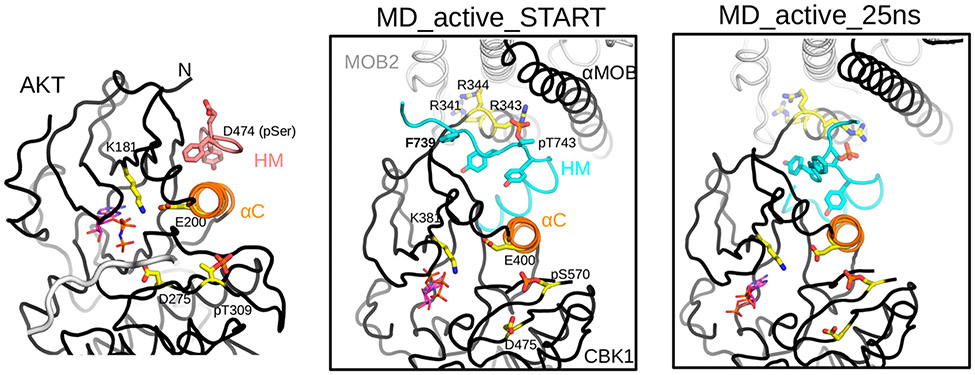
Figure 7.
Mechanistic model of activation of the Cbk1–Mob2 complex by the HM and activation loop phosphorylation. The crystal structure of active AKT/PKB (PDB entry 1O6K) highlights the central role of αC in assembling a catalytically competent AGC kinase active site. Glu-200 on αC together with Lys-181 from the N-lobe of the kinase orients the phosphate moiety of ATP (shown in sticks where carbon atoms are colored magenta) and aligns it for substrate phosphorylation involving Asp-275. (The backbone of the substrate peptide bound in the substrate-binding pocket is colored gray.) Activated HM binding in the PIF pocket and activation loop (AL) phosphorylation at Thr-309 buttress αC and promote the proper ATP alignment. The middle panel shows the structural model of the activated Cbk1–Mob2 complex (MD_active_START). The model, which had αC built on the basis of the active AKT/PKB crystallographic model shown in the left panel (but lacked long flexible regions invisible in the crystal structure, e.g., the linker connecting the HM to the kinase domain core or most of the long AL), was created using the revised crystal structure of the nonphosphorylated Cbk1–Mob2 complex. The panel shows the energy-minimized phosphorylated Cbk1–Mob2 structural model and highlights the same key residues in the AGC kinase domain as shown in the AKT/PKB structure on the left. Note that Arg-343 is well-positioned to coordinate phosphoThr-743 on the HM, and this together with phosphoSer570 at the AL affects salt bridge formation between Glu-400 and Lys-381, which in turn could influence active site assembly as in AKT/PKB. The right panel shows the active Cbk1–Mob2 complex after a 25 ns molecular dynamics simulation (MD_active_25 ns).