Abstract
Skin repair is a significant aspect of human health. While the makeup of healthy stratum corneum and epidermis is generally understood, the mobilization of molecular components during skin repair remains largely unknown. In the present work, we utilize multi-modal, in-situ, mass spectrometry and immunofluorescence imaging for the characterization of newly formed epidermis following an initial acute wound for the first 96h of epithelization. In particular, TOF-SIMS and confirmatory MALDI FT-ICR MS (/MS) analysis permitted the mapping of several lipid classes including phospholipids, neutral lipids, cholesterol, ceramides, and free fatty acids. Endogenous lipid species were localized in discrete epidermal skin layers, including the stratum corneum (SC), stratum granulosum (SG), stratum basale (SB), and dermis. Experiments revealed that healthy re-epithelializing skin is characterized by diminished cholesterol sulfate signal along the stratum corneum towards the migrating epithelial tongue. The spatial distribution and relative abundances of cholesterol sulfate are reported and correlated with the healing time. The multi-modal imaging approach enabled in-situ high-confidence chemical mapping based on accurate mass and fragmentation pattern of molecular components. The use of post-analysis immunofluorescence imaging from the same tissue confirmed the localization of endogenous lipid species at high spatial resolution (~ few microns).
Graphical Abstract
Introduction
Skin tissue repair and barrier restoration is a significant aspect of human health. The outer epidermal skin layer functions as a barrier, which protects against pathogens and prevents trans-epidermal water loss.1 Cutaneous wound repair is a multifaceted process which involves a sequence of biological events, characterized by an inflammatory response followed by proliferation and subsequent remodeling of the wounded tissue. In order for wounds to heal properly, temporal regulation of inflammatory, proliferative and remodeling phases of wound healing are essential, where disruption to any stage of this progression leads to complications and formation of non-healing chronic wounds.2 Non-healing chronic wounds impact ~26 million people with diabetes mellitus (DM) worldwide, with 1 of 3 DM patients developing a foot ulcer during their lifetime; resulting in an approximate cost of $58 billion annually in the US alone3–5. Moreover, wounds that achieve closure can be perceived as in remission, since up to 2 of every 3 ulcers re-occur within 5 years5, which can be associated with ineffective barrier restoration. Keratinocytes, as major cellular component of epidermis, require tight regulation during wound healing and undergo multiple phenotypic changes including migratory, where keratinocytes must first migrate to cover the wound surface, and then proliferate and differentiate into a stratified protective epithelium to restore barrier.2, 6
Historically, mouse models have been used to study wound healing for potential therapeutic applications.7 Despite the large overlap between molecular and cellular mechanisms, a body of literature addressed significant anatomic and physiologic divergence found between human and the widely used rodent models2, 4, 8–12 The wound healing process between the two species is biomechanically and biologically different, from hair pattern and cycle, thickness of dermal and epidermal layers, to speed and mechanisms of wound closure (rodent skin heals by contraction primarily, whereas human skin heals by re-epithelialization). As a result, many potential therapeutic agents have shown efficacy in animal models but not in clinical trials, which impeded development of effective therapies.2, 8–10, 12 There has been a notable increase in utilization of ex vivo human wound models. The translatability of human ex vivo wound model is very high as documented by recent studies ranging from infection and immunity, scar formation, angiogenesis, hair follicle neogenesis, wound re-epithelialization,13–17 as well as for cosmetological application, such as penetration of active components into human skin.18–20
It has been shown that SC lipids are comprised of ceramides, free fatty acids, and cholesterol.21–23 Changes in skin metabolites with age have also been documented, supporting the idea that de-regulation of lipids may lead to loss of skin elasticity or anti-oxidant capability.24 While the makeup of healthy stratum corneum and epidermis is mostly understood, the role of lipids involved in skin repair has not been extensively studied. In atopic eczema patients, decreased free fatty acid (FFA) chain length has been observed in the SC of non-lesional skin but especially in lesional regions.25 Recently, it was demonstrated that long chain fatty acids and cholesterol were correlated with faster recovery from transepidermal water loss and SC hydration.26 While bulk methods such as Raman spectroscopy and liquid chromatography have shown promises in the study of the wound healing research,27–28 they lack the specificity to identify and map molecular components during the skin healing process. On the other hand, Mass Spectrometry Imaging (MSI) has been shown to be very informative, generating molecular maps with high spatial resolution (e.g., using TOF-SIMS19–20) and the distribution of a wide variety of molecular components (e.g., using matrix-assisted laser desorption-ionization, MALDI29–30). When complemented with traditional fluorescence microscopy for secondary confirmation31–33, MSI have been successfully applied to the study of biological problems combining spatial localization and high mass resolution measurements.34 Recently, MALDI was applied for the study of a living skin equivalent model with a focus on incisional wound healing.30 Several authors have demonstrated the suitability of the SIMS platform for applications in skin research including the penetration of topically applied ceramides,20 fatty acids,19 pharmaceuticals,35 and age-related SC lipids.36
In the present work, for the first time we evaluate the molecular distribution as a function of the epidermis healing for an ex-vivo human skin acute wound using multi-modal imaging. Specifically, we show the unique advantages of in-situ endogenous lipid markers and secondary immunofluorescence confirmation for the localization of cholesterol sulfate as a function of the epithelization time with high spatial (TOF-SIMS) and high mass resolution (MALDI-FT-ICR MS).
Experimental Section
Ex-vivo human skin wound protocol
Healthy human skin specimens were obtained as discarded tissue from reduction surgery procedures in accordance to institutional approvals as previously described.37 University of Miami Institutional Review Board (IRB) determined that such protocol does not constitute Human Subject Research per 45 CFR46.101.2. Human skin specimens from these reduction surgery were used to generate acute wounds as previously described.37–38 Briefly, human skin was obtained following an abdominoplasty procedure from age matched healthy male donors. Skin was excised to remove the subcutaneous layer before excising a circular disk using an 8 mm biopsy punch. A smaller, 3 mm medial punch in the center was used to induce an acute wound, corresponding to removal of epidermis. Tissues were grown on air-liquid interface and supplemented daily with growth media containing Dulbecco’s Modified Eagle Medium (DMEM) supplemented with Fetal Bovine Serum (FBS), antibiotic/antimycotic (Figure 1 a and b). Tissues were harvested by flash freezing at the time of wound induction (0h), or 48h, and 96h after wound induction. A cryotome was used to section tissue perpendicular to the initial acute wound, resulting in 12 μm thick slices thaw-mounted onto conductive Indium-tin oxide (ITO) coated glass slides. Alternating tissue slices were taken for Hematoxylin and Eosin staining and TOF-SIMS analysis, respectively.
Figure 1.
