Abstract
EEGLAB signal processing environment is currently the leading open-source software for processing electroencephalographic (EEG) data. The Neuroscience Gateway (NSG, nsgportal.org) is a web and API-based portal allowing users to easily run a variety of neuroscience-related software on high-performance computing (HPC) resources in the U.S. XSEDE network. We have reported recently (Delorme et al., 2019) on the Open EEGLAB Portal expansion of the free NSG services to allow the neuroscience community to build and run MATLAB pipelines using the EEGLAB tool environment. We are now releasing an EEGLAB plug-in, nsgportal, that interfaces EEGLAB with NSG directly from within EEGLAB running on MATLAB on any personal lab computer. The plug-in features a flexible MATLAB graphical user interface (GUI) that allows users to easily submit, interact with, and manage NSG jobs, and to retrieve and examine their results. Command line nsgportal tools supporting these GUI functionalities allow EEGLAB users and plug-in tool developers to build largely automated functions and workflows that include optional NSG job submission and processing. Here we present details on nsgportal implementation and documentation, provide user tutorials on example applications, and show sample test results comparing computation times using HPC versus laptop processing.
2. Introduction
Despite the explosion in the use and diversification of research and applications using electroencephalography (EEG), other than one current privately financed project (Alexander et al., 2017; O’Connor et al., 2017), significant recent brain biomarker discovery initiatives have not involved EEG-derived measures (Pedroni, Bahreini, & Langer, 2019). Other neuroimaging techniques, chiefly MRI and fMRI, are the focus of leading initiatives such as the IMAGEN study (Schumann et al., 2010), the U.K. Biobank project (Sudlow et al., 2015), the Human Connectome Project (Van Essen et al., 2012), the Autism Brain Imaging Data Exchange (Di Martino et al., 2014) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (Mueller et al., 2005). Three major factors are hindering the large-scale data collection, analysis, and meta-analysis of EEG data.
Physiologic and experimental variability.
The first factor is the native high inter-subject signal variability in EEG signals themselves. Brain anatomic and physiological (as well as cognitive and behavioral) differences between individuals, produce differences in their EEG signals that may severely affect the performance of models and preprocessing techniques meant to generalize across many subjects (Pedroni et al., 2019). Circumventing issues arising from this variability requires major efforts both in data standardization and in data analysis (Bigdely-Shamlo, Makeig, & Robbins, 2016; Bigdely-Shamlo, Mullen, Kothe, Su, & Robbins, 2015; Gabard-Durnam, Mendez Leal, Wilkinson, & Levin, 2018; Pedroni et al., 2019). Differences in number and placement of the recording electrodes across studies, and difficulty in co-registering the electrode positions to the head and underlying functional EEG brain sources add further complexity.
Format variability and lack of specificity.
A second factor hindering development of large scale EEG data analysis and meta-analysis is the lack of agreement within researchers and system manufacturer communities regarding EEG data formats. Lack of standardization in this area has negatively impacted the growth of EEG data sharing. Inconsistent and often insufficient documentation of data collection and analysis procedures have made it difficult to reproduce results across laboratories and limited the possibility of bringing together data recorded in different formats using varying experimental paradigms and recording systems and parameters.
The Brain Imaging Data Structure (BIDS) specification initiative is attempting to establish a common set of standards for, first, MRI and fMRI (Gorgolewski et al., 2016) and most recently MEG (Niso et al., 2018), iEEG (Holdgraf et al., 2019), and EEG (Holdgraf et al., 2018) data. The emerging BIDS standards are sets of interrelated research community-developed specifications for organizing, archiving, sharing, and easily analyzing brain imaging data collected within and across studies and laboratories. The new BIDS standards, now being rapidly adopted by neuroinformatics projects in the US and in Europe, address the issue of heterogeneity in the structure of archived data across studies. Importantly for EEG data, the root BIDS standard supports the Hierarchical Event Descriptor (HED) system for annotating the precise nature of experimental events recorded in the data (Bigdely-Shamlo, Cockfield, et al., 2016), thereby promoting greater depth of detail and improved consistency across labs in the descriptions of experimental events recorded in the data.
Need for high-performance computing.
A third factor hindering the development of large-scale EEG analysis is that, given the success of applications of machine learning methods to very large data archives in an increasing number of fields, and the relatively large amount of EEG data that could potentially be made available for such analyses, there is an increasing need for using high-performance computing (HPC) resources. However, there are few readily available tools for solving difficult EEG signal processing problems such as EEG source localization using realistic electrical head models (Akalin Acar & Makeig, 2010; Brunet, Murray, & Michel, 2011; Wolters, Kuhn, Anwander, & Reitzinger, 2002); time-frequency (for example, (S Makeig et al., 2002; Onton, Delorme, & Makeig, 2005; Pfurtscheller & Da Silva, 1999)) and cross-frequency coupling analysis (Canolty & Knight, 2010; Martínez-Cancino et al., 2019; Tort, Komorowski, Eichenbaum, & Kopell, 2010); stochastic analyses including independent component analysis (ICA) (Artoni, Menicucci, Delorme, Makeig, & Micera, 2014; Scott Makeig, Jung, Bell, Ghahremani, & Sejnowski, 1997); and a growing variety of machine learning methods (see examples in (Lotte et al., 2018; Scott Makeig et al., 2012)). Thus, for many EEG researchers lack of access to sufficient well-documented and formatted data, readily applicable analysis tools, and connected computing resources are major current obstacles to applying new large-scale analysis and meta-analysis methods to EEG and related data
HPC science gateways.
Publicly available high-performance computing (HPC) resources do exist at national academic supercomputer centers, but access by neuroscientists to these resources is limited by both administrative and technical barriers including the steep learning curve required to understand HPC hardware, software, usage policies, user environments and to install and run applications on HPC resources (Delorme et al., 2019). These obstacles are being partially eliminated through the creation of science gateways to publicly-funded HPC resources (Wilkins-Diehr, Gannon, Klimeck, Oster, & Pamidighantam, 2008). Each science gateway provides a customized set of compute-intensive applications to researchers in a specific scientific field, making them available for use via a simple to use web portal and/or an application programming interface (API). These gateways allow scientists to access HPC resources without dealing with the complexities associated with the machine environment. A gateway has been developed for the neuroscience community, the Neuroscience Gateway (Sivagnanam et al., 2013; Sivagnanam, Yoshimoto, Carnevale, & Majumdar, 2018). Here we present a solution linking the most widely used signal processing environment for EEG research, EEGLAB (Delorme & Makeig, 2004), to the U.S. HPC Extreme Science and Engineering Discovery Environment (XSEDE) computer network (xsede.org) via the Neuroscience Gateway (NSG).
The Neuroscience Gateway (NSG)
(Sivagnanam et al., 2018) is a National Science Foundation (NSF) funded project focused on providing HPC resources to neuroscience researchers. In doing so, NSG aims to lower or eliminate the administrative and technical barriers that currently make it difficult for investigators to use these resources. The NSG (nsgportal.org) gateway offers free computer time to neuroscientists on the XSEDE computer network, providing a ready means to use HPC resources for popular neuroscience tools, pipelines, data processing, and software including NEURON (Hines & Carnevale, 2008), GENESIS (Bower & Beeman, 2012), MOOSE (Ray, Deshpande, Dudani, & Bhalla, 2008), NEST (Gewaltig & Diesmann, 2007), Brian (Goodman & Brette, 2009) and PyNN (Davison et al., 2009), as well as software for analyzing and visualizing structural and functional brain imaging data, such as Freesurfer (Fischl, 2012) and MRtrix (Tournier, Calamante, & Connelly, 2012). NSG leverages access to resources including the supercomputers Comet at the University of California San Diego and Stampede2 at University of Texas Austin, and the Jetstream cloud resource at the University of Indiana.
The EEGLAB signal processing environment.
Resources for analyzing EEG data are increasingly freely available as open-source software. Leading examples are the EEGLAB (Delorme & Makeig, 2004), Fieldtrip (Oostenveld, Fries, Maris, & Schoffelen, 2011), Brainstorm (Tadel, Baillet, Mosher, Pantazis, & Leahy, 2011), and MNE (Gramfort et al., 2014) tool environments. In 2011, EEGLAB was reported to be the most widely used by the cognitive neuroscience community (Hanke & Halchenko, 2011), a report consistent with counts of more recent research publications reporting the use of these respective environments (see Inline Supplementary Figure 1). EEGLAB (Delorme & Makeig, 2004), first introduced in 2002 organizing tools first released by Makeig and colleagues at the Salk Institute in 1997, comprises a large set of tools for performing ICA decomposition, time/frequency analysis, and effective source-level (or scalp channel-level) data visualization. EEGLAB combines a simple but powerful graphical user interface (GUI) for data exploration with an easy transition to command line scripting for custom analysis, exploiting the richness of the MATLAB environment (The Mathworks, Inc., Natick, Massachusetts, United States) on which it runs. EEGLAB is also compatible with the open source, cross-platform Octave application, and is readily extensible through its increasing library of more than 100 plug-in extensions contributed by many laboratories, extensions that appear as easy-to-launch items in the EEGLAB window menu of users who download them. These plug-ins can take advantage of the wide range of easily accessible core MATLAB graphics and other libraries.
Using EEGLAB on high-performance computing resources.
In a recent publication (Delorme et al., 2019), we introduced the Open EEGLAB Portal (OEP) software framework enabling interaction between EEGLAB and the Neuroscience Gateway (NSG, nsgportal.org) to offer free use of the high-performance computing (HPC) resources of the U.S. XSEDE network for analysis of EEG and related data from the EEGLAB environment. The OEP makes use of NSG to provide ready access to HPC resources without the burden of complications typically associated with using HPC services.
Earlier, in (Delorme et al., 2019), we previewed the EEGLAB functions supporting the OEP and discussed future development, e.g., a programmatic user interface from EEGLAB to NSG. Here, we introduce the EEGLAB plug-in, nsgportal (github.com/sccn/nsgportal) that fulfills this purpose -- to give cognitive neuroscientists free and easy access to HPC resources from an EEGLAB session running on any lab or personal computer running MATLAB1. The current manuscript is organized as follows: In Section 3, we expand on the structure and capabilities of the OEP, ways of accessing it and the characteristics and structure of the computational jobs. In Section 4, we introduce the nsgportal plug-in and discuss its architecture, its GUI structure, and its set of MATLAB command line tools. In Section 5, we present three nsgportal applications, the first illustrating the use of the nsgportal GUI, the second its MATLAB command line tools, and the third showing use of these tools to incorporate HPC access into new or existing EEGLAB plug-ins, culminating with building nsgportal capabilities into the compute-intensive RELICA (for ‘RELiable ICA’) plug-in of Artoni et al. (Artoni et al., 2014). Finally, in Sections 6 and 7, we outline current and future developments and present conclusions from our work to date.
3. The Open EEGLAB Portal
3.1. NSG and Open EEGLAB Portal structure and capabilities.
The Neuroscience Gateway software is based on the Workbench Framework (WF) (Miller, Pfeiffer, & Schwartz, 2010, Miller, 2011 #23), a software development kit designed to deploy analytical jobs and database searches to a generic set of computational resources and databases. The WF has been used in building successful science gateways such as the CIPRES phylogenetic gateways (Miller et al., 2010) and the Portal for Petascale Life science Applications and Research (PoPLAR) gateway (Rekapalli, Giblock, & Reardon, 2013). Among these, CIPRES is the most successful gateway, handling about 40,000 unique users over a period of ten years (2009 – 2019). This speaks of the maturity of the WF. To create NSG, the CIPRES-WF was modified to hide all the complexities associated with accessing and using HPC resources for various neuroscience tools made optimally available on HPC resources (for more detail, see (Sivagnanam et al., 2015)).
The Open EEGLAB Portal (OEP) is built upon the NSG framework, making use of its well-established and tested features, e.g., user interface, account management system, documentation, a ticketing system for bug reports and resolution, a mechanism for user support as well as policies to equitably distribute HPC time. Figure 1 (above) presents an overview of OEP workflow. The OEP architecture connects three main components: The user’s local computer, the NSG gateway interface, and the NSG backend HPC resources. NSG resources may be engaged either through the NSG web interface (nsgportal.org) or through its API, NSGR, operating in the Representational State Transfer (REST) environment. Here we focus on the interface between EEGLAB and NSG-R through the NSG REST API.
Figure 1. High-level overview of the Open EEGLAB Portal (OEP) workflow.

The OEP architecture has three main components: The EEGLAB user local computer resource (in blue), the Neuroscience Gateway (NSG) (in orange), and the NSG back-end HPC resources (in green). NSG can be accessed via the NSG web interface or through the NSG applications programming interface (API) operating in the Representational State Transfer (REST) environment (Miller et al., 2015). The EEGLAB plug-in nsgportal exploits the latter access point to provide seamless cross-communication between an EEGLAB session and NSG. Once jobs are submitted to NSG, scripts to execute and data to operate on are deployed to the back-end HPC resources for processing (red arrow). Once job processing is finished, the NSG working directory is first returned to the NSG server (green arrow) and then compressed. Following, the user is notified of the job completion via email. Status of jobs submitted directly from EEGLAB using the API-based nsgportal tools can also be monitored using plug-in tools and features. The results from the computation can be retrieved manually via the NSG web interface, or automatically through the nsgportal plug-in GUI running within the same or a new EEGLAB session.
