Abstract
Convolutional neural networks, the state of the art for image segmentation, have been successfully applied to histology images by many computational researchers. However, the translatability of this technology to clinicians and biological researchers is limited due to the complex and undeveloped user interface of the code, as well as the extensive computer setup required. We have developed a plugin for segmentation of whole slide images (WSIs) with an easy to use graphical user interface. This plugin runs a state-of-the-art convolutional neural network for segmentation of WSIs in the cloud. Our plugin is built on the open source tool HistomicsTK by Kitware Inc. (Clifton Park, NY), which provides remote data management and viewing abilities for WSI datasets. The ability to access this tool over the internet will facilitate widespread use by computational non-experts. Users can easily upload slides to a server where our plugin is installed and perform the segmentation analysis remotely. This plugin is open source and once trained, has the ability to be applied to the segmentation of any pathological structure. For a proof of concept, we have trained it to segment glomeruli from renal tissue images, demonstrating it on holdout tissue slides.
Keywords: WSI segmentation, plugin, cloud based analysis, glomeruli
I. INTRODUCTION
Recent advancements in machine learning techniques have attained previously unachievable accuracy for image analysis tasks. In particular, convolutional neural networks (CNNs) have great potential for impactful applications for the segmentation of image structures. In the field of pathology, CNNs have been successfully utilized by many research groups for the segmentation of whole slide images (WSIs)1–4. However, thus far tools to segment WSIs have been complex to deploy and use, requiring use of the command line interface and computational expertise. Going beyond development, the target demographic for these tools is the pathologist or biological scientist, who’s clinical workflow or research questions could leverage fast and accurate segmentation of relevant structures. To address this gap, we have developed a