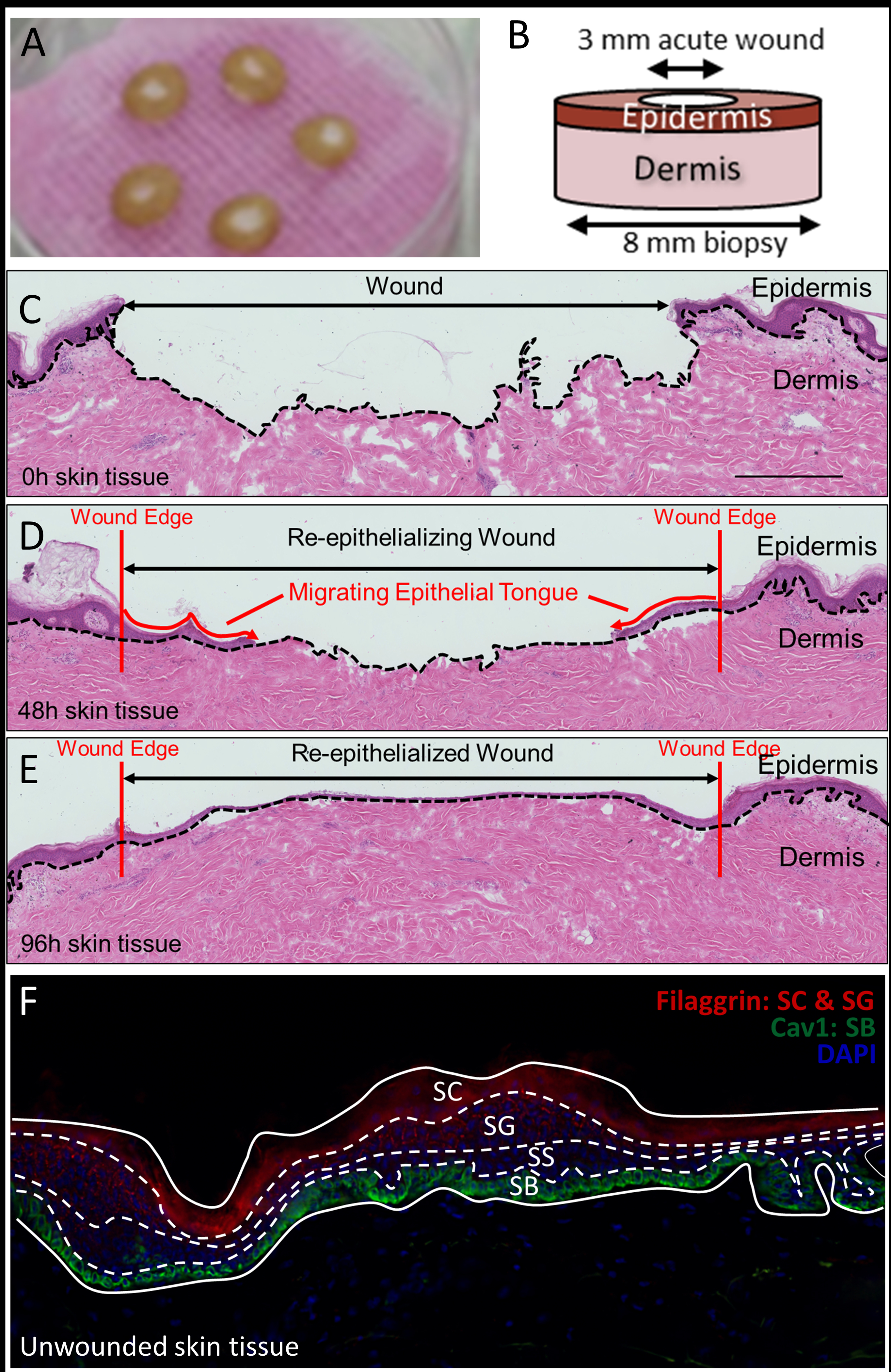
(A) Tissue disks incubating in an air-liquid interface. (B) Schematic representation of biopsy punch and resulting disk. (C-E) H&E staining of parallel tissue sections. (F) Fluorescence microscopy using Filaggrin, Cav-1, and DAPI to delineate the stratum corneum, granulosum, spinosum, and basale of the epidermis.
TOF-SIMS imaging
Slides containing tissue sections were kept frozen at −80 °C until dehydration. Tissues mounted on ITO slides were freeze-dried over dry ice in a custom-built vacuum drier equipped with a cold trap for 2 hours. Samples were allowed to warm to room temperature before transferring into the analytical chamber of the mass spectrometer. TOF-SIMS experiments were carried out on a TOF-SIMS5 instrument (ION-TOF, Münster, Germany), fitted with a bismuth NanoProbe column operated at 25 keV, selected for Bi3+ clusters,39–40 and an electron flood gun to reduce charging during analysis. The TOF-SIMS instrument was operated in the “high current bunched” (HCBU) mode, described elsewhere.33, 41 A typical spatial resolution of < 2.6 μm (20% to 80% of maximum on tissue) and a resolving power of m/Δm ~8,000 at a m/z of 465 were observed. Experiments were carried out at primary ion doses of 3 × 1012 primary ions•cm−2 over selected 400 × 400 μm regions of re-epithelialized tissue, directed by parallel H&E staining and post-analysis fluorescence microscopy. Data were collected and processed using the ION-TOF SurfaceLab 6.4 (Münster, Germany).
Rehydration and Immunofluorescence labeling
Following TOF-SIMS analysis, samples were fixed in ice cold acetone for 2 min and air dried prior to staining. Samples were blocked for 1hr with 2.5% normal goat serum in Phosphate Buffered Saline supplemented with 0.01% Tween (PBSt), prior to incubating with Rabbit anti-Cav1 (1:100, Sigma Aldrich, St. Louis, MO, USA, cat # HPA049326) and mouse anti-filaggrin antibodies (1:200 Abcam, Cambridge, UK, cat# ab3137) overnight at 4oC and then with either Alexa 488 anti-rabbit or Alexa 594 anti-mouse secondary antibodies (Life technologies, Carlsbad, CA, USA) and mounted using Prolong Gold Antifade with DAPI (Life Technologies). Immunofluorescent and phase contrast images were obtained using a Keyence BZ-X700 inverted microscope (Osaka, Japan) and compared to TOF-SIMS-based imaging.
MALDI FT-ICR MS Imaging
Parallel skin tissue samples were prepared on ITO coated slides and dehydrated similar to TOF-SIMS samples described above. Cholesterol sulfate standard (Sigma-Aldrich) was prepared in methanol and used to obtain reference spectra. Samples were then coated with 9-aminoacridine (Sigma-Aldrich) via sublimation (200 × 152 mm apparatus, p/n Z258717, Sigma-Aldrich). Briefly, 300 mg of 9-aminoacridine (9-AA) was added to the bottom of the sublimation chamber. Next, the sample slide was fixed to the cold finger using double-sided carbon tape for heat transfer. The apparatus was then assembled and the cold finger filled with an ice slush. Sublimation was carried out for 9 minutes at a sand bath temperature of 190 °C and pressure of 6 mbar.
MALDI imaging was carried out in a SolariX 7.0T FT-ICR MS instrument (Bruker Daltonics Inc., Billerica, MA). A Smartbeam Nd:YAG laser was operated at 500 Hz at a laser power of 40% and focus set to ‘minimum.’ A total of 800 shots were collected at each spectrum, with a step size of 50 μm for tissue imaging experiments. For MS2 confirmation of the cholesterol sulfate standard, an isolation window of 1 m/z and a CID energy of 40 eV was used.
