Abstract
Base editors are an innovative addition to the genome editing toolbox that introduced a new genome editing strategy to the field. Instead of using double-stranded DNA breaks, base editors use nucleobase modification chemistry to efficiently and precisely incorporate single nucleotide variants (SNVs) into the genome of living cells. Two classes of DNA base editors currently exist: deoxycytidine deamination-derived editors (CBEs, which facilitate C•G to T•A mutations) and deoxyadenosine deamination-derived base editors (ABEs, which facilitate A•T to G•C mutations). More recently, the development of mitochondrial base editors allowed the introduction of C•G to T•A mutations into mitochondrial DNA as well. Base editors show great potential as therapeutic agents and research tools, and extensive studies have been carried out to improve upon the original base editor constructs to aid researchers in a variety of disciplines. Despite their widespread use, there are few publications that focus on elucidating the biological pathways involved during the processing of base editor intermediates. Because base editors introduce unique types of DNA damage products (a U•G mismatch with a DNA backbone nick for CBEs, and an I•T mismatch with a DNA backbone nick for ABEs) to facilitate genome editing, a deep understanding of the DNA damage repair pathways that facilitate or impede base editing represents an important aspect for the further expansion and improvement of the technologies. Here, we first review canonical deoxyuridine, deoxyinosine, and single-stranded break repair. Then, we discuss how interactions among these different repair processes can lead to different base editing outcomes. Through this review, we hope to promote thoughtful discussions on the DNA repair mechanisms of base editing, as well as help researchers in the improvement of the current base editors and the development of new base editors.
1. Introduction
Single nucleotide variants (SNVs) account for over 96% of currently identified human genetic variation.[1] While the majority of observed genetic variants (over 99%) currently lack a clinical interpretation, of those known to cause genetic diseases, roughly 55% are SNVs. [2] Therefore, the ability to precisely and efficiently introduce SNVs into the genome of living cells holds great potential for the treatment of genetic diseases, as well as for modelling the impact that currently unclassified genetic variants have on cellular function and human health. The development of CRISPR-associated (Cas) nucleases as genome editing agents has led to a revolution and paved the way for the development of next generation genome editing agents, such as base editors, the subject of this review. [3,4]
Originally functioning as adaptive immune systems in bacteria and archaea, Cas nucleases (such as Cas9 and Cas12) have been repurposed to perform genome editing in a wide variety of cells and organisms. Their ability to create double-stranded DNA breaks (DSBs) with high efficiencies in a programmable, RNA-guided manner has enable their rapid adoption by the genome editing community at large. To direct Cas nucleases to a genomic locus of interest, researchers simply change the spacer sequence of the guide RNA (gRNA). This then facilitates Cas:gRNA-DNA binding via Watson-Crick-Franklin base pairing rules between the spacer sequence of the gRNA and the protospacer sequence of the target locus. The protospacer must also be directly next to a protospacer adjacent motif (PAM). Once bound to its target DNA sequence, the Cas nuclease will introduce a DSB.
However, the introduction of a DSB is only the first step of the genome editing process. The next step involves the cellular response to this DNA damage product. In fact, all current genome editing technologies (including “nontraditional” genome editing agents, such as base editors and prime editors, which are discussed later) employ a two-step process to perform their function: 1) Deliberately introduce a DNA lesion at a user-programmed location in the genome, followed by 2) Manipulate or rely on the endogenous DNA damage repair pathways to process the lesion into a desired editing outcome. In the case of DSB repair (which has been extensively studied and reviewed elsewhere [5,6]), multiple pathways compete with each other to process the lesion, resulting in a mixture of outcomes. In brief, processing by the non-homologous end joining (NHEJ) or microhomology-mediated end joining (MMEJ) pathways results in insertions, deletions, translocations, and other rearrangements at the site of the break, which are particularly useful for gene disruption studies.[7,8] Alternatively, precise DNA edits can be installed via the homology-directed repair (HDR) pathway through the use of an exogenously supplied DNA donor template. This donor template is designed to contain the desired sequence modification flanked by homology arms; the HDR machinery will use this as a template to repair the DSB and in the process incorporate the modified sequence into the genome.[9] HDR-mediated genome editing can be used to install any type of sequence modification of interest, including SNVs, but it is highly cell cycle dependent (and only active during G2 and S phases), and usually outcompeted by NHEJ for the repair of DSBs. [10,11]
DNA base editing is a recent technology specifically developed to combat these challenges inherent to DSB-reliant genome editing methods. [12,13] Base editors facilitate the efficient and precise introduction of SNVs using non-DSB DNA damage intermediates. DNA base editors are generally protein fusion constructs consisting of a catalytically impaired Cas nuclease (dCas9, dCas12, or Cas9n) linked to a single-stranded DNA (ssDNA)-specific nucleobase modifying enzyme (Figure 1A). Upon localization of the Cas protein to its target DNA sequence, hybridization of the gRNA to its complementary DNA sequence displaces and exposes a small stretch of ssDNA at the PAM-distal region of the non-complementary strand. This exposure allows the ssDNA modifying enzyme to gain access to and chemically modify its target base within this ssDNA window. To date, by harnessing hydro-deamination chemistry performed by deaminase enzymes, two major classes of base editors have been developed. Cytosine base editors (CBEs) catalyze the transition mutation of C•G base pairs to T•A base pairs via the deamination of deoxycytidine to deoxyuridine as the mutagenic intermediate (Figure 1B). Adenine base editors (ABEs) catalyze the transition mutation of A•T base pairs to G•C base pairs via the deamination of deoxyadenosine to form deoxyinosine as the mutagenic intermediate (Figure 1C). Because both deoxyuridine and deoxyinosine are noncanonical DNA nucleotides, they are recognized as DNA lesions by the cell and repaired by cellular DNA repair mechanisms distinct from those utilized in DSB-reliant methods. To improve base editing efficiencies, the catalytically dead dCas9 protein can be changed to the nickase version, Cas9n, to introduce a nick on the non-modified DNA strand. This nick serves to bias the cellular repair mechanisms to preferentially replace this strand and use the mutagenic intermediate as a template for repair.
Figure 1: Overview of DNA base editing technologies.
A) DNA base editors are generally protein fusion constructs consisting of a catalytically impaired Cas nuclease (dCas9, dCas12, or Cas9n) linked to a ssDNA-specific nucleobase modifying enzyme. (1) Binding of a base editor to its target locus forms a DNA/RNA hybrid R-loop, which exposes the target nucleotide X in a stretch of ssDNA outside of the Cas protein. (2) The ssDNA nucleobase modifying enzyme now chemically modifies its target base X into X*, a noncanonical nucleotide that serves as a mutagenic intermediate. (3) X* is further processed by cellular repair or replication machinery, resulting in its conversion to P. For CBEs, X•Z, X*•Z and P•Q are C•G, U•G, and T•A, respectively. For ABEs, they are A•T, I•T, and G•C, respectively. B) The ssDNA nucleobase modifying enzymes used in CBEs are cytidine deaminases, which chemically modify deoxycytidine to deoxyuridine via hydro-deamination chemistry. C) Adenosine deaminases are used in ABEs, which chemically modify deoxyadenosine to deoxyinosine via hydro-deamination chemistry. D) The unique DNA damage intermediate harnessed for base editing, featuring a modified nucleotide(X*)-containing mismatch and a nick located 3 to 14 nucleotides away in the 5’ direction from the mismatched site on the unmodified strand. E) DdCBEs are constructed by fusing two halves of a dsDNA cytidine deaminase DddA to separate TALE proteins that bind at neighboring “half sites”.
Base editors therefore use unique types of DNA damage: a mutagenic, noncanonical DNA nucleotide on one strand, and a DNA backbone nick on the other strand, 5’ upstream from the modified nucleotide (Figure 1D). A variety of comprehensive reviews have been reported elsewhere on the development of base editing technologies, including the many modifications of the original systems. [14-18] Therefore, here we focus on the potential DNA repair mechanisms of base editing. We hope to aid researchers in the improvement of current base editors and promote thoughtful discussion on the identification of potential intermediate to use for new base editors.
