Abstract
Due to the coronavirus disease 2019 (COVID-19) pandemic, wastewater surveillance has become an important tool for monitoring the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) within communities. In particular, reverse transcription-quantitative PCR (RT-qPCR) has been used to generate large datasets aimed at detecting and quantifying SARS-CoV-2 RNA in wastewater. Although RT-qPCR is rapid and sensitive, there is no standard method yet, there are no certified quantification standards, and experiments are conducted using different assays, reagents, instruments, and data analysis protocols. These variations can induce errors in quantitative data reports, thereby potentially misleading interpretations, and conclusions. We review the SARS-CoV-2 wastewater surveillance literature focusing on variability of RT-qPCR data as revealed by inconsistent standard curves and associated parameters. We find that variation in these parameters and deviations from best practices, as described in the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines suggest a frequent lack of reproducibility and reliability in quantitative measurements of SARS-CoV-2 RNA in wastewater.
Keywords: RT-qPCR, assay validity, standard curve, quality assurance, quality control, wastewater surveillance
Graphical abstract
1. Introduction
The screening of untreated wastewater and primary solids for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA has emerged as an effective means of tracking coronavirus disease 2019 (COVID-19) at the population level (Ahmed et al., 2021a; Medema et al., 2020a; Peccia et al., 2020; D'Aoust et al., 2021; Li et al., 2021). SARS-CoV-2 RNA wastewater surveillance is used as a tool to monitor COVID-19 alongside traditional clinical monitoring in over 50 countries across hundreds of organizations and sites (https://arcg.is/1aummW; Bivins et al., 2020; Naughton et al., 2021). However, SARS-CoV-2 RNA surveillance in wastewater is a complex process that involves a sequence of steps including wastewater sampling, transport, virus concentration (if required), RNA extraction, reverse-transcription quantitative PCR assays (RT-qPCR) and data interpretation (Ahmed et al. 2021b). Unless validated and optimized, these steps can produce erroneous results that may cause inaccurate evaluations of COVID-19 status within communities. This is particularly important when reporting quantitative data. While recent reviews have highlighted the limitations and bottlenecks of wastewater surveillance (Medema et al., 2020b; Michael-Kordatou et al., 2020; Ahmed et al., 2020; Ahmed et al. 2021b; Zhu et al., 2021), the reliability, accuracy, and relevance of quantitative data generated by RT-qPCR-based wastewater testing have not been examined.
RT-qPCR quantification is based on the quantification cycle (Cq). This value is usually obtained by defining a baseline cycle range, below which amplification is not recorded, to establish a threshold line that, upon crossing the amplification curves, generates Cq values. The Cq is inversely proportional to the initial template concentration. Samples are quantified by comparing their Cq values relative to assay-specific standard curves. Standard curves are constructed using a dilution series of standard control materials, typically four to six 10-fold dilution points with at least three replicates per point and a defined number of target molecules.
The standard curve is created by plotting the resultant Cq values against log-transformed gene copy (GC) quantities from control materials by fitting a linear trend line to the data (y = mx + b). The precision of the quantitative data is strongly influenced by the quality of the standard curve, as reflected by its slope (m), PCR efficiency, linearity (r2 value) and y-intercept (b). PCR efficiency can be calculated from the slope (m) of the trendline, with 100% efficiency (doubling of the PCR product after each cycle) characterized by a slope of −3.32. Although amplification efficiencies between 90% and 110% are considered acceptable, a small change in PCR efficiency from 100 to 97% over 30 cycles causes a 57% difference in the estimated input DNA, while a change from 100 to 90% results in a 365% difference (Boulter et al., 2016). The r 2 and the standard error of the estimated amplification efficiency can be used to evaluate the quality of the efficiency determination. While the intercept of the standard curve on the y-axis gives a theoretical sensitivity of the assay, which denotes the number of cycles required for detecting a single unit of measurement, the quantities of target sequence in all samples can be calculated by comparing the respective Cq measurement to the established log-linear relationship between GC and Cq, as indicated by the standard curve. A standard curve can be constructed using various control materials produced in the laboratory or purchased from commercial vendors, including plasmid DNA constructs, PCR amplicons, synthetic RNA or DNA, genomic DNA, cDNA, and RNA or DNA from biological samples. Such templates may be single-stranded (synthetic oligonucleotides) or double-stranded (plasmids, gene fragments). Quantities of such materials may comprise certified, reference, or information values (Bustin et al., 2009; Hou et al., 2010).
Adding to the variability within the standard curve parameters, RT-qPCR assays are characterized by a dynamic range, which is the log concentration range of the control material over which the standard curve is confirmed to be linear. Finally, RT-qPCR assays are also characterized by a limit of detection (LOD) and a limit of quantification (LOQ). While the definitions of these terms may vary, they are generally used to describe the smallest concentration of target molecules that can be reliably detected and quantified via the assay. Bustin et al. (2009) described the LOD as the gene copy quantity that yields a 95% probability of detection in a single reaction but did not provide a precise quantitative definition of the LOQ.
Although RT-qPCR is a sensitive and specific technique for the quantification of nucleic acid targets, the data reproducibility and reliability are critical parameters and must be established for each assay. Variations in protocols, reagents, sample quality, instruments, operators, analysts, data analysis, and interpretation within and the across laboratories can lead to the production of inaccurate and unreliable quantitative data. To circumvent these issues, Bustin and colleagues (2009) recommended a set of protocols and guidelines, “The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) for conducting and reporting qPCR data.” The guidelines describe experimental stringency and uniformity practices to ensure the production of reproducible and scientifically defensible quantitative data within and between laboratories and been applied in the field of environmental microbiology (de Bruin et al., 2011; Hughes et al., 2017). Nevertheless, there remain concerns regarding the quality of RT-qPCR results in the published literature (Bustin and Nolan, 2017; Taylor et al., 2019), particularly in the emerging wastewater surveillance literature (Ahmed et al., 2020).