Results and Discussion
Re-epithelialization of Ex-vivo human skin
Ex -vivo human tissue sections were obtained such that each slice reveals epidermis, dermis, and regions of re-epithelialized skin. As demonstrated by Figure 1 (C–E), the epithelial tongue migrates inward from the edge of the initial ~3 mm wound crater as a function of incubation time in the air-liquid interface previously described. Using Immunofluorescence (IF) staining, the stratum corneum (SC), stratum granulosum (SG), stratum spinosum (SS), and stratum basale (SB) of the epidermis can be clearly observed (Figure 1 F). Antibodies against Filaggrin (red), a significant epidermal protein, primarily stained the lipid envelope of corneocytes of the SC, and to a lesser extent, the SG. DAPI (blue), an AT-region DNA fluorescent dye, was observed to stain nuclei of keratinocytes of the SG and SS. Antibodies against Cav-1 (green), a scaffolding protein, were observed to stain cells of the SB.37
Upon wounding, basal keratinocytes proximal to the wound need to first start proliferating and then migrating into the wound bed in order for the wound to close efficiently. These migrating keratinocytes are what is referred to as the migrating epithelial tongue. In order for these cells to move, they need to reorganize their cytoskeletal architecture and change expression pattern of key intermediate filaments and integrins, as well as expression and activation status of key microfilaments and microtubules.42 Briefly, resting basal keratinocytes express keratins 5 and 14, which upon wounding become activated by release of pro-inflammatory cytokine IL-1 at the wound edge and start expressing keratins 5 and 16.42 This results in a hyperproliferative and migratory keratinocyte phenotype which maintain their active status by secreting various growth factors and cytokines (including TNF-alpha and TGF-alpha). When the wound becomes closed, keratinocytes again change their cytoskeletal architecture and start expressing keratin 17 in order to contract and reorganize the provisional basement membrane and recruit fibroblast to the wound site to produce TGF-beta and start producing more collagen as part of the remodeling phase of wound healing, which also signals to the keratinocytes to change their architecture again and start expressing their resting keratins 5 and 14. Interestingly, we have previously shown that Caveolin-1 negatively regulates both proliferation as well as migration of keratinocytes and thus its expression needs to be spatially (at the wound edge) and temporally (during inflammatory & proliferative stages of wound healing) downregulated, thus allowing keratinocytes to both divide and migrate into the wound bed.37 Expression of caveolin-1 needs to be restored to homeostatic levels to prevent development of scar tissue, as well as numerous hyperproliferative skin disorders.
Control experiments on skin dehydration/rehydration
Taken together, phase contrast and IF provide an avenue for verifying the sample integrity following a freeze-drying procedure, while also allowing for epidermal layers to be referenced in relation to endogenous markers. It has been shown that freeze-drying can prevent the loss of diffusible ions and endogenous lipids.43 Since MSI sample preparation are tissue specific, control experiments were performed to evaluate the viability of this freeze-drying process for the skin sections.
TOF-SIMS and immunofluorescence images are shown from the same tissue slice (Figure 2); note that in-situ analysis implies that immunofluorescence images correspond to sections hydrated/stained after TOF-SIMS analysis. Fatty acids FA 24:0, 25:0, and 26:0 [M-H]- ions co-localize with Filaggrin in the SC. In addition, adenine [M-H]- ions (m/z 134.1) also colocalize with DAPI in region of the nuclei in the epidermis. A comprehensive list of negative mode secondary ions is shown in Table 1. Considering the good correspondence between the images, this freeze-drying was considered effective for preserving the epidermis.
Figure 2.
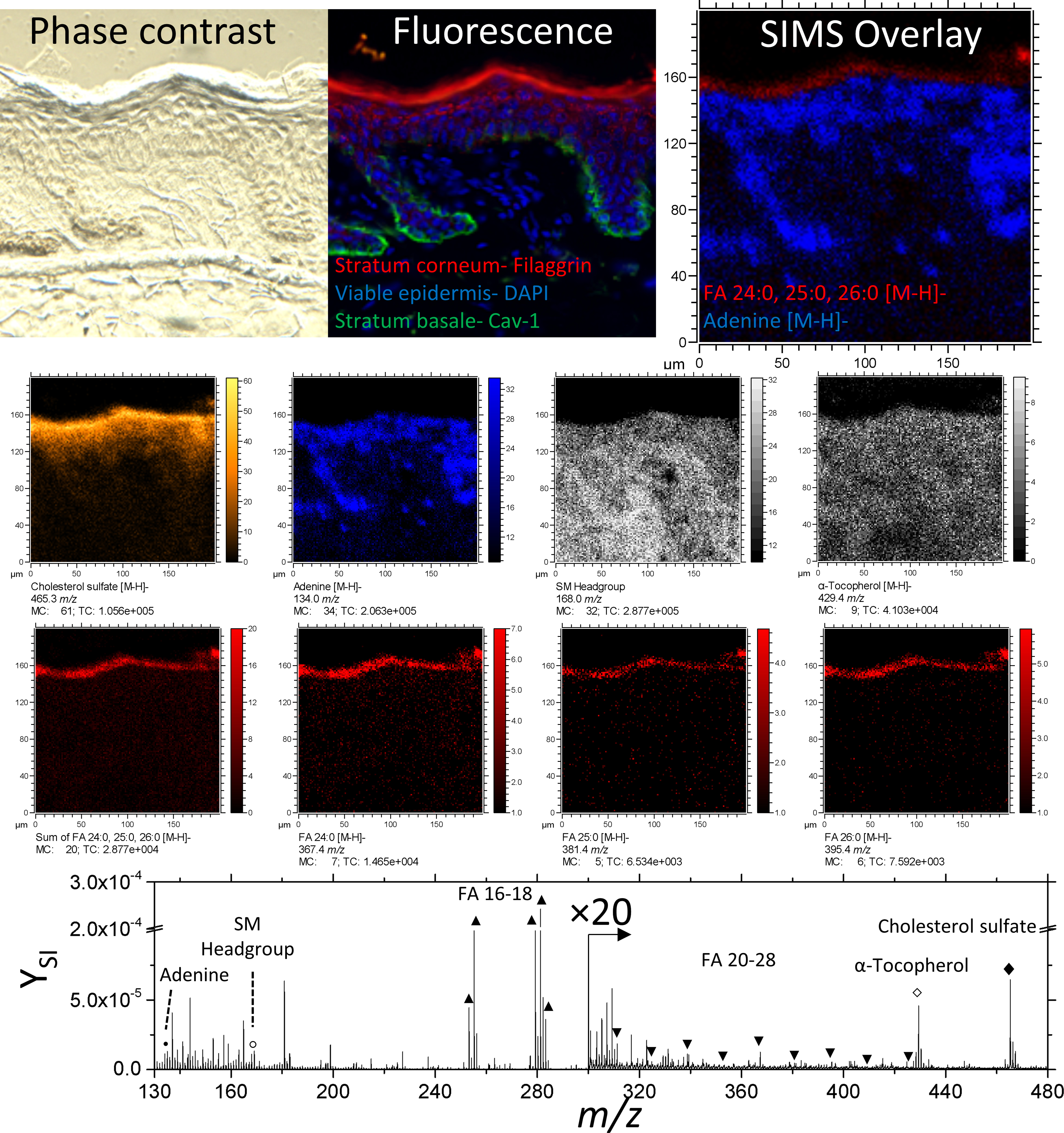
Phase contrast, fluorescence, and secondary ion images of an unwounded region of skin tissue. Negative ion images of cholesterol sulfate, adenine, α-Tocopherol, FA 24:0, 25:0, 26:0 are of the form [M-H]-. The ion at 168.0 m/z is used here to represent the Sphingomyelin headgroup. A comparison between fluorescence and SIMS overlay demonstrates the fidelity of endogenous lipids as biomarkers for epidermal layers.
Table 1.