2. Deoxycytidine Deamination-Derived Base Editors: C•G to T•A Base editors (CBEs), C•G to G•C Base Editors (CGBEs) and mitochondrial C•G to T•A Base editors (DdCBEs)
The first DNA base editor utilized a naturally occurring ssDNA-specific cytidine deaminase, rAPOBEC1, fused to dCas9 to convert target deoxycytidines to deoxyuridines. As deoxyuridine has the base-pairing properties of deoxythymidine, this base editor was engineered to introduce C•G to T•A base pair conversions in a programmable manner. [12] Further optimizations led to the use of Cas9n and the addition of the uracil glycosylase inhibitor (UGI) peptide. The UGI component served to prevent excision of the uridine intermediate by the cellular DNA repair machinery (discussed in more detail below). Extensive optimizations of the original CBE construct have created a plethora of different CBEs, which are reviewed elsewhere. [14,15,19]
While CBEs were expected to cleanly introduce C•G to T•A SNVs, concomitant C•G to non-T•A conversions (such as C•G to G•C or A•T) at the target deoxycytidine were commonly observed. Knock-out studies demonstrated these conversions required the base excision repair enzyme uracil N-glycosylase (UNG).[20] CBE constructs lacking the UGI component have been used for targeted random mutagenesis, as they display elevated rates of C•G to non-T•A conversions compared to constructs containing UGI. [21,22] Additionally, C•G to G•C base editors (CGBEs) have been developed by fusing the UNG enzyme instead of UGI to the CBE architecture. These CGBEs promote the generation of abasic sites at the target cytidine, facilitating the C•G to G•C mutations in a sequence-dependent manner.[23,24] More recently, XRCC1 (X-ray repair cross-complementing protein 1), a scaffolding protein for base excision repair (BER), was fused to the CBE construct, which also lead to an increase in C•G to G•C editing efficiencies. [25]
Mitochondrial DNA (mtDNA) genome editing has long been elusive due to the challenges associated with using mitochondrial DSB repair pathways for editing, and a lack of methods for delivering nucleic acids (and thus CRISPR gRNAs) into the mitochondrion. [26] However, CRISPR-free mtDNA CBEs (DdCBEs) were recently reported that perform C•G to T•A base editing in mtDNA. [27] In contrast to CRISPR-based CBEs, DdCBEs are constructed by fusing two halves of a dsDNA cytidine deaminase (DddA) to separate TALE (transcription activator-like effector) proteins that have been engineered to bind at neighboring “half sites” (Figure 1E). DdCBEs demonstrated for the first time efficient and precise genome editing in human mtDNA, using deoxyuridine intermediates.
3. Deoxyadenosine Deamination-Derived Base Editors: A•T to G•C Base Editors (ABEs)
One year after the publication of the first CBE, the first ABE was developed.[13] Unlike cytidine deaminases, there are no naturally occurring adenosine deaminases capable of using DNA as a substrate. The TadA* (mutant TadA) enzyme used to produce the final ABE construct (ABE7.10) was developed from seven rounds of directed evolution on the E. coli TadA enzyme that naturally deaminates adenosines in tRNA. Later studies reported ABE8 variants (engineered through additional rounds of directed evolution), which demonstrate enhanced editing activity and compatibility with Cas9 and Cas12a homologs. [28,29] Unlike the C•G to non-T•A conversions observed with CBEs, ABEs produce more precise editing outcomes, with no A•T to non-G•C conversions and minimal indel formation.
4. Processing of Base Editing Intermediates by DNA Repair Pathways
The most innovative aspect of CBEs, DdCBEs, and ABEs were their utilization of new types of DNA damage as genome editing intermediates; previous tools had almost exclusively relied on DSBs and single-stranded breaks (SSBs) as intermediates. CBEs and DdCBEs on the other hand generate a U•G mismatch intermediate at the site of interest, and ABEs utilize an I•T mismatch intermediate. In addition, the most commonly used CBE and ABE variants employ a Cas9n to introduce an SSB on the opposite strand from the deoxyuridine or deoxyinosine. Depending on the location of the target deoxycytidine or deoxyadenosine within the protospacer, this nick is usually located 3 to 14 nucleotides away in the 5’ direction from the mismatched site (Figure 1D). Here, we review canonical deoxyuridine, deoxyinosine, and SSB repair pathways individually, and analyze how the combination of these types of damage can result in different base editing outcomes.
4.1. Deoxyuridine Repair
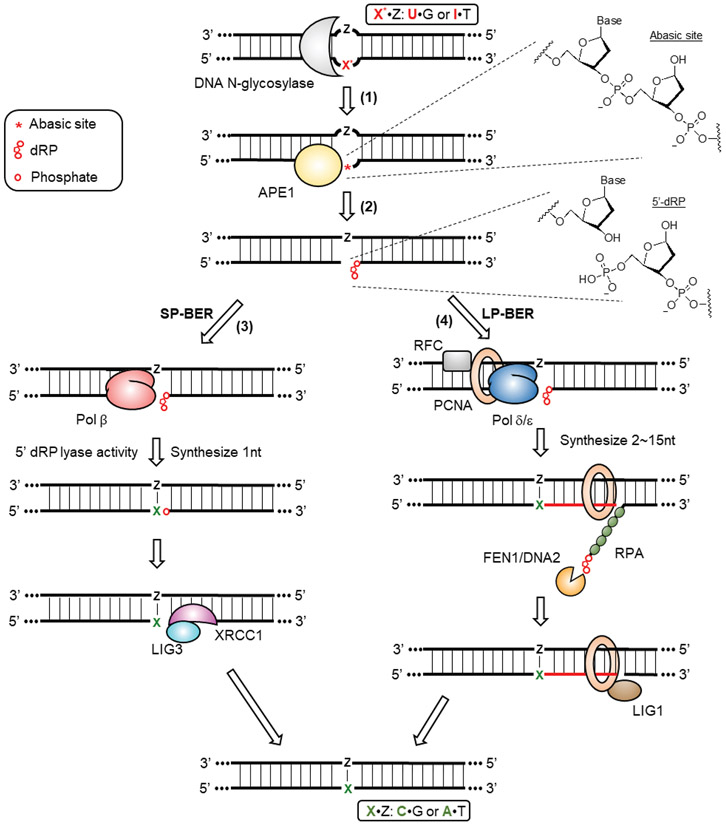
Deoxyuridine is a common DNA damage product caused by the spontaneous deamination of deoxycytidine, and its repair has been studied extensively.[6,30,31] Deoxyuridine in DNA is efficiently recognized by the glycosylase enzyme UNG (Uracil N-Glycosylase) as the first step of the base excision repair (BER) pathway (Figure 2). [32] There are two splice variants produced from the UNG gene, UNG1, which is mitochondrially-localized, and UNG2, which is localized to the nucleus.[33] In addition to UNG, mammalian cells have at least three additional glycosylases, SMUG1 (Single-strand selective monofunctional uracil DNA glycosylase), TDG (G/T mismatch-specific thymine DNA glycosylase) and MBD4 (Methyl-CpG-binding domain protein 4) that also have the ability to recognize and remove deoxyuridine from DNA. [34-36] Upon deoxyuridine recognition, all four of these glycosylases hydrolyze the N-glycosidic bond between the uracil nucleobase and the 2-deoxyribose sugar backbone to generate an abasic site (Figure 2, step 1). The abasic site is then recognized by an apurinic/apyrimidinic endonuclease, most likely APE1 (Apurinic/Apyrimidinic Endonuclease 1) but potentially APE2 as well, which cleaves the sugar phosphate backbone immediately 5’ to the abasic site to produce a 3’-hydroxyl (3’-OH) terminus and a 5’-terminal 2-deoxyribose-5-phosphate (5’-dRP, Figure 2, step 2).[37,38] Following this incision, there are two sub-pathways of BER available to further process the nicked, abasic site-containing strand: short-patch (Figure 2, step 3) and long-patch BER (Figure 2, step 4).[39-41] Short-patch BER (SP-BER) involves the replacement of only the abasic site-containing nucleotide, which is first mediated by the DNA polymerase Pol β. Pol β removes the 5’ dRP moiety through its dRP lyase activity and then incorporates the correct nucleotide onto the 3’-hydroxyl end via its polymerase activity, using the intact strand as a template. The final ligation step is performed by the XRCC1/LIG3 (DNA Ligase 3) complex. Alternatively, long-patch BER (LP-BER) can process the nicked, abasic site-containing intermediate by replacing 2-15 nucleotides on this strand. This process relies on enzymes that normally participate in DNA replication, such as the DNA polymerases Pol δ or Pol ε, RFC (Replication factor C), and PCNA (Proliferating cell nuclear antigen). The newly synthesized strand displaces the old strand to form a 5’ flap, which is then digested by FEN1 (Flap endonuclease 1) or DNA2 (DNA replication helicase/nuclease 2). Instead of XRCC1/LIG3, the final ligation step in LP-BER is carried out by the PCNA/LIG1 (DNA ligase 1) complex. How the cell chooses between SP- and LP-BER is not currently well-understood, but several factors are thought to influence this decision including the type of damaged base, the stage of the cell cycle, and the cell differentiation state.[42] Repair of uracil through both SP- and LP-BER have been observed.[39,43]
Figure 2: Base excision repair.