3.2. Registering EEGLAB as an NSG application
To use NSG-R, the EEGLAB toolbox itself has been registered to use NSG UMBRELLA authentication, whereby jobs can be submitted to NSG on behalf of multiple registered users. The EEGLAB NSG identifier and key are specified within the nsgportal authentication function pop_nsginfo (see Inline Suppl. Figure 2) and should not need to be changed by the user.
3.3. User interaction with the OEP
The first step to using the Open EEGLAB Portal is to create a user NSG account on the NSG home page (www.nsgportal.org). Obtaining an account first requires providing identity information to allow NSG administrators to verify user authenticity. This information comprises name, email, responsible principal investigator, and institution. Any researcher associated with a not-for-profit institution may apply. Receiving an account credential typically requires two days.
Any job submitted to NSG must be in the form of a compressed .zip file of a folder including raw or pre-processed data files (typically, EEGLAB-formatted), an EEGLAB analysis script to run on MATLAB (The Mathworks, Inc.), plus any custom MATLAB functions not in the main EEGLAB distribution and used in the analysis script. Code for built-in MATLAB and MATLAB toolbox functions, as well as functions in the current EEGLAB release, are already available via NSG and do not have to be provided in the job file.
The version of EEGLAB available in NSG, always the current stable version, includes the most popular EEGLAB data processing plug-in extensions as well as all of the current EEGLAB data import plug-ins. If a toolbox, plug-in, or function used in the job analysis script is not built-in by default in MATLAB, in the EEGLAB release, or in one of the NSG-installed plug-ins, the user must include the corresponding files in the job .zip file. (Note: We may add to the installed NSG base other EEGLAB plug-ins of general interest; please email requests to eeglab@ucsd.edu).
NSG job submission.
User input files can be provided to NSG by using its two main entry ports (see Fig. 1): (1) The user can upload through the simple NSG web-based interface (nsgportal.org) a job .zip file containing the data and the Matlab code to execute, plus further submission parameters (Section 4.4 and 4.5). Else, (2) the user can submit, monitor, and retrieve NSG jobs through tools using the NSG REST API (Section 4.2).
Job execution.
Once the job files and relevant settings are submitted to NSG, independent of the interface used NSG assigns a unique identifier to the job (Unique_job_identifier) and generates a job status object with a handle in the following format:
The NSG front end then sends the input job files with a job request to the remote HPC server for processing. Once the job completes, its working directory is compressed (by zip) along with the original job input and any output files. Additional files generated by NSG to report HPC scheduler processing (scheduler_stderr.txt and scheduler_stdout.txt) or job errors (stderr.txt), as well as the MATLAB command window output record (stdout.txt) are included in this directory. The resulting .zip file is transferred to the NSG file system storage (Sivagnanam et al., 2015).
Once the results are ready for retrieval, an email is sent to the user to notify them of the job completion. The user can then download the results contained in the .zip output file. Results retrieval must be done through the same NSG access mode used for job submission (the NSG web portal or the NSG REST API).
4. The EEGLAB nsgportal plug-in
Here we formally introduce nsgportal, a plug-in interface between EEGLAB and NSG using the REST API.
4.1. Accessing NSG through the plug-in nsgportal
Nsgportal capitalizes on the availability of the NSG REST API to provide EEGLAB users an nsgportal GUI plus MATLAB command line tools allowing them to manage and interact with NSG jobs directly from the MATLAB and EEGLAB interfaces. (Alternatively, programmatic access using open source curl commands allows the user to perform the same tasks outside the EEGLAB environment, directly from the operating system command line, though doing this requires detailed familiarity with operating system and curl syntax).
4.2. Implementation, architecture, and dependencies
The nsgportal plug-in, implemented and running in MATLAB, calls curl command line tools that provide an interface to the NSG REST service. As a consequence, to run nsgportal the user must have MATLAB and curl/libcurl installed on their computer. Currently, most macOS and Windows (Windows10 and later) releases have curl installed by default, while Linux users can easily install curl from their software repository. Since MATLAB and curl command line tool code are compatible with macOS, Windows and Linux, the plug-in nsgportal will run without modification in MATLAB on any of these platforms. As a plug-in to EEGLAB, nsgportal also requires the EEGLAB environment functions to be in the MATLAB path and must be installed as per the standard EEGLAB plug-in installation instructions (available online under sccn.ucsd.edu/wiki at EEGLAB_Extensions_and_plug-ins).
Functions included in the nsgportal plug-in are designated by mimicking the function name hierarchy used in EEGLAB. Nsgportal has two main sets of functions, designated by the prefixes pop_ and nsg_. When called with fewer than the required arguments, pop_ functions open a parameter entry window, else run directly without opening a window. The second class of nsgportal functions, with prefix nsg_, can be called directly from MATLAB command line or from other MATLAB scripts or functions. These functions perform lower-level tasks than the pop_ functions. A main plug-in function (eegplugin_nsgportal) effects the inclusion and manages the appearance of the nsgportal item(s) in the main EEGLAB GUI window menu.
The NSG interface in nsgportal is implemented in the functions nsg_jobs, nsg_run and nsg_delete. They each format a curl command as per NSG REST API specifications, as a MATLAB string variable using the inputs received as arguments. The formatted curl command is then issued for execution using the MATLAB system command call. All communication with NSG through the REST interface occurs within an SSL (Secure Sockets Layer) encrypted session, ensuring that the information is transmitted safely. The code samples in Appendix A show typical curl calls to NSG-R in nsgportal for (A.1) Submitting a job, (A.2) Checking job status, (A.3) Retrieving job results, and (A.4) Deleting a job. Here the submitter must be a registered NSG user (here credited with symbolic credentials your_username and your_password). These examples do not demonstrate the full scope of the NSG REST API. For more information, see the online API guide (nsgportal.org/guide.html).
Successful NSG job submission through the REST interface returns a job status XML object. In nsgportal, this object is parsed into a MATLAB job structure containing job information, parameters, and settings. To manage this interaction, the output from the curl instructions used to submit the job and to request job status are saved in a temporary file with extension .TXT or .XML. These files are parsed into a structure by xml2struct.m within nsgportal and are then deleted. Similar workflows apply to other nsgportal interactions with NSG (job creation and polling). Details of the XML object returned after submitting or managing an NSG job can be found in the NSG REST API user guide (nsgportal.org/guide.html). To download job output results only, the curl command line tool syntax (A.3) is quite similar but there is no need to parse the output .zip file, and deleting a job does not return a job object.
Job structures contain information on current job status, results and errors at a more detailed level of specificity and organization. To assist nsgportal users in troubleshooting, an error-reporting heuristic has been implemented: this categorizes job status as either Completed (job processed), Processing (job processing in progress), MATLAB Error encountered, or NSG Error encountered. A successful NSG job should progress through a series of states (listed in Appendix B), the last (Completed) indicating that the job results are available for download. Note that these four NSG status categories are not the same as the internal nsgportal status states, although they are related, as we will show next.
To check NSG job status, nsgportal first checks for the latest job message issued by NSG. If it is Completed, it checks output file stderr.txt to look for error messages issued by MATLAB (typically these are in the first line of the file, enclosed in curly brackets). If an error is found, nsgportal sets its internal job status to MATLAB Error, or if not, to Completed.
If the last message issued for the job is not Completed, nsgportal checks the Failed field in the job structure. If its value is 1, signifying an NSG failure in processing the job, nsgportal sets its internal job status to NSG Error. If not, it sets its internal status to Processing to indicate that the job is still being processed. This internal categorization is available only within the nsgportal GUI.
4.3. Preparing nsgportal for use
Installation.
All EEGLAB plug-ins including nsgportal can be installed by following the procedure described (EEGLAB extensions/plug-ins under https://sccn.ucsd.edu/wiki/EEGLAB) in the EEGLAB documentation. For the following, we assume the plug-in nsgportal has been added to the EEGLAB installation.
Setting NSG credential and other options.
Although nsgportal uses UMBRELLA authentication within NSG-R, curl commands issued from EEGLAB to interface NSG employ username and password while sharing the common application key ‘EEGLAB_TG’. These commands also use settings that have to be defined by the user, e.g., the NSG URL (typically ‘https://nsgr.sdsc.edu:8443/cipresrest/v1’). Storing NSG credentials and managing other nsgportal settings is performed by pop_nsginfo. This function belongs to the group of pop_ functions introduced in the previous section; we can therefore invoke its GUI by calling the function without any argument (e.g., by simply entering pop_nsginfo on the MATLAB command line) or by selecting EEGLAB menu item ‘Tools > NSG Tools > NSG account info’ (see Inline Supplementary Figure 2).
The user-provided credentials and options are stored in file nsg_info.m in the root of the home folder, or if provided in the path designated for EEGLAB options (see EEGLAB script eeg_options.m). Alternatively, the same process of setting up the plug-in, performed in the pop_nsginfo GUI (shown in Inline Supplementary Figure 2), can be accomplished by calling pop_nsginfo from the MATLAB command window and providing the required inputs as key-value pairs, as in the following code sample:
Code sample 1: Generic command line call to pop_nsginfo to store NSG user credential information.
pop_nsginfo(‘nsgusername’,’[your_username]’, … ‘nsgpassword’,’[your_password]’, … ‘nsgkey’,’TestingEEGLAB-BCE8EC90088F4475AE48190A1B87EF8D’, … ‘nsgurl’,’https://nsgr.sdsc.edu:8443/cipresrest/v1’, … ‘outputfolder’,’/data/tmp’);
The EEGLAB default ‘nsgkey’ and ‘nsgurl’ values shown in the code sample should not be altered. After registering for and obtaining an NSG account, as described above, the install process described in this section should be performed only once to install the plug-in.
4.4. The main plug-in GUI: pop_nsg
The nsgportal GUI.
The nsgportal graphical user interface (GUI) is controlled by the function pop_nsg which, through its associated window and command line interface, is intended to be the primary point of user interaction with NSG within EEGLAB. The main functionalities of nsgportal, comprise the submission, managing, deletion, and retrieval of NSG jobs. The functionalities supported by the pop_nsg GUI allow users to: (1) Submit an EEGLAB job to NSG and set NSG-R options. (2) Perform test runs of NSG jobs on the local computer (typically using a reduced instruction set). (3) Delete jobs from the user’s NSG account. (4) Download NSG job results. (5) Load NSG job results into EEGLAB. (6) Visualize NSG job error and intermediate job logs. (7) Access pop_nsg help documentation. The pop_nsg GUI (Fig. 2) can be invoked by executing ‘pop_nsg’ on the MATLAB command windows or by selecting menu item ‘Tools > NSG Tools > Manage NSG jobs’ from EEGLAB menu.
Figure 2. The pop_nsg.m graphical user interface (GUI) window.
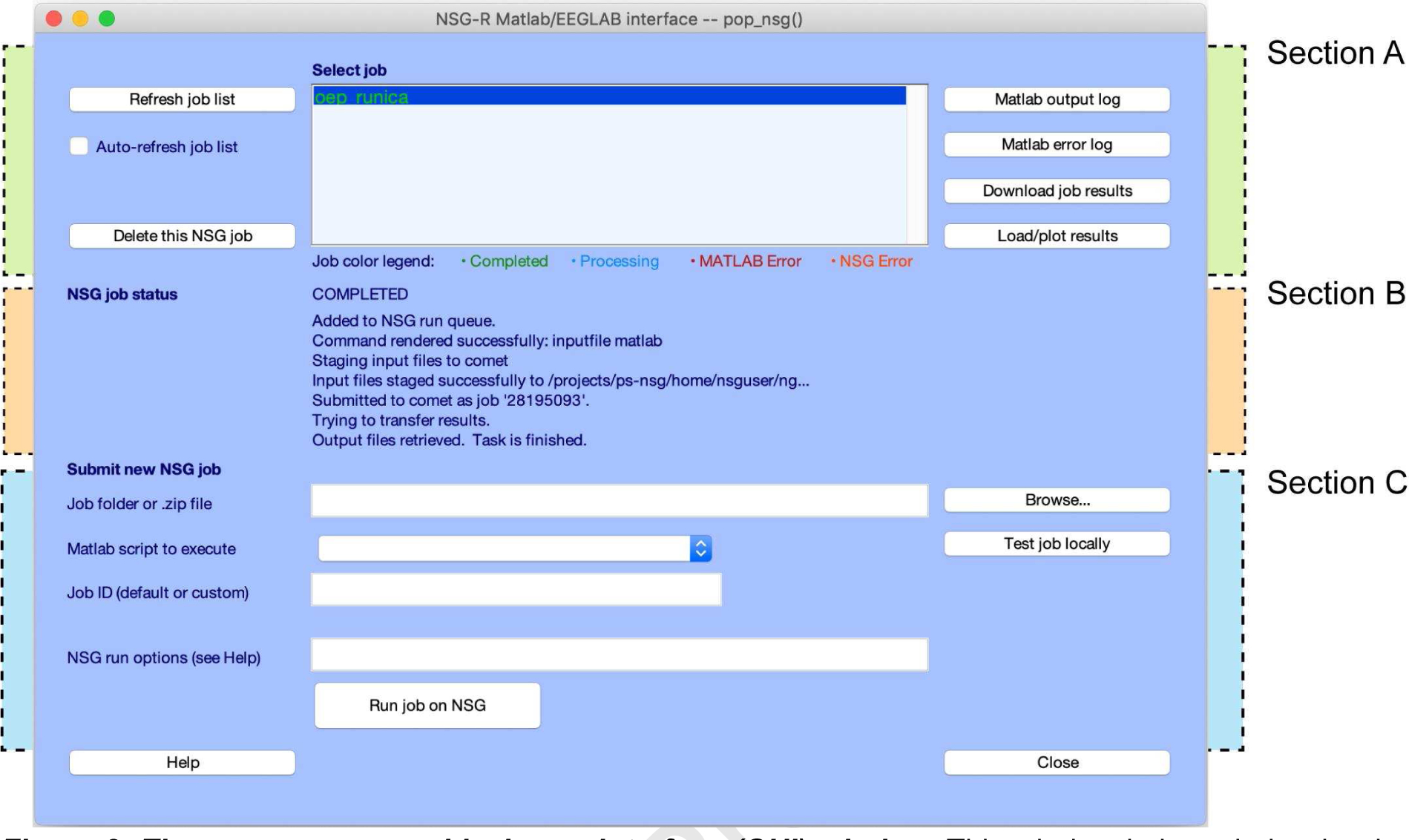
This window is intended to be the primary point of user interaction with NSG within EEGLAB graphical environment. From this interface the user can handle the submission, managing, deletion, and retrieval of jobs. The pop_nsg GUI comprises three main sections, highlighted by dashed lines in the figure: Top section A (see green backing) allows user to interact with jobs already submitted under their own personal credentials. A color-coded scheme (see its legend in the bottom of the display window) reveals to the user the status of the job selected. Middle section B (orange backing) shows status and other messages associated with the job selected in Section A. Section C (cyan backing) manages user job submission.