Herein, we review the SARS-CoV-2 wastewater surveillance literature to assess the appropriate use of RT-qPCR calibrators and associated performance parameters. All publications included in the analysis are compiled in an open access database, as detailed below.
2. Screening the SARS-CoV-2 wastewater surveillance literature
We screened 125 preprint and peer-reviewed publications concerning wastewater surveillance for SARS-CoV-2 RNA, as listed on the COVID-19 Wastewater-Based Epidemiology Collaborative (WBEC) publication map (https://www.covid19wbec.org/publication-map). These publications were compiled through alerts via Google Scholar using the keyword string (“SARS-CoV-2” AND (“wastewater” OR “sewage”)) from April 2020 to 15 May 2021. Of those publications, 44 were excluded from further analysis due to the use of platforms other than RT-qPCR for SARS-CoV-2 RNA quantification (e.g., digital PCR), reporting qualitative results (e.g., positive/negative or Cq values), or genomic rather than quantitative results. The remaining 81 publications (46 peer-reviewed and 35 pre-prints) reported quantitative GC results as measured by RT-qPCR. Where available, we extracted the following information for each RT-qPCR assay reported: SARS-CoV-2 gene target, one-step or two-step RT-qPCR protocol, standard curve parameters (i.e., y-intercept, slope, r 2, and efficiency), LOD, LOQ, the dynamic range, control material used for standard curve (i.e., plasmid DNA, gBlock, PCR amplicon, etc.), the vendor of the control material, pre-treatment of the control material (if applied), and if any independent quantification was performed prior to use. The studies were evenly screened by three co-authors. Relevant data were extracted with a duplicated review of 5 to 10 randomly selected entries by each author. The resulting database was independently reviewed by each author, and discrepancies were remedied via discussion to form a consensus. The consensus database was then independently reviewed and audited by a fourth author. The compiled database used for the analysis can be found at https://osf.io/q7dnp/ doi: 10.17605/OSF.IO/Q7DNP. The purpose in conducting this review is not to identify individual laboratories with questionable practices but to highlight the importance of reporting standard curve parameters and to assess the performance of the entire wastewater surveillance community. Hence, citations were not provided when discussing practices within individual publications.
3. RT-qPCR and standard curve reporting
From the 81 selected publications, we extracted details pertinent to the assays used, standard curve parameters, and positive control materials for 208 separate quantitative assays (i.e., an average of 2.6 assays per publication). Only 26% of the total 208 RT-qPCR assays reported in the literature included all essential standard curve parameters, with 41% (86 of 208) reporting at least one standard curve parameter, i.e., 30, 33, 35, and 40% of studies detailing y-intercept, slope, r 2 values, and efficiency data, respectively. Among the 208 RT-qPCR assays, 130 targeted the N gene, 25 targeted ORF1, 23 targeted the E gene, 19 targeted RdRp, and 10 targeted the S gene, while one did not report any assay target. For assays targeting the N gene of SARS-CoV-2, the US CDC N1 assay was applied most frequently (39%), followed by the US CDC N2 assay (32%). Collectively, these two assays accounted for 45% of the RT-qPCR assays reportedly used to quantify SARS-CoV-2 RNA in wastewater.
RT‐qPCR assays are performed using two approaches: a separate RT reaction (i.e., cDNA synthesis) followed by qPCR (two‐step) or a combined RT and qPCR reaction in the same tube (one‐step). The two-step approach often uses random hexamer primers during RT followed by target-specific primers, with the qPCR step offering more flexibility to optimize amplification conditions (Bustin and Nolan, 2017). However, the two-step approach requires additional sample handling, potentially inducing greater measurement variability and risk of contamination. A potential advantage of the two-step protocol is its ability to reverse transcribe RNA to cDNA immediately after extraction to avoid RNA degradation during freezing and storage. The one-step approach utilizes gene-specific primers and minimizes sample handling by carrying out the RT and qPCR steps in the same microtube, reducing bench time and the risk of contamination at the expense of less flexibility for assay optimization. However, the one-step protocol may also lead to RNA degradation during storage or freezing and thawing if extracts are stored before further analysis.
The majority (83%) of the reported RT-qPCR assays utilized the one-step protocol and 13% of RT‐qPCR assays were performed as two-step assays. The remaining 4% did not report whether a one-step or two-step protocol was used. The performances of one-step and two-step RT-qPCR assays were compared in a previous study, and the results indicated significant variation in quantification of the targets between the two protocols (Bustin and Nolan, 2017). However, in an earlier study (Wackerd and Godard, 2005), both one-step and two-step protocols produced similar standard curves with RT-qPCR efficiencies close to 100%, suggesting that discrepancies may be protocol- and assay-specific.