(1) Base excision repair is initiated by the recognition of modified nucleotides (X*) by DNA N-glycosylases. Deoxyuridine is recognized by four DNA N-glycosylases, including UNG, SMUG1, TDG and MBD4. Deoxyinosine can be recognized by MPG. DNA N-glycosylases hydrolyze the N-glycosidic bond between the nucleobase and the 2’-deoxyribose, leading to the formation of abasic sites. (2) APE1 recognizes abasic sites and incises the DNA sugar backbone to result in a SSB containing a 3’-OH and a 5’-dRP terminus. Further processing of the SSB diverts into two potential pathways. (3) SP-BER involves Pol β removing the 5’ dRP moiety and incorporating the correct nucleotide X back using Z as a template. The final ligation is performed by the XRCC1/LIG3 complex. (4) LP-BER involves the recruitment of DNA replication factors to the site of the SSB, including RFC, PCNA and Pol δ/ε. Synthesis of a new DNA strand (colored in red) displaces the old strand to form a 5’-flap, which is later digested by FEN1 or DNA2. The final ligation is performed by the PCNA/LIG1 complex.
Deoxyuridine in mitochondrial DNA is processed through both LP- and SP-BER as well. [44-47] Some nuclear BER factors, including APE1, FEN1, DNA2, LIG3, are imported into the mitochondrion, while others have distinct mitochondrial counterparts. Instead of UNG2, uracil in mitochondria is excised by UNG1. Following incision of the abasic site by APE1, DNA synthesis in both SP- and LP-BER is mediated by the mitochondrial-specific replicative polymerase Pol γ, and the ligation is then performed by LIG3 alone without XRCC1.[48]
4.2. Deoxyinosine Repair
In mammalian cells, deoxyinosine is also mainly repaired through BER (Figure 2). Hypoxanthine (the nucleobase of deoxyinosine) is recognized and excised by MPG (N-methylpurine-DNA glycosylase, also known as AAG), resulting in the generation of an abasic site.[49,50] Similar to the repair of deoxyuridine, this abasic site is then processed by APE1, followed by either short-patch or long-patch DNA synthesis and ligation. Recently, an alternative excision repair (AER) pathway, initiated by EndoV (endonuclease V), was found to play a major role in deoxyinosine repair in E. coli instead of BER. [51] EndoV, unlike DNA glycosylases, recognizes deoxyinosine and cleaves the DNA sugar phosphate backbone at the second phosphodiester bond 3’ to the lesion. In in vitro experiments, the deoxyinosine can then be excised by the E. coli DNA polymerase Pol I through its 3’-to-5’ proofreading exonuclease activity.[52] The resulting gap is filled through DNA synthesis and ligation. The human homologue of EndoV, ENDOV, was recently identified and found to exhibit deoxyinosine 3’-endonuclease activity on ssDNA. [53,54] However, this AER pathway has yet to be confirmed in vivo in humans.
4.3. Cas-induced Nick Processing and Strand Replacement
4.3.1. SSB repair (SSBR)
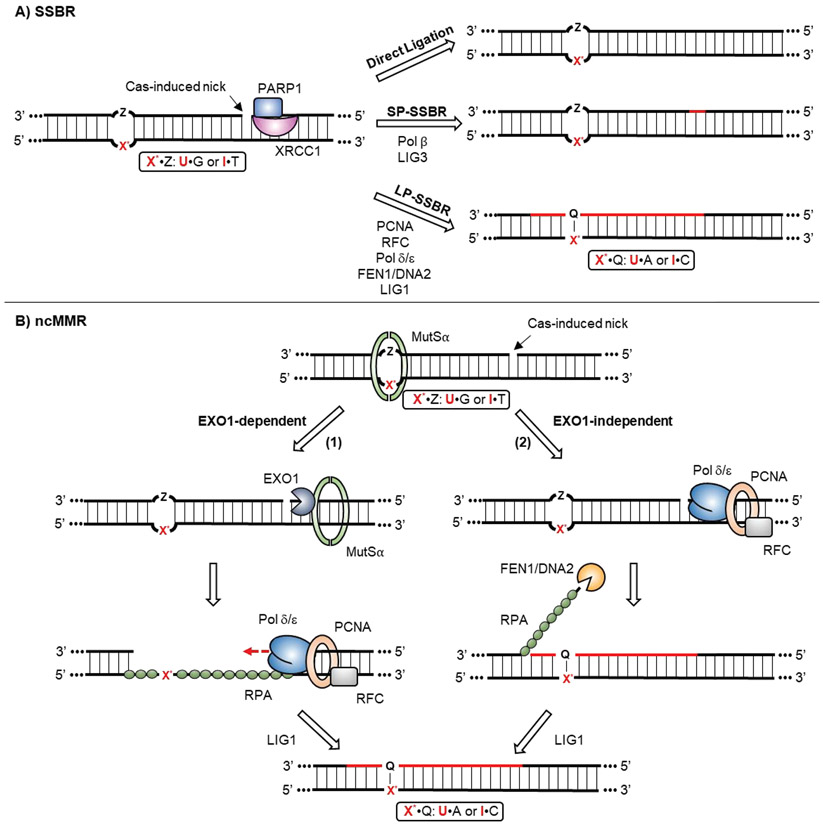
Naturally, SSBs are produced by certain DNA-damaging agents, including most commonly by endogenous reactive oxygen species (ROS). Because SSBs are intermediates in the BER pathway, SSBR and BER have certain machinery in common, but additional factors are required for SSB detection and processing (Figure 3A).[42,55] SSBR is initiated when the SSB is recognized by PARP1 (Poly [ADP-Ribose] Polymerase 1), which then recruits the remainder of the SSBR machinery to the site of the break via its interactions with the scaffolding protein XRCC1. [56,57] Damaged 3’ ends (such as 3’-phosphate and 3’-phosphoglycolate) and 5’ ends (such as 5’-hydroxyl and 5’-adenosine monophosphate) are then further processed by enzymes such as PNKP (Bifunctional polynucleotide phosphatase/kinase), TDP1 (Tyrosyl-DNA phosphodiesterase 1) and APTX (Aprataxin).[58-60] The next DNA strand replacement and ligation processes are identical to the BER pathway, which can be carried by either a short-patch (SP-SSBR) or a long-patch mechanism (LP-SSBR). However, SSBs introduced by Cas nucleases have “clean” 3’-hydroxyl and 5’-phosphate ends, and thus do not need to be end-processed.[61,62] Cas-induced SSBs therefore can potentially be re-ligated directly by either the XRCC1/LIG3 or PCNA/LIG1 complexes.[63]
Figure 3: Cas-induced nick processing and strand replacement.
A) SSBR is initiated by detection of the Cas-induced nick by the XRCC1/PARP1 complex. Then, either direct ligation, SP-SSBR or LP-SSBR can be involved to re-seal the nick. Newly synthesized strands are colored in red. B) ncMMR is initiated by detection of the X*•Z mismatch by MutSα. Subsequently, there are two potential pathways that can process the Cas-induced nick: the EXO1-dependent and -independent pathways. (1) EXO1 is directly recruited and activated at the site of the nick by MutSα. It digests the nicked strand in a 5’-to-3’direction across from the mismatch, leading to formation of a gap across from the X*-containing strand, which becomes coated by RPA. DNA replication factors, including RFC, PCNA and Pol δ/ε, are then recruited to fill in the gap, using the X*-containing strand as a template. (2) In the EXO1-independent pathway, the DNA replication complex is directed recruited to the nick to carry out strand replacement synthesis. The displaced 5’-flap is then digested by FEN1 or DNA2. In both pathways, the final ligation is performed by LIG1, with the X*•Z mismatch being turned into a X*•Q base-pair intermediate.
