Abstract
Teichoic acids (TA) are anionic polymers comprised of polyol phosphate repeat units that are abundant in the cell wall of Gram-positive bacteria. Both wall teichoic acid (WTA) and lipoteichoic acid (LTA) play important roles in regulating cell wall remodeling as well as conferring antibiotic resistance. To analyze TA, we describe a polyacrylamide gel electrophoresis (PAGE) method for both WTA and LTA. To extract crude WTA, the peptidoglycan sacculus is first isolated and WTA is then liberated by hydrolysis. LTA is extracted by 1-butanol and pre-treated with lipase to prevent aggregation and improve single-band resolution by PAGE. Crude extracts of both TAs are then subjected to PAGE followed by Alcian blue and silver staining. These protocols are easily adoptable by laboratories interested in rapidly analyzing TAs and can be used determine the relative abundance, relative polymer length and whether TAs are glycosylated. More detailed TA structural and compositional information can be obtained using the described purification protocols by nuclear magnetic resonance (NMR) and electrospray ionization mass spectrometry (ESI-MS) analysis.
Keywords: Wall teichoic acid, Lipoteichoic acid, PAGE, Cell wall, Staphylococcus aureus, NMR, ESI-MS, Gram-positive bacteria
Background
The hallmark of Gram-positive bacteria is a thick cell wall comprised of a peptidoglycan matrix embedded with anionic polyol phosphate polymers collectively called teichoic acids (TA) (Weidenmaier and Peschel, 2008; Silhavy et al., 2010 ; Brown et al., 2013 ; Percy and Gründling, 2014). Many Gram-positive bacteria have both wall teichoic acid (WTA) and lipoteichoic acid (LTA) whereby WTA is covalently linked to the peptidoglycan layer and LTA is anchored to the cell membrane via a diacylglycerol moiety. TAs regulate numerous cell envelope-associated processes, including autolysin activity and confer resistance to antibiotics such as daptomycin and β-lactams ( Winstel et al., 2014 ). TA function can be further modulated by a variety of side chain modifications, including D-alanylation (D-ala) and glycosylation ( Kovács et al., 2006 ; Mechler et al., 2016 ; Rismondo et al., 2018 ). In Staphylococcus aureus, WTA is comprised of poly-ribitol phosphate repeats whereas its LTA contains polyglycerol phosphate repeats ( Swoboda et al., 2010 ; Percy and Gründling, 2014; Kho and Meredith, 2018). Both polymers are modified with D-ala and α/β-GlcNAc residues, in a manner that can vary between S. aureus strains and environmental culture conditions (Percy and Gründling, 2014; Kho and Meredith, 2018). Methods to rapidly monitor changes in TA structure and identify modifications are thus useful in understanding cell envelope physiology.
Although the S. aureus TA polymer backbones are structurally similar and common methods can be used to analyze their composition, WTA and LTA isolation requires specific extraction methods and care must be taken to prevent cross contamination between polymers. WTA is isolated from the crude peptidoglycan sacculus after thorough washing with detergent to remove LTA. Hydrolysis with either sodium hydroxide or trichloroacetic acid is then used to liberate water soluble WTA from peptidoglycan ( Meredith et al., 2008 ). It is often advisable to extract parallel WTA samples using both methods, as acid/base labile side chain modifications may be inadvertently cleaved during isolation and subsequently overlooked. For instance, D-ala esters are readily cleaved under moderately basic conditions ( Morath et al., 2001 ). To obtain membrane-bound LTA, cells are counter extracted with 1-butanol to disrupt the phospholipid bilayer ( Morath et al., 2001 ; Draing et al., 2006 ; Gründling and Schneewind, 2007). WTA remains insoluble in the cell remnant pellet, while more hydrophobic phospholipids partition into the organic phase. Once clarified, the aqueous phase is enriched with highly charged polyanionic LTA.
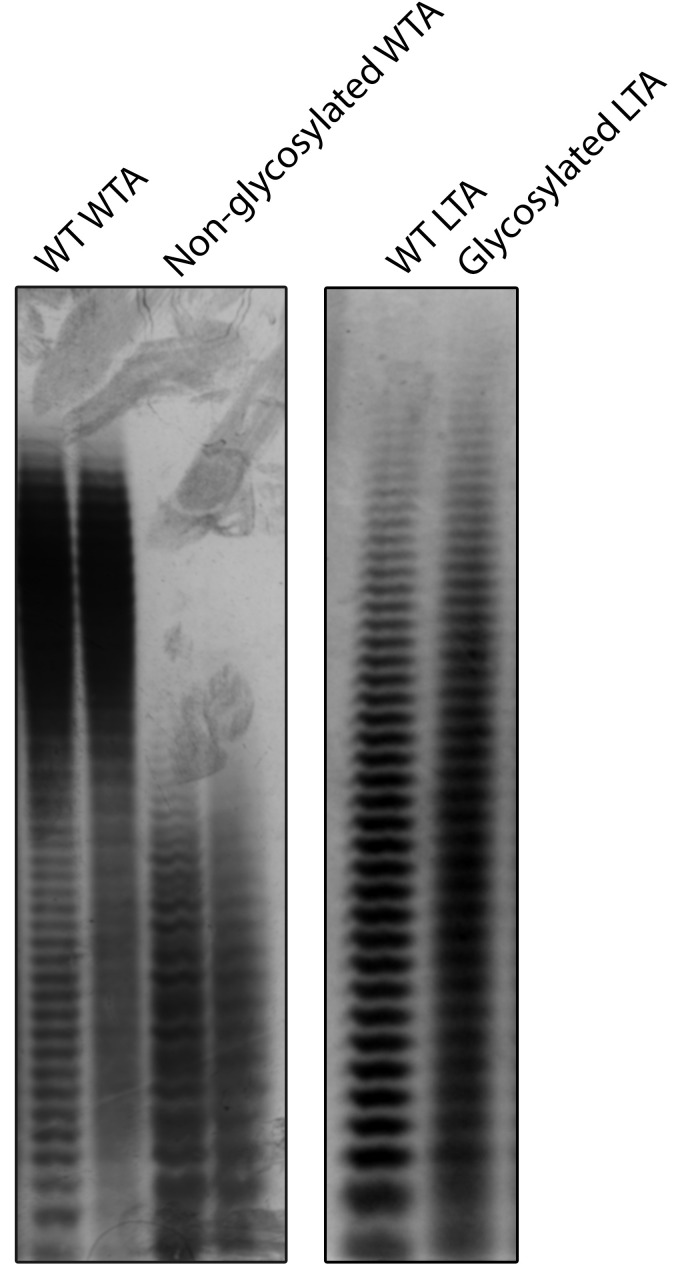
Either crude WTA or LTA extract can then be initially analyzed by PAGE ( Wolters et al., 1990 ; Pollack and Neuhaus, 1994). While WTA can be loaded directly, LTA PAGE resolution benefits from pretreatment with lipases to deacylate the diacylglycerol anchor (Kho and Meredith, 2018). The distinctive “laddering” of bands in TA-PAGE results from TA chain lengths whereby each subsequent band represents a polymer chain length of n + 1 (Figure 1). The relative abundance and polymer length profile can be evaluated, as well as the presence of heterogeneous side chain modifications which induce an ill-defined smear upon TA visualization. TA can be visualized by immunoblotting with α-TA antibodies ( Wörmann et al., 2011 ), or directly imaged using the cationic dye Alcian blue coupled with modified silver staining for enhanced sensitivity ( Meredith et al., 2008 ; Kho and Meredith, 2018).
Figure 1. Sample results for TA-PAGE.
(Left) S. aureus WTA is normally glycosylated. Knockout of the redundant WTA glycosylation genes tarM and tarS results in lower molecular weight WTA. (Right) Wild-type S. aureus LTA has very low levels of glycosylation. Overexpression of the LTA glycosylation pathway non-uniformly modifies LTA resulting in the loss of single-band resolution. Gel images are adapted from published figures in Kho and Meredith (2018).