4. The US CDC N1 and N2 standard curves
To assess the impact of heterogeneity in the reported RT-qPCR standard curves used for SARS-CoV-2 RNA quantification in wastewater, the reported y-intercepts, slopes, r 2 values, and efficiencies for the two most frequently used RT-qPCR assays, the US CDC N1 and N2 were analyzed (Fig. 1 ). Quantitative analysis of other RT-qPCR assays was precluded by the low number of studies reporting standard curve metrics, for example, ORF1ab (n = 4), RdRp (n = 4), S gene (n = 1), N gene (n = 3), and E gene (n = 3). For this review, the copy numbers of unknown samples were calculated by using the y-intercept and the slope of the standard curve as shown in equation (1).
| (1) |
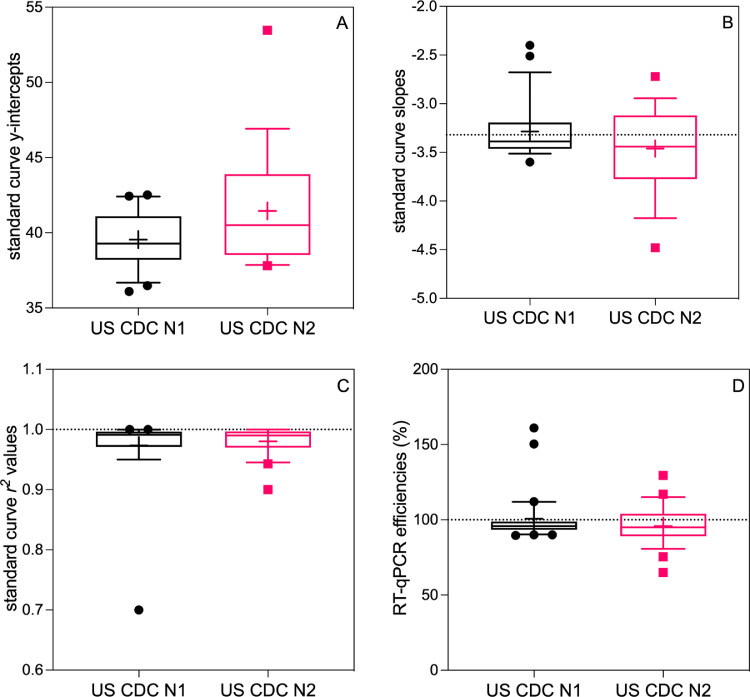
Fig. 1.
SARS-CoV-2 US CDC N1 and N2 standard curve parameters, as reported in the literature: (A) y-intercept values, (B) slopes, (C) r2 values, and (D) RT-qPCR efficiencies. The boxplots display the 10th and 90th percentiles (whiskers), interquartile range (box), median (line), and mean (+). The dashed line on panel (B) depicts an ideal slope of −3.32, on panel (C) depicts an r2 value of 1.00, and on panel (D) depicts an ideal efficiency of 100%.
4.1. Y-intercept values
The reported y-intercepts for CDC N1 (n = 21) and N2 (n = 18) standard curves (Figure 1A) ranged from 36.1 to 42.5 and 37.8 to 53.5, respectively, with 30 and 24 publications not reporting this value. These intercepts indicate that anywhere from 36 to 43 RT-qPCR, thermal cycles would be required to detect a single GC by the CDC N1 assay and 38 to 54 for the CDC N2 assay. However, these values were obtained using various reference materials from different vendors, and the y-intercept value depended on the concentration or copy number associated with that reference material. Therefore, starting with a nominally high copy number of a standard could result in a lower y-intercept value compared with the same reference material that has been ascribed a lower copy number value. The most common source for standard material was plasmid from IDT (n = 13 and 11 for N1 and N2, respectively), and reported y-intercepts for this control material ranged from 36.1 to 42.5 (N1) and 37.8 to 53.5 (N2). The wide range of reported intercepts derived from the same standard material, which was presumably quality-controlled prior to shipment, draws scrutiny to the handling of this reference material during RT-qPCR.
4.2. Slope values and RT-qPCR efficiencies
The qPCR efficiency is derived from the slope of a standard curve as shown in equation (2).
| (2) |
While efficiency values (E) often vary between templates, they are typically highly reproducible for the same template (A-Z of Quantitative PCR, 2004). The ideal slope of a standard curve is −3.32, which indicates 100% RT-qPCR efficiency, although a range from −3.1 (110%) to −3.58 (90%) is typical for an optimized probe-based assay (A−Z of Quantitative PCR, 2004). Slope is also typically more reproducible between laboratories and instruments than the y-intercept (A−Z of Quantitative PCR, 2004).
Fig. 1B shows the reported RT-qPCR slopes for the US CDC N1 and N2 assays. For the US CDC N1 assay, the mean standard curve slope was −3.29, which is within the typical interval; however, the reported slopes ranged from −3.60 (90%) to −2.40 (161%); outside the acceptable range of -3.1 (110%) to -3.58 (90%). A similar pattern was observed for the US CDC N2, with a mean slope of −3.46 (95%) and a range from −4.48 (67%) to −2.72 (133%).
Fig. 1D shows the reported and calculated RT-qPCR efficiencies (calculated with the reported slope if not explicitly described in the publication) for the CDC N1 and N2 assays. For the reported CDC N1 standard curves, the mean reported efficiency was 101% (median 95.8%) and ranged from 89.6 to 161%. For CDC N2 assays, the reported range was from 65 to 129%, with a mean reported efficiency of 95.8% (median 95%).
4.3. r2 values
Standard curves should demonstrate strong linear fits, with r2 values ranging from 0.980 to 1.00 (A−Z of Quantitative PCR, 2004). The r2 value of a standard curve is influenced by the precision of replicate standard material Cq measurements. Lower r2 values indicate contributions to variations from sources other than the control material copy number (e.g., pipetting error, variable reaction conditions, standard dilution preparation error, or instrument variation). For CDC N1 and N2 assays as shown in Fig. 1C, r2 values ranged from 0.70 to 1.00 and 0.90 to 1.00, respectively. While means were comparable (N1 = 0.973, N2 = 0.989), the CDC N1 assay exhibited a much wider range of reported r2 values. The lowest reported r2 values across all SARS-CoV-2 RT-qPCR assays were 0.70 for the CDC N1 standard curve. This value is well below 0.980 and strongly suggests variation attributable to sources other than the standard material GC number.
