follows:
for all classes as follows:
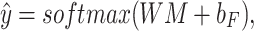 |
(7) |
where 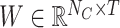 denotes the weight matrix of the fully connected network and
denotes the weight matrix of the fully connected network and 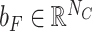 denotes the bias.
denotes the bias.
ARG-InterPro: logistic regression model using InterPro features
The InterPro database integrates protein domains, families, and functional sites from multiple resources (29). First, we ran InterProScan (20) on the sequences to obtain their functional information against the InterPro database. Then, we generated n signatures according to the InterProScan output of the training data, and each signature (InterPro ID) corresponds to a protein domain, family, or functional site. For protein pi, the binary feature vector 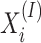 is as follows:
is as follows:
 |
(8) |
where 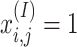 means pi has the j-th signature. We then used the feature vectors of the sequences for multi-class logistic regression.
means pi has the j-th signature. We then used the feature vectors of the sequences for multi-class logistic regression.
ARG-KNN: KNN (k-nearest neighbor) method using BLAST alignment results as input
We aligned the sequences against the training database generated from the training data with BLAST (e-value ≤1e − 3; max_target_seq: 30) to find homologous sequences, as suggested in (30) that protein:protein alignments with expectation values < 1e-3 can reliably be used to infer homology. If the lowest e-value is >1e-3, we say that the protein sequence cannot be aligned to the training database. For each query sequence, the alignment results of the k proteins in the training database with the highest bit scores are kept for classification. For a given query sequence pq, the score for the ARG class Ci, S(Ci, pq) is defined as Equation (9).
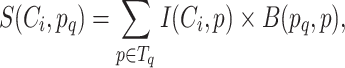 |
(9) |
where Tq denotes the set of proteins having the top k bit scores for pq identified by BLAST, p denotes any protein in Tq, I(Ci, p) is a binary indicator that shows whether p belongs to the ARG class Ci and B(pq, p) is the bit score of the alignment between protein pq and p. The normalized values of S(, pq) by softmax for the ARG classes are used for subsequent integration. The ARG class with the highest score is the result of ARG-KNN.
The ensemble module
ARG-SHINE: an ensemble method for antibiotic resistance class prediction
After generating prediction scores of the three component methods for the antibiotic resistance classes, we used LambdaMART (31), an advanced LTR algorithm, to rank all the antibiotic resistance classes for each sequence. For the sequences that could not be aligned to the training database, ARG-KNN did not provide prediction values, and we used the LTR model trained by the predictions of ARG-CNN and ARG-InterPro to make the prediction. For any other sequence, its identity score is defined as the highest identity score against the database according to the BLAST output in ARG-KNN. An identity score of 50% is the usual cutoff used in the best hit approach (10). For the sequences with high identity scores (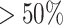 ) with some sequences in the training database, we used the LTR model trained by the identity scores and the prediction scores of ARG-CNN and ARG-KNN to make the prediction. For the sequences with low identity scores (
) with some sequences in the training database, we used the LTR model trained by the identity scores and the prediction scores of ARG-CNN and ARG-KNN to make the prediction. For the sequences with low identity scores (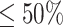 ), we used the LTR model trained by the identity scores and the prediction scores of ARG-CNN, ARG-InterPro and ARG-KNN to make the prediction. The strategy of the ensemble model is determined by its performance on the COALA90 validation data and more details are given in the Supplementary Material.
), we used the LTR model trained by the identity scores and the prediction scores of ARG-CNN, ARG-InterPro and ARG-KNN to make the prediction. The strategy of the ensemble model is determined by its performance on the COALA90 validation data and more details are given in the Supplementary Material.
Competing methods and implementation details
We compared our methods with several methods for ARG classification using the protein sequences or genes as input. The benchmark datasets were curated from multiple databases. Most existing alignment-based methods are developed based on one specific database, which limits their performance in our experiments. We chose RGI (23) as a representative method, which predicts resistomes based on the CARD database (23,32).
BLAST best hit
BLAST (21) is one of the most powerful tools for sequence alignment, which is used by most of the alignment-based methods. We ran it with ‘-max_target_seqs 1’ and different e-value cutoffs as the representatives of the best hit approach.
DIAMOND best hit
DIAMOND (22) is another widely used sequence alignment tool. We ran it with ‘–max-target-seqs 1’ and different e-value cutoffs in the ‘sensitive’ mode.