Tentative SI identifications.
| SIMS m/z | Tentative ID | Tentative Chemical Formula | Reference | MALDI FT-ICR MS error (ppm) | Region (sub-layer) | Ion form | Symbol |
|---|---|---|---|---|---|---|---|
| 134.1 | Adenine | C5H4N4- | SIMS 51 | Epidermis (SG,SS) | [M-H]- | • | |
| 168.0 | SM Headgroup | C4H11NO4P- | SIMS 52 | Dermis | - | ○ | |
| 253.2 | FA 16:1 | C16H29O2- | SIMS 18–20, 53 LC-MS 22 |
Dermis, Epidermis | [M-H]- | ▲ | |
| 255.2 | FA 16:0 | C16H31O2- | SIMS 18–20, 53 LC-MS 22 |
Dermis, Epidermis | [M-H]- | ▲ | |
| 279.2 | FA 18:2 | C18H31O2- | SIMS 18–20, 53 LC-MS 22 |
Dermis, Epidermis | [M-H]- | ▲ | |
| 281.2 | FA 18:1 | C18H33O2- | SIMS 18–20, 53 LC-MS 22 |
Dermis, Epidermis | [M-H]- | ▲ | |
| 283.3 | FA 18:0 | C18H35O2- | SIMS 18–20, 53 LC-MS 22 |
Dermis, Epidermis | [M-H]- | ▲ | |
| 311.3 | FA 20:0 | C20H39O2- | LC-MS 22 | Epidermis (SC) | [M-H]- | ▼ | |
| 325.3 | FA 21:0 | C21H41O2- | SIMS 20 LC-MS 22 |
Epidermis (SC) | [M-H]- | ▼ | |
| 339.3 | FA 22:0 | C22H43O2- | SIMS 20, 53 LC-MS 22 |
Epidermis (SC) | [M-H]- | ▼ | |
| 353.3 | FA 23:0 | C23H45O2- | SIMS 20, 53 LC-MS 22 |
Epidermis (SC) | [M-H]- | ▼ | |
| 367.4 | FA 24:0* | C24H47O2- | SIMS 20, 36, 53 LC-MS 22 |
0.81 | Epidermis (SC) | [M-H]- | ▼ |
| 381.4 | FA 25:0 | C25H49O2- | SIMS 20, 53 LC-MS 22 |
Epidermis (SC) | [M-H]- | ▼ | |
| 395.4 | FA 26:0* | C26H51O2- | SIMS 20, 36, 53 LC-MS 22 |
−1.42 | Epidermis (SC) | [M-H]- | ▼ |
| 409.4 | FA 27:0 | C27H53O2- | SIMS 20 LC-MS 22 |
Epidermis (SC) | [M-H]- | ▼ | |
| 426.4 | FA 28:0 | C28H55O2- | SIMS 20 LC-MS 22 |
Epidermis (SC) | [M-H]- | ▼ | |
| 429.4 | α-Tocopherol | C29H49O2- | SIMS 20 | Dermis | [M-H]- | ■ | |
| 465.3 | Cholesterol sulfate** | C27H45SO4- | SIMS 20, 36, 53 | 0.53 | Epidermis (SS,SG,SC) | [M-H]- | ♦ |
Lipids denoted with a * were confirmed by complimentary MALDI FT-ICR MS analysis.
Cholesterol sulfate (**) was additionally confirmed using MS2.
TOF-SIMS imaging
Positive mode TOF-SIMS analysis of ex-vivo human skin reveals a structured assembly of lipids, including cholesterol, ceramides, and sphingosine and phosphocholine (Table S1). Negative mode TOF-SIMS reveals short-chain fatty acids (≤ C18), long-chain fatty acids (≥ C20), cholesterol sulfate, α-tocopherol, and sphingomyelin (Table 1). The SC, SG, corneocytes of the viable epidermis, and the dermis can be represented by long-chain fatty acids, cholesterol sulfate, adenine, and sphingomyelin, respectively (Figure 3). Complementary analysis of the same field of view is shown in supplementary Figure S2, where the cholesterol sulfate signal is detected.
Figure 3.
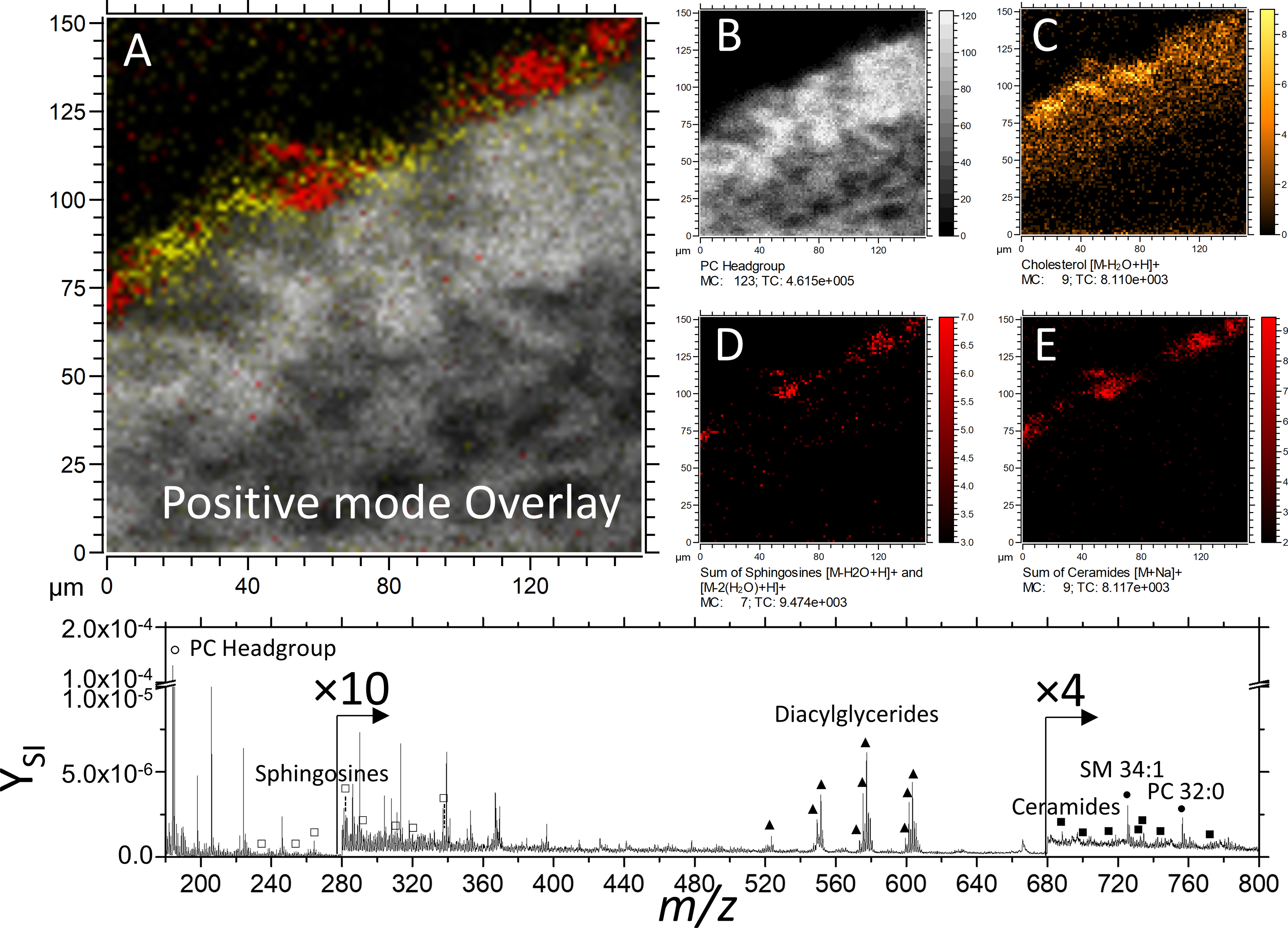
TOF-SIMS imaging of unwounded skin tissue in positive mode detection. (A) Composite of the m/z channels shown in panels B-D. The m/z of 184 is shown to represent PC headgroup (B). For (C) cholesterol [M-H2O+H]+ ion is shown. Images D and E are constructed as the sum of various channels of the same lipid class, shown in Supplementary Figure S1 and listed in detail in Supplemental Table 1.