The pop_nsg GUI has three main sections (highlighted in Fig. 2 by backing green, orange and cyan areas; see labels on the upper right side of Fig. 2). Top Section A allows the user to interact with their own jobs after submission to NSG. A list of all the active jobs under the user account is displayed in the central text box. There, users can select a job with a mouse click, either to then delete it from the queue, to examine its error logs, or to download and display its results. Here, a font color code with four categories (see display window legend) is used to display job status. Middle Section B displays job status and other messages associated with the job selected in Section A. The first line of this information is one of the job status indicators (Appendix B). The lower Section C allows users to submit jobs to NSG. Here the user can specify the job files (as a named .zip file or folder). The MATLAB data processing script to run must also be defined here, as well as other NSG processing options (e.g., requested maximum run time). Another important feature allows the user to run a (user-prepared) downscaled version of the job on their local computer to test the integrity of the script. To use this option, the user must prepare and identify a limited version of the processing script, for example replacing its major processing loop variable (e.g., N = all_files_to_process) with the quicker-running (e.g., N = 1)’. For a detailed explanation of the elements of this GUI and their functionality, see Appendix D.
4.5. Nsgportal Matlab-interface command line tools
Nsgportal command line tools allow users to largely automate their workflow and make the NSG job submission, job status monitoring, and job results retrieval processes simple for users --and/or EEGLAB functions and plug-ins -- to accomplish. Main nsgportal functionalities --submitting, monitoring, deleting jobs, and/or retrieving job results without GUI interaction -- can be also performed directly using command line calls to function pop_nsg providing all its required input arguments. Other useful functions include pop_nsginfo (to set up and record user NSG credentials), nsg_jobs (to return the current list of user NSG jobs) and nsg_recurspoll (to monitor the status of an NSG job). See their function help messages for usage examples.
Setting up user NSG credentials.
After installing the nsgportal plug-in in EEGLAB, function pop_nsginfo can be used to specify the user NSG credential. This function was introduced in Section 4.3, where an example of its command line call was presented in the Code sample 1.
Submitting and managing NSG jobs.
Function pop_nsg is the workhorse of the nsgportal command line tools. It enables users to: (1) create and run an NSG job (‘run’ option); (2) test a short form of the job on your local computer (‘test’ option); (3) retrieve NSG job results (‘output’ option), and (4) delete an NSG job (‘delete’ option). The function pop_nsg must be called with key-value input pairs as in the following pseudo-code:
Code sample 2: Generic key-value input pairs used to invoke pop_nsg.
NSGJobStructure = pop_nsg(‘option_name’, ‘option_value’);
For options ‘run’ and ‘test’, the second argument must always be the pathname of the job .zip file or the folder containing the job to be submitted (or locally tested). Using any of these two options also requires a second pair of compulsory arguments (first argument, ‘filename’) to identify the .m processing script file to be run in the user test or NSG run. For instance to run a job located in path/to/my/job/folder running MATLAB script my_job_script.m:
Code sample 3: Example command line call running an NSG job from the MATLAB command line using pop_nsg.
[NSGJobStructure, AllNSGJobStructure] = … pop_nsg(‘run’,’path/to/my/job/folder’, … ‘filename’, ‘my_job_script.m’);
When pop_nsg is executed with any of the four mode parameters (‘run’, ‘test’, ‘output’, or ‘delete’), the function returns two outputs: (1) a job structure containing all relevant information of the submitted or managed job, and (2) a cell array listing all NSG jobs currently in the user NSG account. In the example code above, these two outputs are called NSGJobStructure and AllNSGJobStructure respectively. These output objects use the same NSG job structure introduced in Section 4.2.
When pop_nsg is called with options ‘output’ or ‘delete’, users can pass the job ID, NSG job structure, or the job URL (see Section 4.2) as the second argument. To use either of these two options, the referenced NSG job must exist in the user’s NSG REST account; to use the ‘output’ option, the job must have completed. This flexibility in invoking pop_nsg is supported by the nsgportal functions nsg_getjobid and nsg_flindclinetjoburl, which respectively translate job URLs to job IDs and vice versa. Function nsg_jobs retrieves information about the jobs associated with the user’s NSG credential. Users can request a recursive check of the status of a submitted job using nsg_recurspoll (see Appendix A.2). The call below sets the polling interval to 2 minutes (120 seconds).
Code sample 4: MATLAB command line call to nsg_recurspoll.
NSGJobStructure = nsg_recurspoll(‘My_Job_ID’, ‘pollinterval’, 120);
Recursive polling in nsg_recurspoll is implemented using the MATLAB timer class to create an object that manages a recursive execution of the sub-function nsg_poll, which in turn executes nsg_jobs repeatedly to check job status by pulling the job status object.
4.6. Nsgportal documentation
A comprehensive nsgportal wiki2 linked to the nsgportal repository3, as well as function help messages contained in nsgportal functions document its use.
The nsgportal wiki.
The wiki begins with a quick introduction to the Open EEGLAB Portal and details how to register as an NSG user and use its web portal. Details on installation and use of the nsgportal plug-in are provided. To represent the workflow within nsgportal, we have generated a plug-in function-calling scheme that may ease future modifications and maintenance4. Finally, a section of the wiki presents detailed examples of how to use the nsgportal plug-in both from the EEGLAB GUI and from the MATLAB command line. Additional examples are given illustrating how to use nsgportal command line tools to equip new or existing EEGLAB plug-ins with HPC optional capability.
Nsgportal function help messages.
Each main function of nsgportal begins with a detailed help message in EEGLAB format: a brief description of the function, a usage example, a table of compulsory and optional input parameters, and a description of the outputs.
5. Applications and examples
Below, we show three applications using the nsgportal plug-in, applied to a representative EEG dataset. The first two applications illustrate the submission, managing, results retrieval, and deletion of jobs from the nsgportal GUI and MATLAB command line, respectively. The third application demonstrates the use of nsgportal command line tools to implement a computationally intense EEGLAB plug-in that gives the user the option to perform the computation on HPC resources via NSG. We first demonstrate this capability by implementing a simple plug-in using nsgportal functions, and then by extending the more complex RELICA plug-in (Artoni et al., 2014 21) to include the option to run its computationally demanding processing on NSG resources. Finally, we report the results of testing the NSG-enabled RELICA on actual EEG data.
5.1. The test data and simple job example Test data description and preprocessing.
To demonstrate the use of nsgportal, we use a single dataset from the EEG/MEG study reported in (Wakeman & Henson, 2015) and made publicly available on OpenNeuro.org (accession number: ds000117). For information on the experimental paradigm and a description of the preprocessing steps applied to these data, see Appendix C. The preprocessed data are a set of 132 EEG 3-sec data epochs time-locked to presentations of visual stimuli and recorded from 70 EEG channels at a sampling rate of 250 Hz, saved as an EEGLAB dataset.
The test job script.
Electric potentials recorded with EEG are the summation of the underlying activity of multiple brain activity sources. Under favorable circumstances, the activity from these sources can be separated by ICA (S Makeig et al., 2002). However, ICA computation may be lengthy and computationally expensive. Here we show how to build a job script to use NSG to decompose the data into maximally independent components (ICs) by applying Extended Infomax Independent Component Analysis (ICA) using the EEGLAB function runica.
First, we create a job folder (runica_on_nsg) containing the test data (files wh_sub11_preproc.set and wh_sub11_preproc.fdt) plus a simple MATLAB script (runica_on_nsg.m) that executes the decomposition via NSG and plots some results. The script (Code sample 5 below) loads the dataset, runs the decomposition, plots results, then saves the figure file and output EEGLAB dataset (now including the ICA mixing and unmixing matrices) to return to the user. The input dataset is (optionally) deleted. The figure (IC_scalp_maps.jpg) shows scalp maps (scalp projection patterns) of the largest 20 ICs (arranged in a 4-by-5 grid).
Code sample 5: Job script ‘runica_on_nsg.m’.
% Launch EEGLAB eeglab; % Load the sample EEGLAB dataset EEG = pop_loadset(‘wh_sub011_proc.set’); % Decompose the data into independent component (IC) processes EEG = pop_runica(EEG, ‘icatype’, ‘runica’); % Plot the scalp maps of the first (and largest) 20 ICs pop_topoplot(EEG, 0, [1:20] ,’EEG Data epochs’,[4 5] ,0,’electrodes’,’on’); % Save the figure as a JPEG file print(‘-djpeg’, ‘IC_scalp_maps.jpg’); % Save the dataset, now including the ICA decomposition matrix pop_saveset(EEG, ‘filename’, ‘wh_sub11_proc_output.set’); % Delete the input dataset to reduce the output file size delete(‘wh_sub011_proc.set’); delete(‘wh_sub011_proc.fdt’);
Assuming we have obtained an NSG account and have installed the nsgportal plug-in, we can then zip the job folder, log in to the NSG website (www.nsgportal.org), and submit the job by identifying the .zip file containing the job folder. Else, we can do this and more using the nsgportal GUI.
Building a job test script for local execution.
A second, ‘test’ version of the job script (‘test_runica_on_nsg.m’) in the job folder, differs from runica_on_nsg.m in only one way: it reduces its computational complexity by asking for a much smaller number of ICA training steps (here, it sets the runica option ‘maxsteps’ to only 5 instead of the default 512). The nsgportal GUI can then be used to run the test script on the user’s computer to verify the accuracy of the syntax in the rest of the script, thereby avoiding wasting user and NSG time and resources attempting to run scripts that still need debugging. For jobs involving many data files (for different participants and/or conditions) and/or surrogate data analysis, this might also involve severely limiting the number of datasets and/or loop iterations to minimize compute time while allowing MATLAB, running on the user’s local computer, to find any coding mistakes before launching the much longer-running full job script on NSG.
5.2. Submitting, managing and retrieving the job using the nsgportal GUI
After we have installed the nsgportal plug-in (Section 4.3), we launch the EEGLAB GUI (command, eeglab) and then the pop_nsg GUI -- either by entering pop_nsg on the MATLAB command line or by selecting item ‘Tools > NSG Tools > Manage NSG jobs’ from the EEGLAB GUI menu. To submit the sample job for processing via NSG from the nsgportal GUI (Figure 2 above), we first browse and select the (compressed or uncompressed) job file runica_on_nsg.zip using the ‘Browse’ button. After selecting the job file, the edit box ‘Job folder or .zip file’ and the drop-down menu ‘Matlab script to execute’ are populated with the path to the zip file and the names of the scripts runica_on_nsg.m and test_runica_on_nsg.m. The field ‘Job ID’ is also populated with a default job name that we choose to change to the more recognizable ‘runica_on_nsg_test’.
Running the test script locally.
Field ‘Matlab script to execute’ has the two .m file entries: runica_on_nsg.m and test_runica_on_nsg.m. To test the script syntax on our local computer, we first select the test script (test_runica_on_nsg.m) and press button ‘Test job locally’. This script is the light version of the job script runica_on_nsg.m. After the job completes its local processing without any error, we are sure the job script syntax at least contains no syntax errors. Testing the jobs script locally is an optional step not required to submit a job for processing in NSG. However, this check may save a lot of time in troubleshooting syntax errors that may delay and interfere with the completion of the job.
Submitting the job to NSG.