4.4. Within-Study Variation of Standard Curve Parameters
MIQE guidelines recommend reporting the confidence interval or standard error of the RT-qPCR efficiency but do not deem this practice essential (Bustin et al., 2009). Standard deviations were reported for all standard curve parameters for only seven RT-qPCR assays in three publications of SARS-CoV-2 quantification in wastewater. Ranges for at least one standard curve parameter were reported for another 12 assays, bringing the total to 9% of RT-qPCR assays with some reporting of variation. While there is no standard for allowable variation, the ideal quantification platform should minimize such variation. None of the studies specified whether the reported variation was a measure of repeatability (within-run variation) or reproducibility (between-run variation). Additionally, variation in standard curves can be dependent on the type of standard curve used for quantification - single curves, master calibration curves, or more sophisticated pooled and mixed models (Sivaganesan et al., 2010). However, standard curve methodologies were seldom reported in the included publications.
5. Standard curve control materials
Performance of RT-qPCR depends on the standard materials used to produce the standard curve and good laboratory practices (A−Z of Quantitative PCR, 2004). Careful, application-specific optimization is required to maximize the performance of the standard materials and the subsequent quantification of genetic targets in samples.
In many cases, commercially available control materials were employed. Often, these materials are provided at a vendor-specified titer, which is assumed to increase the likelihood of producing reliable standard curves. However, it is clear from the variability described above that the dilutions or pipetting steps are highly variable and operator dependent as reported previously (Bustin, 2002).
5.1. Control material reporting
A description of the control material was reported for 78% of the SARS-CoV-2 RT-qPCR assays in the wastewater surveillance literature. The description was sometimes ambiguous, including terms such as “genes encoding nucleocapsid protein,” “synthetic oligonucleotide,” and “cDNA standards,”. Such ambiguous descriptions can be especially problematic if the vendor is not specified, which was the case for 28% of the reported assays screened for this review. Using broad classifications, control materials included plasmids for 59 assays, synthetic cDNA for 29, synthetic RNA for 36, various forms of transcripts for 19, ambiguous or unclear for 19, and not reported for 46 RT-qPCR assays. Plasmids and synthetic oligonucleotides (single-stranded cDNA or RNA) accounted for the majority of reported control materials, with a nearly even split between the two (28 and 31%, respectively).
Few published studies have mentioned adjusting estimated quantities to account for the difference in double-stranded DNA control materials (e.g., plasmids or gene fragments) and the single-stranded SARS-CoV-2 RNA genome. Importantly, unlike RNA control materials, DNA controls would not be subjected to the RT step; therefore, quantifications produced using DNA as control material would not account for RT variability. Pre-treatment of control materials was reported for 7 RT-qPCR assays (heat inactivation of isolated strain in two and linearization of plasmid in the remaining five instances). Independent quantification of control materials was only reported for 6% (12 of 208) of the RT-qPCR assays.
5.2. Effects of control material type on standard curve parameters
Previous studies have reported significant variations in Cq of standard curves produced using non-linearized plasmids compared to linear control materials (synthetic cDNA or RNA) (Hou et al., 2010; Lin et al., 2011; Werling et al., 2015; Beinhauerova et al., 2020), although these are not universal findings (Oldham and Duncan, 2012). Chik et al. (2021) reported up to one order of magnitude of difference between laboratories using linear vs. plasmid standards, with an 8.4 Cq difference observed in one instance between linearized vs. non-linearized plasmids. Gerrity et al. (2020) reported a droplet digital PCR (ddPCR)-measured copy number 5-fold lower than the vendor-specified titer for a circular plasmid control and noted heteroscedasticity in the variance caused by the non-linearized plasmid control material, with a 9.4-fold bias at 40 Cq and a 5.5-fold bias at 25 Cq. Others noted that circular DNA plasmids may require linearization to avoid impacts on PCR efficiency, leading to subsequent bias in qPCR measurements (Hou et al., 2010). Among the publications included in this review, linearization was only reported for 8% of the RT-qPCR assays, where a plasmid was used as the control material for standard curves.