The distribution of fatty acids and cholesterol sulfate is expected, as the stratum corneum is known to be comprised of mostly ceramides, cholesterol, and free fatty acids.1 In the positive polarity, cholesterol and various ceramides are observed as [M-H2O+H]+ and [M+Na]+, respectively. Cholesterol sulfate, which is secreted from keratinocytes and migrates toward the lamellar membrane of the SC,1 is here observed in the SG and SC in agreement with previous reports.44
MALDI FT-ICR MS Imaging
The localization and TOF-SIMS assignment of the cholesterol sulfate were confirmed with MALDI FT-ICR MS imaging (accurate mass) and fragmentation studies using a standard. MALDI FT-ICR MS imaging from a 96 h epithelialized skin tissue section showed the localization of cholesterol sulfate, as well as endogenous markers FA 24:0 and 26:0 of the stratum corneum (Supplementary Figure S3), in good agreement with the TOF-SIMS observations. The MS/MS analysis of the cholesterol sulfate standard showed the typical ion fragment ion SO4H- (Supplementary Figure S4 and S5), in good agreement with the on- tissue analyses MALDI-FT-ICR MS/MS analysis and TOF-SIMS observations (Supplementary Figure S4). Cholesterol sulfate was also previously observed and in human skin epidermis using MALDI-TOF imaging and confirmed using post-source decay MS/MS.45
TOF-SIMS imaging of re-epithelialized skin
Using a Cav-1 stain, the wound edge of re-epithelialized skin tissue can be precisely determined.37 From left to right, the disruption of IF labeled basal keratinocytes indicates where the initial biopsy occurred (Figure 4).
Figure 4.
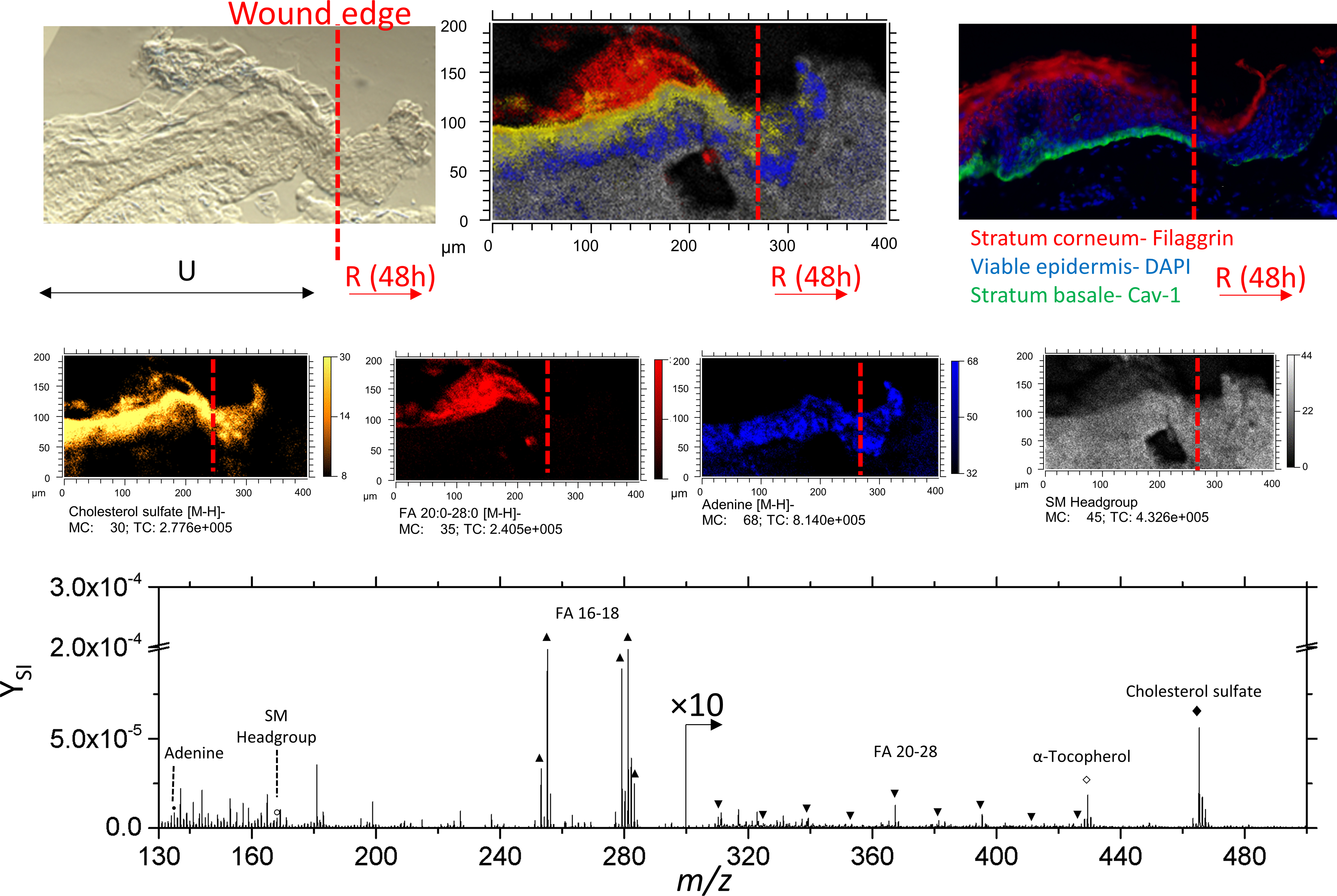
Optical, SIMS imaging, and fluorescence imaging of re-epithelializing skin tissue. The ‘U’ and ‘R’ denote unwounded and re-epithelialized areas of 48h incubated skin, respectively. Individual secondary ion images comprising the SIMS overlay are shown in the middle row. Secondary ion images of sphingomyelin is represented as the of ion at 168.0 m/z. Here, signal of fatty acids 20:0–28:0 were combined and shown as a summed secondary ion image.
In addition, it can be observed from TOF-SIMS secondary ion images that stratification of characteristic lipids of each epidermal layer is disrupted at the migrating epithelial tongue.
Cholesterol sulfate distribution in acute wounds
Cholesterol sulfate is a precursor excreted by keratinocytes of the SG layer, which becomes transformed by steroid sulfatase into cholesterol as it migrates toward the SC as a vital component of the permeability barrier.1, 44 When a tissue region is analyzed in both TOF-SIMS polarities, cholesterol sulfate (negative ion mode) and cholesterol (positive ion mode) are co-localized in the epithelium (Figure 3).
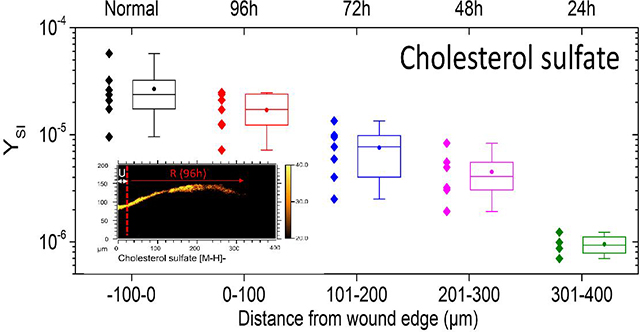
From the TOF-SIMS analysis, cholesterol sulfate density in the epidermis appears to peak between the SG and SC. In Figure 5, cholesterol sulfate is mapped across a 400 μm area of skin tissue near the site of the initial acute wound. In areas of unwounded epidermis (e.g., 0h tissue section shown in Figure 5), cholesterol sulfate signal is relatively uniform. Along the epithelial tongue, however, cholesterol sulfate expression is diminished as a function of distance from the initial wound edge. In order to determine trends in cholesterol sulfate across various SIMS analyses, analyzed tissue areas were virtually divided in 100 μm segments, roughly equivalent to the distance epithelialized every 24h of incubation. The secondary ion yield (YSI) of the m/z of 465.3 was integrated per segments and summarized as a boxplot in Figure 5. Data from multiple replicate analyses from 48h and 96h epithelialized tissue are considered. As a general trend, diminished cholesterol sulfate towards newly formed epithelial tissue is observed. One implication of such epithelialization dynamics may be related to the incorporation of caveolin-1 into the membranes of the basal layer of the epidermis.