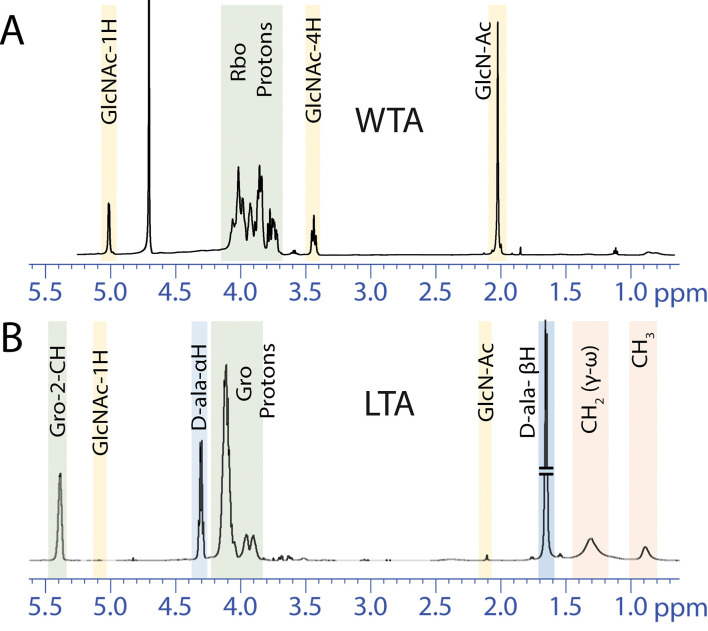
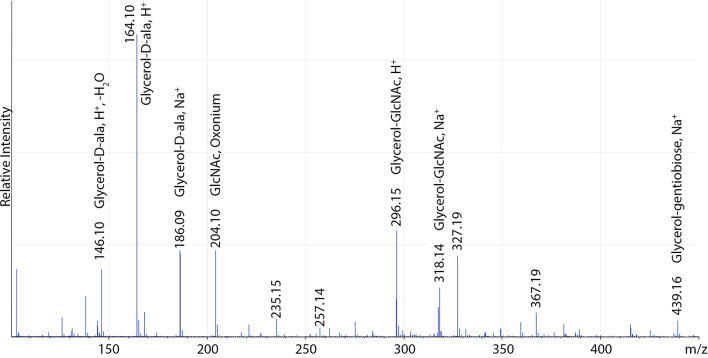
To elucidate the TA structure, composition and degree of modifications, extraction protocols can be scaled up and nuclear magnetic resonance (NMR) and electrospray ionization mass spectrometry (ESI-MS) used for subsequent characterization ( Bernal et al., 2009 ; Eugster and Loessner, 2011; Shen et al., 2017 ). Although purity is often adequate for NMR analysis of hydrolyzed WTA at this stage (Figure 3), hydrophobic interaction chromatography is used for further purification of LTA ( Morath et al., 2001 ). Anion exchange chromatography is a convenient method for further purification of either polymer ( Fischer et al., 1993 ; Kim et al., 2005 ; Eugster and Loessner, 2011). Purified and intact LTA can be analyzed directly by NMR (Figure 3) but ESI-MS analysis of TA monomers is a particularly sensitive approach for detection of low abundance modifications. Hydrofluoric acid (HF)-catalyzed TA depolymerization through dephosphorylation has been developed for WTA (Eugster and Loessner, 2011; Shen et al., 2017 ), but can also be utilized to analyze LTA (Figure 4; Kho and Meredith, 2018).
Figure 3. Sample 1H NMR spectra of WTA and LTA.
A. WTA was liberated from peptidoglycan of wild-type S. aureus by NaOH hydrolysis causing loss of D-ala modifications. This WTA spectrum was obtained using the ZG pulse sequence without water suppression. B. LTA purified from wild-type S. aureus has very low levels of GlcNAc glycosylation but is highly D-alanylated. This LTA spectrum was obtained using the ZGPR pulse sequence with water suppression. These NMR spectra are adapted from published figures in Kho and Meredith (2018). Rbo = Ribitol; Gro = Glycerol.
Figure 4. Sample positive-ion mode ESI-MS of S. aureus LTA overexpressing the LTA glycosylation system.
A full list of assigned masses can be found in Kho and Meredith (2018).
Although the methods described here are routinely used to analyze the TA of Staphylococcus aureus, it should be noted they are most suitable for TA chemotypes harboring phosphodiester repeating units. In contrast, the cell wall polymers of some Gram-positive bacteria such as Streptococcus pyogenes (Group A Streptococcus) are comprised of rhamnose repeating units ( Mistou et al., 2016 ), and may lack the polyanionic charge necessary for certain methods such as TA-PAGE.
Protocols:
Part I: Extraction and analysis of teichoic acids by PAGE
Crude wall teichoic acid (WTA) extraction for PAGE
Crude lipoteichoic acid (LTA) extraction for PAGE
-
Teichoic acid PAGE and staining
Polyacrylamide gel electrophoresis (PAGE)
Alcian blue–silver staining
Part II: Extraction and analysis of teichoic acids by 1H NMR and ESI-MS
Wall teichoic acid (WTA) precipitation for nuclear magnetic resonance
-
Lipoteichoic acid (LTA) purification by hydrophobic interaction chromatography
Extraction of crude LTA
Hydrophobic interaction chromatography
Phosphate content analysis of LTA fractions
-
1H NMR and ESI-MS analysis of teichoic acids
1H Nuclear magnetic resonance
Dephosphorylation and monomerization of LTA for ESI-MS
Electrospray ionization mass spectrometry (ESI-MS)
Part I: Extraction and analysis of teichoic acids by PAGE
Materials and Reagents
Crude wall teichoic acid (WTA) extraction for PAGE
Pipette tips
50 ml centrifuge tubes (Fisher Scientific, catalog number: 14-955-240)
Inoculation loops (VWR, catalog number: 12000-810)
2 ml microcentrifuge tubes (VWR, catalog number: 87003-298)
1.7 ml microcentrifuge tubes (VWR, catalog number: 87003-294)
Staphylococcus aureus or other Gram-positive bacteria (Approximately 100 μl of wet cell pellet)
MilliQ dH2O
Tryptic soy broth (TSB) (BD, catalog number: 211822)
2-(N-morpholino)ethanesulfonic acid (MES) (Fisher Bioreagents, catalog number: BP300-100)
Sodium dodecyl sulfate (SDS) (BP, catalog number: 166-500)
Sodium chloride (NaCl) (Sigma, catalog number: S5886)
Tris (Fisher Bioreagents, catalog number: BP152-5)
Proteinase K (Amresco, catalog number: 0706-100mg)
Sodium hydroxide (NaOH) (EMD Millipore, catalog number: 1064621000)
Hydrochloric acid (HCl) (Sigma, catalog number: 258148-500ML)
Trichloroacetic acid (TCA) (BDH, catalog number: BDH9310-500G)
Buffer 1 (50 mM MES, pH 6.5) (see Recipes)
Buffer 2 (4% [wt/vol] SDS, 50 mM MES, pH 6.5) (see Recipes)
Buffer 3 (2% NaCl, 50 mM MES, pH 6.5) (see Recipes)
Digestion buffer (20 mM Tris-HCl pH 8.0, 0.5% [w/v] SDS) (see Recipes)
Proteinase K solution (2 mg/ml) (see Recipes)
Crude lipoteichoic acid (LTA) extraction for PAGE
Pipette tips
50 ml centrifuge tubes (Fisher Scientific, catalog number: 14-955-240)
Inoculation loops (VWR, catalog number: 12000-810)
2 ml microcentrifuge tubes (VWR, catalog number: 87003-298)
1.7 ml microcentrifuge tubes (VWR, catalog number: 87003-294)
PCR tubes (VWR, catalog number: 20170-004)
pH strips (EMD Millipore, catalog number: 700181-2)
Staphylococcus aureus or other Gram-positive bacteria (Approximately 100 μl of wet cell pellet)
MilliQ dH2O
Tryptic soy broth (TSB) (BD, catalog number: 211822)
Citric acid monohydrate (Sigma-Aldrich, catalog number: C7129)
Sodium hydroxide (NaOH) (EMD Millipore, catalog number: 1064621000)
Zirconia/silica beads (0.1 mm; Biospec Products, catalog number: 11079101z)
1-butanol (Thermo Scientific, catalog number: A399)
Tris (Fisher Bioreagents, catalog number: BP152-5)
Resinase HT (Strem Chemicals, catalog number: 06-3125)
20x Citrate buffer (see Recipes)
Resuspension buffer (see Recipes)
Teichoic acid PAGE and staining
Pipette tips
Gel loading tips (VWR, catalog number: 53509-015)