powerful tool for the segmentation of WSIs and deployed it as an easy to use plugin in a cloud based WSI viewer. Our code is open-source and easily deployable on a remote server for use by the community over the web. While the plugin is agnostic to tissue type or structure of interest, we test this pipeline by training a CNN model for the segmentation of glomeruli from renal tissue images.
II. RESULTS
Using a network model trained to detect glomeruli, we deployed our plugin on the web. Currently the network model is still being developed but the plugin is fully functional. It produces glomerular annotations which are automatically displayed on top of the slide using the HistomicsTK interface5. The user can pan and zoom the slide as well as interact with the annotations, removing or adding regions. See Figs. 1 and 2 for details about this graphical user interface.
Fig. 1. The graphical user interface of our plugin running in HistomicsTK available via the web.
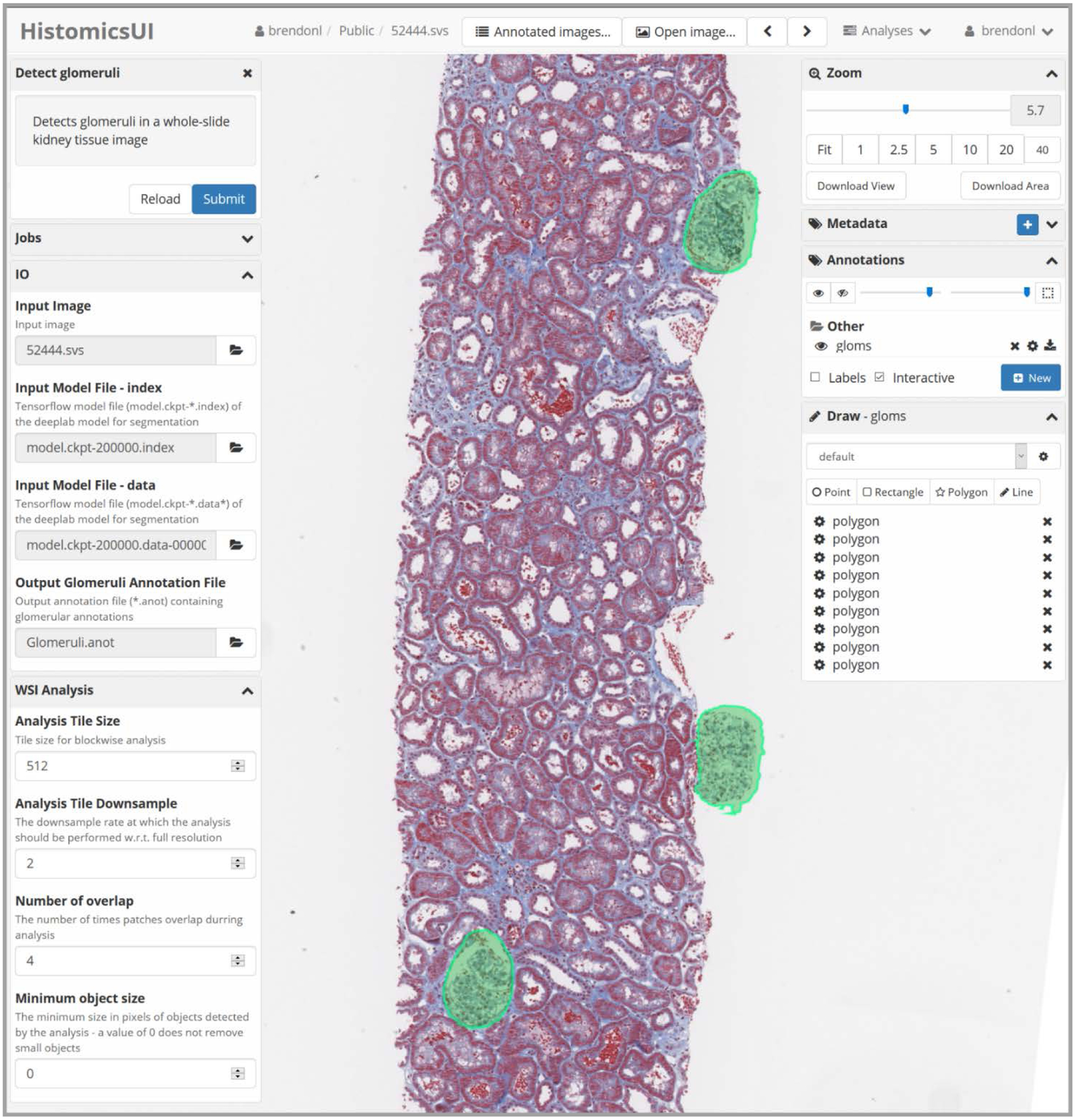
The left Detect Glomeruli column shows the controls for the plugin: IO is required arguments and WSI Analysis contains optional parameters. The right column shows the WSI viewer controls and annotations created by our plugin. The green annotations on the holdout slide are predicted by our plugin and are easily editable by the user. Slides are easily analyzed by clicking the submit button in the top left corner, which submits a segmentation job, running the Deeplab network on the remote server (where HistomicsTK is installed).
Fig. 2. Examples of the HistomicsTK user interface.
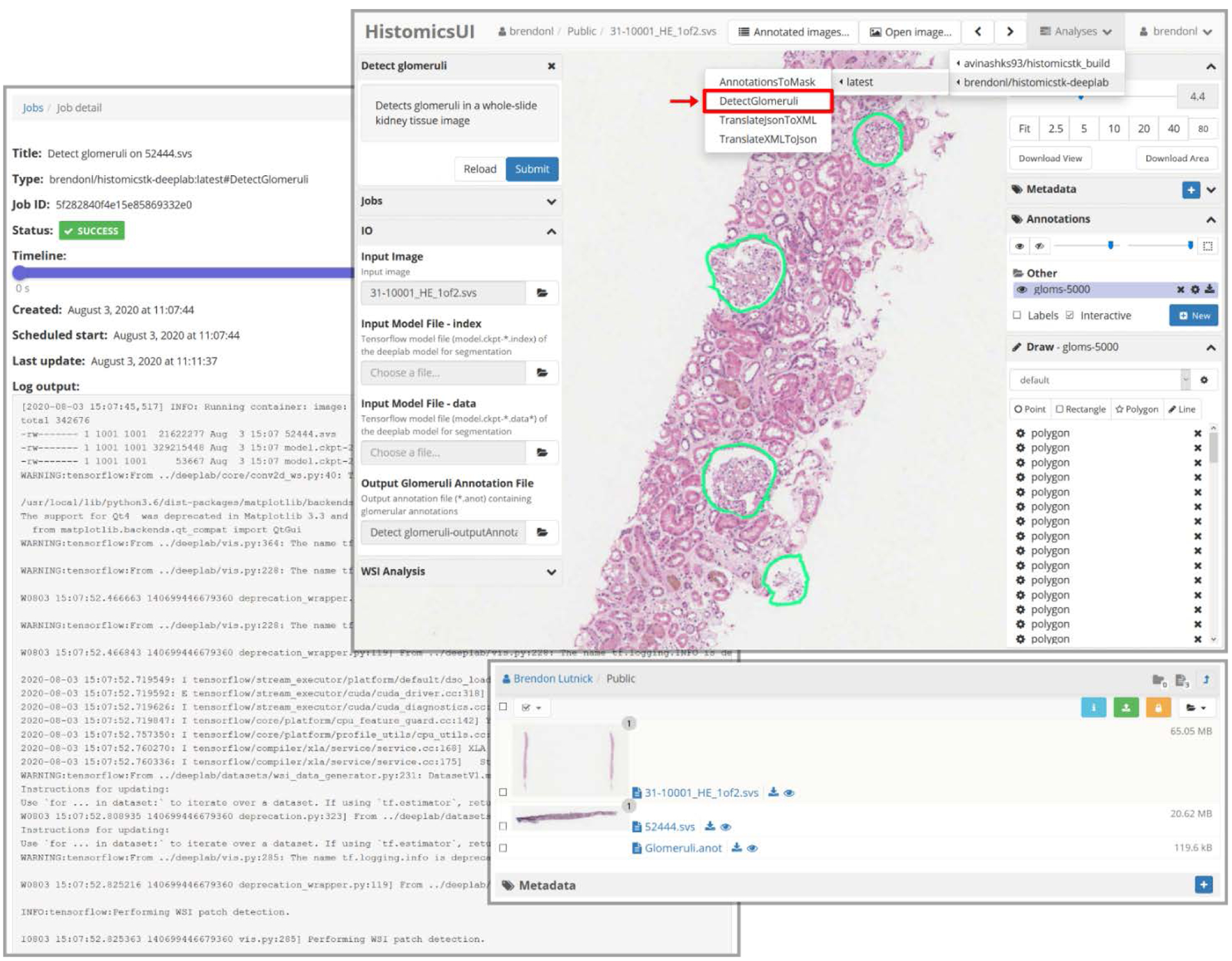
The left panel shows the “Jobs” page where users can check the status of a running segmentation job. This displays the command line output of the running plugin that has been abstracted away from the user. The top right panel shows how the plugin is accessed from the user interface (highlighted in red) as well as another example glomeruli segmentation by the plugin on a holdout WSI. The bottom right panel shows an example of file management using HistomicsTK. Here users can upload and manage slides and annotations on the remote server.