Figure 5.
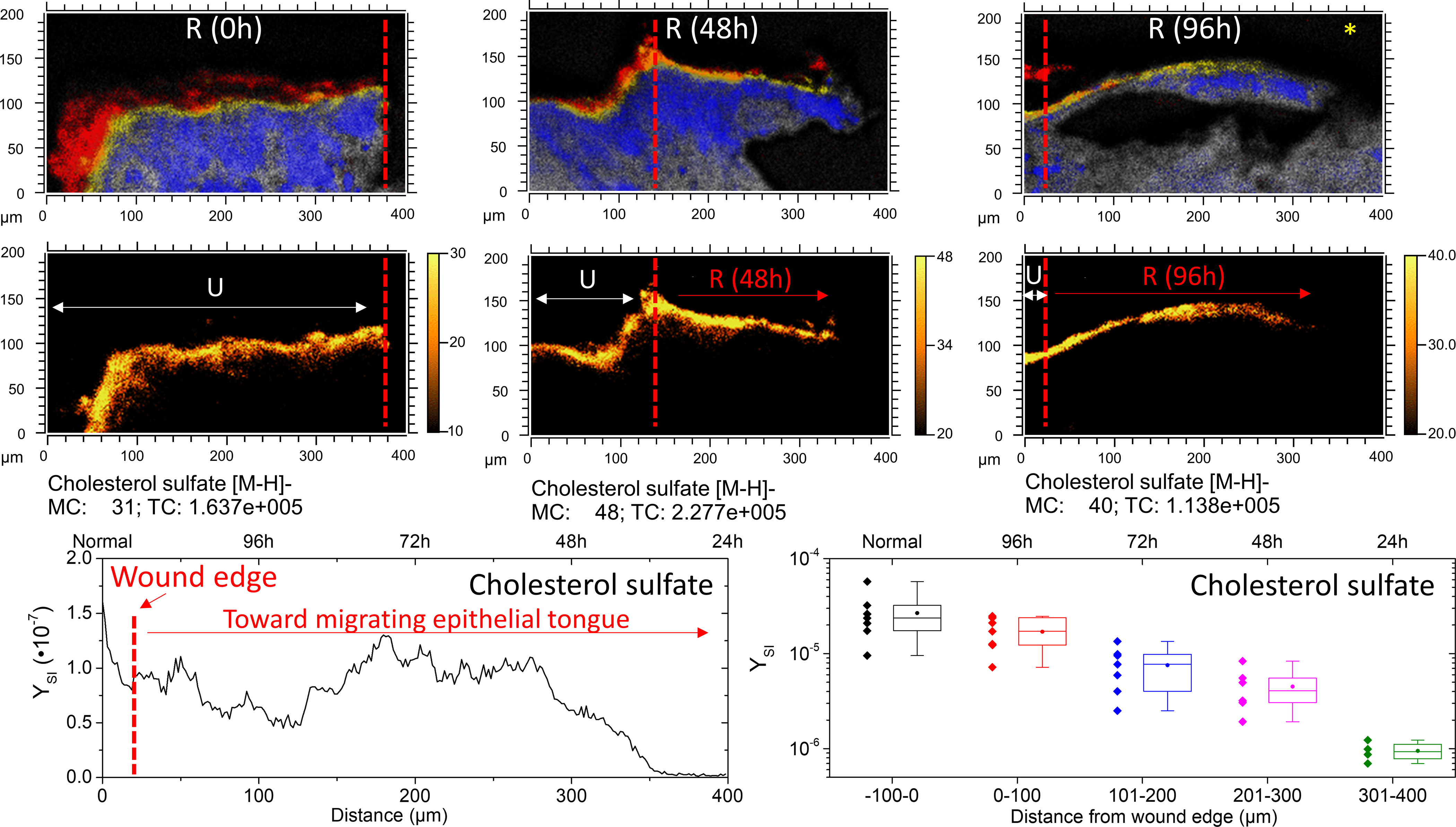
Cholesterol sulfate progression in re-epithelializing tissue. The top row shows SIMS overlays of FA 20:0–28:0 (Red), cholesterol sulfate (yellow), adenine (blue), and sphingomyelin (grey). The red dashed line denotes the wound edge, as corroborated by microscopy. A typical linescan plot of the 96h re-epithelialized tissue (denoted with an *) in the lower left shows the integrated y-area intensity of cholesterol sulfate as a function of distance. Profiles were divided into quarters roughly equivalent to 24 h of re-epitheliazation, approximately 100 μm segments. In the lower right, a boxplot shows the minimum, maximum, lower quartile, upper quartile, median (line) and mean (circle) of observations pooled from 48h and 96h incubated tissues according to distance from the initial wound. N=7 for −100–200 μm; N=6 for 201–300; N=4 for 301–400 μm.
Caveolin-1 is the primary structural component of caveolae, which are specialized lipid rafts, found in areas of the cell membrane that are enriched in cholesterol (and sphingomyelins). However, caveolin’s stability is tied to its localization within caveolae and thus levels of cholesterol.46–47 Caveolin-1 has been demonstrated to function as a negative regulator of proliferation,48–49 and elevated caveolin-1 levels have been reported to be characteristic of reduced corneal re-epithelialization capacity.50 Previous studies have shown that cholesterol depleting agents (including methyl-b-cyclodextrin, statins and cyclosporins) disrupt formation of caveolae by preventing caveolin-1 from binding to cholesterol (and sphingomyelin) rich regions in the membrane, causing caveolin-1 oligomers to depolymerize into monomers which then get poly-ubiquitinated and proteasomally degraded.37 Whether HMG-CoA reductase, or other enzymes responsible for synthesis of cholesterol are also downregulated during wound re-epithelialization remains to be answered, however that is not the scope of the current study. Still, attenuation of cholesterol should lead to attenuation of caveolin-1 in the basal layer, thereby facilitating the rate of re-epithelialization.
