Abstract
Correction for ‘Fragment-based covalent ligand discovery’ by Wenchao Lu et al., RSC Chem. Biol., 2021, DOI: 10.1039/d0cb00222d.
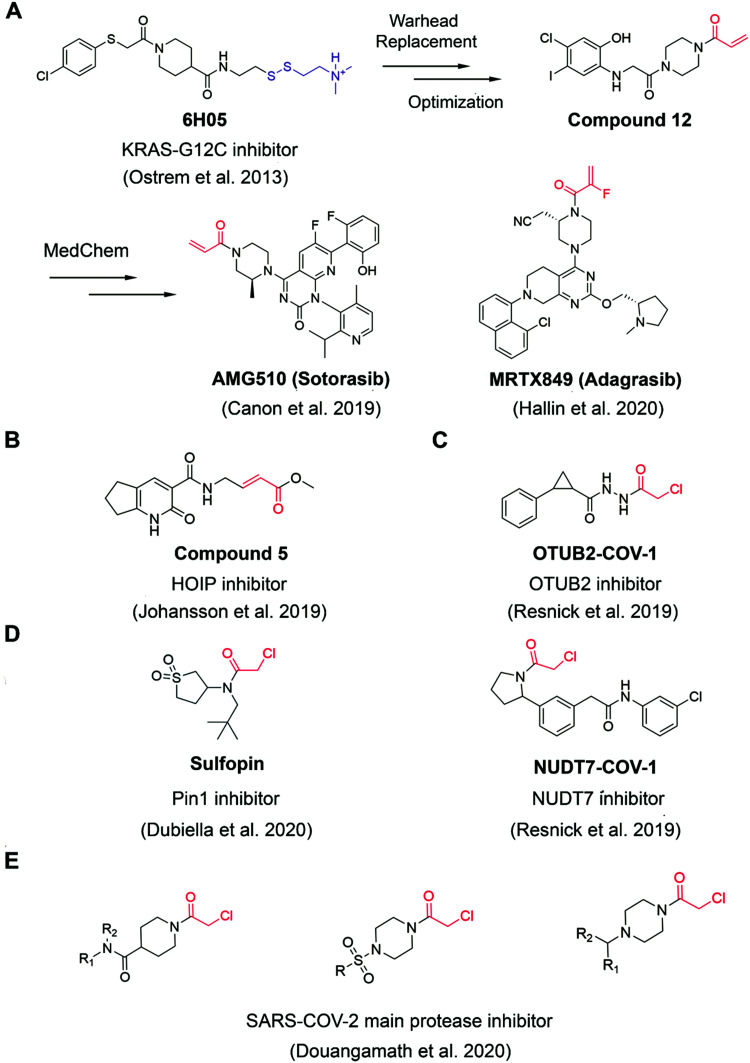
The authors regret that an incorrect version of Fig. 2 was included in the original article, where the structure of Sulfopin in Fig. 2D was incorrectly shown. The correct version of Fig. 2 is presented below.
Fig. 2. The structures of representative well-characterized electrophilic fragments identified from target-based screening strategies in recent years. (A) KRAS-G12C allele-specific covalent fragment (6H05) identified from tethering screen, which was further elaborated to compound 12.31 This inspired numerous groups to develop further optimized inhibitors, within which AMG51033 and MRTX84936 successfully entered clinical trials. (B) Compound 5 targets the active cysteine (C885) of HOIP.37 (C) OTUB2-COV-1 targets the active cysteine (C51) of OTUB2 and NUDT7-COV-1 target C73 of NUDT7.38 (D) Sulfopin targets the active cysteine of Pin1 (C113).39 (E) Representative covalent fragment scaffolds target the active cysteine (C145) of SARS-COV-2 main protease (Mpro).40.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.