The purpose of this abstract is not the performance of our glomerular segmentation algorithm, but the demonstration of its usefulness when deployed as an online plugin. We qualitatively evaluate this algorithm using holdout WSIs, and expect similar performance as achieved in our earlier work4. We note that at the time of writing the network has not been fully trained. Examples of these holdout segmentations, displayed in the HistomicsTK interface are visualized in Figs. 1 and 2.
A video overview of this tool is available here: https://buffalo.box.com/s/w9ao5p1qs9o3lgyqk8ioih6grklb9p9i.
III. METHODS
With the goal of developing a tool with class leading WSI segmentation accuracy as well as easy accessibility to computational non-experts, we have integrated the popular semantic segmentation network Deeplab V3+6 with Digital Slide Archive5 the open-source cloud-based histology management program. Specifically, we have created an easy to use plugin using HistomicsTK, an application programing interface of the Digital Slide Archive for running Python codes. This plugin efficiently runs the Deeplab network for native segmentation of WSIs, making testing new slides accessible through the HistomicsTK graphical user interface.
Software:
This plugin, built using open-source software, is available to the digital pathology community for use and further development. Our modified HistomicsTK-Deeplab codebase is available on GitHub and also as a prebuilt Docker image for easy installation: https://hub.docker.com/repository/docker/brendonl/histomicstk-deeplab. This software is deployed in the cloud and is accessible via the web, making it easily accessible to the community (Fig. 1).
Functionality:
Our plugin outputs predictions as a series of image contours which we format to a JSON format for display in HistomicsTK. We have made our code modular with the ability to handle multi-class segmentation, and included optional prediction parameters for advanced users. We have also included functionality for conversion to the XML format used to display contours in Aperio ImageScope, a popular WSI viewer, as well as export annotated regions as image masks (Fig. 2).