Conclusions
The use of multi-modal, in-situ mass spectrometry and immunofluorescence imaging allowed for the study of the ex-vivo acute wound healing epithelization by correlating cholesterol, cholesterol sulfate, FA and lipid signals with the high selectivity of immunofluorescence staining for discrete epidermal layers (SC, SG, SS, SB) from the same tissue section. The high spatial resolution MSI images allowed for precise visualization of skin layers and provides a useful tool for localizing key molecular components during the process of human skin repair. For example, the potential of endogenous markers (e.g., fatty acids ≤ FA 20:0, adenine m/z 134.1, and sphingomyelin m/z 687.5 C38H76N2PO6-) to differentiate layers of the skin is shown. Moreover, accurate mass and tandem MS/MS from tissue slices allowed for the confirmation of TOF-SIMS assignments of molecular signals (e.g., cholesterol sulfate in skin sections). Experiments demonstrated that cholesterol sulfate signal is diminished in the SC from the wound edge towards the epithelial tongue. These results are in good agreement with previous observations of attenuation of caveolin-1 in the basal layer and suggest the possibility of using cholesterol sulfate as a biomarker by which human skin epithelization may be evaluated.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH grant No. R21AI135469-01A1 to FFL, PhRMA Foundation Grant and Medline Wound Healing Foundation Innovation Grant to IJ, AR060562, NR015649, and NR013881 to MTC, and Dwoskin family gift to the Dr Phillip Frost Department of Dermatology and Cutaneous Surgery. AC was fully supported by NSF grant HRD-1547798 through a Fellowship offered by FIUs Centers of Research Excellence in Science and Technology (CREST) program over the course of this study. This is contribution number ### from the Southeast Environmental Research Center in the Institute of Water and Environment at Florida International University.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Positive mode TOF-SIMS imaging of sphingosine fragment ions and ceramides; Positive and Negative mode TOF-SIMS spectra of cholesterol and cholesterol sulfate standards; Negative mode MALDI-FT-ICR MS analysis of a cholesterol sulfate standard and confirmation of cholesterol sulfate on tissue; Positive mode TOF-SIMS tentative lipid assignments. (PDF)
REFERENCES
- 1.Feingold KR, Thematic review series: Skin Lipids. The role of epidermal lipids in cutaneous permeability barrier homeostasis. Journal of Lipid Research 2007, 48 (12), 2531–2546. [DOI] [PubMed] [Google Scholar]
- 2.Eming SA; Martin P; Tomic-Canic M, Wound repair and regeneration: mechanisms, signaling, and translation. Science Translational Medicine 2014, 6 (265), 265sr6–265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexiadou K; Doupis J, Management of diabetic foot ulcers. Diabetes Ther 2012, 3 (1), 4–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong DG; Boulton AJM; Bus SA, Diabetic Foot Ulcers and Their Recurrence. New England Journal of Medicine 2017, 376 (24), 2367–2375. [DOI] [PubMed] [Google Scholar]
- 5.Nussbaum SR; Carter MJ; Fife CE; DaVanzo J; Haught R; Nusgart M; Cartwright D, An Economic Evaluation of the Impact, Cost, and Medicare Policy Implications of Chronic Nonhealing Wounds. Value Health 2018, 21 (1), 27–32. [DOI] [PubMed] [Google Scholar]
- 6.Pastar I; Stojadinovic O; Yin NC; Ramirez H; Nusbaum AG; Sawaya A; Patel SB; Khalid L; Isseroff RR; Tomic-Canic M, Epithelialization in Wound Healing: A Comprehensive Review. Advances in Wound Care 2014, 3 (7), 445–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galiano RD; Michaels VJ; Dobryansky M; Levine JP; Gurtner GC, Quantitative and reproducible murine model of excisional wound healing. Wound Repair and Regeneration 2004, 12 (4), 485–492. [DOI] [PubMed] [Google Scholar]
- 8.Stojadinovic O; Ito M; Tomic-Canic M, Hair Cycling and Wound Healing: To Pluck or Not to Pluck? Journal of Investigative Dermatology 2011, 131 (2), 292–294. [DOI] [PubMed] [Google Scholar]
- 9.Ansell DM; Holden KA; Hardman MJ, Animal models of wound repair: Are they cutting it? Experimental Dermatology 2012, 21 (8), 581–585. [DOI] [PubMed] [Google Scholar]
- 10.Shay T; Jojic V; Zuk O; Rothamel K; Puyraimond-Zemmour D; Feng T; Wakamatsu E; Benoist C; Koller D; Regev A; ImmGen C, Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (8), 2946–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliot S; Wikramanayake TC; Jozic I; Tomic-Canic M, A Modeling Conundrum: Murine Models for Cutaneous Wound Healing. Journal of Investigative Dermatology 2018, 138 (4), 736–740. [DOI] [PubMed] [Google Scholar]
- 12.Pastar I; Wong LL; Egger AN; Tomic-Canic M, Descriptive vs mechanistic scientific approach to study wound healing and its inhibition: Is there a value of translational research involving human subjects? Experimental Dermatology 2018, 27 (5), 551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corzo-León DE; Munro CA; MacCallum DM An ex vivo Human Skin Model to Study Superficial Fungal Infections Front Microbiol [Online], 2019, p. 1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon DJ; Fregoso DR; Nguyen D; Chen V; Strbo N; Fuentes JJ; Tomic-Canic M; Crawford R; Pastar I; Isseroff RR, A tractable, simplified ex vivo human skin model of wound infection. Wound Repair and Regeneration 2019, 27 (4), 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkers T; Boiten WA; Absalah S; van Smeden J; Lavrijsen APM; Bouwstra JA, Compromising human skin in vivo and ex vivo to study skin barrier repair. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2019, 1864 (8), 1103–1108. [DOI] [PubMed] [Google Scholar]
- 16.Hu AC; Hong EM; Toubat O; Sivoraphonh R; Barnes C; Moy WJ; Krasieva TB; Wong BJF, Multiphoton Microscopy of Collagen Structure in Ex Vivo Human Skin Following Electrochemical Therapy. Lasers in Surgery and Medicine 2019. [DOI] [PubMed] [Google Scholar]
- 17.Egu DT; Sigmund AM; Schmidt E; Spindler V; Walter E; Waschke J, A new ex vivo human oral mucosa model reveals that p38MAPK inhibition is not effective to prevent autoantibody-induced mucosal blistering in pemphigus. British Journal of Dermatology 2019, 0 (ja). [DOI] [PubMed] [Google Scholar]
- 18.Kezutyte T; Desbenoit N; Brunelle A; Briedis V, Studying the penetration of fatty acids into human skin by ex vivo TOF-SIMS imaging. Biointerphases 2013, 8 (1), 3. [DOI] [PubMed] [Google Scholar]
- 19.Čižinauskas V; Elie N; Brunelle A; Briedis V, Fatty acids penetration into human skin ex vivo: A TOF-SIMS analysis approach. Biointerphases 2017, 12 (1), 011003. [DOI] [PubMed] [Google Scholar]
- 20.Sjövall P; Skedung L; Gregoire S; Biganska O; Clément F; Luengo GS, Imaging the distribution of skin lipids and topically applied compounds in human skin using mass spectrometry. Scientific Reports 2018, 8 (1), 16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weerheim A; Ponec M, Determination of stratum corneum lipid profile by tape stripping in combination with high-performance thin-layer chromatography. Archives of Dermatological Research 2001, 293 (4), 191–199. [DOI] [PubMed] [Google Scholar]
- 22.van Smeden J; Boiten WA; Hankemeier T; Rissmann R; Bouwstra JA; Vreeken RJ, Combined LC/MS-platform for analysis of all major stratum corneum lipids, and the profiling of skin substitutes. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 2014, 1841 (1), 70–79. [DOI] [PubMed] [Google Scholar]
- 23.Norlén L; Plasencia I; Bagatolli L, Stratum corneum lipid organization as observed by atomic force, confocal and two-photon excitation fluorescence microscopy. International Journal of Cosmetic Science 2008, 30 (6), 391–411. [DOI] [PubMed] [Google Scholar]