DeepARG
In DeepARG (10), sequences are represented by their bit scores to known ARGs by aligning them against the known ARGs using DIAMOND. A deep learning model is then used for ARG classification. We retrained DeepARG-LS 1.0.1 (model for long sequences) using our training data. The retrained DeepARG-LS model could not achieve good performance using default parameters, so we changed several parameters. For the predictions with low probability, DeepARG-LS may report more than one ARG class as the results, and other methods can also report more than one ARG class. Therefore, we kept the ARG class with the highest predicted probability for comparison.
RGI
RGI (23) predicts resistome based on homology. RGI analyzes sequences under three paradigms according to the bit score: Perfect, Strict and Loose (low bit score). We report the results with the Loose hits and the results without the Loose hits.
HMMER
HMMER (24) can be used for searching sequence homologs against HMM profiles. The training sequences from each class were aligned using MAFFT v7.475 (33) and the alignments were used to build HMM profiles with HMMER 3.3.2 (24) using ‘hmmbuild’. The testing sequences were classified with the HMM profiles using ‘hmmsearch’ with parameters ‘-E 1000, –domE 100 –max’ as done in (34).
TRAC
The transfer-learning based model, TRAC (11), contains three training stages: (i) the general-domain language model; (ii) the target task language model and (iii) the target task classifier. We retrained their fine-tuned language model using the sequences in the datasets that are not included in the dataset they used and retrained the classifier using our training data. In their paper (11), they trained ten classifiers, ran ten classifiers, and built a soft majority voting classifier for predictions. To make a fair comparison with other baselines and our component methods, we only trained one classifier for predictions. The training process reproduces the strategies mentioned in (11) as much as possible. We used the AdamW (35) optimizer and the label smoothing cross entropy for training.
Evaluation metrics
We used prediction accuracy and f1-score to evaluate the performance. F1-score is the harmonic mean of the precision and recall. The metrics were defined as universal definitions and were detailed in DeepARG (10). We also reported the macro-average results and weighted-average results. Macro-average means that the average performance of each class, and weighted-average means the average performance of each class weighted by the number of sequences.
RESULTS
ARG-SHINE outperforms other available ARG classification tools on the COALA90 dataset
We compared the performance of ARG-SHINE with currently available ARG classification methods including BLAST-best hit (21), DIAMOND-best hit (22), DeepARG (10), RGI (23), HMMER (24) and TRAC (11). In this section, we reported the results of the competing methods using the parameters that achieve the best performance on the validation data. Supplementary Tables S3 and S4 show the prediction accuracy of BLAST, DIAMOND and DeepARG with the different parameters on the COALA90 data. Parameter settings of the compared methods and our methods are given in Supplementary Table S5.
Overall performance
Table 2 shows the accuracy, macro-average f1-score and weighted-average f1-score of the various methods on the COALA90 test data. Among the stand-alone methods, ARG-CNN performs the best in terms of accuracy. Compared with TRAC, which achieves the best performance among the available methods, ARG-CNN increases the accuracy from 0.8115 to 0.8467. Among the alignment-based methods, ARG-KNN and BLAST best hit achieve better performance than DIAMOND best hit and DeepARG. ARG-SHINE performs better than the component methods and other compared methods in terms of all the metrics as shown in Table 2. Compared with the best performer among the available methods, ARG-SHINE increases the value from 0.8115 to 0.8614 in accuracy, from 0.8258 to 0.8555 in macro-average f1-score and from 0.8423 to 0.8591 in weighted-average f1-score. There is no one-to-one correspondence between the ARG classes of RGI and that in our study, so we do not report its average f1-score. To investigate the robustness of ARG-SHINE, we further compared its performance with two well-performing tools, BLAST best hit and TRAC, and our component methods using 5-fold cross-validation. The results and more details about the experiments are given in Table 3 and Supplementary Material. The main findings are consistent with those shown in Table 2.
Table 2.