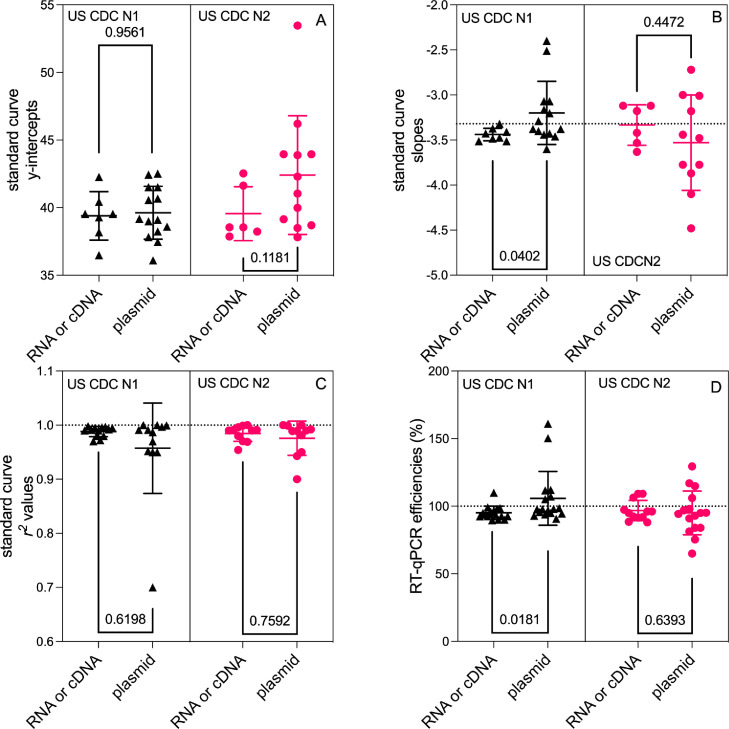
We considered the effects of control material type, linear materials (cDNA or RNA) versus plasmid materials (mostly circular) on reported RT-qPCR standard curves for the CDC N1 and N2 assays across the wastewater surveillance literature. Through the screening of publications, we were able to extract standard curve parameters for 21 CDC N1 assays (7 linear control materials and 14 plasmids) and 18 CDC N2 assays (six linear control materials and 12 plasmid). Fig. 2 shows the scatter plots of each standard curve parameter for CDC N1 and CDC N2 assays stratified by the control material type. Standard curve parameters based on synthetic RNA or cDNA and plasmid materials were evaluated using a Mann−Whitney U test (Mann and Whitney, 1947). A significant difference was observed only between the standard curve slopes (p = 0.0402; Fig. 2B) and efficiencies (p = 0.0181; Fig. 2D) for the CDC N1 assays indicating a potential source of bias.
Fig. 2.
Dot plots of reported RT-qPCR standard curve parameters for CDC N1 and N2 assays: (A) y-intercepts, (B) slopes, (C) r2 values, and (D) efficiencies stratified by linear synthetic RNA or cDNA versus plasmid control material. Individual values, mean, and standard deviation are displayed. The dashed line on panel (B) depicts an ideal slope of −3.32, on panel (C) depicts an r2 value of 1.00, and on panel (D) depicts an ideal efficiency of 100%.
Although most of the statistical tests imply no significant differences, the scatter plots in Fig. 2 suggest that plasmid control materials consistently exhibit a broader range of variation across all standard curve parameters for both the CDC N1 and N2 assays. A coefficient of variation (CV = standard deviation divided by mean) was calculated for each standard curve parameter and standard material group combination for both CDC N1 and N2 assays; CV values were greater for curves produced using plasmid materials (slope CVs 11–15%; efficiency CVs 17–19%) compared to linearized materials (slope CVs 2–7%; efficiency CVs 5–8%), further supporting this observation. Differences were particularly pronounced for standard curve slopes, r 2 values, and RT-qPCR efficiencies. For the CDC N2 assay, the use of plasmid control materials resulted in a roughly 2-fold increase in the CV of the standard curve slope, r 2 value, and efficiency. Whereas, for the CDC N1 assay, the plasmid control materials resulted in a 5-, 9-, and 4-fold increase in CV for the slope, r 2, and efficiency, respectively.
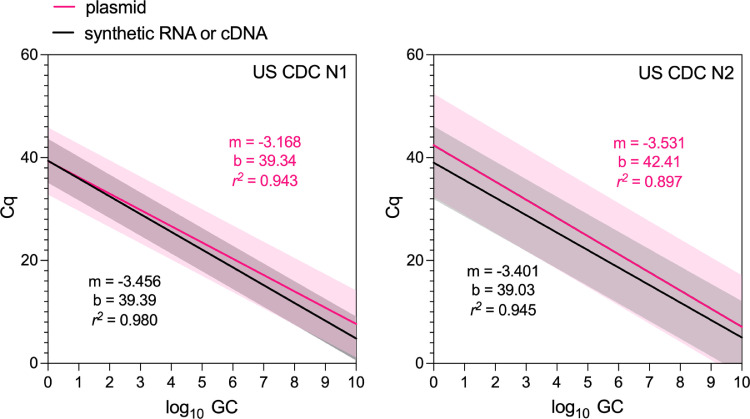
These observations suggest increased variability and, lower reproducibility, among CDC N1 and N2 standard curves produced using non-linearized plasmid control materials compared to linear RNA or cDNA materials, with the effect being particularly pronounced for the CDC N1 RT-qPCR assay. From Fig. 3 , we further visualized the effect of this variation by fitting log-linear regressions to reported RT-qPCR standard curves for the CDC N1 and N2 assays stratified by the control material, treating each curve as a replicate. While the mean slopes (m) and y-intercepts (b) are comparable, the increased width of the shaded area illustrates the increased 99th percentile prediction interval for the standard curves produced from plasmids compared to synthetic RNA or cDNA. The r 2 values for both CDC N1 and N2 plasmid-based standard curves (0.943 and 0.897, respectively) were lower than the r 2 values for RNA/cDNA curves and are well below the recommended 0.980 lower bound. These decreased r 2 values suggest an increased proportion of the variation in experiments performed with plasmids is attributable to a source other than the variability in standard dilution GC quantities. Our observations corroborate with those of Chik et al. (2021) and Gerrity et al. (2020) and further suggest a systematic deficiency in the protocols and procedures used for plasmid control materials in RT-qPCR quantification of SARS-CoV-2 RNA in wastewater.
Fig. 3.