After testing the downscaled version of the job locally, we selected the script to execute in NSG (runica_on_nsg.m) in the field ‘Matlab script to execute’. Next, in edit box ‘NSG run options’, we set the job maximum time allocation (here) to one hour by typing ‘runtime’, 1. The unit here is hours, and the maximum setting is 48 hours. Note: do not set this to less time than the job will require! Then, we click the button ‘Run job on NSG’ to submit the job for processing. Upon successful job submission, the Job ID we assigned (‘runica_on_nsg_test’), is shown in the list of job records NSG associates with our NSG user credential. The current status of the job (‘INPUTSTAGING’) is displayed in the job status panel (see Appendix B for status name interpretations).
Monitor job status.
After the job is submitted, to ask pop_nsg to check periodically for its status check the checkbox ‘Auto-refresh job list’. Messages giving the current NSG status of the selected job are then printed (by default, every 30 seconds) on the MATLAB command line. The text color used to display the job ID and job status in the nsgportal GUI, will also be updated to show the current job status.
Retrieving the MATLAB command window output of the job.
The MATLAB command window output issued in the NSG MATLAB session during the job processing can be checked from pop_nsg GUI. To retrieve and display this information in the MATLAB browser, we used the button ‘Matlab output log’. It is possible that this information is not available at the time of the request; in this case the user will be notified of this by a MATLAB pop-up window.
Retrieving and loading job results.
When the job has completed, an email to this effect is sent to the registered user. At this point, the job status in the GUI also reads ‘COMPLETED’. We can then download the job results by clicking on ‘Download job results’. The names of the downloaded and unzipped files are then printed on the MATLAB command window. The following code sample resembles the message printed on the MATLAB command window after downloading the results of the runica_on_nsg.zip test job.
Code sample 6: The code above shows the MATLAB command window output after retrieving job results using the ‘Download job results’ button in the pop_nsg GUI. Lines 1–5 (in green) show the files generated during the processing. The input job data files were (optionally) removed by our test script, so are not contained in the downloaded results.
1>> ./runica_on_nsg/ 2>>./runica_on_nsg/IC_scalp_maps.jpg 3>>./runica_on_nsg/runica_on_nsg.m 4>>./runica_on_nsg/wh_sub11_proc_output.set 5>>./runica_on_nsg/wh_sub11_proc_output.fdt 6>>./scheduler_stderr.txt 7>>./scheduler_stdout.txt 8>>./stderr.txt 9>>./stdout.txt 10>>Done. 11>>File downloaded and decompressed in the output folder specified in the settings 12>>Accessing job: “https://nsgr.sdsc.edu:8443/cipresrest/v1/job/my_useraname/NGBW-JOB-EEGLAB_TG-9FB3515E932C4CA8AADAEF6D40167F0D” on NSG… 13>>Done.
Here, we can see that both results and files submitted (in green) are in the downloaded file, saved in the path defined previously in pop_nsginfo.
To explore the output files, we use the file explorer implemented in the button Load/plot results. This application can load datasets into EEGLAB as well as display a wide range of image formats generated during a job processing in NSG. From this file explorer, we can navigate to (by double-clicking into) the downloaded folder runica_on_nsg and access the result files. For instance, after navigating to the folder with the results of the job in this example, if the file wh_sub11_proc_outpout.set is selected and opened by using the button Load/plot, the main EEGLAB window will pop up, showing that the selected data has been loaded and that the ICA decomposition has been performed (the ICA weights field in the GUI reads ‘yes’).
Deleting a job.
After retrieving the results, we may proceed to delete the job from our NSG account, here by selecting the job ‘runica_on_nsg_test’ in the pop_nsg job list and clicking the button ‘Delete this NSG job’. This functionality can be used either on finished jobs or jobs being currently processed. The deletion of jobs from NSG is accomplished by calling nsg_delete from pop_nsg.
5.3. Submitting, managing and retrieving a job using nsgportal command line tools.
Next, we show how to create, manage, and retrieve NSG jobs using nsgportal command line tools to automate a data analysis pipeline. For this purpose, we will use the same runica test job and data introduced in Section 5.1. The nsgportal command line tools were introduced in Section 4.5; there their key components are described in more detail. Here, we again assume we have obtained an NSG account and have installed the nsgportal plug-in (see Section 4.3).
Submitting a job to NSG.
The code sample below uses function pop_nsg to submit the job runica_on_nsg.zip for processing via NSG. Here we store the path to the job .zip file in a variable; this path can also be passed directly as a string to the function:
Code sample 7: Using pop_nsg for submitting the runica_on_nsg.zip job for processing at NSG.
% The job zip file is located in home directory path2zip = ‘Users/eeglabHome/runica_on_nsg.zip’; % Execute the job currentjob = pop_nsg(‘run’,path2zip,’filename’, ‘runica_on_nsg.m’, … ‘jobid’,’runica_on_nsg_test’);
Notice that three pairs of key-value parameters were used here (‘run’, ‘filename’ and ‘jobid’). The ‘filename’ argument is compulsory when using the option ‘run’ to specify the top-level script run in the job. Default options in pop_nsg assign a randomly generated ID to the job. However, we encourage users to specify a meaningful job ID for their NSG jobs, as this will help in later job management, especially when the number of user submitted jobs is large. In the code sample above, we specify the job ID (‘runica_on_nsg_test’) using the input parameter jobid. After executing the code above, the function pop_nsg returns a MATLAB NSG job structure for the submitted job (currentjob) that we will use as an input for the next processing steps.
Although not shown here, users can also specify other job parameters while submitting a job by providing key-value pair arguments to the function call. These optional arguments include the following: 1) ‘outfile’: the local folder to which the NSG results are to be downloaded. The default value for this parameter is the jobname and job ID plus prefix ‘nsgresults_’ (for example, nsgresults_ testjob_1234). 2) ‘runtime’: the maximum time (in wall clock hours) to allocate for computation of the job on NSG. The default value is 0.5 hours (the maximum value is 48 hours). Note, again, that if the job takes longer to complete than asked for here, it will be stopped! This maximum time estimate is used by the job scheduler to maximize XSEDE resources. ‘subdirname’: Name of the sub-directory containing the script file to run on NSG if the script file is not in the job’s root folder. The options available in pop_nsg are not restricted to those shown here, as the list may grow based on the users needs. The changes and additions are always made available through the nsgportal documentation (Section 4.6).
Monitoring job status periodically.
After the job is submitted to NSG it will be processed on the XSEDE network. We can check the status of the job periodically by calling function nsg_recurspoll, providing as arguments the job structure obtained when submitting the job (job ID, URL or structure are valid inputs), currentjob, and the polling interval in seconds. This function is particularly handy for running functions that operate on the output of a submitted NSG job. Below, we specify the job structure currentjob as a first argument and 120 seconds (2 minutes) as the polling interval:
Code sample 8: Command line call initiating recursive polling of the job status of job ‘currentjob’.
currentjob = nsg_recurspoll(currentjob, ‘pollinterval’, 120);
Once initiated, function nsg_recurspoll exits when the NSG job has completed and its output is ready to be accessed by the user. The function also returns a job structure containing the status of the specified job. This structure is basically the same as that provided as input, but with the job status field updated.
Retrieving job results.
After the job completes and function nsg_recurspoll exits, job results can be retrieved using pop_nsg by providing the job structure following the first argument ‘output’.
Code sample 9: Command line call to pop_nsg to retrieve results of job ‘currentjob.’.
pop_nsg(‘output’, currentjob);
The input currentjob contains the NSG job structure of the job we want to retrieve. As in the example using nsportal GUI, the output files are downloaded to the location defined by the user using pop_nsginfo (see Section 4.3).
Deleting an NSG job.
Below, we use the output structure of nsg_recurspoll to delete the job from the NSG record associated with the user’s NSG credential.
Code sample 10: Command line call to delete the job whose job structure is currentjob.
NSGjobstruct = pop_nsg(‘delete’, currentjob);
The output here is the NSG job structure (NSGjobstruct) of the job deleted. Note that when this command is executed the job is deleted from the user’s NSG account and can no longer be accessed.
In this sub-section, we have shown a proof of concept example automating the use of NSG resources using nsgportal command line tools. With minor changes to the job script, the example here might be scaled up to process data from many subjects within a single job, etc.
The tools framed here can also be used to implement HPC access offered by NSG from within any EEGLAB function or plug-in. This is the focus of the next section.
5.4. Accelerating EEGLAB plug-in performance using nsgportal tools
EEGLAB plug-in extensions allow users to build and publish new or customized data processing and visualization functions using EEGLAB data structures, functions, and conventions. Plug-in functions can be called conveniently by selecting the new menu item(s) they introduce into the EEGLAB menu of users who have downloaded and installed them. In this Section, we use nsgportal command line tools to implement access to NSG HPC capabilities in: (1) an example plug-in that performs the same processing as in the sample job shown in Section 5.1 and, (2) an example adding nsgportal command line calls to an existing plug-in, RELICA (Artoni et al., 2014).
5.4.1. Example plug-in: RUNICA_NSG
Here we describe a simple yet fully functional EEGLAB plug-in (RUNICA_NSG) exemplifying the application of nsgportal command line tools to implement HPC computation from within EEGLAB plug-ins. To create this plug-in, we modified the simple runica_on_nsg.m script presented above (Section 5.1) by turning it into a function eeg_runica_nsg.m that accepts inputs that 1) point to the data matrix to decompose, and 2) select the ICA algorithm to use in the decomposition. To allow the sample plug-in to be called directly from the EEGLAB GUI menu, two additional functions are needed: function eegplugin_runica_nsg manages the listing of the plug-in in the EEGLAB menu, while pop_runica_nsg manages the inputs, first popping up a parameter entry window if its two required input arguments are unspecified, and then redirecting computation either to the local computer resource or to NSG. The example RUNICA_NSG plug-in function code is available online, both from the EEGLAB Extension Manager5 and from its GitHub repository6.
The workflow of RUNICA_NSG is purposely simple. When its main pop_window function pop_runica_nsg is invoked either from the EEGLAB menu or MATLAB command line, the data matrix in the currently loaded EEG dataset (EEG.data) is selected for stochastic decomposition. Two further parameters are required to specify: (1) whether the computation should be performed on the user’s computer or on XSEDE network HPC resources via the user’s NSG account, and (2) which ICA algorithm to use (here, given only two choices: ‘runica’ and ‘jader’). If the user chooses to perform the computation on their local computer (second argument ‘local’) pop_runica_nsg will perform the processing locally (by calling eeg_runica_nsg.m), returning the input EEG dataset structure, now containing the computed ICA decomposition parameters.
If the user chooses to perform the computation through NSG (second argument ‘nsg’), the function pop_runica_nsg will: (1) create a temporary folder, (2) save the EEG data passed as input in this folder, and (3) generate and save in this folder a MATLAB script (runica_nsg_job.m) including a call to the RUNICA_NSG plug-in function eeg_runica_nsg including the user-specified parameters. Then pop_runica_nsg will proceed to submit this job folder to NSG using nsgportal command line tools, identifying runica_nsg_job.m as the main script to execute (first argument ‘filename’). After submitting the job, eeg_runica_nsg will exit, returning the NSG job ID assigned to the task.
The user can provide this job ID to pop_nsg to monitor the job progress and then, when it completes, use it to retrieve its results through pop_runica_nsg. The process of job submission and retrieval, as implemented using nsgportal command line functions, is shown in Fig. 3, in which the nsgportal functions used in the process are shown in green boxes.
Figure 3. Workflow of the example plug-in RUNICA_NSG.