ARG-SHINE outperforms existing ARG classification programs and the component methods in terms of classification accuracy, macro-average F1-score and weighted-average F1-score on the COALA90 test data
| Methods | Accuracy | Macro-average F1-score | Weighted-average F1-score |
|---|---|---|---|
| BLAST best hit | 0.8092 | 0.8258 | 0.8423 |
| DIAMOND best hit | 0.7986 | 0.8103 | 0.8423 |
| DeepARG | 0.7810 | 0.7303 | 0.8419 |
| RGI | 0.0528 | - | - |
| (perfect+strict) | |||
| RGI | 0.5584 | - | - |
| (perfect+strict+loose) | |||
| HMMER | 0.4938 | 0.4499 | 0.4916 |
| TRAC | 0.8115 | 0.7399 | 0.8097 |
| ARG-CNN | 0.8467 | 0.8167 | 0.8427 |
| ARG-InterPro | 0.8197 | 0.7382 | 0.8151 |
| ARG-KNN | 0.8115 | 0.8047 | 0.8457 |
| ARG-SHINE | 0.8614 | 0.8555 | 0.8591 |
The best results among all the methods and best results among the stand-alone methods are in bold. The ARG classes we used are different from that used by RGI and thus RGI’s macro-average F1-scores and weighted-average F1-scores are not shown.
Table 3.
ARG-SHINE outperforms existing ARG classification programs and the component methods in terms of classification mean accuracy, macro-average F1-score and weighted-average F1-score on the COALA90 dataset based on 5-fold cross-validation
| Methods | Accuracy | Macro-average F1-score | Weighted-average F1-score |
|---|---|---|---|
| BLAST best hit | 0.8045 (±0.0047) | 0.8414 (±0.0163) | 0.8452 (±0.0042) |
| TRAC | 0.8075 (±0.0150) | 0.7615 (±0.0374) | 0.8042 (±0.0158) |
| ARG-CNN | 0.8402 (±0.0059) | 0.8405 (±0.0279) | 0.8373 (±0.0058) |
| ARG-InterPro | 0.8244 (±0.0069) | 0.7703 (±0.0345) | 0.8211 (±0.0078) |
| ARG-KNN | 0.8065 (±0.0035) | 0.8381 (±0.0140) | 0.8472 (±0.0030) |
| ARG-SHINE | 0.8557 (±0.0055) | 0.8595 (±0.0230) | 0.8534 (±0.0057) |
The best results among all the methods and best results among the stand-alone methods are in bold. Standard deviation of accuracy is shown in the brackets.
The three component methods synergistically contributed to the results
We next investigated the contributions of each component method to ARG-SHINE. Figure 2 A shows the venn diagram for the sequences in the COALA90 test data correctly classified by the three component methods. The three component methods complement each other in classification. Among all the 1703 test sequences, 43, 25 and 17 sequences are uniquely correctly classified by ARG-CNN, ARG-InterPro and ARG-KNN, respectively. There are 1259 sequences that are correctly classified by all the three component methods. Venn diagram for the three component methods and ARG-SHINE is shown in Figure 2 B. It reflects that all the component methods contribute to the final results, especially for ARG-CNN, which provides 31 unique correctly classified sequences for ARG-SHINE. We further analyzed the specific genes that only ARG-SHINE could discover. Our component methods classify both sequences incorrectly but generate a relatively high probability for the correct class. For example, one of the sequences is from the TETRACYCLIN class with 38.78% identify score against the training database. ARG-KNN and ARG-InterPro classify it as the BETA-LACTAM class, but they give the second-highest probability for the TETRACYCLIN class. ARG-CNN classifies it as the FSI class (prediction probability: 0.2968) but with a relatively high probability for the TETRACYCLIN class (prediction probability: 0.1014). ARG-SHINE integrates the prediction probabilities for all the classes of the component methods and the identify score, and classifies the sequence into the correct class.
Figure 2.
The Venn diagrams for the sequences in the COALA90 test data that are correctly classified by the three component methods and ARG-SHINE. (A) Venn diagram for the three component methods. (B) Venn diagram for the three component methods and ARG-SHINE.