4.3.2. Mismatch repair (MMR)
As mentioned previously, CBEs and ABEs generate a unique type of DNA damage, which can be thought as of a lesion-containing “mismatch” accompanied by a nick on the opposite strand, 5’ downstream from the mismatched site (Figure 1D). This damage resembles the intermediates processed by the replication-uncoupled, noncanonical mismatch repair (ncMMR) pathway.[31,64] Canonical MMR (cMMR) is associated with DNA replication, and therefore its substrates are mismatched base pairs that consist of two canonical, or undamaged, nucleotides produced by replication errors.[65] In contrast, ncMMR operates independently of replication (predominantly during the G1-phase of the cell cycle), and recognizes mismatches containing damaged nucleotides that have been formed as a result of deamination, oxidation, and alkylation of DNA bases.[64,66] Although ncMMR was initially identified as a player in somatic hypermutation in B cells, later studies demonstrated its presence in other cell types, including nondividing cells.[67] Base editors have been shown to be compatible with many cell types, indicating that ncMMR may play an important role in processing base editing intermediates.[68] During ncMMR, the “mismatch” is recognized by the mismatch recognition complex MutSα (which is comprised of MSH2 and MSH6, Figure 3B). Conventionally, MutSα recruits the MutLα complex (comprised of MLH1 and PMS2) to mismatched sites to introduce a nick on either the 5’ or 3’ side of the mismatch.[69] Unlike cMMR, which utilizes pre-existing strand discontinuities (such as Okzaki fragment termini) to direct the repair to the nascent strand during DNA replication, ncMMR lacks strand discrimination signals. In fact, MutLα can nick either strand during ncMMR, and not necessarily the lesion-containing strand.[64,70,71] In the case of base editing, the strand discrimination signal is potentially provided by the nick introduced by Cas9n on the non-lesion-containing strand, and therefore downstream repair factors could be directly recruited to the nick
Nick-containing mismatches can be processed through an EXO1 (exonuclease 1)-dependent (Figure 3B, route 1) or an EXO1-independent pathway (Figure 3B, route 2).[72] In the EXO1-dependent pathway, MutSα directly recruits EXO1 to the site of the nick and activates it.[73] EXO1 is a 5’-to-3’ exonuclease and will degrade the nicked strand, creating a ssDNA gap of up to 150nt. It is important to note that in the case of base editing intermediates, the directionality of this exonuclease activity will generate a gap across from the deoxyuridine or deoxyinosine. DNA resynthesis then fills in the gap via normal DNA replication factors, including RPA (Replication protein A, which binds to the ssDNA of the gap to protect it from further damage), RFC, PCNA and Pol δ.[74] When DNA resynthesis encounters a replication-blocking lesion, such as an abasic site, DNA translesion synthesis (TLS) factors may be recruited to bypass the lesion (discussed in more details later).[31,64,66,75] It is also worth noting that the ssDNA gap generated by EXO1 may expose additional deoxycytidines within this region to the ssDNA cytidine deaminase component of CBEs.[31] Indeed, recent work that analyzed base editing outcomes in high-throughput revealed statistically significant cytosine editing spanning from the −15nt position 5’ upstream of the protospacer to the +17nt position where the nick is introduced.[76] This observation could be explained by EXO1-dependent strand replacement during MMR being involved in base editing.
The EXO1-independent pathway resembles LP-BER.[77] The MutSα and MutLα complexes can recruit PCNA and Pol δ to the nick to carry out strand displacement synthesis spanning the mismatched site. The displaced 5’ flap is then digested by FEN1 or DNA2, and the resulting nick is sealed by a DNA ligase.
5. CBE Outcomes in Genomic DNA (gDNA): An Interplay Between Deoxyuridine Repair and Cas-induced Nick Processing
Cytosine base editing in genomic DNA (gDNA) can result in four different outcomes: Repair back to the original C•G base pair, C•G to T•A editing, C•G to non-T•A editing, and indel outcomes. We discuss here potential DNA repair mechanisms that can cause these different outcomes (Figure 4).
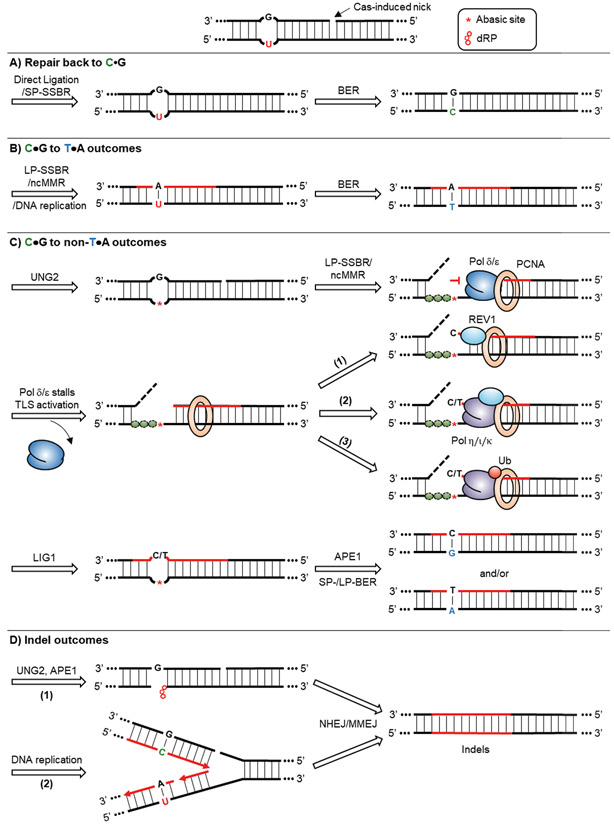
Figure 4: CBE outcomes in gDNA.
A typical CBE intermediate features a U•G mismatch accompanied by a 5’ Cas-induced nick. A) The CBE intermediate can be repaired back to C•G by first re-ligation of the nick through SSBR, followed by the deoxyuridine being processed by BER. B) The CBE intermediate can lead to C•G to T•A outcomes when DNA synthesis occurs across the deoxyuridine, which can be achieved by either the LP-SSBR, ncMMR, or normal DNA replication. The resulting U•A base pair can be resolved into a T•A base pair by BER. C) The CBE intermediate can also result in C•G to non-T•A outcomes, which is mediated by DNA synthesis across abasic sites. Abasic sites are generated by excision of uracil by UNG2. DNA synthesis to replace the nicked strand is then carried out by LP-SSBR or ncMMR. Depending on the pathway involved, either an RPA-coated ssDNA gap or a 5’-flap would form, which are depicted by dashed lines. Replicative polymerases Pol δ and Pol ε stall at abasic sites, leading to a subsequent TLS activation to rescue the stalled DNA synthesis and bypass the lesion. There are three potential downstream routes. (1) REV1 is recruited to PCNA. It can insert a dCMP opposite abasic sites. (2) Alternatively, REV1 can recruit other error-prone polymerases, such as Pol η, Pol ι and Pol κ, to the stalled site. They are capable of incorporating both dCMP and dTMP opposite abasic sites. (3) The REV1-independent pathway depends on the recruitment of error-prone polymerases by monoubiquitylation of PCNA. Ligation of the newly synthesized strand by LIG1 forms an intact abasic site-containing intermediate, which is further processed by APE1 and the rest of the BER machinery. This eventually leads to the formation of C•G to G•C and C•G to A•T outcomes. D) The CBE intermediate can be processed into indels, which we attribute to in situ DSB formation. (1) DSBs can arise from processing of deoxyuridine by UNG2 and APE1 before the re-ligation of the Cas-induced nick. (2) DSBs can also be originated from helicase unwinding of the nicked strand during DNA replication. DSB processing by both NHEJ and MMEJ can lead to indels.
5.1. Repair back to C•G
A U•G, nick-containing intermediate can be repaired back to an intact C•G base pair via sequential processing by SSBR and BER. The nick has to be sealed by SSBR first, followed by BER reverting the U•G base pair back to a C•G base pair (Figure 4A). If the deoxyuridine is processed by BER before the re-ligation of the nick, incision of the abasic site by APE1 in concert with the pre-existing nick would generate a DSB, which could lead to indel formation.[70,78]
5.2. C•G to T•A outcomes
To produce a C•G to T•A outcome, DNA synthesis needs to occur across the deoxyuridine (Figure 4B). This can be done during normal DNA replication (during S-phase of the cell cycle) if the U•G intermediate is still intact, or if Cas-induced nick processing and strand replacement by either LP-SSBR or ncMMR occurs prior to uracil excision by UNG2. However, the strand replacement needs to extend long enough that the polymerase reads across the uracil – 3 to 14 nucleotides long for typical base editing intermediates. Uracil can be readily bypassed by the replicative polymerases Pol δ and Pol ε, thus TLS activation is not required.[79-81] These scenarios would both produce a U•A intermediate, and the deoxyuridine would eventually be replaced by a deoxythymidine by BER.