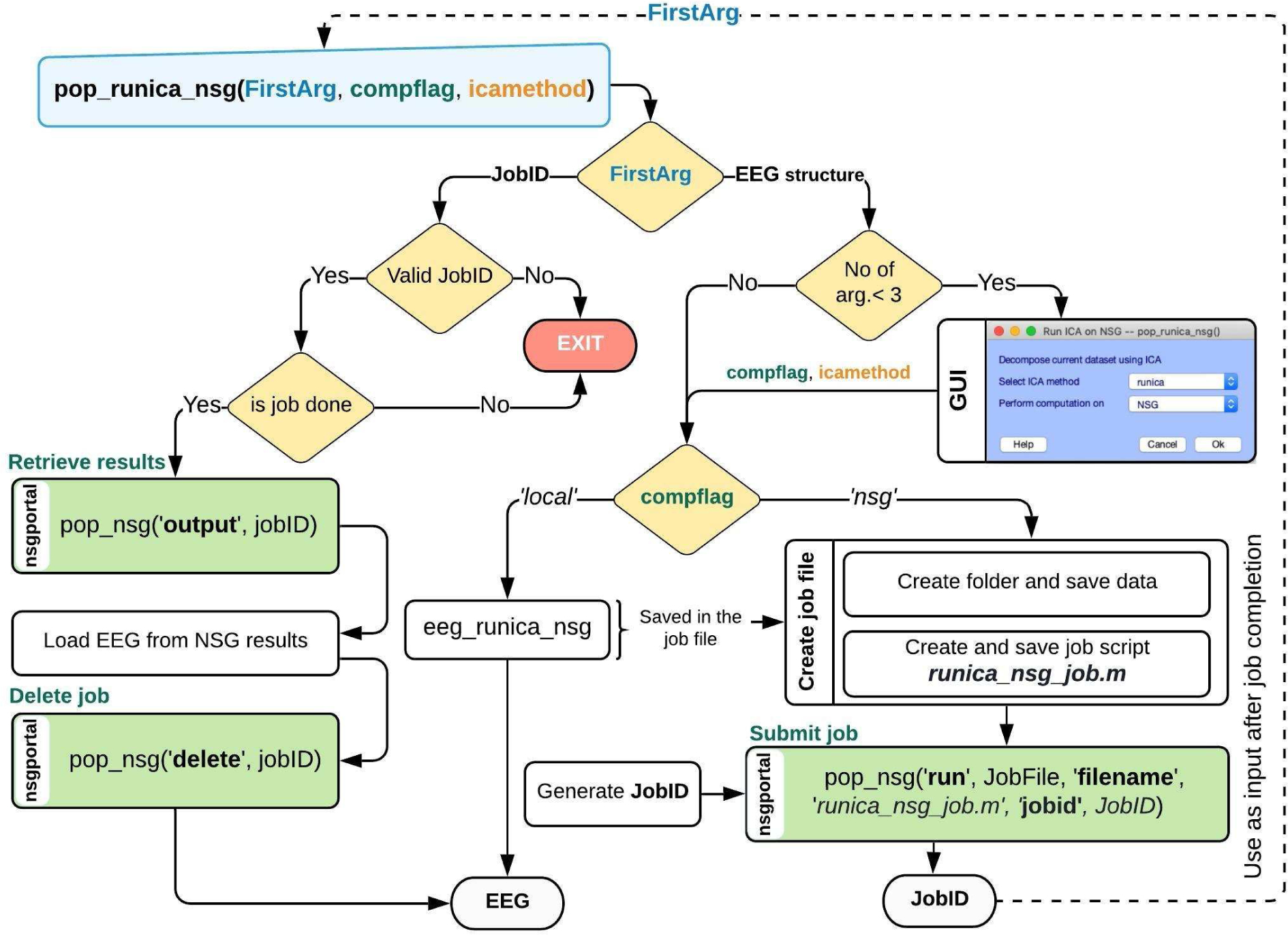
This plug-in illustrates the use of nsgportal command line functions that enable EEGLAB functions and plug-in tools to run on HPC resources via NSG. After installing the plug-in in the eeglab/plugins/ directory, the user can call it via the EEGLAB menu (Tools > Compute ICA via NSG) or equivalently by invoking function pop_runica_nsg from the MATLAB command window. In the second case, an EEG data structure must be provided as input. If invoked from the menu, or when either of its other two arguments, compflag (run locally or via NSG) or icamethod (ICA algorithm to use) are not specified, a MATLAB window will pop up to allow the user to input these parameters. When pop_runica_nsg is invoked from the EEGLAB GUI, the currently loaded EEGLAB dataset (EEG.data) is processed and the pop window is always launched to allow manual entry of arguments compflag and icamethod. When the computation is to be run via NSG, pop_runica_nsg creates a temporary folder and saves the current EEG dataset in it along with function script eeg_runica_nsg.m and a custom script (runica_nsg_job.m) generated to invoke this function using the parameters provided by the user (here, icamethod). This script, along with the data saved, is then submitted to NSG for processing under a user-designated job ID (JobID in the figure). Function pop_nsg with options ‘run’, ‘filename’ and ‘jobid’ performs these steps. Following job submission, pop_runica_nsg exits, providing the submitted jobID as output. Once the NSG job is complete (as flagged by an email message sent to the user from NSG, or via the open pop_nsg GUI), the user must retrieve its results by calling pop_runica_nsg with the JobID as its first and only argument. Internally, pop_runica_nsg will check the validity of the job ID and will then call pop_nsg (with arguments ‘run’ and ‘delete’) to retrieve the results and then delete the job from the user’s NSG account record. Finally, the EEG data structure provided originally as input, including the ICA decomposition parameters, is loaded into EEGLAB by pop_runica_nsg and returned as output.
5.4.2. Adding HPC computation to the RELICA plug-in
RELICA (for ‘RELiable ICA’) is a method that uses a stochastic approach to characterize the reliability of component processes identified by ICA decomposition of a multichannel dataset, by analyzing the stability of the component processes under bootstrap resampling. RELICA thus performs multiple ICA decompositions of bootstrapped versions of the input data, then clusters the ICs from all the decompositions based on similarities between their time courses. By default, the number of clusters corresponds to the number of ICs per decomposition. Each cluster in the analysis is associated with an exemplar IC, selected to be the IC nearest to the cluster centroid in the clustering space. Finally, RELICA computes a measure of the compactness of each bootstrap IC cluster that provides a within-subject measure of IC stability for each IC.
Figure 4 shows a high-level view of the process implemented in the original RELICA implementation by Artoni et al. (Artoni et al., 2014). Most of the computation time here is spent in the sequence of ICA decompositions performed on the bootstrapped data (red area, Fig. 4) . The high computational load required to perform these processes, particularly for large datasets with many channels, makes the RELICA plug-in a good candidate for running on NSG high-performance computing resources using nsgportal tools.
Figure 4. Original RELICA workflow.
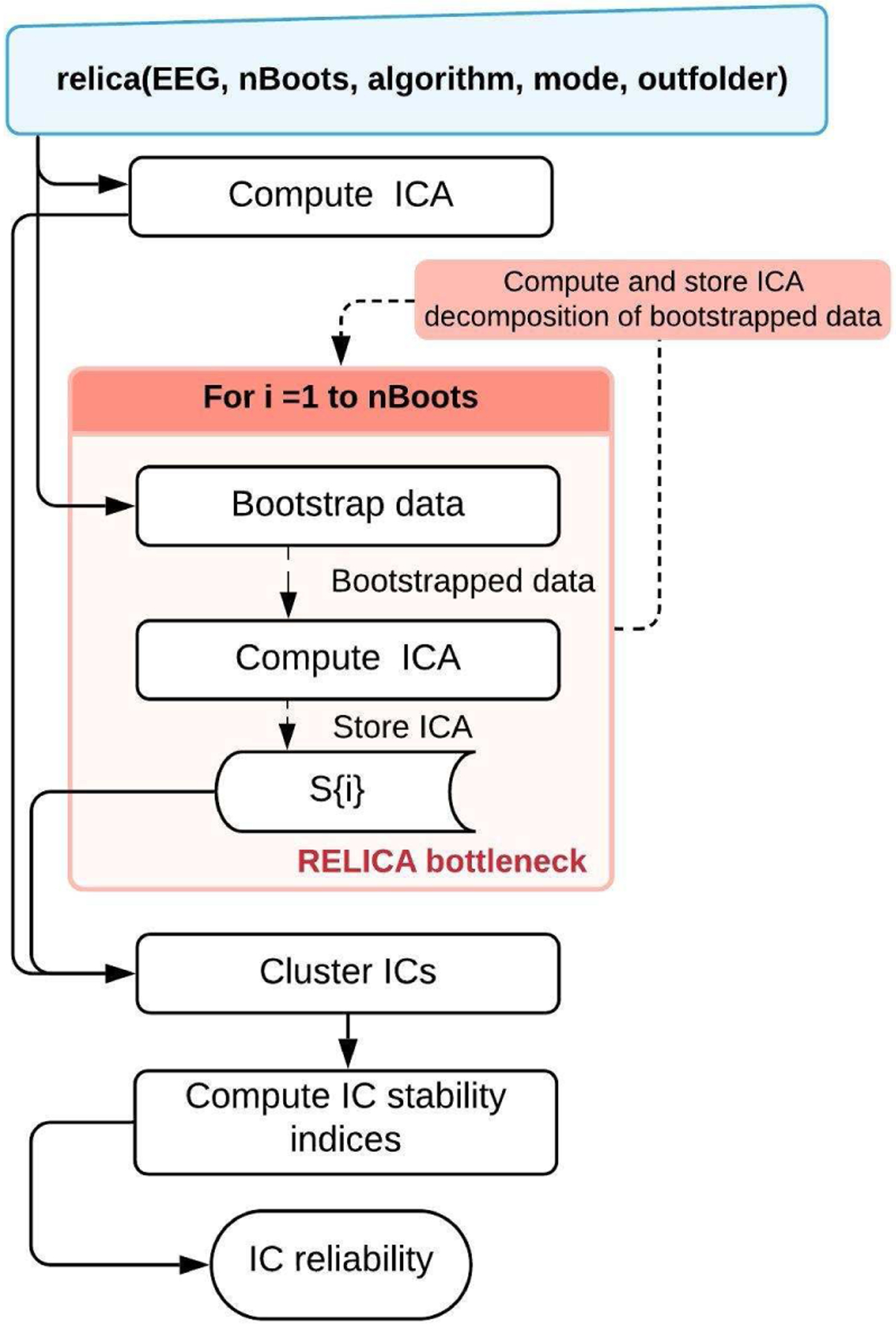
A high-level view of the original implementation of RELICA as proposed by Artoni et al. (Artoni et al., 2014). Given an EEG dataset, relica first performs a reference decomposition of the whole input data, and then multiple ICA decompositions of bootstrapped versions of the same data (red area). The function relica then clusters the identified ICs from all the decompositions and from this computes a measure of the stability of each IC in the reference decomposition.
Here we modified the RELICA implementation to reduce its runtime. First we parallelized the main for loop in the RELICA bottleneck (red area) using parfor from the MATLAB Parallel Computing Toolbox to speed processing on either local (multi-core) or (NSG-accessed) HPC computers. Then we added nsgportal code to enable it to run on XSEDE HPC resources via NSG. Fig. 5 shows a high level view of the resulting NSG-capable RELICA code.
Figure 5. The NSG-capable RELICA plug-in.

A high level view of the NSG-capable RELICA plug-in. Here we have added a last input, compflag, to the original inputs of the main function relica to indicate where the computation should be carried out (‘nsg’ or ‘local’). When relica is called with an EEG set structure and compflag is ‘local’, computation is performed as usual (central yellow panel). If compflag is ‘nsg’, nsgportal code in relica first saves the current EEG dataset in a temporary folder also containing a script (relica_nsg_job.m) generated to run relica on NSG with the parameters provided by the user. This script and the data provided as input are submitted to NSG for processing using pop_nsg with first argument ‘run’ (green panel on right). Additional inputs are provided to indicate the script to run (relica_nsg_job.m), and the designated job ID. This uses pop_nsg options ‘run’, ‘filename’, and ‘jobid’. After the job submission, relica exits, providing the jobID as output. After this, the plug-in processing proceeds as in runica_nsg (Figure 3). Once the NSG job is complete, the user provides the job ID to NSG to retrieve the results by again calling relica with the job ID as the first and only argument. After checking the validity of the submitted job ID, relica uses pop_nsg (with arguments ‘run’ and ‘delete’) to retrieve the results and then delete the job from the user’s NSG account. Finally the submitted EEG dataset plus the RELICA-computed IC reliability measures are provided in the function output.
The user first provides an EEGLAB dataset (in the form of an EEG data structure) for processing by relica. As in Section 5.4.1, by specifying the ‘nsgflag’ argument as ‘local’ or ‘NSG’, the user can direct the computation to the user’s local computer or to NSG resources. In the latter case, relica will create a command line call script to invoke relica on the HPC resource and pass the user-selected options to it. This script plus the input data are saved in a temporary folder and submitted to NSG as a job using pop_nsg with argument ‘run’. If the submission is successful, the (assigned or optionally user-specified) job ID is returned as a text output. This job ID must later be used to retrieve the results when available. Here, the user provides relica with the job ID as first argument.
When relica confirms the validity of the provided job ID and that the job is complete, it downloads the job results (pop_nsg ‘output’), deletes the job from the user’s NSG account (pop_nsg ‘delete’) and loads the output of relica into the EEG dataset structure. The results of RELICA are stored in the EEG dataset structure, allowing seamless integration of NSG computation into RELICA post-processing and results visualization.
5.4.2.1. RELICA performance: HPC versus local computing
To test the performance of RELICA running via NSG on XSEDE cluster Comet, we ran RELICA on the sample EEG dataset referenced in Section 5.1, setting the number of bootstraps to 100 and specifying ICA algorithm ‘runica’ (Extended Infomax ICA) (Bell & Sejnowski, 1995; Lee, Girolami, & Sejnowski, 1999). The two bars on the left in Fig.6 show RELICA processing time on a modern laptop (MacBook Pro, Intel(R) Core(TM) i7–4770HQ 4-core CPU @ 2.20 GHz) and on NSG (1 24-core node of XSEDE network HPC resource Comet). Timestamps for each NSG process were retrieved from the job status XML object introduced in Section 4.2. Despite the relative simplicity of the task, via NSG the RELICA job was performed 11 times faster than on the laptop. A detail of the NSG processing time, magnified x10 in the gray panel on the right side of Fig.6, shows the time the job spent in the different NSG job processing stages including data handling and job scheduling. The scheduling delay depends on the queue wait time of the HPC machine to which NSG sends jobs for processing, so may vary with machine demand intensity.