ARG-SHINE markedly improves the classification accuracy for sequences with relatively low sequence similarity against the training database
When an ARG sequence is highly similar to one of the sequences in the training database, we expect that it is relatively easy to classify the ARG sequence just by BLAST best alignment. However, when an ARG sequence diverges from the sequences in the training database, alignment-based methods will not work well. Therefore, we compared the performance of ARG-SHINE with other available ARG classification methods according to the highest similarity between the query ARG sequence and the sequences in the training database. To assess the methods with different identity cutoffs, we divided the test data into three groups according to their highest identity scores against the training database according to the BLAST output in ARG-KNN. Among the 1703 sequences, 142 of them cannot be aligned to the database using BLAST with e-value no more than 1e-3. Table 4 shows the prediction accuracy of the methods stratified by the identify score against the training database. Among the available methods, BLAST best hit achieves the best performance on the sequences with identity scores. Compared with the BLAST best hit, ARG-SHINE increases the prediction accuracy from 0.6243 to 0.6864 on the sequences with identity scores no more than 50%, and it achieves slightly better performance on sequences with high identity scores. For the sequences that cannot be aligned to the database using BLAST, ARG-SHINE achieves 0.4648 accuracy. Moreover, ARG-CNN and ARG-InterPro achieve better performance than other stand-alone methods on the sequences with identity scores no more than 50%, and the accuracy is 0.6538 and 0.6509, respectively. ARG-SHINE has better performance compared with our component methods. We also present the results with different identity cutoffs using lower e-value cutoffs and the results are presented in Supplementary Table S6. The same conclusions are obtained.
Prediction performance for each antibiotic resistance class
We next evaluated the performance of the different methods for each ARG class and the results are given in Table 5. We analyze the following results based on the f1-score metric. ARG-SHINE performs the best in ten classes among the sixteen ARG classes, and it ties with other methods in five classes of the ten classes. ARG-SHINE also achieves the highest macro-average f1-score (0.8555) and weighted-average f1-score (0.8591). The BLAST best hit, DIAMOND best hit, DeepARG and TRAC perform best in five, five, five and two ARG classes, respectively. Supplementary Table S7 shows the results of our component methods and ARG-SHINE on the COALA90 test data for each antibiotic resistance class. We can find that ARG-SHINE achieves the best performance for the antibiotic resistance classes containing <50 sequences in the training data (the STREPTOGRAMIN class, the RIFAMYCIN class and the MLS class), and this is due to the good performance of ARG-CNN and ARG-KNN on these classes. We suppose that the identity scores of the query sequences against the database affect the accuracy. For example, both testing sequences from the MLS class have over 50% identity scores against the training data. Therefore, BLAST best hit and ARG-SHINE achieve good performance in this class in our experiments. HMMER does not perform well in most classes. This may be due to the low reliability of the multiple sequence alignments (MSAs). To confirm this hypothesis, we calculated the Transitive Consistency Score (TCS) (36,37) of the MSA for each ARG class and compared it with the f1-score based on the HMMER prediction (Supplementary Table S8). The TCS score measures the reliability of MSA. We also observed that the lengths of sequences in each ARG class vary widely, which can markedly impact the reliability of MSA (Supplementary Figure S2A). For example, when the standard deviation of the sequence lengths in an ARG class is less than 50, all the TCS scores are at least 770. However, the TCS scores for all other ARG classes are less than 700. We observed that the f1-score of the HMMER model increases with TCS with a Pearson correlation coefficient of 0.54 (p-value = 0.03) (Supplementary Figure S2B). When TCS > 550, only one out of seven ARG classes has an f1-score less than 0.5. On the other hand, when TCS < 550, seven out of nine ARG classes have an f1-score less than 0.5. The f1-score was also observed to decrease with the standard deviation of the lengths of sequences in the ARG class.
Table 5.