5.3. C•G to non-T•A outcomes
C•G to non-T•A outcomes can only occur in cells with UNG2.[20] Therefore, these outcomes most likely originate from an abasic site-containing intermediate. We believe this occurs via a very nuanced scenario, when nick processing and strand replacement by LP-SSBR or ncMMR occur after uracil excision by UNG2, but before the abasic site has been incised by APE1 (Figure 4C).[31,82,83] While not a C•G to non-T•A outcome, it is worth mentioning that replicative DNA polymerases have the ability to bypass abasic sites, and most likely incorporate a dAMP (deoxyadenosine monophosphate) opposite them. This is known as the “A-rule”, and would result in the desired C•G to T•A outcome.[84] However, this bypass efficiency is low, and replicative DNA polymerases are severely stalled at abasic sites, which leads to the activation of TLS.[81,85]
There are two TLS pathways known to rescue stalled DNA replication machinery and bypass abasic sites: the REV1-dependent and REV1-independent pathways. In the REV1-dependent pathway, REV1 is recruited to a stalled PCNA via its N-terminal domain.[86,87] REV1 is a dCMP (deoxycytidine monophosphate) transferase capable of inserting dCMPs opposite abasic sites, which would lead to a C•G to G•C outcome (Figure 4C, route 1).[88-90] Alternatively, instead of inserting dCMP, the C-terminal domain of REV1 can recruit other error-prone Y-family TLS polymerases, such as Pol η, Pol ι and Pol κ, to the damaged site (Figure 4C, route 2).[86,91] All these TLS polymerases can bypass abasic sites.[84] Notably, Pol η and Pol ι can incorporate a dTMP (deoxythymidine monophosphate) opposite abasic sites in vitro fairly efficiently, which would lead to a C•G to A•T outcome. However, their roles in vivo have not yet been confirmed. On the other hand, the REV1-independent pathway relies on monoubiquitylation of the stalled PCNA (Figure 4C, route 3).[92,93] Monoubiquitylated PCNA (PCNA-Ub) can recruit TLS polymerases directly to the stalled site. A recent study showed that the C•G to G•C outcome of cytosine base editing does not clearly correlate with REV1 expression, suggesting that the REV1-independent pathway may be involved in this process.[76]
Several studies have observed that the deaminase components of CBEs can affect C•G to non-T•A outcomes.[23,76] It has been suggested that these deaminases may possess a previously unrecognized role in directing repair pathways, while others suggest that the dynamics of the deaminase may affect editing outcomes. It is interesting to note that in both studies, rAPOBEC1 and eA3A, the two cytidine deaminase enzymes with the fastest kinetics, facilitated the highest levels of C•G to non-T•A outcomes.[94,95] Additionally, editing of multiple cytidines concurrently within the base editing window has shown to dramatically decrease C•G to non-T•A outcomes.[20] Overall, we believe the kinetics of cytidine deamination impact C•G to non-T•A outcomes due to the unique timing of events as described above required for these outcomes to occur.
5.4. Indel outcomes
We attribute indel outcomes to the generation of DSBs in situ (Figure 4D). A DSB can be generated if incision of the abasic site happens before re-ligation of the nick. Furthermore, if the nick escapes global SSBR and persist until DNA replication, it will be turned into a DSB upon helicase unwinding. [96] Template switching or HDR pathways can repair the DSB back to the original sequence, whereas NHEJ or MMEJ pathways would both lead to the formation of indels.[7,9]
6. Cytosine Base Editing Outcomes in Mitochondrial DNA (MtDNA): Interplays Among Deoxyuridine Repair, MtDNA Replication, and Degradation
MtDNA cytosine base editing is distinct from genomic cytosine base editing in three major aspects. First, each cell contains thousands copies of mtDNA, meaning there are thousands of target sites for the mitochondrial base editor in contrast to typically two target alleles for genomic CBEs.[97,98] Despite this, DdCBEs demonstrated up to 50% editing efficiencies, implying that many uracils may simultaneously be present in mtDNA during editing. This in turn may pose challenges to the mitochondrial uracil repair machinery. Second, the two TALE proteins used in DdCBEs do not introduce DNA breaks of any kind (therefore, no nicks will be present in the intermediate). Third, DddA is a dsDNA cytidine deaminase, resulting in a wider editing window (14-18bp) than CBEs. This can result in deamination of multiple cytidines at a time, and on both strands. SSBs or DSBs can be generated as a result of BER processing of these multi-U intermediates. Despite this, the only observed editing outcomes of DdCBEs are repair back to the original C•G base pair and the C•G to T•A outcome. We will discuss how interactions among deoxyuridine repair, mtDNA replication, and mtDNA degradation can lead to these different outcomes (Figure 5).
Figure 5: Cytosine base editing outcomes in mtDNA.
Unlike CBE in gDNA, a DdCBE intermediate in mtDNA contains a U•G mismatch (or multiple) without any nicks. A) This intermediate can be repaired back to C•G by mitochondrial BER. B) There are several scenarios in which this intermediate can result in DSB formation, and eventually mtDNA degradation. (1) Pol γ stalls at abasic sites generated by UNG1 during mtDNA replication. (2) mtDNA replication encounter SSBs generated by APE1. Note that the leading-strand and the lagging-strand synthesis during mtDNA replication are uncoupled.[116] Hence, the replication forks are depicted with dashed lines. DNA synthesis based on the unmodified strand leads to repair back to C•G. (3) If two (or more) deoxycytidines are deaminated on opposite strands, processing of these deoxyuridines by UNG1 and APE1 can lead to DSBs. C) The U•G intermediate can be resolved into a T•A outcome by DNA synthesis across the deoxyuridine during mtDNA replication, followed by the deoxyuridine being replaced by a deoxythymidine through mitochondrial BER.
6.1. Repair back to C•G
The U•G intermediates of DdCBEs can be repaired back to C•G base pairs by mitochondrial BER in an analogous manner to nuclear BER (Figure 5A). However, we believe additional repair mechanisms may be at play here as well for two reasons. First, there is a lack of C•G to non-T•A and indel outcomes observed with DdCBEs despite the sheer number of uracils they are introducing into mtDNA.[27] Furthermore, TLS and NHEJ/MMEJ, which we hypothesize to be responsible for C•G to non-T•A and indel outcomes, respectively, with genomic CBEs, exist in human mitochondria.[44,99-101]
The DNA degradation pathway is a prevalent DNA repair pathway in mitochondria, which we believe contributes to the high precision of DdCBEs.[101-103] While DNA degradation is not an option for genomic DNA, a significant number of mtDNA molecules can be degraded without compromising mitochondrial functions, as there are typically thousands copies of mtDNA within each cell. MtDNA degradation is triggered by DSBs, and there are three possible scenarios during the processing of mitochondrial base editing intermediates that can produce DSBs (Figure 5B). First, Pol γ could stall at a UNG1-generated abasic site during mtDNA replication. Although Pol γ can bypass abasic sites in vitro, this activity in vivo is low (Figure 5B, route 1).[101] Second, DSBs can be formed when mtDNA replication encounters a SSB (which would be produced by APE1 incision of abasic sites) (Figure 5B, route 2). And third, if two uracils (or more) are on the opposing strands and are both processed by UNG1 followed by APE1, this would result in a DSB (Figure 5B, route 3). Once DSBs form, rapid degradation of mtDNA is mediated by the 5’-to-3’ exonuclease MGME1 (Mitochondrial Genome Maintenance Exonuclease 1), the 3’-to-5’ exonuclease activity of Pol γ, and the mitochondrial replicative DNA helicase TWNK (Twinkle protein).[104] A decrease in mtDNA copy number was not observed during base editing with DdCBE, but mtDNA replication can quickly rescue the loss of mtDNA.
6.2. C•G to T•A outcomes
The target C•G to T•A outcome is most likely achieved by Pol γ incorporating an A opposite the U during mtDNA replication (Figure 5C). In rare cases in which Pol γ bypasses abasic sites, it obeys the A-rule, which would lead to C•G to T•A outcomes as well.[101]
7. Adenine Base Editing Outcomes: An Interplay between Deoxyinosine Repair and Cas-induced Nick Processing
ABEs feature highly precise editing profiles as well, with mostly A•T to G•C outcomes and repair back to A•T observed (Figure 6). Indel outcomes are minimal but do sometime occur. Notably, despite ABE8 variants producing much higher editing efficiencies than ABE7.10, their indel formation is not significantly increased.[28,29] Because ABEs and CBEs generate very similar base editing intermediates, the high product purity of ABEs indicates a lack of TLS or NHEJ/MMEJ activation, which we attribute to inefficient deoxyinosine excision. Indeed, MPG, the DNA glycosylase responsible for excising of a wide range of damaged bases including hypoxanthine, has a kchem (the rate constant for hydrolysis of the N-glycosidic bond) of 0.033 s−1.[105] This is orders of magnitude slower than the kchem Of UNG (115 s−1).[106,107] Deoxyinosine formation in DNA occurs much less often than deoxyuridine, as spontaneous deoxyadenosine deamination occurs at only 2-3% of the rate of deoxycytidine deamination.[108] Therefore, a much lower selection pressure occurred during the evolution of an enzyme for deoxyinosine repair. This observation in turn suggests that the development of future base editors would benefit from using more rarely occurring DNA damaged intermediates.