Dataset:
For the sake of this abstract, we have constrained our testing of this pipeline to the segmentation of glomeruli from renal tissue WSIs. At the time of writing our glomerular segmentation Deeplab model has limited training time. Currently this model is trained using 24 human biopsies and has not yet reached convergence, therefore, the output segmentations it produces are of limited quality. With our plugin established we plan to focus on the further training of our model, aiming for a training set of at least 1000 PAS, H&E, Trichrome, and Silver stained renal slides. We expect to achieve this extensive training as well as testing during the time of the SPIE 2021 conference.
Computational model:
The Deeplab V3+ network was trained for 300000 steps using cropped patches of size 512×512 pixels, a batch size of 12, and learning rate of 1e−3. This model used the Xception network backbone with an output stride of 16 as described in the Deeplab paper6. Fig. 2 gives an example of the command line output of this network running when a job is submitted using the plugin.
Human data:
Biopsy samples from human diabetic nephropathy patients were obtained from Kidney Translational Research Center at Washington University School of Medicine directed by Dr. Sanjay Jain and from Vanderbilt University Medical Center via collaborator Dr. Agnes Fogo. Description of these samples are available in our recent publication7. We also used human transplant biopsy samples provided by Dr. Kuang-Yu Jen from University of California at Davis. Human data collection procedure followed a protocol approved by the Institutional Review Boards at University at Buffalo, Vanderbilt University Medical Center, and University of California at Davis. Ground-truth annotations of glomeruli were preformed using the annotation tools provided by HistomicsTK and Aperio ImageScope, where computational predictions were corrected as described in our previous work4.
Imaging and data preparation:
Human tissue slices of 2–5 μm were stained using diverse histological stains, and imaged using a whole-slide imaging scanner (Aperio VERSA 200, Leica Biosystems, Buffalo Grove, IL). We followed similar imaging protocol as described in our previous work8.
IV. CONCLUSION AND FUTURE WORK
We have developed a user friendly plugin for cloud based segmentation of WSI. To show the utility of this method, we trained a model for the segmentation of glomeruli from renal tissue slides which we demonstrate on holdout slides. At the time of writing, this plugin is running on our server, a demonstration is available in the video linked in the results section. This plugin is open source and packaged as a pre-built Docker image which is easy to setup on a server. The software is plug-and-play, allowing users to upload data in the Digital Slide Archive, run segmentation via HistomicsTK, and correct segmentation error via HistomicsTK web-viewer. We designed the plugin with flexibility in mind, it was designed to efficiently run segmentation on WSIs of any tissue type. Segmentation of new structures of interest is possible by retraining the Deeplab network used for segmentation. We are currently working to build a larger training set for glomerular segmentation, aiming for 1000 human biopsy slides. When this network is trained we plan to open our server to the community for glomerular detection. In the future we plan to extend this plugin for the segmentation of multiple kidney structures.
REFERENCES
- 1.Santo BA, Rosenberg AZ & Sarder P Artificial intelligence driven next-generation renal histomorphometry. Current opinion in nephrology and hypertension 29, 265–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginley B et al. Computational segmentation and classification of diabetic glomerulosclerosis. 30, 1953–1967 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronneberger O, Fischer P & Brox T in International Conference on Medical image computing and computer-assisted intervention. 234–241 (Springer; ). [Google Scholar]
- 4.Lutnick B et al. An integrated iterative annotation technique for easing neural network training in medical image analysis. Nature Machine Intelligence 1, 112–119, doi: 10.1038/s42256-019-0018-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutman DA et al. The digital slide archive: A software platform for management, integration, and analysis of histology for cancer research. Cancer research 77, e75–e78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L-C, Zhu Y, Papandreou G, Schroff F & Adam H in Proceedings of the European conference on computer vision (ECCV). 801–818. [Google Scholar]
- 7.Ginley B et al. Computational segmentation and classification of diabetic glomerulosclerosis. Journal of the American Society of Nephrology : JASN (To appear). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginley B, Tomaszewski JE, Yacoub R, Chen F & Sarder P Unsupervised labeling of glomerular boundaries using Gabor filters and statistical testing in renal histology. Journal of Medical Imaging 4, 021102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