The f1-scores of the compared methods and ARG-SHINE on the COALA90 test data for each class
| ARG Class | BLAST best hit | DIAMOND best hit | DeepARG | HMMER | TRAC | ARG-SHINE |
|---|---|---|---|---|---|---|
| MULTIDRUG | 0.8791 | 0.8696 | 0.8889 | 0.4268 | 0.7327 | 0.8842 |
| AMINOGLYCOSIDE | 0.8783 | 0.8929 | 0.8571 | 0.5382 | 0.8789 | 0.9099 |
| MACROLIDE | 0.9612 | 0.9612 | 0.9764 | 0.8552 | 0.9394 | 0.9767 |
| BETA-LACTAM | 0.8335 | 0.8299 | 0.8355 | 0.4518 | 0.8424 | 0.8584 |
| GLYCOPEPTIDE | 0.8284 | 0.8353 | 0.8451 | 0.6048 | 0.7637 | 0.8416 |
| TRIMETHOPRIM | 0.9630 | 0.9625 | 0.9434 | 0.6527 | 0.9068 | 0.9419 |
| FSI | 0.8578 | 0.8732 | 0.8683 | 0.1412 | 0.7443 | 0.8565 |
| TETRACYCLINE | 0.7923 | 0.7932 | 0.8241 | 0.6617 | 0.7824 | 0.8069 |
| SULFONAMIDE | 1.0000 | 1.0000 | 1.0000 | 0.8333 | 0.9836 | 1.0000 |
| FOSFOMYCIN | 0.7826 | 0.7826 | 0.9524 | 0.2778 | 0.7692 | 0.8696 |
| PHENICOL | 0.5366 | 0.5500 | 0.5823 | 0.2000 | 0.4675 | 0.5946 |
| QUINOLONE | 0.9804 | 0.7805 | 0.4375 | 0.8136 | 0.9600 | 0.9804 |
| STREPTOGRAMIN | 0.5000 | 0.5000 | 0.0000 | 0.0645 | 0.0000 | 0.5000 |
| BACITRACIN | 0.9524 | 1.0000 | 0.9524 | 0.5405 | 1.0000 | 1.0000 |
| RIFAMYCIN | 0.6667 | 0.6667 | 0.5000 | 0.0385 | 0.6667 | 0.6667 |
| MLS | 0.8000 | 0.6667 | 0.2222 | 0.0976 | 0.4000 | 1.0000 |
| Macro-average | 0.8258 | 0.8103 | 0.7303 | 0.4499 | 0.7399 | 0.8555 |
| Weighted-average | 0.8423 | 0.8423 | 0.8419 | 0.4916 | 0.8097 | 0.8591 |
‘Macro-average’ means that we average the f1-score of each individual class; ‘Weighted-average’ means that we average the f1-score of each individual class weighted by the number of sequences. The best values of f1-score for per class are in bold.
Prediction performance on the COALA100 dataset
To investigate the performance of ARG-SHINE on the dataset containing more sequences with high identity scores against the training data than that based on COALA90, we further compared its performance with two well-performing tools, BLAST best hit and TRAC, and our component methods on the COALA100 dataset. Table 6 shows that ARG-KNN and ARG-CNN perform the best among the stand-alone methods in general. BLAST best hit outperforms TRAC. Compared with BLAST best hit, ARG-SHINE increases the accuracy 0.9066 to 0.9286, the macro-average f1-score from 0.9131 to 0.9225, and the weighted-average f1-score from 0.9221 to 0.9276. The prediction accuracy of the different methods on the sequences of the COALA100 test data stratified by identity scores against the training database is shown in the Supplementary Table S9. The table shows that the improvement in prediction accuracy mainly comes from sequences with relatively low identify scores against the training data.
Table 6.
ARG-SHINE outperforms existing ARG classification programs and the component methods in terms of classification mean accuracy and weighted-average F1-score on the COALA100 test data
| Methods | Accuracy | Macro-average F1-score | Weighted-average F1-score |
|---|---|---|---|
| BLAST best hit | 0.9066 | 0.9131 | 0.9221 |
| TRAC | 0.9043 | 0.8963 | 0.9020 |
| ARG-CNN | 0.9219 | 0.9176 | 0.9206 |
| ARG-InterPro | 0.8689 | 0.8135 | 0.8654 |
| ARG-KNN | 0.9112 | 0.9215 | 0.9266 |
| ARG-SHINE | 0.9286 | 0.9225 | 0.9276 |
The best results among all the methods and best results among the stand-alone methods are in bold.
Analysis of the motifs identified by ARG-CNN
We further analyzed the functional information of the motifs identified by ARG-CNN to explain why this method works well. We took the SULFONAMIDE class that ARG-InterPro achieves the best performance on the COALA90 test data (Supplementary Table S7) for analysis. For each protein belonging to the SULFONAMIDE class in the COALA90 test data, we selected the five most important S-length (S = 20) fragments according to their attention scores for each of the r attention sets. We obtained 877 fragments from 30 protein sequences. We then ran InterProScan on these fragments and obtained seven ARG-CNN identified InterPro signatures. As shown in Supplementary Table S10, the important fragments generated by ARG-CNN can identify six out of nine InterPro signatures relevant to the SULFONAMIDE class in ARG-InterPro. These results show that our ARG-CNN can find motifs that cover sequence functional information.