50 ml centrifuge tubes (Fisher Scientific, catalog number: 14-955-240)
14 ml tubes (Corning, catalog number: 352057)
Large glass dish for staining (Pyrex, catalog number: 1085782)
MilliQ dH2O
Tris (Fisher Bioreagents, catalog number: BP152-5)
Hydrochloric acid (HCl) (Sigma, catalog number: 258148-500ML)
40% acrylamide solution (Fisher Bioreagents, catalog number: BP1402-1)
Bis-acrylamide (EMD Millipore, catalog number: 8059680250)
Tricine (Fisher Bioreagents, catalog number: BP315-100)
TEMED (Fisher BioReagents, catalog number: BP150-20)
Ammonium persulfate (APS) (Fisher BioReagents, catalog number: BP179-25)
70% ethanol (Fisher BioReagents, catalog number: BP8201500)
Glycerol (Fisher Bioreagents, catalog number: BP229-4)
Bromophenol blue (Sigma, catalog number: 114391-5G)
Alcian blue 8GX (Alfa Aesar, catalog number: J60122)
Glacial acetic acid (EMD, catalog number: AX0073-9)
Potassium dichromate (JT Baker, catalog number: 3090-01)
Nitric acid (Amresco, catalog number: 0697-100ML)
Silver nitrate (Fisher Bioreagents, catalog number: BP2546-100)
Sodium carbonate (VWR, catalog number: 0585-500G)
Formaldehyde, 37% (Sigma, catalog number: F1635-500ml)
Gel buffer (see Recipes)
10% ammonium persulfate (APS) (see Recipes)
10x Tris-Tricine buffer (see Recipes)
Running buffer (see Recipes)
Resolving acrylamide solution (30% T, 6% C) (see Recipes)
Stacking acrylamide solution (30% T, 2.6% C) (see Recipes)
4x loading dye (see Recipes)
Alcian blue solution (1 mg/ml, 3% acetic acid) (see Recipes)
Oxidizing solution (see Recipes)
Silver nitrate solution (see Recipes)
Developer (see Recipes)
Acetic acid, 0.1 M (see recipes)
Equipment
Crude wall teichoic acid (WTA) extraction for PAGE
Shaking incubator (Thermo Scientific, catalog number: SHKE6000)
Table-top centrifuge (Eppendorf, catalog number: 022621807)
Micropipettes
Hot plate (VWR, catalog number: 97042-698)
Beaker
Thermomixer (Eppendorf, catalog number: 022670107)
Crude lipoteichoic acid (LTA) extraction for PAGE
Shaking incubator (Thermo Scientific, catalog number: SHKE6000)
Centrifuge (Eppendorf, catalog number 022623508)
Rotor (Eppendorf, catalog number: 022638009)
Table-top centrifuge (Eppendorf, catalog number: 022621807)
Micropipettes
MagNA Lyser (Roche, catalog number: 03358968001)
Thermomixer (Eppendorf, catalog number: 022670107)
Thermocycler (Bio-Rad, catalog number: 1861096)
Teichoic acid PAGE and staining
PROTEAN® II xi 2-D Tube Gel Cell (Bio-Rad, catalog number: 1651931)
Beakers
Stir plate (VWR, catalog number: 97042-590)
Stir bars
Pipette controller (BrandTech Scientific, catalog number: 26330)
Micropipettes
Shaker (Alkali Scientific, catalog number: RS7061)
Procedure
Crude wall teichoic acid (WTA) extraction for PAGE
Transfer 20 ml of TSB into a 50 ml centrifuge tube and if necessary, add the required antibiotics for selection.
Using a sterile inoculation loop, inoculate the media from a single Staphylococcus aureus colony.
Incubate the bacterial culture at 37 °C for 16-20 h with shaking at 250 rpm.
-
Heat the hot plate to boil 800 ml of water in a beaker.
Note: It is critical to obtain a rolling boil to ensure thorough cell breakage.
Centrifuge the cell culture at 5,000 × g for 5 min.
Resuspend the cell pellet in 30 ml of Buffer 1.
Centrifuge cells at 5,000 × g for 5 min.
Resuspend the cell pellet in 30 ml of Buffer 2.
Put the centrifuge tube containing the cell resuspension into the boiling water bath for 1 h.
-
Remove the centrifuge tubes from the boiling water bath and allow it to cool at room temperature for 30 min or until it is cool to touch.
Note: Do not cool on ice as SDS will precipitate.
Centrifuge at 5,000 × g for 5 min.
-
Resuspend in 1 ml of Buffer 1 and transfer into a 2 ml microcentrifuge tube.
Note: An additional wash with SDS-containing Buffer 1 is critical to remove LTA and other contaminating lipids.
Centrifuge at 16,000 × g for 1 min.
Resuspend in 2 ml of Buffer 2 and centrifuge as in Step 13.
Resuspend in 2 ml of Buffer 3 and centrifuge as in Step 13.
Resuspend in 2 ml of Buffer 1 and centrifuge as in Step 13.
Resuspend in 1 ml of Digestion buffer.
Add 10 μl of 2 mg/ml proteinase K solution (20 μg)
Incubate on a shaking heat block at 50 °C for 4 h at 1,400 rpm.
Centrifuge at 16,000 × g for 1 min.
Resuspend in 2 ml of Buffer 3 and centrifuge as in Step 20.
Resuspend in 2 ml of dH2O and centrifuge as in Step 20.
Repeat dH2O washes for a total of 3 washes to remove SDS.
Resuspend thoroughly in 1 ml of 0.1 M NaOH. See Notes for alternative TCA hydrolysis.
Incubate on a thermomixer for 16 h at room temperature at 1,400 rpm.
Centrifuge at 16,000 × g for 1 min.
Transfer the supernatant into a new 1.7 ml microcentrifuge tube.
Add 250 μl of 1 M Tris-HCl, pH 7.8 to neutralize.
Store at -20 °C or proceed with TA-PAGE.
Crude lipoteichoic acid (LTA) extraction for PAGE
Transfer 20 ml of TSB into a 50 ml centrifuge tube and if necessary, add the required antibiotics for selection.
Using a sterile inoculation loop, inoculate the media from a single Staphylococcus aureus colony.
Incubate the bacterial culture at 37 °C for 16-20 h with shaking at 250 rpm.
Centrifuge cells at 5,000 × g for 5 min.
Resuspend cell pellet in 2 ml of resuspension buffer.
Transfer cell suspension into a 2 ml screw cap microcentrifuge tube.
Centrifuge cells at 16,000 × g for 1 min and discard supernatant.
Add ~700 μl of zirconia/silica beads.
Fill the tube with resuspension buffer to minimize headspace.
Chill the sample on ice for 5 min.
-
Lyse cells with MagNA-lyser
Run for 30 s at 7,000 rpm.
Chill on ice for 5 min.
Repeat 3 more times.
Pellet the cell membranes at 20,000 × g, 4 °C for 1 h.