In a second test, we explored the reliability of the RELICA computation performed via NSG vis a vis two trial runs performed on the same laptop. To do this, we computed pairwise correlations between the IC cluster exemplar maps obtained from RELICA computed via NSG and from the two runs on the laptop using relica options as in Section 5.4.2. Results of this analysis are shown in Fig. 7, in which the sorted best-matching IC-pair correlations between the cluster exemplar IC maps obtained from the NSG and first laptop run (NSG, Laptop1), NSG, and the second laptop run (NSG, Laptop2), and between the two laptop runs (Laptop1, Laptop2), are shown in blue, green and orange traces respectively. As we can see, in all the output IC-pair comparisons (NSG versus Laptop1, NSG versus Laptop2, and Laptop1 versus Laptop2) at least 60 of the 70 IC pairs are near-perfectly replicated. The small remaining variability results from the stochasticity of the ICA computation itself.
Figure 7.
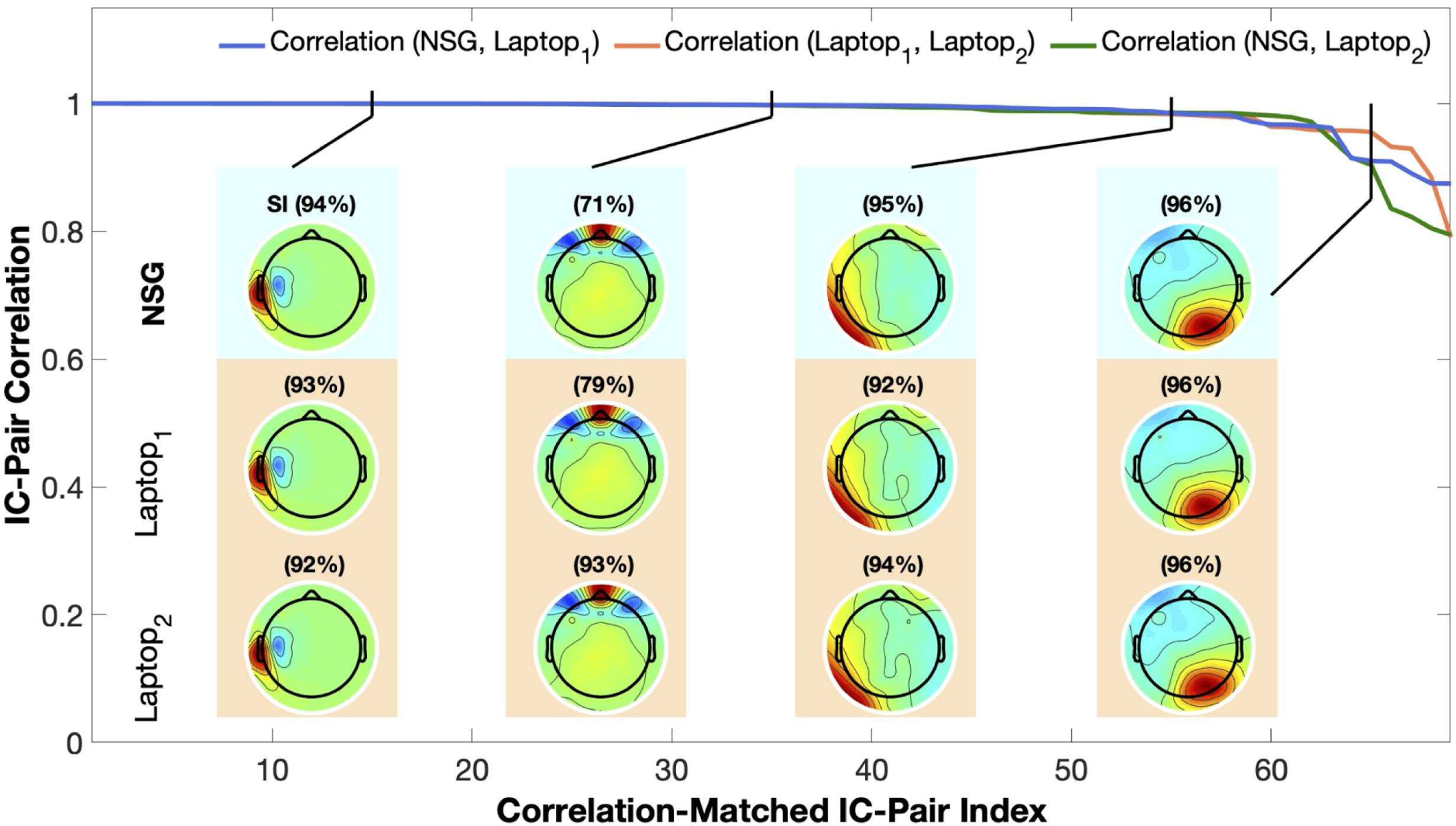
Cluster exemplar scalp map correlations for best-matching IC pairs in the output of the same RELICA decomposition run (twice) on a laptop and (once) on XSEDE network cluster Comet via the Neuroscience Gateway using nsgportal. Top blue trace: correlations of best-matching IC scalp map pairs, sorted by decreasing correlation. Scalp maps: Scalp maps of some best-matching ICs from the three decompositions. Black traces point to the mean correlation rank of these ICs in the three pairwise comparisons among the three maps. Numbers above the scalp maps give the stability index reported by RELICA for these ICs.
Fig. 7 also shows four sets of best-matching IC maps from the three runs. Black lines show the mean correlation-sorted rank of the three correlated IC cluster exemplar maps. Numbers above the maps give the stability index reported by RELICA for these ICs. The high similarity of the maps (even for the rightmost pair), demonstrate the high level of consistency of RELICA output across runs and computing resources.
6. Discussion
Recently (Delorme et al., 2019) we introduced the Open EEGLAB Portal (OEP) framework providing free use of high-performance computing (HPC) resources from the EEGLAB environment through the Neuroscience Gateway (NSG, nsgportal.org). Here, we report the release of an EEGLAB plug-in, nsgportal, implementing an interface between NSG and EEGLAB running on MATLAB on any Web-connected computer. The nsgportal plug-in uses the capabilities of the NSG REST API to enable optional HPC processing of computationally intensive tasks directly from any internet-connected EEGLAB GUI, or from any suitably adapted EEGLAB function or the MATLAB command line. The nsgportal features a flexible and user friendly GUI that allows users to directly submit, manage and retrieve jobs running on the U.S XSEDE network of HPC resources from within any EEGLAB session. The set of nsgportal command line tools supporting the functions of its GUI, enables users to create their own HPC computation-enabled functions, function pipelines, and EEGLAB plug-ins, while making the NSG job submission and results retrieval process as simple as possible. Detailed documentation and wiki pages are available to support nsgportal use and maintenance.
Here we have described nsgportal functions and interfaces, and pointed to online documentation to support their use, maintenance and further development. We have presented three examples to illustrate the use of nsgportal and its potential applications. The first two examples demonstrated how to use nsgportal to create, manage, retrieve results from, and then delete an NSG job within the nsgportal GUI and from the EEGLAB session command line. Here we used a simple script to compute an ICA decomposition of an EEG dataset, save the results, and plot images of the resulting IC scalp maps. The structured approach and guidelines used in these examples can be easily modified to accomplish more computationally intensive tasks.
We then demonstrated (Section 5.4.2), through a concrete example, how to use nsgportal command line functions to implement direct HPC access from within any EEGLAB function or plug-in. For this purpose, we modified the job presented in Section 5.1 by turning the earlier example script into a function accepting inputs pointing to the data matrix to decompose and selecting the ICA algorithm to use in the decomposition. The resulting simple example plug-in (RUNICA_NSG) is available from the EEGLAB plug-in manager and includes abundant comments and documentation to support implementing HPC access using nsgportal within other EEGLAB functions and plug-ins. Please note that EEGLAB plug-ins can be run in NSG processing scripts without including any nsgportal functions; many commonly-used plug-ins are available in the EEGLAB NSG distribution -- any that are not yet available directly within NSG can be uploaded to NSG, along with the data and any other custom EEGLAB functions. Using the nsgportal functions within EEGLAB functions and plug-ins (as in Sections 5.4.1 and 5.4.2) is a convenient option for authors of new or existing plug-ins to consider, as using nsgportal functions they can provide users an NSG-mediated computation option within the plug-in itself.
To show this application operating in an existing EEGLAB plug-in in, we chose to use the more computationally demanding RELICA plug-in of Artoni (Artoni et al., 2014). The addition of nsgportal tools to RELICA was presented in Section 5.4.2.
To explore the impact of these changes on RELICA performance, we ran a RELICA task on NSG and on a modern laptop (MacBoook Pro, Intel(R) Core(TM) i7–4770HQ 4-core CPU @ 2.20GHz). The NSG computation was eleven times faster than on the laptop, and the resulting output across three runs (one via NSG and two on the laptop) were practically indistinguishable (Section 5.4.2.1). NSG computation time included data handling and scheduling delays. These times can vary depending on the system load at the time of submission. To probe variability in the times required for NSG data handling and job scheduling, we submitted the ICA decomposition job (Section 5.1) to NSG repeatedly over a month, and recorded computation times and scheduling delays. Total computation time averaged 31 ± 3 min (mean ± standard deviation). Of this, job scheduling accounted for 2 ± 4 min (with a non-normal distribution). These results provide a rough idea of the stability of NSG regarding scheduling delay, here under relatively modest user demand.
Job file upload and download durations may depend heavily on user Internet bandwidth and job size. Data upload and download times for the RELICA test job were negligible compared to the required length of the computation. However, we must warn users that data I/O speed may be an important factor to consider when weighing the advantages of running NSG jobs requiring very large data file uploads. NSG data I/O speeds vary greatly depending on the bandwidth and then-existing traffic load of each link in the user connection to UCSD.
Lack of adequate computational power currently acts as a kind of ‘compute horizon’ boundary on scientific imagination itself -- enhancing computational power can thus, at its best, both prompt and enable new scientific vision and discovery. However, before submitting EEGLAB jobs to run via NSG using nsgportal, we encourage users to consider the time cost/benefit ratio of submitting a job for processing to NSG versus using a local compute resource. There may be instances in which the use of a local resource and/or alternate analysis method are more efficient than using NSG -- for example, in cases in which uploading the data to NSG might itself require more time than performing the computation locally or where the applied algorithm may itself be easily optimized to achieve the desired computation speed. Use of the substantial compute resources available via NSG should be exercised thoughtfully, particularly in these times in which reducing the human carbon footprint appears key to preserving human civilization in its current form.
Another important aspect to consider before submitting a job to NSG is whether and to what extent the task itself can take good advantage of parallel computing resources. In some (extreme) cases, the overhead involved in setting up and maintaining parallel processing might require more time than the actual computation. Jobs running iterative processes each requiring a long computation time (such as optimal electrical head modeling or ICA decomposition of large datasets), or jobs applying intensive processing to a large amount of data (applying time/frequency analysis to a large EEG study, for example) may best take advantage of NSG resources. Other EEGLAB scripts, functions, and pipelines that use vectorized code already optimized for local computation may not benefit from being sent to NSG for batch processing. The latter could only be justified when the data handled by the pipeline are too large for a personal computer to manage. Finally, jobs that require human interaction (e.g., manual data cleaning) cannot take useful advantage of batch-mode HPC processing.
Some current limitations: Currently, on the Comet supercomputer running (at present) 24-core nodes (note: the planned successor to Comet will use 64-core nodes), and NSG can assign a maximum of one node (24 cores) to each EEGLAB job, though this limitation will soon be relaxed with the planned introduction on Comet of the MATLAB Parallel Server™ library. Another current limitation is the lack of NSG support for data retention and shareability within user NSG accounts. Under a current NeuroElectroMagnetic data Archive and tools Resource (NEMAR) project operating under the OpenNeuro data archive umbrella (openneuro.org), we are developing a solution that will ultimately allow joint use within NSG of personal and/or public data published on OpenNeuro.
7. Conclusions
Here we have described nsgportal, an EEGLAB plug-in toolbox providing neuroscientists and others using the EEGLAB signal processing environment (sccn.ucsd.edu/eeglab) free access to HPC resources of the U.S. XSEDE high-performance computing network directly from the EEGLAB GUI or session MATLAB command line through a programmatic interface to the Neuroscience Gateway (nsgportal.org). We also showed how to use the nsgportal command line tools to equip EEGLAB functions and plug-in toolboxes with direct HPC run capabilities. Finally, we performed a proof-of-concept test of nsgportal performance on a suitable EEG data processing task, and confirmed the equivalence of the obtained results.
The ease and flexibility of the nsgportal tools can accelerate any neuroelectromagnetic data research in need of higher computational power. In particular, dynamic modeling and machine learning approaches that are now being applied to human electrophysiological data can require unprecedented computing power. Limitations in available computing resources might be said to constitute a ‘compute horizon’ beyond which researchers cannot or dare not explore or imagine. Extending the compute horizon for electrophysiological imaging research using nsgportal tools may hopefully stimulate researchers to use new research methods built on richer data models and thereby obtain new results stimulating new insights into brain function and health.
Supplementary Material
Figure 6. RELICA processing time on NSG versus a laptop.