Validation on Novel ARGs
Campbell et al. (38) used functional metagenomics to probe for novel ARGs. They used 15 antibiotics or antibiotic combinations to functionally screen the 16 functional libraries they created and generated the annotation of 332 ARGs (GenBank: MK935708–MK936039). Most of them are from the BETA-LACTAM, TETRACYCLINE, AMINOGLYCOSIDE and PHENICOL classes. We utilized EMBOSS Transeq (39) to translate the nucleic acid sequences to their corresponding peptide sequences. Ten of the sequences have low identity scores (<50%, BLAST) against our COALA90 training database. ARG-SHINE correctly classifies 328 of the 332 sequences. For the other four sequences, three of them are annotated as ‘efflux transporter-like’, which does not match the ARG class labels of our database, and our database does not contain the category of another sequence.
Willms et al. (40) used function-based metagenomic library screening to discover novel sulfonamide and tetracycline resistance genes in soil samples, and identified eight unknown ARGs (GenBank: MK159018 to MK159025). Three of the eight sequences have low identity scores (<50%, BLAST) against our COALA90 training data (database). ARG-SHINE correctly classifies seven of eight novel ARGs into the SULFONAMIDE class or the TETRACYCLINE class. ‘pLAEG3_tet01’ (GenBank: MK159022) is classified into the PHENICOL (AMPHENICOL) class by ARG-SHINE. As shown in Table 6 of (40), the gene also influences the effect of lincomycin antibiotic, belonging to the LINCOSAMIDE class. As presented in Figure 1A of (7), antibiotics in the LINCOSAMIDE class, and antibiotics in the AMPHENICOL class share the same target site. The COALA90 database does not contain the LINCOSAMIDE class. Therefore, it is understandable that ARG-SHINE classifies the pLAEG3_tet01 (GenBank: MK159022) into the PHENICOL class.
DISCUSSION
In this paper, we developed an ensemble method, ARG-SHINE, for ARG classification based on LTR. ARG-SHINE consists of two modules for extracting different features and achieves better performance compared to existing methods. The ‘Component module’ contains three component methods, and they are applied for ARG classification. The component methods are then integrated using stat-of-the-art LTR ensemble method. To the best of our knowledge, this is the first time that protein domains/families/motifs are used for ARG classification and they are effectively integrated with alignment-based and amino acid representation-based deep learning methods. We evaluated ARG-SHINE and other methods on two benchmark datasets, COALA100 and COALA90, and showed that ARG-SHINE outperforms other available ARG classification methods including DeepARG and TRAC. Moreover, compared with BLAST best hit, our final model can achieve much better performance on the sequences with low identity score (<50%) against the training database, and slightly better performance on the sequences with high identity score against the database (Table 4). Note that two of the component methods, ARG-CNN and ARG-InterPro, can also achieve better performance compared with other published methods. Furthermore, ARG-KNN and BLAST best hit usually have better performance than the DIAMOND best hit and DeepARG-LS in our experiments.
Without changing any parameters of the model trained by the COALA90 dataset, we also tested ARG-SHINE on some novel ARGs identified by functional metagenomics. Among the eight novel ARGs from the SULFONAMIDE class or the TETRACYCLINE class, ARG-SHINE correctly classified seven ARGs, with the eighth classification sharing the same antibiotic target site with the true class. Among the 332 ARGs annotated by Campbell et al. (38), ARG-SHINE correctly classified 328 ARGs.
Despite the successes of ARG-SHINE for ARG classification, it has several limitations. First, we only focus on ARG classification using ARG-like protein sequences. If users want to apply ARG-SHINE on the genes, including the genes that are not ARGs, DIAMOND or other methods need to be used to select the ARG-like sequences like DeepARG does. Suppose users would like to use this tool on the assembled contigs from a metagenomic data set. We recommend using Prodigal (41) to predict the ORFs and to obtain the translated sequences first. Next, DIAMOND can be used to align these translated sequences against the ARG database. Sequences meeting the e-value and identity requirement are considered ARG-like sequences and can be further classified using our tool. The choice of the parameters for passing the ARG-like sequences is a trade-off between false positives and false negatives. If users would like to discover more novel ARG genes at the cost of higher false-positive rates, the parameters should be loose, and ARG-SHINE can find more novel ARGs with a looser threshold. Second, the ARG-like sequences cannot be correctly classified if they are not from the ARG classes included in the database. Third, the classification accuracy of ARG sequences for sequences with low identity score with known ARGs is still relatively low and needs to be further increased.