-
Suspend the cell membrane in 350 μl of resuspension buffer.
Note: The pellet is hard to resuspend so scrap it with a pipet tip and pipet mix as well as possible. Freezing the pellet at -20 °C prior to resuspending can help.
Add 350 μl of 1-butanol.
Incubate on shaking heat block at 37 °C for 45 min at 1,400 rpm.
Centrifuge at 20,000 × g, 4 °C for 45 min to pellet insoluble debris and for phase separation.
-
Extract the lower aqueous layer into a 1.7 ml microcentrifuge tube.
Note: If there are still cell debris, centrifuge at 20,000 x g, 4 °C for 20 min and extract the supernatant into another 1.7 ml microcentrifuge tube.
-
In a PCR tube, set up the lipase reaction to hydrolyze the LTA lipids
50 μl of crude LTA extract
50 μl of 50 mM Tris free base
2 μl of 1 M NaOH
Mix well and ensure that the pH is around pH 8.5 with a pH strip.
Add 1 μl of Resinase HT and mix.
Incubate at 50 °C for 16-18 h in a thermocycler.
Store at -20 °C or proceed with TA-PAGE.
Teichoic acid PAGE and staining
Polyacrylamide gel electrophoresis (PAGE)
Fill the gel cooling core with dH2O and chill at 4 °C.
Chill running buffer at 4 °C.
Assemble the gel glass plates.
-
Make the resolving gel (20% T, 6% C final concentration) in a 50 ml tube.
13.3 ml of gel buffer (3 M Tris-HCl, pH 8.5)
26.67 ml of resolving acrylamide solution (30% T, 6% C)
400 μl of 10% APS
40 μl of TEMED
Pipet 35 ml between the gel plates, leaving enough space for the stacking gel.
-
Layer the surface with 70% ethanol.
Note: It is better to insert the comb here to properly space the glass plates.
Allow the gel to solidify for at least 30 min.
Pour off the ethanol and rinse with dH2O.
Dab off the excess water with a paper towel.
-
Make the stacking gel (3% T, 2.6% C) in a 15 ml tube
2 ml of MilliQ dH2O
1 ml of gel buffer (3 M Tris-HCl, pH 8.5)
333 μl of stacking acrylamide solution
33 μl of 10% APS
3.3 μl of TEMED
Fill the headspace with the stacking gel and carefully insert the comb while wiping away the gel overflow with a paper towel.
Allow the gel to solidify.
Gently remove the gel comb and straighten the wells with a gel loading tip if necessary.
Assemble the gel apparatus by first wetting the gasket and locking the gel in position.
-
Fill the top chamber with running buffer.
Note: Ensure that there are no leaks. If it is leaking, remove the gel and reassemble the apparatus.
Pour the remaining running buffer into the gel tank.
Prior to loading the wells, wash out the wells with a 1,000 μl pipet by pipetting running buffer into wells.
Mix 15 μl of each sample with 5 μl of 4x loading dye.
Load 15 μl of samples per well.
Run at 40 mA per gel, 4 °C for 20-22 h with constant stirring of the buffer in the gel tank.
Alcian blue–silver staining
Carefully disassemble the glass plates and remove the stacking gel.
Fill a large Pyrex dish with some MilliQ dH2O and carefully transfer the resolving gel into the dish.
Gently agitate to wash the gel.
Decant the water.
Wash the gel briefly twice more with MilliQ dH2O.
Gently agitate gel in Alcian blue solution for at least 3 h or overnight on a shaker.
Decant the dye solution.
Wash the gel briefly 5 times with MilliQ dH2O.
Destain the gel in MilliQ dH2O with several water changes for several hours or overnight with shaking until the dye front is no longer visible.
Decant the water.
Soak the gel in oxidizing solution and gently agitate for 7 min.
Wash 5 times with MilliQ dH2O.
-
Soak the gel in 12 mM silver nitrate solution with gentle agitation under bright illumination for 24 min.
Note: The illumination in a laminar flow hood is sufficient.
Wash 5 times quickly with MilliQ dH2O.
-
Develop the gel with 3 brief treatments of developer
Wash it briefly until the solution turns brown and decant it.
Repeat the washing a second time.
Soak the gel in the developer and gently agitate it until the bands intensify. Let the bands develop sufficiently but decant the solution before the background darkens. See Figure 1.
Immediately flood the gel with 0.1 M acetic acid to stop the development process and let it sit in the acetic acid for at least 15 min.
Decant the acetic acid and store the gel in dH2O prior to imaging.
Image the gel using a gel imager or a camera.
Notes
Crude wall teichoic acid (WTA) extraction for PAGE
If necessary, instead of using 1 ml of 0.1 M NaOH, WTA hydrolysis can be performed in 1 ml of 5% TCA at 4 °C for 45 h before neutralization with 36.5 μl of 10 M NaOH and 100 μl of 1 M Tris-HCl, pH 7.8. Although this might yield slightly “cleaner” WTA, D-alanine modifications might remain intact and affect PAGE resolution.
WTA hydrolysis incubation periods with either acid or base may have to be adjusted for a particular strain of interest in order to minimize TA chain hydrolysis (seen as time-dependent faster-migrating bands in TA-PAGE) while retaining acceptable WTA yield.
This protocol should yield at least 0.5 μg of crude WTA if lyophilized.
Crude lipoteichoic acid (LTA) extraction for PAGE
10 OD/ml of exponentially growing cells can be used instead.
Enzymatic lysis (with lysozyme/lysostaphin/mutanolysin) should be avoided to minimize LTA cross contamination (see as faster migrating material by TA-PAGE).
While bead beating is not absolutely essential, LTA yields are enhanced with prior mechanical disruption.
If you don’t have access to a refrigerated microcentrifuge, a regular centrifuge will suffice.
More concentrated LTA preparations can be achieved by suspending the cell membrane in 200 μl of resuspension buffer with 200 μl of 1-butanol.
It is important that the lipase reaction is carried out at pH 8.5 to promote base-catalyzed hydrolysis of D-ala modifications on LTA which can improve single-band resolution on PAGE by increasing LTA homogeneity.
This protocol should yield at least 50 μg of crude LTA based on PAGE comparison with purified LTA.
Teichoic acid PAGE and staining
The bis-acrylamide in the resolving acrylamide solution (30% T, 6% C) can precipitate during extended storage and may require mixing prior to use.
Alcian blue 8GX should be dissolved in MilliQ dH2O with 3% acetic acid to maintain dye solubility. High purity dH2O should be used with all Alcian blue 8GX solutions.
Alcian blue staining alone may be sufficient to visualize TA bands if the sample is concentrated.
Gently handle the gel by the edges when staining and washing. Any indentations/prints will be silver stained and potentially obscure the bands.