Log-linear regressions fit to the reported standard curves for CDC N1 assays (left; n = 22) and CDC N2 assays (right; n = 19) in the wastewater surveillance literature. Regressions fit to data from synthetic RNA or cDNA control materials are denoted in black, while those fit to plasmid control materials are denoted in pink. The shaded region displays the 99th percentile for each. The slope (m), y-intercept (b), and r2 values for the linear fit to each set of data are shown in the color corresponding to the regression line.
6. Reported LODs and LOQs
The MIQE guidelines list “evidence for LOD” and “Cq variation at lower limit” (i.e., LOQ) as of essential importance for reporting RT-qPCR experiments. Operational definitions of LOD and LOQ varied among the studies we reviewed and were frequently unspecified. The LOD values were reported for only 34% of RT-qPCR assays, ranging between one and 500 GC per reaction (the most frequently used unit). LOD values were also reported in volumetric units (i.e., µL, mL, etc.), but the source of this volume (e.g., sewage, extract eluate, and reaction mixture) was often unspecified. A method definition for the LOD determination was rarely provided, and in several cases, the reported LOD values were below the 95% LOD theoretical limit of 3 GC/reaction derived from the Poisson distribution (Bustin et al., 2009). An LOQ value was reported for only 19% of the RT-qPCR assays, about half as frequently as an LOD. LOQ values were also most frequently reported in GC per reaction, with values ranging from 5 to more than 10,000. Similar to LOD, when volumetric units were used to report LOQ, the source of the relevant volume was often ambiguous. Furthermore, when multiple RT-qPCR assays were used in a single publication, the reported LOD and LOQ values were often not attributed to a specific assay.
7. Discussion
The quantitative potential of RT-qPCR assays has resulted in the broadening of its use from the qualitative detection of SARS-CoV-2 to the quantification of viral load in patients or environmental samples. While a binary result demands reliability, specificity, and sensitivity, quantitative analysis sets a much higher bar regarding assay quality, as conveyed by reproducibility, accuracy, precision, and comparability. These parameters were established using optimized assay-specific standard curves generated using appropriate and validated control materials (A−Z of Quantitative PCR, 2004). As mentioned above, some variability, both within and between laboratories, is expected during RT-qPCR experiments due to variations in protocols, reagents, sample quality, instruments, operators, data analysis, and interpretation.
The MIQE guidelines, published 11 years ago, were intended to guide investigators and reviewers in their generation of reliable and reproducible RT-qPCR data (Bustin et al., 2009). Such reliability is even more critical when the data produced are used to guide responses to an outbreak or pandemic. In response to the ongoing COVID-19 pandemic, many laboratories, some with limited or no environmental microbiology experience, quickly pivoted and adopted various assays, protocols, materials, and techniques, including RT-qPCR, to detect and quantify SARS-CoV-2 RNA in wastewater. While such efforts are commendable and shortcomings under the duress of standing up entire surveillance programs on short notice are understandable, this review suggests a neglect of best practices concerning to RT-qPCR experiments in the SARS-CoV-2 wastewater surveillance literature. Consequently, there is significant potential for inaccurate and misleading reporting of SARS-CoV-2, with wide-ranging implications for developing policies and guidelines. Furthermore, the departure from best practice in this important process has serious implications for environmental monitoring of high priority biothreat agents in general.
Basic and essential information for RT-qPCR assays including the standard curve parameters of y-intercept, slope and/or efficiency, and r2 value, were reported for only 26% of the RT-qPCR assays used for wastewater surveillance. Variation in these parameters is even more rarely reported, being published for only 9% of assays. The reported standard curve data exhibit broad heterogeneity that, in turn, limit the reproducibility and reliability of RT-qPCR data for SARS-CoV-2 wastewater surveillance. For example, y-intercepts ranged from 36.1 to 53.5; slopes ranged from -2.4 to -4.5; and r2 values were as low as 0.700 with reported efficiencies ranging from 65 to 161%. Many of these are well outside of the expected range for optimized RT-qPCR assays. Variation in RT-qPCR performance seems further exacerbated by the prevalent use of plasmid control material without linearization. This practice is likely to produce increased variation across all standard curve parameters, with a particularly pronounced increase in the widely used CDC N1 RT-qPCR assay.
The published SARS-CoV-2 wastewater surveillance literature also contains other scenarios that reinforce the need for higher standards in practice and reporting. In some publications, “standard curves” were generated using one to three standard dilutions, while others made GC concentration estimates in samples falling well outside the confirmed dynamic range. Many publications provided no standard curve information at all, while one publication stated that the quantitative data were generated by a commercial laboratory without providing any pertinent RT-qPCR information, not even the assay used to generate the data. In a few cases, sophisticated epidemiological models such as susceptible-exposed-infectious-recovered (SEIR), vector auto-regression, and Monte Carlo simulations, were applied to quantitative data from RT-qPCR experiments without reporting any of the standard curve parameters, making it essentially impossible to validate the utility of these approaches. Conversely, at least two SARS-CoV-2 wastewater surveillance studies explicitly followed the MIQE guidelines, although they are studies using digital PCR (Graham et al., 2021; Wolfe et al., 2021).