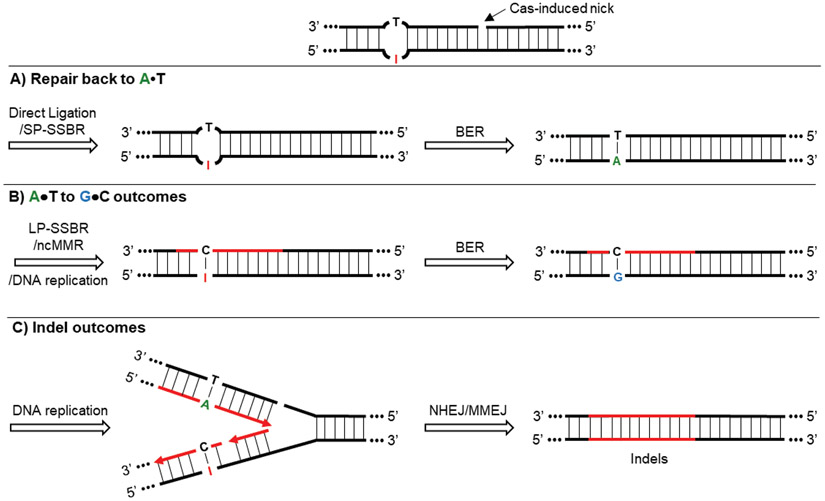
Figure 6: ABE outcomes.
A typical ABE intermediate contains an I•T mismatch with a 5’ Cas-induced nick. A) The ABE intermediate can be repaired back to an intact A•T base pair by sequential processing of nick re-ligation and BER. B) The ABE intermediate can be resolved into a T•A outcome when DNA synthesis occurs across the deoxyinosine during LP-SSBR, ncMMR, or normal DNA replication. The resulting I•C base pair can be converted to a G•C base pair by BER. C) Indel outcomes are attributed to helicase unwinding of the nicked strand during DNA replication, followed by processing of the DSB through NHEJ/MMEJ.
7.1. Repair back to A•T
Similar to CBEs, an I•T, nick-containing intermediate can be repaired back to an intact A•T base pair by SSBR-mediated nick re-ligation followed by MPG-initiated BER (Figure 6A). However, ABE experiments performed in MPG knock-out cells did not exhibit a significant increase in editing efficiency compared to wild-type controls. We therefore think this process is quite inefficient.[13]
7.2. A•T to G•C outcomes
The A•T to G•C outcome requires DNA synthesis extended across the deoxyinosine. This could be achieved through strand replacement of the nicked, unedited strand by LP-SSBR, ncMMR, or normal DNA replication analogous to the corresponding CBE outcome (Figure 6B).
7.3. Indel outcomes
As excision of hypoxanthine by MPG is inefficient, and indel rates do not scale with increasing base editing efficiencies, we believe ABE-mediated indels are more likely a result of DSBs formation due to helicase unwinding of the Cas9n-induced nick during DNA replication (Figure 6C).
8. Conclusion
Base editors demonstrated a new genome editing strategy of harnessing non-DSB DNA damage intermediates. Extensive engineering efforts from many researchers have been devoted to tackle the limitations of base editors, including off-target DNA editing, off-target RNA editing, and bystander editing.[14,15] An additional key limitation of base editors was their inability to introduce transversion mutations. Researchers have begun to chip away at this limitation through the manipulation of the DNA repair pathways involved in cytosine base editing. This highlights the importance of understanding the DNA repair mechanisms that produce different base editing outcomes. We anticipate that the development of additional new base editors will require exploiting new nucleic acid modification chemistries and/or DNA repair manipulation strategies.
In addition to base editors, prime editors are another class of editing agents that use non-DSB intermediates.[109] Prime editors can mediate the incorporation of insertions, deletions, and all possible SNVs in mammalian cells by using a 5’-flap-containing mismatch or bulge intermediate. These intermediates are introduced by a reverse transcriptase-Cas9n fusion construct using a 3’-extension on the gRNA as a template for the reverse transcriptase. Although the mechanisms involved in the processing of prime editor intermediates are outside the scope of this review, prime editors highlight the value of using new DNA damage products as genome editing intermediates for the development of new genome editing tools.
Base editors showcase great potential as therapeutics and research tools due to their high efficiency, precision, and compatibility with many cell types. In fact, base editing technologies have already been applied in the ongoing development of treatments for various genetic diseases and cancer immunotherapy.[14,68] However, they are not without room for improvement. As first proposed by Thuronyi and co-workers, while increasing base editor expression levels and deaminase activity led to improvements in overall editing efficiencies, maximum editing levels reach saturation at a certain limit imposed by editor-independent factors, which could include cell cycle states, local chromatin structures, and DNA repair processes.[28,29,110-112] Specifically, some sites seem to be intrinsically difficult to edit (even when highly active deaminases are used), potentially due to low saturation limits at these sites. Raising the saturation limits at these sites will require modulating editor-independent factors, which can be achieved through an expanded understanding of the mechanisms by which base editors work (Figure 7). For example, a recent study demonstrated that using small molecule inhibitors of histone deacetylases can increase base editing efficiencies by artificially inducing an open chromatin state and facilitating better base editor access to target loci.[113] Similarly, while nucleosomes impede DNA binding and cutting by Cas enzymes in vitro and in vivo, the addition of nucleosome remodeling (sliding) factors restore activity at previously occluded target sites in vitro.[114,115] Due to the fact that Cas enzymes were not evolved to explore the complex, chromatin-bound eukaryotic genomes, fusion of chromatin remodelers to base editors represents an unexplored avenue to raise the saturation limit at poorly edited sites. In addition, previous studies have already demonstrated that DNA repair processes can be modulated to raise the saturation limits. Base editing outcomes are the result of an equilibrium among competing DNA repair pathways, and tilting the balance towards certain pathways can favor certain desired outcomes. As for CBEs, the inhibition of UNG-initiated BER by fusing UGI peptides to the editor suppresses the major repair pathways that would lead to repair back to C•G and to undesired C•G to non-T•A or indel outcomes, thus greatly promoting the C•G to T•A outcome (Figure 4). For both CBEs and ABEs, we hypothesize that inhibiting the direct reversal of the Cas-induced nick (direct ligation and SP-SSBR) while promoting the processing of the Cas-induced nick by LP-SSBR or ncMMR could further boost the C•G to T•A and A•T to G•C efficiency, respectively. For CGBEs, just as fusing UNG boosted transversion efficiencies, suppressing the incision of abasic sites (APE1) while facilitating the recruitment of error-prone polymerases may increase the efficiency of C•G to G•C outcomes. Future research that illuminates the detailed biological pathways involved in base editing will facilitate the continued optimization of base editors to maximize their efficiencies, targeting abilities, and safety. Research that directly studies the mechanisms of base editing remains to be an indispensable building block for the further expansion and improvement of these technologies.
Figure 7: A conceptual model of base editing.
Editing efficiency reaches a maximum at the optimal target base position. At certain poorly edited sites, the observed base editing efficiencies are low even when a highly active base editor is used. We reason that the low saturation limits of those sites are imposed by editor-independent factors, such as cell states, chromatin structures, and DNA repair processes. Raising the limits by modulation of these editor-independent factors can potentially increase editing efficiencies at these sites.
10. Acknowledgments
A.C.K. and S.G. are partially funded by NIH grant R35 GM138317. Q.T.C. is supported by the Molecular Biophysics Training Grant, NIH Grant T32 GM008326.