Recipes
Crude wall teichoic acid (WTA) extraction for PAGE
-
Buffer 1 (50 mM MES, pH 6.5)
Weigh out 2.665 g of MES (213.2 g/mol)
Dissolve in 175 ml MilliQ dH2O
Adjust pH to 6.5 with NaOH
Top with MilliQ dH2O to 250 ml
Store at room temperature
-
Buffer 2 (4% [wt/vol] SDS, 50 mM MES, pH 6.5)
Dissolve 4 g of SDS in 100 ml of Buffer 1
Store at room temperature
-
Buffer 3 (2% NaCl, 50 mM MES, pH 6.5)
Dissolve 1 g of NaCl in 50 ml of Buffer 1
Store at room temperature
-
Digestion buffer (20 mM Tris-HCl pH 8.0, 0.5% [w/v] SDS)
Dissolve 0.1214 g of Tris in 40 ml of MilliQ dH2O
Adjust to pH 8.0 with HCl
Add 0.25 g of SDS and dissolve
Top with MilliQ dH2O to 50 ml
Store at room temperature
-
Proteinase K solution (2 mg/ml)
Dissolve 2 mg of proteinase K in 1 ml of MilliQ dH2O
Store at 4 °C
Crude lipoteichoic acid (LTA) extraction for PAGE
-
20x Citrate buffer (1 M sodium citrate, pH 4.7)
Weigh out 21.014 g of citric acid monohydrate (MW 210.14)
Dissolve in 70 ml of MilliQ dH2O
Adjust to pH 4.7 with sodium hydroxide pellets
Top with MilliQ dH2O to 100 ml
Store at room temperature
-
Resuspension buffer (50 mM sodium citrate, pH 4.7)
Dilute 25 ml of 20x citrate buffer into 500 ml of MilliQ dH2O
Adjust to pH 4.7 with NaOH if necessary
Store at room temperature
Teichoic acid PAGE and staining
-
Gel buffer (3 M Tris-HCl, pH 8.5)
Dissolve 36.342 g Tris (MW = 121.14 g/mol) in 90 ml of MilliQ dH2O
Adjust to pH 8.5 with concentrated HCl
Top with MilliQ dH2O to 100 ml
Store at 4 °C
-
10% ammonium persulfate
Dissolve 0.1 g APS in 1 ml of MilliQ dH2O
Store at -20 °C
-
10x Tris-Tricine buffer (1 M Tris, 1 M Tricine, pH 8.2)
Weigh out 121.2 g Tris (MW = 121.14 g/mol)
Weigh out 179.2 g Tricine (MW = 179.17 g/mol)
Dissolve powders in 700 ml MilliQ dH2O
Top with MilliQ dH2O to 1 L
Store at room temperature
Do not adjust the pH
-
Running buffer
Dilute 300 ml of 10x Tris-tricine buffer into 3 L with MilliQ dH2O
-
Resolving acrylamide solution (30% T, 6% C)
Dissolve 1.8 g of Bis-acrylamide in 70.5 ml of 40% acrylamide solution
Top with MilliQ dH2O to 100 ml
Store at 4 °C
-
Stacking acrylamide solution (30% T, 2.6% C)
Dissolve 0.78 g of Bis-acrylamide in 73.1 ml of 40% acrylamide solution
Top with MilliQ dH2O to 100 ml
Store at 4 °C
-
4x loading dye
Mix 5 ml of glycerol with 5 ml of running buffer
Add a trace of bromophenol blue and mix well
Store at -20 °C
-
Alcian blue solution (1 mg/ml, 3% acetic acid)
Make this solution fresh
Dissolve 0.2 g of Alcian blue in 194 ml of MilliQ dH2O
Add 6 ml glacial acetic acid
-
Oxidizing solution (3.4 mM potassium dichromate, 3.2 mM nitric)
Make this solution fresh
Weigh out 0.2 g of potassium dichromate (294.185 g/mol)
Dissolve in 200 ml MilliQ dH2O
Add 40 μl of nitric acid
-
Silver nitrate solution (12 mM)
Make this solution fresh
Weigh out 0.408 g of silver nitrate (MW = 169.87 g/mol)
Dissolve in 200 ml MilliQ dH2O
-
Developer (0.28 M sodium carbonate, 1.85 % formaldehyde)
Make this solution fresh
Weigh out 14.84 g sodium carbonate (MW = 105.99 g/mol)
Dissolve in 500 ml MilliQ dH2O
Add 250 μl 37% formaldehyde
-
0.1 M acetic acid
Dilute 1.44 ml glacial acetic acid into 250 ml with MilliQ dH2O
Part II: Extraction and analysis of teichoic acids by 1H NMR and ESI-MS
Materials and Reagents
Wall teichoic acid (WTA) precipitation for nuclear magnetic resonance
14 ml tubes (Corning, catalog number: 352057)
Inoculation loops (VWR, catalog number: 12000-810)
50 ml centrifuge tubes (Fisher Scientific, catalog number: 14-955-240)
50 ml Oak Ridge tubes (Nalgene, catalog number: 3119-0050)
2 ml microcentrifuge tubes (VWR, catalog number: 87003-298)
Staphylococcus aureus or other Gram-positive bacteria (At least 5 ml of wet cell pellet)
Tryptic soy broth (TSB) (BD, catalog number: 211822)
Sodium dodecyl sulfate (SDS) (Fisher Scientific, catalog number: BP166-500)
Sodium chloride (NaCl) (Sigma, catalog number: S5886)
Sodium phosphate dibasic anhydrous (Na2HPO4) (EMD Millipore, catalog number: SX0720-1)
Potassium chloride (KCl) (Sigma, catalog number: P9541-500G)
Potassium phosphate monobasic (KH2PO4) (Fisher Bioreagents, catalog number: BP362-500)
Tris (Fisher Bioreagents, catalog number: BP152-5)
Trypsin (Amresco, catalog number: M150-1G)
Sodium hydroxide (NaOH) (EMD Millipore, catalog number: 1064621000)
Hydrochloric acid (HCl) (Sigma, catalog number: 258148-500ML)
Trichloroacetic acid (TCA) (BDH, catalog number: BDH9310-500G)
Sodium acetate trihydrate (Sigma, catalog number: S8625-500G)
Glacial acetic acid (EMD Millipore, catalog number: AX0073-9)
95% ethanol (Koptec, catalog number: V1101)
Dry ice
10x phosphate buffered saline (see Recipes)
1x phosphate buffer saline (PBS) (see Recipes)
2 M NaCl (see Recipes)
1 M Tris-HCl, pH 7.0 (see Recipes)
1 M Tris-HCl, pH 7.0 containing 1 M NaCl (see Recipes)
0.5% SDS (see Recipes)
10% trichloroacetic acid (see Recipes)
3 M NaOAc, pH 5.1 (see Recipes)
Trypsin solution (see Recipes)
Lipoteichoic acid (LTA) purification by hydrophobic interaction chromatography
50 ml centrifuge tubes (Fisher Scientific, catalog number: 14-955-240)
Inoculation loops (VWR, catalog number: 12000-810)
50 ml Oak Ridge tubes (Nalgene 3119-0050)
2 ml microcentrifuge tubes (VWR, catalog number: 87003-298)
Rubber stopper (VWR, catalog number: 59586-167)
96-well plate (Greiner, catalog number: 655185)
14 ml tubes (Corning, catalog number: 352057)
Scintillation vial (VWR, catalog number: VW74511-20)
Kimwipes (Kimberly-Clark, catalog number: 34120)
Vacuum tubing
0.45 μm PES syringe filter (VWR, catalog number: 28145-503)
5 ml syringe (Fisher Scientific, catalog number: 14-829-45)
HiPrep Octyl FF 16/10 column (GE Healthcare, catalog number: 28936548)
Elastic band
Staphylococcus aureus or other Gram-positive bacteria (At least 20-25 ml of wet cell pellet)
Tryptic soy broth (TSB) (BD, catalog number: 211822)
Citric acid monohydrate (Sigma-Aldrich, catalog number: C7129)
Sodium hydroxide (NaOH) (EMD Millipore, catalog number: 1064621000)
0.1 mm zirconia/silica beads (BioSpec Products, catalog number: 11079101z)
1-butanol (Thermo Scientific, catalog number: A399)
Sodium phosphate, monobasic dihydrate (NaH2PO4·2H2O) (Acros Organics, catalog number: 271750010)
Sulfuric acid (Sigma, catalog number: 258105-500ML)
70% Perchloric acid (BDH, catalog number: 4550-500ML)
Ammonium molybdate tetrahydrate (Sigma, catalog number: A7302-100G)
Sodium acetate trihydrate (Sigma, catalog number: S8625-500G)
L-Ascorbic acid sodium salt (VWR, catalog number: 0561-100G)