This review has several limitations. First and foremost, this is not a formal systematic review; instead, relevant publications were identified from a curated collection of SARS-CoV-2 wastewater surveillance publications from April 2020 to May 2021. Although this review did not consider every SARS-CoV-2 RT-qPCR publication, the inclusion of 125 studies provided sufficient information to identify multiple deficiencies in standard curve performance, assays and reporting practices. Second, this review relied on self-reported data from pre-print and published scientific literature, which could be subject to bias. Additionally, there are sources of variation that have not been explicitly analyzed in this review, such as time between extraction and RT or RT-qPCR (frequently unreported), and differences between instruments and master mixes. Importantly, variation between RT-qPCR replicates, experimental runs, instruments, and laboratories is expected to a degree, and is well documented for various SARS-CoV-2 RT-qPCR assays (Bustin et al., 2021). While it is not possible to independently account for this expected variation, the variability observed in this review often exceeded the recommend best practices, strongly suggesting that the wastewater surveillance community must make a concerted effort to improve both the reporting of RT-qPCR parameters and the optimization of SARS-CoV-2 RT-qPCR assays.
8. Conclusions and recommendations
This brief literature review describing the use of RT-qPCR assays for wastewater surveillance has revealed significant variation in the standards of the reported results, although the paucity of quality control data makes it difficult to ascertain which reports are likely to be the most credible. Therefore, there is an urgent need to improve the transparency of reporting and disclose more information about the characteristics of the assays being used. We propose the following measures:
-
•
A starting point for improvement would be the adoption of the MIQE guidelines for reporting RT-qPCR (and digital PCR) experiments performed for wastewater surveillance purposes (Bustin et al., 2009; Huggett et al., 2013). Comprehensive reporting of all experimental details including RT-qPCR formats (one-step vs. two-step), instrument and reagent manufacturers, and protocols is critical to account for all potential sources of variation in quantitative results. Therefore, it is critical that journal editors and reviewers should consider requiring the submission of the pertinent MIQE checklist (https://rdml.org/miqe.html) not only for SARS-CoV-2 wastewater surveillance publications but also for others.
-
•
To produce reliable data especially for public health decision-making, the wastewater surveillance community should, at a minimum, aspire to achieve 100% reporting of all standard curve performance parameters (y-intercept, slope/efficiency and, r2 value). If the standard curve parameters do not fall within general data acceptance ranges, additional actions must be undertaken such as optimization, calibration of pipets and/or thermal cycle optimization, among others until the appropriate performance is achieved for the intended application.
-
•
Because the reproducibility of RT-qPCR data relies, in part, on the standard curves produced by the control materials, the type and vendor for such materials should be transparently and unambiguously reported. Any pre-treatment or manipulation of these control materials prior to quantification should also be documented, especially for plasmid controls. Appropriate adjustments for the use of double-stranded control materials to quantify single-stranded RNA should also be reported. Caveat emptor must be the motto when relying upon commercially available control materials.
-
•
It would also be useful to independently confirm vendor-reported titers to ensure that materials were not degraded during shipment or due to mishandling in the laboratory.
In summary, to maximize the utility of wastewater surveillance for public health, it is time for wastewater surveillance practitioners to hold themselves to the highest quality control standards and “walk the walk” (Bustin and Nolan, 2017).
Disclaimer
Information has been subjected to U.S. EPA peer and administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the official positions and policies of the U.S. EPA. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This study was partially supported by NSF award 2038087.
References
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K.V., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimization and quality control are crucial for generating reliable public health information. Curr Opin Environ Sci Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S., Bertsch P., Bibby K., Bivins A., Blackall L., Bofill-Mas S., Bosch A., Brandao J., Choi P., Ciesielski M., Donner E., D'Souza N., Farnleitner A., Gerrity D., Gonzalez R., Griffith J., Gyawali P., Haas C., Hamilton K., Hapuarachchi C., Harwood V., Haque R., Jackson G., Khan S., Khan W., Kitajima M., Korajkic A., La Rosa G., Layton B., Lipp E., McLellan S., McMinn B., Medema G., Metcalfe S., Meijer W., Mueller J., Murphy H., Naughton C., Noble R., Payyappat S., Petterson S., Pitkanen T., Rajal V., Reyneke B., Roman F., Rose J., Rusinol M., Sadowsky M., Sala-Comorera L., Setoh Y.X., Sherchan S., Sirikanchana K., Smith W., Steele J., Sabburg R., Symonds E., Thai P., Thomas K., Tynan J., Toze S., Thompson J., Whiteley A., Wong J., Sano D., Wuertz S., Xagoraraki I., Zhang Q., Zimmer-Faust A., Shanks O. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Preprints. 2021 doi: 10.20944/preprints202104.0481.v1. 2021040481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinhauerova M., Babak V., Bertasi B., Boniotti M.B., Kralik P. Utilization of digital PCR in quantity verification of plasmid standards used in quantitative PCR. Front Mol Biosci. 2020;7:155. doi: 10.3389/fmolb.2020.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Gurol Z.C., Chakraborty C., Costa F., Curcio S., de los reyes F.L., III, Delgado Vela J., Farkas K., Fernazdez-Casi X., Gerba C., Gerrity D., Girones R., Gonzzalez R., Haramoto E., Harris A., Holden P.A., Ispam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., Mclellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wiggington K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bustin S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J.F., Kubista M., Mueller R.D., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Bustin S., Nolan T. Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research. Eur. J. Clin. Invest. 2017;47:756–774. doi: 10.1111/eci.12801. [DOI] [PubMed] [Google Scholar]