Footnotes
Competing Interests
A.C.K. is a member of the SAB of Pairwise Plants, and is an equity holder for Pairwise Plants and Beam Therapeutics. A.C.K.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies
9. References
- [1].The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 2016;44:D862–8. 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pickar-Oliver A, Gersbach CA. The next generation of CRISPR–Cas technologies and applications. Nat Rev Mol Cell Biol 2019;20:490–507. 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jiang F, Doudna JA. CRISPR–Cas9 Structures and Mechanisms. Annu Rev Biophys 2017;46:505–29. 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- [5].Scully R, Panday A, Elango R, Willis NA. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol 2019;20:698–714. 10.1038/s41580-019-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Altieri F, Grillo C, Maceroni M, Chichiarelli S. DNA Damage and Repair: From Molecular Mechanisms to Health Implications. Antioxid Redox Signal 2008;10:891–938. 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- [7].Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol 2017;18:495–506. 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 2018;36:765–71. 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jasin M, Rothstein R. Repair of Strand Breaks by Homologous Recombination. Cold Spring Harb Perspect Biol 2013;5:a012740–a012740. 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yeh CD, Richardson CD, Corn JE. Advances in genome editing through control of DNA repair pathways. Nat Cell Biol 2019;21:1468–78. 10.1038/s41556-019-0425-z. [DOI] [PubMed] [Google Scholar]
- [11].Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. ELife 2014;3:e04766. 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533:420–4. 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017;551:464–71. 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Porto EM, Komor AC, Slaymaker IM, Yeo GW. Base editing: advances and therapeutic opportunities. Nat Rev Drug Discov 2020;19:839–59. 10.1038/s41573-020-0084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 2020;38:824–44. 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- [16].Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 2018;19:770–88. 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ranzau BL, Komor AC. Genome, Epigenome, and Transcriptome Editing via Chemical Modification of Nucleobases in Living Cells. Biochemistry 2019;58:330–5. 10.1021/acs.biochem.8b00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Komor AC, Badran AH, Liu DR. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017;168:20–36. 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Evanoff M, Komor AC. Base editors: modular tools for the introduction of point mutations in living cells. Emerg Top Life Sci 2019;3:483–91. 10.1042/ETLS20190088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 2017;3:eaao4774. 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 2016;353:aaf8729–aaf8729. 10.1126/science.aaf8729. [DOI] [PubMed] [Google Scholar]
- [22].Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 2016;13:1029–35. 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- [23].Kurt IC, Zhou R, Iyer S, Garcia SP, Miller BR, Langner LM, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol 2020. 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhao D, Li J, Li S, Xin X, Hu M, Price MA, et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol 2020. 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- [25].Chen L, Park JE, Paa P, Rajakumar PD, Prekop H-T, Chew YT, et al. Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat Commun 2021;12:1384. 10.1038/s41467-021-21559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gammage PA, Moraes CT, Minczuk M. Mitochondrial Genome Engineering: The Revolution May Not Be CRISPR-Ized. Trends Genet 2018;34:101–10. 10.1016/j.tig.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mok BY, de Moraes MH, Zeng J, Bosch DE, Kotrys AV, Raguram A, et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 2020;583:631–7. 10.1038/s41586-020-2477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol 2020;38:883–91. 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gaudelli NM, Lam DK, Rees HA, Solá-Esteves NM, Barrera LA, Born DA, et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat Biotechnol 2020;38:892–900. 10.1038/s41587-020-0491-6. [DOI] [PubMed] [Google Scholar]
- [30].Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA – occurrence, consequences and repair. Oncogene 2002;21:8935–48. 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- [31].Pilzecker B, Jacobs H. Mutating for Good: DNA Damage Responses During Somatic Hypermutation. Front Immunol 2019;10:438. 10.3389/fimmu.2019.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Olsen LC, Aasland R, Wittwer CU, Krokan HE, Helland DE. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J 1989;8:3121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, et al. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res 1997;25:750–5. 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nilsen H Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J 2001;20:4278–86. 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hardeland U, Bentele M, Lettieri T, Steinacher R, Jiricny J, Schär P. Thymine DNA glycosylase. Prog. Nucleic Acid Res. Mol. Biol, vol. 68, Elsevier; 2001, p. 235–53. 10.1016/S0079-6603(01)68103-0. [DOI] [PubMed] [Google Scholar]
- [36].Bellacosa A Role ofMED1 (MBD4) Gene in DNA repair and human cancer. J Cell Physiol 2001;187:137–44. 10.1002/jcp.1064. [DOI] [PubMed] [Google Scholar]
- [37].Wilson DM. Properties of and substrate determinants for the exonuclease activity of human apurinic endonuclease Ape1. J Mol Biol 2003;330:1027–37. 10.1016/s0022-2836(03)00712-5. [DOI] [PubMed] [Google Scholar]
- [38].Demple B, Sung J-S. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair 2005;4:1442–9. 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- [39].Dianov G, Price A, Lindahl T. Generation of single-nucleotide repair patches following excision of uracil residues from DNA. Mol Cell Biol 1992;12:1605–12. 10.1128/MCB.12.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, Cox LS, et al. Two Pathways for Base Excision Repair in Mammalian Cells. J Biol Chem 1996;271:9573–8. 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- [41].Klungland A Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO J 1997;16:3341–8. 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fortini P, Dogliotti E. Base damage and single-strand break repair: Mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair 2007;6:398–409. 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- [43].Dianov GL, Prasad R, Wilson SH, Bohr VA. Role of DNA Polymerase β in the Excision Step of Long Patch Mammalian Base Excision Repair. J Biol Chem 1999;274:13741–3. 10.1074/jbc.274.20.13741. [DOI] [PubMed] [Google Scholar]
- [44].Kazak L, Reyes A, Holt IJ. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol 2012;13:659–71. 10.1038/nrm3439. [DOI] [PubMed] [Google Scholar]
- [45].Stierum RH, Dianov GL, Bohr VA. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts#. Nucleic Acids Res 1999;27:3712–9. 10.1093/nar/27.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. Long Patch Base Excision Repair in Mammalian Mitochondrial Genomes. J Biol Chem 2008;283:26349–56. 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair 2008;7:605–16. 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- [48].Lakshmipathy U Mitochondrial DNA ligase III function is independent of Xrcc1. Nucleic Acids Res 2000;28:3880–6. 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Alseth I, Dalhus B, Bjørås M. Inosine in DNA and RNA. Curr Opin Genet Dev 2014;26:116–23. 10.1016/j.gde.2014.07.008. [DOI] [PubMed] [Google Scholar]
- [50].Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proc Natl Acad Sci U S A 1994;91:5873–7. 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yao M, Hatahet Z, Melamede RJ, Kow YW. Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3’ endonuclease, from Escherichia coli. J Biol Chem 1994;269:16260–8. [PubMed] [Google Scholar]
- [52].Lee C-C, Yang Y-C, Goodman SD, Yu Y-H, Lin S-B, Kao J-T, et al. Endonuclease V-mediated deoxyinosine excision repair in vitro. DNA Repair 2010;9:1073–9. 10.1016/j.dnarep.2010.07.007. [DOI] [PubMed] [Google Scholar]
- [53].Fladeby C, Vik ES, Laerdahl JK, Gran Neurauter C, Heggelund JE, Thorgaard E, et al. The Human Homolog of Escherichia coli Endonuclease V Is a Nucleolar Protein with Affinity for Branched DNA Structures. PLoS ONE 2012;7:e47466. 10.1371/journal.pone.0047466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mi R, Alford-Zappala M, Kow YW, Cunningham RP, Cao W. Human endonuclease V as a repair enzyme for DNA deamination. Mutat Res 2012;735:12–8. 10.1016/j.mrfmmm.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet 2008;9:619–31. 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- [56].Chalmers AJ. Poly(ADP-ribose) polymerase-1 and ionizing radiation: sensor, signaller and therapeutic target. Clin Oncol R Coll Radiol G B 2004;16:29–39. 10.1016/s0936-6555(03)00223-1. [DOI] [PubMed] [Google Scholar]
- [57].London RE. The structural basis of XRCC1-mediated DNA repair. DNA Repair 2015;30:90–103. 10.1016/j.dnarep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 2001;104:107–17. 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- [59].Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 1999;286:552–5. 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- [60].Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 2006;443:713–6. 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- [61].Zuo Z, Liu J. Structure and Dynamics of Cas9 HNH Domain Catalytic State. Sci Rep 2017;7:17271. 10.1038/S41598-017-17578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yoon H, Zhao LN, Warshel A. Exploring the Catalytic Mechanism of Cas9 Using Information Inferred from Endonuclease VII. ACS Catal 2019;9:1329–36. 10.1021/acscatal.8b04324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mortusewicz O Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res 2006;34:3523–32. 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Peña-Diaz J, Bregenhorn S, Ghodgaonkar M, Follonier C, Artola-Borán M, Castor D, et al. Noncanonical Mismatch Repair as a Source of Genomic Instability in Human Cells. Mol Cell 2012;47:669–80. 10.1016/j.molcel.2012.07.006. [DOI] [PubMed] [Google Scholar]