Dry ice
20x citrate buffer (see Recipes)
Resuspension buffer (see Recipes)
Equilibration buffer (see Recipes)
Elution buffer (see Recipes)
Ashing solution (2 M H2SO4, 0.44 M HClO4) (see Recipes)
Reducing solution (3 mM ammonium molybdate, 0.25 M sodium acetate, 1% ascorbic acid) (see Recipes)
0.1 M phosphate buffer (see Recipes)
Phosphate standards (see Recipes)
1H NMR and ESI-MS analysis of teichoic acids
1.7 ml microcentrifuge tube (VWR, catalog number: 87003-294)
NMR tube (Wilmad, catalog number: 528-PP-7)
50 ml centrifuge tube (Fisher Scientific, catalog number: 14-955-240)
Purified teichoic acid
99.8% D2O (Acros Organics, catalog number: 166300100)
100% D2O (Acros Organics, catalog number: 320700075)
48% hydrofluoric acid (Acros Organics, catalog number: 223330250)
OmniTop Assemblies, Pre-Sterilized Single-Use Sample Tubes® (PAW BioScience, catalog number: 50CTA-CF-X-00)
Tubing (VWR, catalog number: 89068-512)
Potassium hydroxide (Sigma, catalog number: 484016)
Sodium carbonate, anhydrous (Amresco, catalog number: 0585-500G)
Ammonium bicarbonate (Sigma, catalog number: 09830-500G)
Ammonium hydroxide solution (Sigma, catalog number: 320145)
Acetonitrile (Thermo Scientific, catalog number: 51101)
Saturated sodium carbonate solution (see Recipes)
1 M ammonium bicarbonate (see Recipes)
5 mM ammonium bicarbonate (see Recipes)
Dilute ammonium hydroxide (see Recipes)
Equipment
Wall teichoic acid (WTA) precipitation for nuclear magnetic resonance
2 L baffled flask (Kimble, catalog number: 25630-2000)
Centrifuge bottles (Thermo Fisher, catalog number: 3120-0500)
Shaking incubator (Thermo Scientific, catalog number: SHKE6000)
BeadBeater (BioSpec Products)
Floor centrifuge (Beckman, catalog number: 369001)
Rotor (Beckman, catalog number: 369687)
Rotoflex tube rotator (Argos Technologies, catalog number: R2000)
Freeze-dryer (Labconco, catalog number: 7670520)
Lipoteichoic acid (LTA) purification by hydrophobic interaction chromatography
Shaking incubator (Thermo Scientific, catalog number: SHKE6000)
2 L baffled flasks x 4 (Kimble, catalog number: 25630-2000)
250 ml baffled flask (Kimble, catalog number: 25630-250)
Nalgene centrifuge bottles (Thermo Fisher, catalog number: 3120-0500)
Floor centrifuge (Beckman, catalog number: 369001)
Rotor (Beckman, catalog number: 369687)
BeadBeater (BioSpec Products)
Freeze-dryer (Labconco, catalog number: 7670520)
1 L Büchner flasks (Kimble, catalog number: 27060-1000)
Vacuum pump (Buchi, catalog number: 11V100000)
Stir bars
Stir plate (VWR, catalog number: 97042-590)
ÄKTA pure chromatography system (GE Healthcare)
5 ml sample loop (GE Healthcare, catalog number: 18114053)
Heat block (VWR, catalog number: 12621-088)
Plate reader (Molecular Devices, catalog number: SpectraMax M2e)
Rotary evaporator (Buchi, catalog number: 11100V202)
Float-A-Lyzer G2 Dialysis Device CE, 0.5-1.0 kD MWCO (Spectrum Laboratories, catalog number: G235051)
1H NMR and ESI-MS analysis of teichoic acids
Freeze-dryer (Labconco, catalog number: 7670520)
Drying apparatus (see Figure 2)
OmniTop assembly (VWR, catalog number: 75840-774)
Drying tube (VWR, catalog number: 46600-054)
Bruker Avance III 600
Waters Q-TOF Premier quadrupole/time of flight mass spectrometer
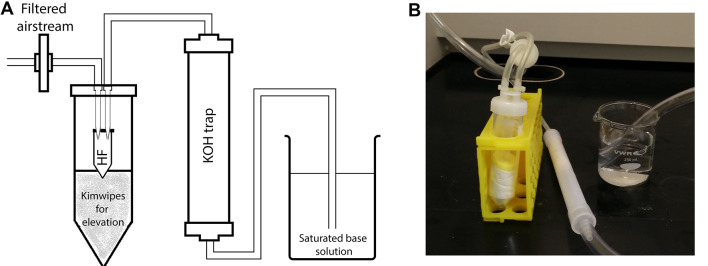
Figure 2. Drying apparatus.
A. Schematic illustration of the filtered airstream setup using a gentle airflow within the chemical fume hood. Preassembled OmniTop assembly tubes fitted with pipette tips into the interior of the lid can be conveniently converted into the diagrammed HF removal apparatus. The microcentrifuge tube containing the sample is inserted into a 50 ml centrifuge tube as a secondary container. The air (or nitrogen) stream is then passed through the trap filled with KOH pellets and further bubbled through a saturated base solution to neutralize HF. B. A picture of the actual setup.
Procedure
Wall teichoic acid (WTA) precipitation for nuclear magnetic resonance
Transfer 2 ml of TSB into a 14 ml tube and add antibiotics for selection if necessary.
Using a sterile inoculation loop, inoculate the media from a single Staphylococcus aureus colony.
Incubate the culture at 37 °C with shaking at 250 rpm over the course of the day until it is turbid.
Inoculate 1 L of TSB in a baffled flask (with antibiotic if necessary) with the pre-grown culture.
Incubate the cultures at 37 °C with shaking at 200 rpm overnight.
Decant cell culture into centrifuge bottles.
Centrifuge at 6,000 × g for 10 min.
Suspend cell pellet in 10 ml of 2 M NaCl.
Fill the small BeadBeater chamber with one-third zirconium/silica beads.
Decant the cell suspension into the BeadBeater chamber.
Fill the chamber with 2 M NaCl to minimize headspace and securely cap it.
Tightly screw into the cooling chamber filled with ice water and let it chill for at least 5 min.
Beat 4 cycles at 2.5 min each with at least 5-10 min cooling in between.
Allow the beads to settle and carefully pipet the lysate into as many 50 ml Oak Ridge tubes as necessary.
Add 10 ml of 2 M NaCl to the beads and pulse for 10 s to wash the beads.
Combine the wash with the lysate in the Oak Ridge tubes.
Centrifuge at 14,000 × g for 15 min to pellet cell wall fragments.
Wash with 10 ml of 1x PBS and centrifuge.
Resuspend the pellet in 30 ml of 0.5% SDS and centrifuge.
Repeat twice more.
Wash 3 times with MilliQ dH2O by resuspending and centrifuging.
Suspend in 30 ml of MilliQ dH2O.
Incubate at 60 °C for 30 min with shaking at 250 rpm to remove non-covalently bound components.
Centrifuge at 14,000 × g for 15 min to pellet cell wall and aspirate the supernatant containing the non-covalently bound components.
Wash the cell wall pellet with 30 ml MilliQ dH2O.
Suspend in 20 ml of trypsin solution.
Incubate at 37 °C for 18 h with constant mixing at 250 rpm.
Centrifuge at 14,000 × g for 15 min.
-
Wash sequentially with 30 ml of the following by resuspending and centrifuging at 14,000 × g for 15 min after each wash:
1 M Tris-HCl pH 7.0
1 M Tris-HCl, pH 7.0 containing 1 M NaCl
1 M Tris-HCl, pH 7.0
3x MilliQ dH2O
Suspend in 10 ml of 0.1 M NaOH. See Notes for alternative TCA hydrolysis.
Incubate at room temperature for 16 h on a tube rotator or with shaking at 250 rpm.
Centrifuge at 5,500 × g for 45 min.
Transfer the supernatant containing WTA into a new tube and discard the peptidoglycan pellet.
Add 0.1 volume of 3 M NaOAc, pH 5.1 and 3 volumes of 95% ice-cold ethanol to precipitate WTA.