- Bustin S., Mueller R., Shipley G., Nolan T. COVID-19 and diagnostic testing for SARS-CoV-2 by RT-qPCR- Facts and Fallacies. Int. J. Mol. Sci. 2021;22:2459. doi: 10.3390/ijms22052459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter N., Suarez F.G., Schibeci S., Sunderland T., Tolhurst O., Hunter T., Hodge G., Handelsman D., Simananien U., Hendriks E., Duggan K. A simple, accurate and universal method for quantification of PCR. BMC Biotechnol. 2016;16:27. doi: 10.1186/s12896-016-0256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chik A.H.S., Glier M.B., Servos M., Mangat C.S., Pang X.-L., Qiu Y., D'Aoust P.M., Burnet J.-B., Delatollea R., Dorner S., Geng Q., Giesy J.P., Jr., McKay R.M., Mulvey M.R., Prystajecky N., Srikanthan N., Xie Y., Conant B., Hrudey S.E. Canadaian SARS-CoV-2 Inter-laboratory Consortium. Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: Results and implications from a collaborative inter-laboratory study in Canada. J. Environ. Sci. (China). 2021;107:218–229. doi: 10.1016/j.jes.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R., Srikanthan N., MacKenzie M., Figeys D., Manuel D., Jüni P., MacKenzie A.E., Delatolla R. Catching a resurgence: Increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin A., de Groot A., de Heer L., Bok J., Wielinga P.R., Hamans M., van Rotterdam B.J., Janse I. Detection of Coxiella burnetti in complex matrices by using multiplex quantitative PCR during a major Q fever uutbreak in the Netherlands. Appl. Environ Micrbiol. 2011;77(18):6516–6523. doi: 10.1128/AEM.05097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: Methodology, occurrence, and incidence/prevalence considerations. Water Res X. 2020;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Grijalva L.M.M., Roldan-Hernandez L., Langenfeld K., Wigginton K., Boehm A.B. SARS-CoV-2 RNA in Wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55 doi: 10.1021/acs.est.0c06191. 488–398. [DOI] [PubMed] [Google Scholar]
- Hou Y., Zhang H., Miranda L., Lin S. Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene. PLoS One. 2010;5:e9545. doi: 10.1371/journal.pone.0009545. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J.F., Foy C.A., Benes V., Emslie K., Garson J.A., Haynes R., Hellemans J., Kubista M., Mueller R.D., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T., Bustin S.A. The digital MIQE Guidelines: Minimum Informantion for publication of quantitative digital PCR experiments. Clin. Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- Hughes B., Beale D.J., Dennis P.G., Cook S., Ahmed W. Cross-comparison of human wastewater-associated molecular markers in relation to fecal indicator bacteria and enteric viruses in recreational beach waters. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00028-17. e00028–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Di D.Y.W., Saingam P., Jeon M.K., Yan T. Fine-scale temporal dynamics of SARS-CoV-2 RNA abundance in wastewater during a COVID-19 Lockdown. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., Chen Y.C., Pan T.M. Quantification bias caused by plasmid DNA conformation in quantitative real-time PCR assay. PLoS One. 2011;6(12):e29101. doi: 10.1371/journal.pone.0029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H.B., Whitney D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Statist. 1947;18(1):50–60. [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8(5) doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C.C., Roman F.A., Jr., Alvarado A.G.F., Tariqi A.Q., Deeming M.A., Bibby K., Bivins A., Rose J.B., Medema G., Ahmed W., Katsivelis P., Allan V., Sinclair R., Zhang Y., Kinyua M.N. Show us the data: Global COVID-19 wastewater monitoring efforts, equity, and gaps. MedRXiv. 2021 doi: 10.1101/2021.03.14.21253564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham A.L., Duncan K.E. Similar gene estimates from circular and linear standards in quantitative PCR analyses using the prokaryotic 16S rRNA gene as a model. PLoS One. 2012;7(12):e51931. doi: 10.1371/journal.pone.0051931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Warren J.L., Weinberger D.N., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaganesan M., Haugland R.A., Chern E.C., Shanks O.C. Improved strategies and optimization of calibration models for real-time PCR absolute quantification. Water Res. 2010;44(16):4726–4735. doi: 10.1016/j.watres.2010.07.066. [DOI] [PubMed] [Google Scholar]
- Taylor S.C., Nadeau K., Abbasi M., Lachance C., Nguyen M., Fenrich J. The Ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37(7):761–774. doi: 10.1016/j.tibtech.2018.12.002. [DOI] [PubMed] [Google Scholar]
- Wacker M.J., Godard M.P. Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J. Biomol. Tech. 2005;16(3):266–271. [PMC free article] [PubMed] [Google Scholar]
- Werling N.J., Satkunanathan S., Thorpe R., Zhao Y. Systematic comparison and validation of quantitative real-time PCR methods for the quantitation of adeno-associated viral products. Hum Gene Ther Methods. 2015;26(3):82–92. doi: 10.1089/hgtb.2015.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe M.K., Archana A., Catoe D., Coffman M.M., Dorevich S., Graham K.E., Kim S., Grijalva L.M., Roldan-Hernandez L., Silverman A.I., Sinnott-Armstrong N., Vugia D.J., Yu A.T., Zambrana W., Wigginton K.R., Boehm A.B. Scaling of SARS-CoV-2 RNA in settled solids from multiple wastewater treatment plants to compare incidence rates of laboratory-confirmed COVID-19 in their sewersheds. Environ. Sci. Technol. Lett. 2021;8(5):398–404. doi: 10.1021/acs.estlett.1c00184. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Oishi W., Maruo C., Saito M., Chen R., Kitajima M., Sano D. Early warning of COVID-19 via wastewater-based epidemiology: potential bottlenecks. Sci. Total Environ. 2021;767 doi: 10.1016/j.scitotenv.2021.145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A-Z of Quantitative PCR. In: Bustin Stephen A., editor. Volume 5. Calif International University Line La Jolla; 2004. Editor. [Google Scholar]