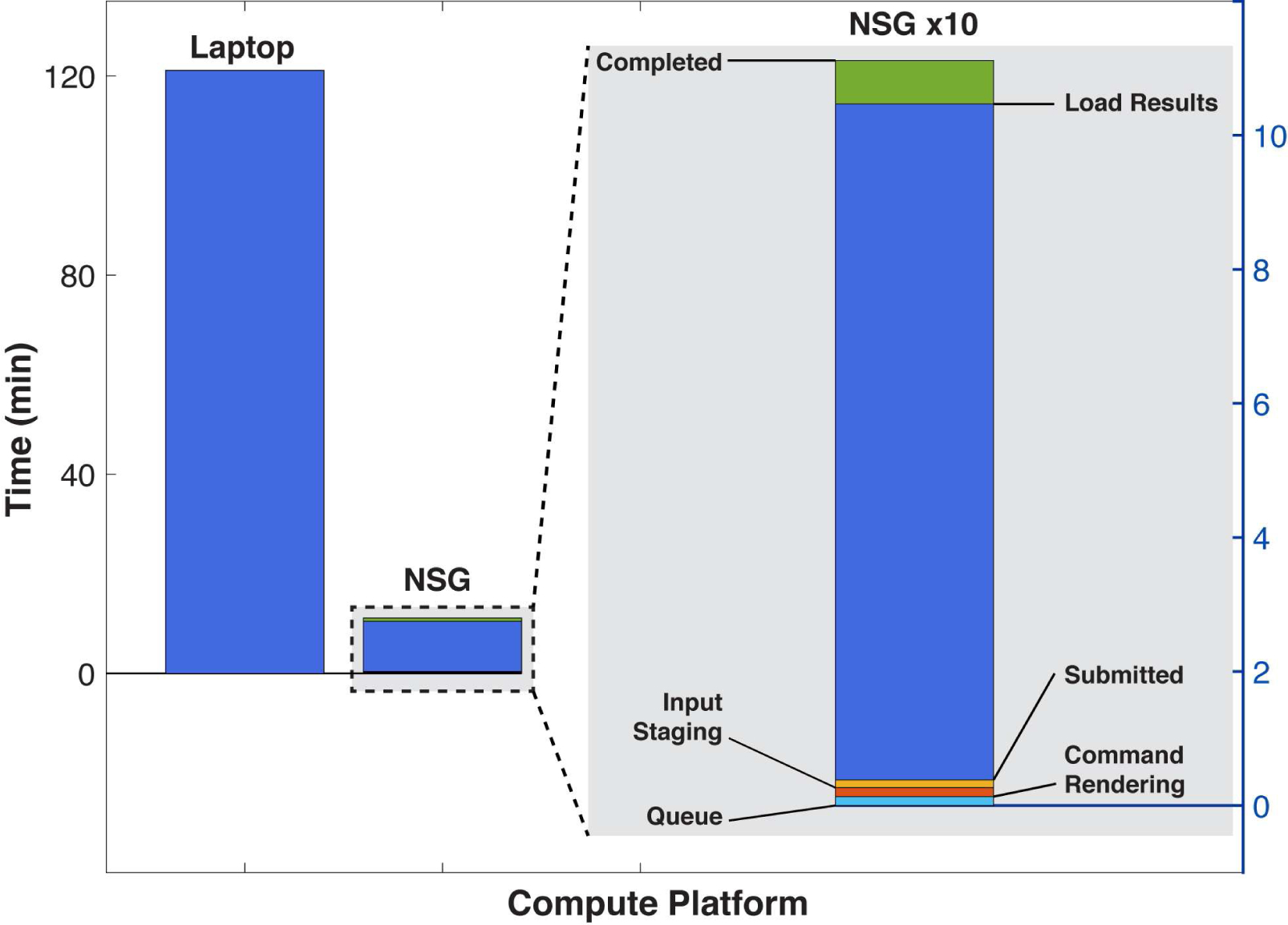
The two leftmost bars represent recorded processing time running RELICA on the sample data using a laptop (MacBoook Pro, Intel(R) Core(TM) i7–4770HQ 4-core CPU @ 2.20GHz) and via NSG (1 node, all 24 cores of the XSEDE network cluster Comet) using 100 iterations of ICA decomposition using ‘runica’ (Scott Makeig et al., 1997). A detail of NSG processing time magnified by 10 is shown in the right gray panel. Here the time has been split to show the time the job spent on each of the NSG stages. Here the initialization stages (bottom of right bar) produced only a small delay, though this may vary with current system load. Data upload and download times (not shown) here required about as much time as the 3 job initialization stages (right column bottom), but these delays are still negligible compared to the time saved by performing the computation using NSG versus on the laptop. Upload and download times depend heavily on the Web bandwidth and traffic load between the user machine and UCSD.
8. Funding sources
This work was supported by the National Institutes of Health, U.S.A. (R01-NS047293, R24-MH120037, R01-MH123231) and the National Science Foundation (#1458840), and by a gift from The Swartz Foundation (Old Field, NY).
Acknowledgements
Figure 1 uses elements designed by macrovector/Freepik and Freepik (freepik.com). The authors also thank all nsgportal beta users for their encouragement and valuable feedback and to Dr. Johanna Wagner for helping with the references.
Appendices
Appendix A.
The code samples below show typical curl calls to the NSG REST API used in nsgportal for: submitting a job, checking job status, retrieving job results, and deleting a job. Lines in the code samples have been numbered for tutorial purposes (they are not present in the actual code). These examples assume the user has been registered in NSG and obtained a valid username and password, designated as your_username and your_password respectively. The application key and NSG URL (nsgr.sdsc.edu:8443/cipresrest/v1) are designated as $KEY and $URL respectively.
A.1. Submitting a job
Code sample A1.
1 curl -s -u your_username:your_password \ 2 -H cipres-appkey:$KEY \ 3 $URL/job/your_username \ 4 -F tool=EEGLAB_TG \ 5 -F input.infile_=@”/data/TestingEEGLABNSG.zip” \ 6 -F vparam.filename_=run_ica_nsg.m 7 -F metadata.clientJobId=TestingEEGLABNSG \ 8 -F vparam.outputfilename_=“nsgresults_TestingEEGLABNSG” \ 9 -F vparam.number_nodes_=1 \
The code sample above (Code sample A1) shows a curl command following NSG REST API specification for submitting a job to NSG using EEGLAB as a tool (see Line 4). The job zip file is specified with the parameter input.infile_ in Line 5, and the script to run with the parameter vparam.filename_ in Line 6. A job ID is indicated with the parameter metadata.clientJobId in Line 7. Although not compulsory in NSG REST API specifications, in nsgportal the use of a job ID is mandatory. The rationale for this is to provide meaningful job identifiers to users, rather than a long job url used in NSG to tag and track jobs. A default job ID assigned in nsgportal is created by taking the name of the file in input.infile_ and attaching three randomly generated numbers in its end. This job ID can be modified at the user’s convenience. In the same way, in nsgportal, the name of the output file is assigned a default value created by attaching the prefix ‘nsgresults_’ to the value defined in metadata.clientJobId. This value is set in Line 8 of the code sample above by using the parameter vparam.outputfilename_. Finally, we define the number of nodes requested for computation with the parameter number_nodes_ in Line 9. At this moment, NSG only allows the selection of one node (currently, of 24 cores) for each EEGLAB job. For more details on the MATLAB call of the code above, see nsgportal function nsg_run.
A.2. Checking job status
Code sample A2.
1 curl -s -u your_username:your_password \ 2 -H cipres-appkey:$KEY \ 3 $URL/job/your_username
The code above (Code sample A2) allows checking for the status of all jobs under the user myusername account. The MATLAB call for this code, as well as the one to retrieve job results, presented next in A.3, are implemented in the ngportal function nsg_jobs.
A.3. Retrieving job results
Code sample A3.
1 curl -s -u your_username:your_password \ 2 -H cipres-appkey:$KEY \ 3 $URL/job/your_username/NGBW-JOB-EEGLAB_TG-Unique_job_identifier/output
The code sample above (Code sample A3) is used to retrieve the job files and results (with job url :$URL/job/nsgusername/NGBW-JOB-EEGLAB_TG-Unique_job_identifier)
A.4. Deleting a job
Code sample A4.
1 curl -u your_username:your_password \ 2 -H cipres-appkey:$KEY \ 3 -X DELETE \ 4$URL/your_username/NGBW-JOB-EEGLAB_TG-Unique_job_identifier
The code above (Code sample A4) is used to delete the job specified by the job URL in line 4 from the user’s NSG account. The MATLAB call for this code is implemented in the ngportal function nsg_delete.
Appendix B
A successful NSG job should progress through the following states (NSG):
QUEUE - The job has been validated and placed in the NSG queue.
COMMANDRENDERING - The job has reached the head of the queue, and NSG has created the command line that will run the job on the HPC resource.
INPUTSTAGING - NSG has created a temporary working directory for the job on the execution HPC host and copied the input files over.
SUBMITTED - The job has been submitted to the HPC host.
LOAD_RESULTS - The job has finished running on the HPC host, and NSG has begun to transfer the results back.
COMPLETED - Results have been successfully transferred and are available in the NSG web portal.
Appendix C
The stimuli presented in this experiment comprised two sets of 300 grayscale photographs, half from known people and half from unknown people. Images of known, unknown and scrambled faces were presented for random durations between 800 and 1,000 ms following the appearance of a fixation cross cue with a random duration between 400 and 600 ms. During the 1,700-ms interstimulus interval, a central white circle was presented. The participant was told to fixate centrally throughout the experiment and asked to press one of two keys based on the degree of bilateral symmetry (more or less) of each presented face. EEG data were recorded synchronously with MEG data using an Elekta Neuromag Vectorview 306 system (Helsinki, FI). A 70-channel Easycap (easycap.de/wordpress) cap was used to record the EEG data, with electrode layout conforming to the extended 10–10% system. The EEG electrodes location was digitized by using a 3-D digitizer (Fastrak Polhemus Inc., Colchester, VA, USA). Data were acquired at an 1100-Hz sampling rate with a low pass filter applied below 350 Hz and no high pass filter. The EEG reference electrode was placed on the nose, and the common ground electrode was placed at the left collar bone. Stimuli were presented during six, 7.5-min runs. The protocol is described in more detail in Wakeman and Henson, 2015 (Wakeman & Henson, 2015). Data preprocessing, performed using EEGLAB and custom scripts, written in MATLAB (The Mathworks, Inc.), proceed through the following steps:
Data file from subject sub-11 was downloaded from OpenNeuro at openneuro.org/crn/datasets/ds000117/snapshots/1.0.3/files/sub-11:ses-meg:meg:sub-11_ses-meg_task-facerecognition_run-01_meg.fif
Raw EEG data and event information were imported using FileIO plug-in for EEGLAB. The location of the fiducials were added to the channel location and the channel montage was rotated to match EEGLAB format. Event latencies were corrected by 34 ms as reported by the data authors. Finally, the six sequentially collected task run files were merged and saved.
Downsample data from 1100 Hz to 500 Hz using EEGLAB function pop_resample.
High-pass filter the data above 1 Hz (FIR, Hamming windowed, transition bandwidth 1 Hz).
Remove line noise (50 Hz) by applying a Hamming-windowed (sinc) FIR notch filters centered at the line frequency and its harmonics (e.g., 50 Hz, 100 Hz, 150 Hz, and 200 Hz).
Remove segments in the data containing non-brain artifact (e.g., high-frequency muscle noise and other irregular artifacts) as identified by visual inspection.
Apply common-average reference computation involving all channels
Extract data segments time-locked to the presentation of each of the three main events (known, unknown and scrambled face) and spanning from 1 sec before to 2 sec following stimulus onsets.
Save the dataset in EEGLAB .set format.
Code for the processing described here can be accessed in the GitHub repository7 of supporting material.
Appendix D
This appendix describes in more detail the elements and section of the nsportal main GUI in Figure 2
Section A: Interacting with submitted NSG jobs.
This section contains the following components:
(List box) Select job: lists all existing NSG jobs under the user credentials. Font color coding is used to indicate the job state: completed (Completed, in green), still being processed (Processing, in blue), returning a MATLAB syntax-related job error (MATLAB error, in red) or an NSG-related error (NSG error, in orange). For example, in Figure 2, the job named oep_runica is shown in green, indicating that the job has been completed. Most of the functionalities accessed in this section of the graphical interface will act on the job selected in this list.
(Button) Refresh job list: refreshes the list of existing NSG jobs under the user account.
(Checkbox) Auto-refresh job list: flags the interface to update the user NSG jobs status every thirty seconds. The status of the selected job is shown to the right of ‘NSG job status’ label. This functionality uses a MATLAB timer object to schedule a sequential update of the status of the jobs.
(Button) Delete this NSG job: removes the selected job from the user’s NSG account.
(Button) Matlab output log: downloads and displays the accumulated MATLAB command line output for the selected NSG job.
(Button) Matlab error log: downloads and displays the MATLAB error log for the selected job (if available).
(Button) Download job results: download the job result .zip file from the currently selected job. (Button) Load/plot results: launch a window controlling the browsing, loading, and displaying of job results.
Section B: Checking NSG job status.
This section of the GUI window (beneath ‘NSG job status’) displays the job status and the messages issued by NSG during the submission and processing of the job currently selected in the jobs list. The job-status is displayed in capital letters and reflects the current stage of the job at the time of the last update (see Appendix B for the list of states). These states, while related to the four color-coded categories in the job list, are mostly referring to more fundamental NSG processing stages in the selected job.