Incubate at -20 °C overnight.
Centrifuge at 5,500 × g for 30 min at room temperature.
Suspend the pellet in 2 ml of 95% EtOH and transfer to a 2 ml microcentrifuge tube.
Centrifuge at 16,000 × g for 5 min to collect WTA.
Wash the Oak Ridge tube with of 2 ml of 95% EtOH and combine in the 2 ml microcentrifuge tube to collect residual WTA by centrifuging at 16,000 × g for 5 min.
Wash the WTA pellet by resuspending in 2 ml of 95% ethanol and centrifuging.
Repeat for a total of 5 times.
Allow the ethanol to completely evaporate.
Suspend the pellet in 500 μl of MilliQ dH2O.
Transfer into a pre-weighed 1.7 ml microcentrifuge tube.
Freeze in a dry ice bath or at -80 °C and freeze-dry to yield a white powder.
Weigh and store at -20 °C.
Lipoteichoic acid (LTA) purification by hydrophobic interaction chromatography
Extraction of crude LTA
Transfer 20 ml of TSB into a 50 ml centrifuge tube and add antibiotics for selection if necessary.
Using a sterile inoculation loop, inoculate the media from a single Staphylococcus aureus colony.
Incubate the culture at 37 °C with shaking at 250 rpm during the course of the day until it is turbid.
Inoculate 4 x 1 L flasks of TSB (with antibiotic if necessary) with the pre-grown culture.
Incubate the cultures at 37 °C with shaking at 200 rpm overnight.
Decant cell cultures into centrifuge bottles.
Centrifuge at 6,000 × g for 10 min.
Resuspend and combine the cells in 100 ml of resuspension buffer.
-
Pellet the cells at 6,000 × g for 10 min.
Note: The pellet can be frozen for long-term storage at -80 °C.
Fill the large BeadBeater chamber with 1/3rd zirconium/silica beads.
-
Transfer the bacterial pellet into the BeadBeater chamber using a spatula.
Note: It is important to minimize headspace in the chamber so vortexing to resuspend is not recommended as it will cause foaming.
Wash the bottle with 50 ml of resuspension buffer and decant into the BeadBeater chamber.
Fill the chamber with resuspension buffer to minimize headspace while leaving enough room for the BeadBeater rotor.
Fill the cooling chamber with ice water and let chill for 5 min.
Beat 4 cycles at 2.5 min each with at least 5-10 min cooling in between cycles.
Allow the beads to settle and carefully pipet the lysate into as many 50 ml Oak Ridge tubes as necessary.
Add 50 ml of resuspension buffer to the beads and pulse for 10 s to wash the beads.
Combine the wash with the lysate in the Oak Ridge tubes.
-
Centrifuge at 30,000 × g, 4 °C for 45 min to pellet cell membrane material.
Note: The pellet can be frozen for long-term storage at -80 °C.
-
Thoroughly resuspend and combine the membrane pellets in 60-75 ml total resuspension buffer and pour into a clean 250 ml baffled flask.
Note: The membrane pellet is hard to resuspend, so a combination of scraping with a spatula/pipette and vortex is necessary.
Wash centrifuge tubes with 50 ml of 1-butanol total and decant into in the same flask.
Cover tightly to prevent butanol evaporation and incubate with shaking at 37 °C for 45 min.
-
Split the suspension into 50 ml Oak Ridge tubes.
Note: The same tubes as Steps 19-20 can be used without cleaning.
Centrifuge at 30,000 × g, 4 °C for 45 min to pellet insoluble debris.
Extract the lower aqueous phase containing LTA into 50 ml centrifuge tubes.
Centrifuge at 6,000 × g for 10 min to remove debris.
Extract and combine the supernatants in a freeze-dryer flask.
-
Flash-freeze in a dry ice bath.
Note: Slowly rotate the flask while freezing to coat the walls of the flask. Greater ice surface area will hasten drying.
Freeze-dry overnight or to completion.
Suspend in 5 ml of equilibration buffer.
Split into 3 x 2 ml microcentrifuge tubes.
Centrifuge at 20,000 × g for 10 min to remove insoluble debris.
Combine the supernatant and filter through a 0.45 μm PES syringe filter to remove any residual debris.
Wash the filter with 500 μl of equilibration buffer and combine the filtrate.
-
Draw the solution into a clean 5 ml syringe and proceed with column chromatography.
Notes:
Ensure that the final sample is completely free of debris. Otherwise, centrifuge and/or filter again to clarify.
Ensure that there are no air bubbles. Tap the syringe to dislodge any air bubbles and let it stand upright for several minutes to allow air bubbles to rise.
Hydrophobic interaction chromatography
Transfer 500 ml of equilibration buffer to a 1 L Büchner flask.
Insert a stir bar and cap the flask with a rubber stopper.
Attach one end of a vacuum tubing to the hose barb and the other end to a vacuum pump.
Start the vacuum to de-gas the solution.
Slowly increase the stir rate until it is stirring vigorously, ensuring that the solution does not bubble into the tubing.
De-gas the solution with stirring for 30 min.
Stop the de-gassing by slowly stopping the stirring and then turning off the vacuum.
Repeat the de-gassing procedure with the elution buffer.
Attach the HiPrep Octyl FF 16/10 column to the ÄKTA pure system.
Clean the sample loop with 10 ml of equilibration buffer.
Manually inject the crude LTA extract into the sample loop.
-
Start the column purification run with the following conditions:
Fix the flow rate at 0.5 ml/min or higher depending on the system pressure.
Equilibrate with 2 column volumes (40 ml) of equilibration buffer.
Empty the sample loop with 10 ml of equilibration buffer.
Wash the column with 5 column volumes of equilibration buffer (100 ml).
Elute the column with a linear gradient of equilibration buffer and elution buffer (15% to 65% 1-propanol, 50 mM sodium citrate, pH 4.7) in 100 ml.
Collect 3 ml fractions for approximately 36 total fractions.
Clean the column with 4 column volumes of (80 ml) of elution buffer.
Equilibrate the column with 6 column volumes (120 ml) of equilibration buffer.
Phosphate content analysis of LTA fractions
Transfer 100 μl of phosphate standards and 100 μl of each fraction into 2 ml microcentrifuge tubes.
Work in a chemical fume hood and add 200 μl of ashing solution to each tube.
Leave the microcentrifuge tubes uncapped and incubate on a heat block at 120 °C for 3 h in the chemical fume hood.
Add 1 ml of reducing solution and cap tubes.
Incubate at 45 °C for 2 h with occasional mixing by inversion.
Transfer 250 μl from each tube to a 96-well plate and read at 700 nm.
Combine LTA peak fractions.
Dry on a rotary evaporator with the water bath set to 30 °C.
While drying the fractions, prepare the dialysis device according to manufacturer’s instructions.
Suspend the LTA in a total of 5 ml of MilliQ dH2O and transfer into the prepared dialysis device.
-
Dialyze extensively against MilliQ dH2O at 4 °C
Dialyze at least 7 to 8 rounds for 6 h each in 5 L of MilliQ dH2O.
Stir rigorously to increase dialysis efficiency.
If dialyzing multiple samples at once, add at least 2 more rounds of dialysis.
Transfer dialysate to a pre-weighed scintillation vial.
Carefully freeze the dialysate in a dry ice bath.
Remove the cap and secure a folded piece of Kimwipe in its place with an elastic band.
Freeze-dry overnight or to completion.
Weigh and store at -20 °C.
1H NMR and ESI-MS analysis of teichoic acids
1H Nuclear magnetic resonance
Suspend purified TA in 99.8% D2O.
Transfer to a 1.7 ml microcentrifuge tube.
Incubate at room temperature for 30 min.
Freeze in a dry ice bath or at -80 °C.
Freeze-dry.
Repeat Steps 1-5 without changing tubes.