- 24.Kuehne A; Hildebrand J; Soehle J; Wenck H; Terstegen L; Gallinat S; Knott A; Winnefeld M; Zamboni N, An integrative metabolomics and transcriptomics study to identify metabolic alterations in aged skin of humans in vivo. BMC Genomics 2017, 18 (1), 169–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Smeden J; Janssens M; Kaye ECJ; Caspers PJ; Lavrijsen AP; Vreeken RJ; Bouwstra JA, The importance of free fatty acid chain length for the skin barrier function in atopic eczema patients. Exp. Dermatol. 2014, 23 (1), 45–52. [DOI] [PubMed] [Google Scholar]
- 26.Zouboulis CC; Elewa R; Ottaviani M; Fluhr J; Picardo M; Bernois A; Heusèle C; Camera E, Age influences the skin reaction pattern to mechanical stress and its repair level through skin care products. Mechanisms of Ageing and Development 2018, 170, 98–105. [DOI] [PubMed] [Google Scholar]
- 27.Jain R; Calderon D; Kierski PR; Schurr MJ; Czuprynski CJ; Murphy CJ; McAnulty JF; Abbott NL, Raman spectroscopy enables noninvasive biochemical characterization and identification of the stage of healing of a wound. Analytical Chemistry 2014, 86 (8), 3764–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wijesinghe DS; Warncke UO; Diegelmann RF, Human as the Ultimate Wound Healing Model: Strategies for Studies Investigating the Dermal Lipidome. Curr Dermatol Rep 2016, 5 (4), 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnel D; Legouffe R; Eriksson AH; Mortensen RW; Pamelard F; Stauber J; Nielsen KT, MALDI imaging facilitates new topical drug development process by determining quantitative skin distribution profiles. Analytical and Bioanalytical Chemistry 2018, 410 (11), 2815–2828. [DOI] [PubMed] [Google Scholar]
- 30.Lewis EEL; Barrett MRT; Freeman-Parry L; Bojar RA; Clench MR, Examination of the skin barrier repair/wound healing process using a living skin equivalent model and matrix-assisted laser desorption-ionization-mass spectrometry imaging. International Journal of Cosmetic Science 2018, 40 (2), 148–156. [DOI] [PubMed] [Google Scholar]
- 31.Ostrowski SG; Kurczy ME; Roddy TP; Winograd N; Ewing AG, Secondary Ion MS Imaging To Relatively Quantify Cholesterol in the Membranes of Individual Cells from Differentially Treated Populations. Analytical Chemistry 2007, 79 (10), 3554–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chini CE; Fisher GL; Johnson B; Tamkun MM; Kraft ML, Observation of endoplasmic reticulum tubules via TOF-SIMS tandem mass spectrometry imaging of transfected cells. Biointerphases 2018, 13 (3), 03B409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellanos A; Ramirez CE; Michalkova V; Nouzova M; Noriega FG; Fernández-Lima F, Three dimensional secondary ion mass spectrometry imaging (3D-SIMS) of Aedes aegypti ovarian follicles. Journal of Analytical Atomic Spectrometry 2019, 34 (5), 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desbenoit N; Walch A; Spengler B; Brunelle A; Römpp A, Correlative mass spectrometry imaging, applying time-of-flight secondary ion mass spectrometry and atmospheric pressure matrix-assisted laser desorption/ionization to a single tissue section. Rapid Communications in Mass Spectrometry 2017, 32 (2), 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sjövall P; Greve TM; Clausen SK; Moller K; Eirefelt S; Johansson B; Nielsen KT, Imaging of Distribution of Topically Applied Drug Molecules in Mouse Skin by Combination of Time-of-Flight Secondary Ion Mass Spectrometry and Scanning Electron Microscopy. Analytical Chemistry 2014, 86 (7), 3443–3452. [DOI] [PubMed] [Google Scholar]
- 36.Starr NJ; Johnson DJ; Wibawa J; Marlow I; Bell M; Barrett DA; Scurr DJ, Age-Related Changes to Human Stratum Corneum Lipids Detected Using Time-of-Flight Secondary Ion Mass Spectrometry Following in Vivo Sampling. Analytical Chemistry 2016, 88 (8), 4400–4408. [DOI] [PubMed] [Google Scholar]
- 37.Jozic I; Sawaya AP; Pastar I; Head CR; Wong LL; Glinos GD; Wikramanayake TC; Brem H; Kirsner RS; Tomic-Canic M, Pharmacological and genetic inhibition of Caveolin-1 promotes epithelialization and wound closure. Molecular Therapy 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jozic I; Vukelic S; Stojadinovic O; Liang L; Ramirez HA; Pastar I; Tomic Canic M, Stress Signals, Mediated by Membranous Glucocorticoid Receptor, Activate PLC/PKC/GSK-3β/β-catenin Pathway to Inhibit Wound Closure. J Invest Dermatol 2017, 137 (5), 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brunelle A; Touboul D; Laprevote O, Biological tissue imaging with time-of-flight secondary ion mass spectrometry and cluster ion sources. Journal of Mass Spectrometry 2005, 40 (8), 985. [DOI] [PubMed] [Google Scholar]
- 40.Touboul D; Kollmer F; Niehuis E; Brunelle A; Laprevote O, Improvement of Biological Time-of-Flight-Secondary Ion Mass Spectrometry Imaging with a Bismuth Cluster Ion Source. Journal of the American Society for Mass Spectrometry 2005, 16 (10), 1608. [DOI] [PubMed] [Google Scholar]
- 41.Sodhi RNS, Time-of-flight secondary ion mass spectrometry (TOF-SIMS):-versatility in chemical and imaging surface analysis. Analyst (Cambridge, U. K.) 2004, 129 (6), 483. [DOI] [PubMed] [Google Scholar]
- 42.Freedberg IM; Tomic-Canic M; Komine M; Blumenberg M, Keratins and the Keratinocyte Activation Cycle. Journal of Investigative Dermatology 2001, 116 (5), 633–640. [DOI] [PubMed] [Google Scholar]
- 43.Malm J; Giannaras D; Riehle MO; Gadegaard N; Sjövall P, Fixation and Drying Protocols for the Preparation of Cell Samples for Time-of-Flight Secondary Ion Mass Spectrometry Analysis. Analytical Chemistry 2009, 81 (17), 7197–7205. [DOI] [PubMed] [Google Scholar]
- 44.Strott CA; Higashi Y, Cholesterol sulfate in human physiology: what’s it all about? Journal of Lipid Research 2003, 44 (7), 1268–1278. [DOI] [PubMed] [Google Scholar]
- 45.Enthaler B; Trusch M; Fischer M; Rapp C; Pruns JK; Vietzke J-P, MALDI imaging in human skin tissue sections: focus on various matrices and enzymes. Analytical and Bioanalytical Chemistry 2013, 405 (4), 1159–1170. [DOI] [PubMed] [Google Scholar]
- 46.Li S; Song KS; Lisanti MP, Expression and Characterization of Recombinant Caveolin: PURIFICATION BY POLYHISTIDINE TAGGING AND CHOLESTEROL-DEPENDENT INCORPORATION INTO DEFINED LIPID MEMBRANES. J. Biol. Chem. 1996, 271 (1), 568–573. [PubMed] [Google Scholar]
- 47.Murata M; Peränen J; Schreiner R; Wieland F; Kurzchalia TV; Simons K, VIP21/caveolin is a cholesterol-binding protein. Proceedings of the National Academy of Sciences 1995, 92 (22), 10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bender F; Montoya M; Monardes V; Leyton L; Quest AFG, Caveolae and caveolae-like membrane domains in cellular signaling and disease: Identification of downstream targets for the tumor suppressor protein caveolin-1. Biological Research 2002, 35, 151–167. [DOI] [PubMed] [Google Scholar]
- 49.Gheida SF; Neinaa YME-H; Mohammed DAE-A, Caveolin-1 expression in hyperproliferative skin disorders: A potential predictive marker of disease severity and progression. Dermatologica Sinica 2018, 36 (4), 179–184. [Google Scholar]
- 50.Rhim JH; Kim JH; Yeo E-J; Kim JC; Park SC, Caveolin-1 as a novel indicator of wound-healing capacity in aged human corneal epithelium. Molecular Medicine 2010, 16 (11–12), 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.May CJ; Canavan HE; Castner DG, Quantitative X-ray Photoelectron Spectroscopy and Time-of-Flight Secondary Ion Mass Spectrometry Characterization of the Components in DNA. Analytical Chemistry 2004, 76 (4), 1114–1122. [DOI] [PubMed] [Google Scholar]
- 52.Adams KJ; DeBord JD; Fernandez-Lima F, Lipid specific molecular ion emission as a function of the primary ion characteristics in TOF-SIMS. Journal of Vacuum Science & Technology B 2016, 34 (5), 051804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sjövall P; Greve TM; Clausen SK; Moller K; Eirefelt S; Johansson B. r.; Nielsen KT, Imaging of Distribution of Topically Applied Drug Molecules in Mouse Skin by Combination of Time-of-Flight Secondary Ion Mass Spectrometry and Scanning Electron Microscopy. Analytical Chemistry 2014, 86 (7), 3443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