Section C: Submitting a job to NSG.
The lower section of the GUI (beneath ‘Submit new NSG job’) is dedicated to submitting jobs to NSG as well as for performing testing of the jobs in the user’s local computer. In what follows, we describe and explain the functionality of each component.
(Button) Browse and (edit box) Job folder or zip file: This button launches a window to browse for a .zip file or a folder containing the job to be submitted to NSG (or tested locally). Once the file/folder is selected, its full path is displayed in (edit box) Job folder or zip file. The value in the edit box is used in the NSG-R command line option input.infile_ when submitting a job.
(Drop-down menu) Matlab script to execute: Show all the MATLAB script files (.m) in the job file (.zip file or file folder) selected above. The Matlab script for NSG execution or testing must be selected here. In the case when a folder is selected, the listing of files is done by scanning the folder content with the MATLAB function dir. When a .zip file is selected, the listing of the .m files is done through the nsgportal function listzipcontents.m. This function uses Java objects and methods native to MATLAB to read the content of the .zip file without unzipping it. Both options, for folder and .zip files, are implemented in the callback of (button) Browse in pop_nsg.m. The value in this menu is used in the NSG-R command line option input.filename_ when submitting a job (see examples in Appendix A).
(Button) Test job locally: runs the job indicated by the job file and MATLAB script selected above on the local computer where nsgportal is being executed. Local testing of the jobs is intended to be done by using a downscaled version of the job to be submitted. This can be done, e.g., by using a scaled-down script mimicking the main steps to be performed at NSG, but on a lower scale. Take the example of a job that processes hundreds, say N=200, of subjects in a for loop, the downscaled version of this job may result from the shortening of the loop from 200 to 2. This button is strategically located to the right of (edit box) Matlab script to execute, to emphasize the idea that job testing should be done on a different script from the one intended to run on the HPC resource via NSG.
(Edit box) Job ID (default or custom): displays the unique identifier for the NSG job. The information in this field is generated automatically when a job folder or .zip file is selected in the Browse button above. The default value assigned here is created by taking the name of the job folder or .zip file and attaching three randomly generated numbers at the end. We encourage users to modify this field for easier recall and reference. The value here is used in the optional NSG-R command line option input.clientJobId when submitting a job. This option is compulsory when the job is submitted from nsgportal, since the job ID value is used instead of the long job URL for job management and tracking in the nsgportal GUI machinery.
(Edit box) NSG run options (see Help): allows the user to specify options for the to be submitted job. The options defined here are appended to the curl command when the job is submitted to NSG. Currently, nsgportal includes options for defining the maximum runtime allocation in NSG (runtime), the number of NSG nodes to use in the computation (nnode) (currently, 1), and other options (see the pop_nsg and nsg_run help messages). The list of options will eventually include all NSG-R options.
(Button) Run job on NSG: submits the job to NSG. The button callback calls nsg_run and passes the inputs provided by the user in the GUI. These inputs are formatted in a curl command (see the example in Appendix A), before being issued for execution on the local computer.
(Buttons) Help and Close respectively display the help message of pop_nsg in a browser window and close the pop_nsg GUI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Currently, nsgportal functions cannot be run using Octave.
References
- Akalin Acar Z, & Makeig S (2010). Neuroelectromagnetic forward head modeling toolbox. Journal of neuroscience methods, 190, 258–270. doi: 10.1016/j.jneumeth.2010.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander LM, Escalera J, Ai L, Andreotti C, Febre K, Mangone A, … Kovacs M (2017). The Healthy Brain Network Biobank: An open resource for transdiagnostic research in pediatric mental health and learning disorders. bioRxiv, 149369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artoni F, Menicucci D, Delorme A, Makeig S, & Micera S (2014). RELICA: a method for estimating the reliability of independent components. Neuroimage, 103, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AJ, & Sejnowski TJ (1995). An information-maximization approach to blind separation and blind deconvolution. Neural computation, 7(6), 1129–1159. [DOI] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Cockfield J, Makeig S, Rognon T, La Valle C, Miyakoshi M, & Robbins KA (2016). Hierarchical Event Descriptors (HED): semi-structured tagging for real-world events in large-scale EEG. Frontiers in neuroinformatics, 10, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Makeig S, & Robbins KA (2016). Preparing laboratory and real-world EEG data for large-scale analysis: a containerized approach. Frontiers in neuroinformatics, 10, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Mullen T, Kothe C, Su K-M, & Robbins KA (2015). The PREP pipeline: standardized preprocessing for large-scale EEG analysis. Frontiers in neuroinformatics, 9, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower JM, & Beeman D (2012). The book of GENESIS: exploring realistic neural models with the GEneral NEural SImulation System: Springer Science & Business Media. [Google Scholar]
- Brunet D, Murray MM, & Michel CM (2011). Spatiotemporal analysis of multichannel EEG: CARTOOL. Computational intelligence and neuroscience, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canolty RT, & Knight RT (2010). The functional role of cross-frequency coupling. Trends in cognitive sciences, 14(11), 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AP, Brüderle D, Eppler JM, Kremkow J, Muller E, Pecevski D, … Yger P (2009). PyNN: a common interface for neuronal network simulators. Frontiers in neuroinformatics, 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme A, Majumdar A, Sivagnanam S, Martinez-Cancino R, Yoshimoto K, & Makeig S (2019). The Open EEGLAB portal. Paper presented at the 2019 9th International IEEE/EMBS Conference on Neural Engineering (NER). [Google Scholar]
- Delorme A, & Makeig S (2004). EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. Journal of neuroscience methods, 134(1), 9–21. [DOI] [PubMed] [Google Scholar]
- Di Martino A, Yan C-G, Li Q, Denio E, Castellanos FX, Alaerts K, … Dapretto M (2014). The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular psychiatry, 19(6), 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B (2012). FreeSurfer. Neuroimage, 62(2), 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabard-Durnam LJ, Mendez Leal AS, Wilkinson CL, & Levin AR (2018). The Harvard Automated Processing Pipeline for Electroencephalography (HAPPE): standardized processing software for developmental and high-artifact data. Frontiers in neuroscience, 12, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewaltig M-O, & Diesmann M (2007). Nest (neural simulation tool). Scholarpedia, 2(4), 1430. [Google Scholar]
- Goodman DF, & Brette R (2009). The brian simulator. Frontiers in neuroscience, 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski KJ, Auer T, Calhoun VD, Craddock RC, Das S, Duff EP, … Halchenko YO (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data, 3, 160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramfort A, Luessi M, Larson E, Engemann DA, Strohmeier D, Brodbeck C, … Hämäläinen MS (2014). MNE software for processing MEG and EEG data. Neuroimage, 86, 446–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke M, & Halchenko Y (2011). Neuroscience Runs on GNU/Linux. Frontiers in neuroinformatics, 5(8). doi: 10.3389/fninf.2011.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines ML, & Carnevale NT (2008). Translating network models to parallel hardware in NEURON. Journal of neuroscience methods, 169(2), 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdgraf C, Appelhoff S, Bickel S, Bouchard K, D’Ambrosio S, David O, … Gorgolewski C (2018). BIDS-iEEG: an extension to the brain imaging data structure (BIDS) specification for human intracranial electrophysiology. [DOI] [PMC free article] [PubMed]
- Holdgraf C, Appelhoff S, Bickel S, Bouchard K, D’Ambrosio S, David O, … Foster BL (2019). iEEG-BIDS, extending the Brain Imaging Data Structure specification to human intracranial electrophysiology. Scientific Data, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T-W, Girolami M, & Sejnowski TJ (1999). Independent component analysis using an extended infomax algorithm for mixed subgaussian and supergaussian sources. Neural computation, 11(2), 417–441. [DOI] [PubMed] [Google Scholar]
- Lotte F, Bougrain L, Cichocki A, Clerc M, Congedo M, Rakotomamonjy A, & Yger F (2018). A review of classification algorithms for EEG-based brain–computer interfaces: a 10 year update. Journal of neural engineering, 15(3), 031005. [DOI] [PubMed] [Google Scholar]
- Makeig S, Jung T-P, Bell AJ, Ghahremani D, & Sejnowski TJ (1997). Blind separation of auditory event-related brain responses into independent components. Proceedings of the National Academy of Sciences, 94(20), 10979–10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeig S, Kothe C, Mullen T, Bigdely-Shamlo N, Zhang Z, & Kreutz-Delgado K (2012). Evolving signal processing for brain–computer interfaces. Proceedings of the IEEE, 100(Special Centennial Issue), 1567–1584. [Google Scholar]
- Makeig S, Westerfield M, Jung TP, Enghoff S, Townsend J, Courchesne E, & Sejnowski TJ (2002). Dynamic brain sources of visual evoked responses. Science, 295(5555), 690–694. doi: 10.1126/science.1066168 [DOI] [PubMed] [Google Scholar]
- Martínez-Cancino R, Heng J, Delorme A, Kreutz-Delgado K, Sotero RC, & Makeig S (2019). Measuring transient phase-amplitude coupling using local mutual information. Neuroimage, 185, 361–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, & Schwartz T (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Paper presented at the 2010 gateway computing environments workshop (GCE). [Google Scholar]
- Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, … O’Leary MA (2015). A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evolutionary Bioinformatics, 11, EBO. S21501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, … Beckett L (2005). Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimer’s & Dementia, 1(1), 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niso G, Gorgolewski KJ, Bock E, Brooks TL, Flandin G, Gramfort A, … Moreau JT (2018). MEG-BIDS, the brain imaging data structure extended to magnetoencephalography. Scientific data, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NSG. nsgportal. Retrieved from http://www.nsgportal.org/guide.html
- O’Connor D, Potler NV, Kovacs M, Xu T, Ai L, Pellman J, … Ghosh S (2017). The Healthy Brain Network Serial Scanning Initiative: a resource for evaluating inter-individual differences and their reliabilities across scan conditions and sessions. Gigascience, 6(2), giw011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onton J, Delorme A, & Makeig S (2005). Frontal midline EEG dynamics during working memory. Neuroimage, 27(2), 341–356. [DOI] [PubMed] [Google Scholar]
- Oostenveld R, Fries P, Maris E, & Schoffelen J-M (2011). FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Computational intelligence and neuroscience, 2011, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroni A, Bahreini A, & Langer N (2019). Automagic: Standardized preprocessing of big EEG data. Neuroimage. [DOI] [PubMed] [Google Scholar]
- Pfurtscheller G, & Da Silva FL (1999). Event-related EEG/MEG synchronization and desynchronization: basic principles. Clinical neurophysiology, 110(11), 1842–1857. [DOI] [PubMed] [Google Scholar]
- Ray S, Deshpande R, Dudani N, & Bhalla US (2008). A general biological simulator: the multiscale object oriented simulation environment, MOOSE. BMC Neuroscience, 9(1), P93. [Google Scholar]
- Rekapalli B, Giblock P, & Reardon C (2013). PoPLAR: portal for petascale lifescience applications and research. BMC bioinformatics, 14(9), S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Loth E, Banaschewski T, Barbot A, Barker G, Büchel C, … Gallinat J (2010). The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Molecular psychiatry, 15(12), 1128. [DOI] [PubMed] [Google Scholar]
- Sivagnanam S, Majumdar A, Yoshimoto K, Astakhov V, Bandrowski A, Martone M, & Carnevale NT (2015). Early experiences in developing and managing the neuroscience gateway. Concurrency and Computation: Practice and Experience, 27(2), 473–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivagnanam S, Majumdar A, Yoshimoto K, Astakhov V, Bandrowski A, Martone ME, & Carnevale NT (2013). Introducing the Neuroscience Gateway. Paper presented at the IWSG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivagnanam S, Yoshimoto K, Carnevale NT, & Majumdar A (2018). The Neuroscience Gateway: Enabling Large Scale Modeling and Data Processing in Neuroscience. Paper presented at the Proceedings of the Practice and Experience on Advanced Research Computing. [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, … Landray M (2015). UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS medicine, 12(3), e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadel F, Baillet S, Mosher JC, Pantazis D, & Leahy RM (2011). Brainstorm: a user-friendly application for MEG/EEG analysis. Computational intelligence and neuroscience, 2011, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tort AB, Komorowski R, Eichenbaum H, & Kopell N (2010). Measuring phase-amplitude coupling between neuronal oscillations of different frequencies. Journal of neurophysiology, 104(2), 1195–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, & Connelly A (2012). MRtrix: diffusion tractography in crossing fiber regions. International journal of imaging systems and technology, 22(1), 53–66. [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens T, Bucholz R, … Curtiss SW (2012). The Human Connectome Project: a data acquisition perspective. Neuroimage, 62(4), 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman DG, & Henson RN (2015). A multi-subject, multi-modal human neuroimaging dataset. Scientific data, 2, 150001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins-Diehr N, Gannon D, Klimeck G, Oster S, & Pamidighantam S (2008). TeraGrid science gateways and their impact on science. Computer, 41(11), 32–41. [Google Scholar]
- Wolters CH, Kuhn M, Anwander A, & Reitzinger S (2002). A parallel algebraic multigrid solver for finite element method based source localization in the human brain. Computing and visualization in science, 5(3), 165–177. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