In summary, ARG-SHINE provides a powerful method for ARG classification and integrates three component methods using different features. The component methods utilize sequence homology to known ARGs, protein functional information, or raw sequences, respectively. Users can also choose one of the component methods for ARG classification in specific applications. The methods proposed in this paper can be used for improving the classification of novel ARGs.
DATA AVAILABILITY
COALA dataset is available at https://figshare.com/articles/dataset/COALA_datasets/11413302. Source codes for ARG-SHINE and its component methods are freely available at the https://github.com/ziyewang/ARG_SHINE.
Supplementary Material
Contributor Information
Ziye Wang, School of Mathematical Sciences, Fudan University, Shanghai 200433, China; Department of Pathology and Laboratory Medicine, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Shuo Li, Department of Pathology and Laboratory Medicine, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Ronghui You, School of Computer Science, Fudan University, Shanghai 200433, China.
Shanfeng Zhu, Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai 200433, China; Key Laboratory of Computational Neuroscience and Brain-Inspired Intelligence, Fudan University, Ministry of Education, Shanghai 200433, China; MOE Frontiers Center for Brain Science, Fudan University, Shanghai 200433, China; Zhangjiang Fudan International Innovation Center, Shanghai 200433, China; Institute of Artificial Intelligence Biomedicine, Nanjing University, Nanjing, Jiangsu 210031, China.
Xianghong Jasmine Zhou, Department of Pathology and Laboratory Medicine, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA 90095, USA.
Fengzhu Sun, Quantitative and Computational Biology Department, University of Southern California, Los Angeles, CA 90089, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
S.Z. was supported by National Natural Science Foundation of China (No. 61872094), Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01), ZJ Lab, Shanghai Center for BrainScience and Brain-Inspired Technology and the 111 Project (No. B18015).
Conflict of interest statement. None declared.
REFERENCES
- 1. Chaudhary A.S. A review of global initiatives to fight antibiotic resistance and recent antibiotics discovery. Acta Pharm. Sin. B. 2016; 6:552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gullberg E., Cao S., Berg O.G., Ilbäck C., Sandegren L., Hughes D., Andersson D.I.. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011; 7:e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grumaz S., Stevens P., Grumaz C., Decker S.O., Weigand M.A., Hofer S., Brenner T., von Haeseler A., Sohn K.. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016; 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rizzo L., Manaia C., Merlin C., Schwartz T., Dagot C., Ploy M., Michael I., Fatta-Kassinos D.. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 2013; 447:345–360. [DOI] [PubMed] [Google Scholar]
- 5. Li B., Yang Y., Ma L., Ju F., Guo F., Tiedje J.M., Zhang T.. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015; 9:2490–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wellington E.M., Boxall A.B., Cross P., Feil E.J., Gaze W.H., Hawkey P.M., Johnson-Rollings A.S., Jones D.L., Lee N.M., Otten W.et al.. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013; 13:155–165. [DOI] [PubMed] [Google Scholar]
- 7. Boolchandani M., D’Souza A.W., Dantas G.. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019; 20:356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pham V.H., Kim J.. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012; 30:475–484. [DOI] [PubMed] [Google Scholar]
- 9. Riesenfeld C.S., Goodman R.M., Handelsman J.. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 2004; 6:981–989. [DOI] [PubMed] [Google Scholar]
- 10. Arango-Argoty G., Garner E., Pruden A., Heath L.S., Vikesland P., Zhang L.. DeepARG: a deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome. 2018; 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamid M.N. Transfer learning towards combating antibiotic resistance. 2019; Doctoral Dissertation. Iowa State University. Chapter 3.