- [65].Jiricny J The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 2006;7:335–46. 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- [66].Zlatanou A, Despras E, Braz-Petta T, Boubakour-Azzouz I, Pouvelle C, Stewart GS, et al. The hMsh2-hMsh6 Complex Acts in Concert with Monoubiquitinated PCNA and Pol η in Response to Oxidative DNA Damage in Human Cells. Mol Cell 2011;43:649–62. 10.1016/j.molcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- [67].Rodriguez GP, Romanova NV, Bao G, Rouf NC, Kow YW, Crouse GF. Mismatch repair-dependent mutagenesis in nondividing cells. Proc Natl Acad Sci 2012;109:6153–8. 10.1073/pnas.1115361109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yang B, Yang L, Chen J. Development and Application of Base Editors. CRISPR J 2019;2:91–104. 10.1089/crispr.2019.0001. [DOI] [PubMed] [Google Scholar]
- [69].Kadyrova LY, Kadyrov FA. Endonuclease activities of MutLα and its homologs in DNA mismatch repair. DNA Repair 2016;38:42–9. 10.1016/j.dnarep.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bregenhorn S, Kallenberger L, Artola-Borán M, Peña-Diaz J, Jiricny J. Non-canonical uracil processing in DNA gives rise to double-strand breaks and deletions: relevance to class switch recombination. Nucleic Acids Res 2016;44:2691–705. 10.1093/nar/gkv1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutL endonuclease in mismatch repair. Proc Natl Acad Sci 2010;107:16066–71. 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Goellner EM, Putnam CD, Kolodner RD. Exonuclease 1-dependent and independent mismatch repair. DNA Repair 2015;32:24–32. 10.1016/j.dnarep.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Genschel J, Modrich P. Mechanism of 5’-directed excision in human mismatch repair. Mol Cell 2003; 12:1077–86. 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- [74].Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem 2005;280:39752–61. 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, et al. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J Exp Med 2005;201:637–45. 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Arbab M, Shen MW, Mok B, Wilson C, Matuszek Ż, Cassa CA, et al. Determinants of Base Editing Outcomes from Target Library Analysis and Machine Learning. Cell 2020; 182:463–480.e30. 10.1016/j.cell.2020.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci 2009;106:8495–500. 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and Regulation of Class Switch Recombination. Annu Rev Immunol 2008;26:261–92. 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wardle J, Burgers PMJ, Cann IKO, Darley K, Heslop P, Johansson E, et al. Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya. Nucleic Acids Res 2008;36:705–11. 10.1093/nar/gkm1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fuchs RP, Fujii S. Translesion DNA Synthesis and Mutagenesis in Prokaryotes. Cold Spring Harb Perspect Biol 2013;5:a012682–a012682. 10.1101/cshperspect.a012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sale JE. Translesion DNA Synthesis and Mutagenesis in Eukaryotes. Cold Spring Harb Perspect Biol 2013;5:a012708–a012708. 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 2007;76:1–22. 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- [83].Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol 2006;27:313–21. 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Choi J-Y, Lim S, Kim E-J, Jo A, Guengerich FP. Translesion Synthesis across Abasic Lesions by Human B-Family and Y-Family DNA Polymerases α, δ, η, ι, κ, and REV1. J Mol Biol 2010;404:34–44. 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sale JE. Competition, collaboration and coordination - determining how cells bypass DNA damage. J Cell Sci 2012;125:1633–43. 10.1242/jcs.094748. [DOI] [PubMed] [Google Scholar]
- [86].Vaisman A, Woodgate R. Translesion DNA polymerases in eukaryotes: what makes them tick? Crit Rev Biochem Mol Biol 2017;52:274–303. 10.1080/10409238.2017.1291576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Guo C, Sonoda E, Tang T-S, Parker JL, Bielen AB, Takeda S, et al. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell 2006;23:265–71. 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- [88].Masuda K, Ouchida R, Li Y, Gao X, Mori H, Wang J-Y. A Critical Role for REV1 in Regulating the Induction of C:G Transitions and A:T Mutations during Ig Gene Hypermutation. J Immunol 2009;183:1846–50. 10.4049/jimmunol.0901240. [DOI] [PubMed] [Google Scholar]
- [89].Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 1996;382:729–31. 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- [90].Ross A-L, Sale JE. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol Immunol 2006;43:1587–94. 10.1016/j.molimm.2005.09.017. [DOI] [PubMed] [Google Scholar]
- [91].Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 2012;13:141–52. 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hoege C, Pfander B, Moldovan G-L, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 2002;419:135–41. 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- [93].Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair 2009;8:461–9. 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- [94].Yu Y, Leete TC, Born DA, Young L, Barrera LA, Lee S-J, et al. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat Commun 2020; 11:2052. 10.1038/S41467-020-15887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Doman JL, Raguram A, Newby GA, Liu DR. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat Biotechnol 2020;38:620–8. 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Vrtis KB, Dewar JM, Chistol G, Wu RA, Graham TGW, Walter JC. Single-strand DNA breaks cause replisome disassembly. Mol Cell 2021:S1097276520309606. 10.1016/j.molcel.2020.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol 1988;136:507–13. 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- [98].Reznik E, Miller ML, Şenbabaoğlu Y, Riaz N, Sarungbam J, Tickoo SK, et al. Mitochondrial DNA copy number variation across human cancers. ELife 2016;5. 10.7554/eLife.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res 2009;37:4218–26. 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kasiviswanathan R, Gustafson MA, Copeland WC, Meyer JN. Human mitochondrial DNA polymerase γ exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J Biol Chem 2012;287:9222–9. 10.1074/jbc.M111.306852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kozhukhar N, Spadafora D, Fayzulin R, Shokolenko IN, Alexeyev M. The efficiency of the translesion synthesis across abasic sites by mitochondrial DNA polymerase is low in mitochondria of 3T3 cells. Mitochondrial DNA Part A 2016;27:4390–6. 10.3109/19401736.2015.1089539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Shokolenko IN, Wilson GL, Alexeyev MF. Persistent damage induces mitochondrial DNA degradation. DNA Repair 2013; 12:488–99. 10.1016/j.dnarep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res 2009;37:2539–48. 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Peeva V, Blei D, Trombly G, Corsi S, Szukszto MJ, Rebelo-Guiomar P, et al. Linear mitochondrial DNA is rapidly degraded by components of the replication machinery. Nat Commun 2018;9:1727. 10.1038/s41467-018-04131-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Vallur AC, Maher RL, Bloom LB. The efficiency of hypoxanthine excision by alkyladenine DNA glycosylase is altered by changes in nearest neighbor bases. DNA Repair 2005;4:1088–98. 10.1016/j.dnarep.2005.05.008. [DOI] [PubMed] [Google Scholar]
- [106].Drohat AC, Jagadeesh J, Ferguson E, Stivers JT. Role of Electrophilic and General Base Catalysis in the Mechanism of Escherichia coli Uracil DNA Glycosylase †. Biochemistry 1999;38:11866–75. 10.1021/bi9910878. [DOI] [PubMed] [Google Scholar]
- [107].Schermerhorn KM, Delaney S. A Chemical and Kinetic Perspective on Base Excision Repair of DNA. Acc Chem Res 2014;47:1238–46. 10.1021/ar400275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Karran P, Lindahl T. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry 1980;19:6005–11. 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- [109].Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019;576:149–57. 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol 2018;36:843–6. 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zafra MP, Schatoff EM, Katti A, Foronda M, Breinig M, Schweitzer AY, et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol 2018;36:888–93. 10.1038/nbt.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Thuronyi BW, Koblan LW, Levy JM, Yeh W-H, Zheng C, Newby GA, et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol 2019;37:1070–9. 10.1038/s41587-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Shin HR, See J-E, Kweon J, Kim HS, Sung G-J, Park S, et al. Small-molecule inhibitors of histone deacetylase improve CRISPR-based adenine base editing. Nucleic Acids Res 2021;49:2390–9. 10.1093/nar/gkab052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Horlbeck MA, Witkowsky LB, Guglielmi B, Replogle JM, Gilbert LA, Villalta JE, et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. ELife 2016;5:e12677. 10.7554/eLife.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Isaac RS, Jiang F, Doudna JA, Lim WA, Narlikar GJ, Almeida R. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. ELife 2016;5:e13450. 10.7554/eLife.13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Falkenberg M, Gustafsson CM. Mammalian mitochondrial DNA replication and mechanisms of deletion formation. Crit Rev Biochem Mol Biol 2020;55:509–24. 10.1080/10409238.2020.1818684. [DOI] [PubMed] [Google Scholar]