- 12. Li H. A short introduction to learning to rank. IEICE Trans. Inform. Syst. 2011; 94:1854–1862. [Google Scholar]
- 13. You R., Zhang Z., Xiong Y., Sun F., Mamitsuka H., Zhu S.. GOLabeler: improving sequence-based large-scale protein function prediction by learning to rank. Bioinformatics. 2018; 34:2465–2473. [DOI] [PubMed] [Google Scholar]
- 14. You R., Yao S., Xiong Y., Huang X., Sun F., Mamitsuka H., Zhu S.. NetGO: improving large-scale protein function prediction with massive network information. Nucleic Acids Res. 2019; 47:W379–W387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rawat W., Wang Z.. Deep convolutional neural networks for image classification: A comprehensive review. Neural Comput. 2017; 29:2352–2449. [DOI] [PubMed] [Google Scholar]
- 16. Krizhevsky A., Sutskever I., Hinton G.E.. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems(NeurIPS). 2012; Nevada, USA. [Google Scholar]
- 17. Cai Z., Fan Q., Feris R.S., Vasconcelos N.. A unified multi-scale deep convolutional neural network for fast object detection. European conference on computer vision(ECCV). 2016; Amsterdam, The Netherlands: 354–370. [Google Scholar]
- 18. Kim Y. Convolutional Neural Networks for Sentence Classification. Proceedings of Empirical Methods in Natural Language Processing (EMNLP). 2014; Doha, Qatar: 1746–1751. [Google Scholar]
- 19. LeCun Y., Bengio Y.. Convolutional networks for images, speech, and time series. The handbook of brain theory and neural networks. 1995; Cambridge, Massachusetts: MIT Press; 255–258. [Google Scholar]
- 20. Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G.et al.. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 22. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 23. Alcock B.P., Raphenya A.R., Lau T.T., Tsang K.K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.-L.V., Cheng A.A., Liu S.et al.. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020; 48:D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li W., Godzik A.. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006; 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- 26. Mikolov T., Chen K., Corrado G., Dean J.. Efficient estimation of word representations in vector space. Proceeding of the International Conference on Learning Representations (ICLR) Workshop Track. 2013; Arizona, USA. [Google Scholar]
- 27. Nair V., Hinton G.E.. Rectified Linear Units Improve Restricted Boltzmann Machines Vinod Nair. Proceedings of the 27th International Conference on Machine Learning (ICML). 2010; 27:Haifa, Israel: 807–814. [Google Scholar]
- 28. Lin Z., Feng M., Santos C. N.d., Yu M., Xiang B., Zhou B., Bengio Y.. A structured self-attentive sentence embedding. International Conference on Learning Representations (ICLR). 2017; Toulon, France. [Google Scholar]
- 29. Hunter S., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L.et al.. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009; 37:D211–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pearson W.R. An introduction to sequence similarity (‘homology’) searching. Curr. Protoc. Bioinformatics. 2013; 42:3.1.1–3.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burges C.J. From ranknet to lambdarank to lambdamart: an overview. Learning. 2010; 11:81. [Google Scholar]
- 32. Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N.et al.. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017; 45:D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katoh K., Misawa K., Kuma K., Miyata T.. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002; 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berglund F., Österlund T., Boulund F., Marathe N.P., Larsson D. G.J., Kristiansson E.. Identification and reconstruction of novel antibiotic resistance genes from metagenomes. Microbiome. 2019; 7:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loshchilov I., Hutter F.. Decoupled Weight Decay Regularization. International Conference on Learning Representations (ICLR). 2019; New Orleans, USA. [Google Scholar]
- 36. Chang J.M., Di Tommaso P., Notredame C.. TCS: a new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol Biol Evol. 2014; 31:1625–1637. [DOI] [PubMed] [Google Scholar]
- 37. Chang J.M., Di Tommaso P., Lefort V., Gascuel O., Notredame C.. TCS: a web server for multiple sequence alignment evaluation and phylogenetic reconstruction. Nucleic Acids Res. 2015; 43:3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Campbell T.P., Sun X., Patel V.H., Sanz C.M., Dantas G.. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. ISME J. 2020; 14:1584–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R., Potter S.C., Finn R.D.et al.. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019; 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Willms I.M., Kamran A., Aßmann N.F., Krone D., Bolz S.H., Fiedler F., Nacke H.. Discovery of novel antibiotic resistance determinants in forest and grassland soil metagenomes. Front. Microbiol. 2019; 10:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
COALA dataset is available at https://figshare.com/articles/dataset/COALA_datasets/11413302. Source codes for ARG-SHINE and its component methods are freely available at the https://github.com/ziyewang/ARG_SHINE.