-
Suspend the proton-exchanged TA in 550 μl of 100% D2O.
Note: Wrap a single-use ampule of D2O in a paper towel and carefully snap off the top to prevent injury.
Rinse a clean NMR tube with the remaining 150 μl of D2O in the ampule.
Remove the D2O by gently flicking or using a long tip pipet to aspirate.
Carefully transfer the TA in D2O into the NMR tube.
-
Proceed with NMR using with the following typical settings:
Temperature: 298 K
Pulse sequence: ZG (no water suppression) or ZGPR (water suppression by pre-saturation)
Number of scans: 512
Spectral width: 10 ppm
Acquisition time: 5.45 s
Recycle delay: 5 s
Line broadening: 0.3 Hz
Total experiment time: Approximately 2 h on the Bruker Avance III 600
Calibrate the spectra using the water signal as a reference at 4.7 ppm and assign NMR signals corresponding to S. aureus WTA and LTA as exemplified in Figure 3. See Notes about NMR standard.
Dephosphorylation and monomerization of TA for ESI-MS
Note: Hydrofluoric acid is extremely hazardous. Users should consult standard operating procedures and their safety office prior to use. Work in the chemical fume hood and take all necessary precautions.
Weigh out 1 mg of purified TA in a 1.7 ml microcentrifuge tube.
Suspend the LTA in 100 μl of 48% hydrofluoric acid.
-
Incubate at room temperature for 20 h.
Note: Insert the microcentrifuge tube into a clearly labeled 50 ml centrifuge tube as a secondary container to prevent accidental contact.
Set-up a closed vessel drying apparatus with a filtered airstream (see Figure 2).
-
Pass the air through a KOH trap and insert the outlet tubing into a saturated sodium carbonate solution.
Note: Other saturated base solutions can be used.
Insert the microcentrifuge tube into the closed vessel and evaporate it until there is no residual liquid using a low-velocity airstream.
Resuspend the dried sample in 200 μl of 5 mM ammonium bicarbonate.
Adjust to pH 7 with dilute ammonium hydroxide.
Freeze in a dry ice bath or at -80 °C.
Freeze-dry overnight or to completion.
Store at -20 °C.
Note: Depending on the side chain modifications, the HF incubation time should be adjusted to maximize TA monomerization while minimizing cleavage of side chains.
Electrospray ionization mass spectrometry (ESI-MS)
Note: Perform the mass spectrometry analysis on a Waters Q-TOF Premier quadrupole/time of flight mass spectrometer (Waters Corporation [Micromass, Ltd.], Manchester, UK) operated using MassLynx software version 4.1 (Waters).
Analyze the samples using direct injection analysis using a mobile phase consisting of 50% acetonitrile (LC-MS grade) and 50% aqueous 10 mM ammonium acetate with a flow rate of 0.15 ml/min.
Set the nitrogen drying temperature to 300 °C at a flow of 7 L/min with a capillary voltage of 2.8 kV.
Set the mass spectrometer to scan from 100 to 1,000 m/z in positive- and negative-ion modes using ESI.
Assign calculated masses corresponding to S. aureus LTA as exemplified in Figure 4.
Notes
Wall teichoic acid (WTA) precipitation for nuclear magnetic resonance
If resuspending the pellet becomes difficult, physically scrape the pale/white colored pellet with a pipet tip and vortex to ease resuspension.
The trypsin digest is necessary to remove any cell-wall bound proteins.
NaOH hydrolysis causes the loss of D-alanine modifications on WTA (Figure 3). Alternatively, D-alanine modifications can be preserved by using TCA hydrolysis in 10 ml of 10% TCA (w/v) and incubating at 4 °C for 18 h with constant shaking or inversion.
The protocol typically yields around 5 mg of WTA but it can be strain and species dependent.
Lipoteichoic acid (LTA) purification by hydrophobic interaction chromatography
This protocol typically yields 5-15 mg of LTA and is sufficient quantity for NMR analysis but yields are dependent on the strain.
Even after breaking the membrane pellet apart and extensive vortex prior to 1-butanol extraction, it can still be somewhat chunky but you can proceed with the 1-butanol extraction. Freezing the pellet at -20 °C prior to resuspension can also help.
A control sample of buffer only should be included for ashing assay as many standard laboratory containers will leach and contribute to background absorbance.
It is important to dialyze the LTA extensively to minimize interference from residual citrate signals in NMR and ESI-MS.
1H NMR and ESI-MS analysis of teichoic acids
The Bruker Avance III 600 is routinely used but higher/lower field strength instruments can be used as well.
Depending on the TA concentration and field strength, the number of scans may need to be modified.
Water signal suppression pulse programs should be used with caution if peaks (such as anomeric carbohydrate proton signals) are anticipated.
For initial experiments, an NMR standard such as DSS should ideally be used as the chemical shift of water can vary. The standard can be externally referenced in a coaxial insert to maintain sample purity for downstream applications.
TA can be retrieved after NMR and used for other applications such as mass spectrometry or freeze-dried for long-term storage.
If multiple downstream applications are planned, it is easier to first aliquot into working amounts and then freeze-dry.
Recipes
Wall teichoic acid (WTA) precipitation for nuclear magnetic resonance
-
10x phosphate buffered saline
-
Weigh out the following:
1.44 g of Na2HPO4 (MW = 141.96 g/mol)
8 g of NaCl (MW = 58.44 g/mol)
2 g of KCl (MW = 74.55 g/mol)
0.24 g of KH2PO4 (MW = 136.09 g/mol)
Dissolve in 100 ml of MilliQ dH2O
Store at room temperature
-
-
1x Phosphate buffered saline (PBS)
Dilute 10 ml of 10x PBS into 100 ml with MilliQ dH2O
Store at room temperature
-
2 M NaCl
Weigh out 23.376 g NaCl (MW = 58.44 g/mol)
Dissolve in 200 ml of MilliQ dH2O
Store at room temperature
-
1 M Tris-HCl, pH 7.0
Weigh out 24.228 g Tris (MW = 121.14 g/mol)
Dissolve in 180 ml of MilliQ dH2O
Adjust to pH 7.0
Top to 200 ml with MilliQ dH2O
Store at room temperature
-
1 M Tris-HCl, pH 7.0 containing 1 M NaCl
Weigh out 24.228 g Tris (MW = 121.14 g/mol)
Weigh out 11.688 g NaCl (MW = 58.44 g/mol)
Dissolve in 170 ml of MilliQ dH2O
Adjust to pH 7.0
Top to 200 ml with MilliQ dH2O
Store at room temperature
-
0.5% SDS
Weigh out 0.5 g of SDS (MW = 288.372 g/mol)
Dissolve in 100 ml of MilliQ dH2O
Store at room temperature
-
10% trichloroacetic acid (w/v)
Weigh out 2 g of TCA (MW = 163.38 g/mol)
Dissolve in 20 ml of MilliQ dH2O
Store at 4 °C
-
3 M NaOAc, pH 5.1
Weigh out 40.824 g sodium acetate trihydrate (MW = 136.08 g/mol)
Dissolve in 80 ml of MilliQ dH2O
Adjust to pH 5.1 with glacial acetic acid
Top to 100 ml with MilliQ dH2O
Store at room temperature
-
Trypsin solution (0.2 mg/ml trypsin in 15 mM Tris-HCl, pH 7.0)
Make this solution fresh
Dilute 300 μl of 1 M Tris-HCl, pH 7.0 into 20 ml with MilliQ dH2O
Adjust to pH 7.0 if necessary
Dissolve 4 mg of trypsin
Lipoteichoic acid (LTA) purification by hydrophobic interaction chromatography
-
20x citrate buffer (1 M sodium citrate, pH 4.7)
Weigh out 21.014 g of citric acid monohydrate (MW = 210.14 g/mol)
Dissolve in 70 ml of MilliQ dH2O
Adjust to pH 4.7 with sodium hydroxide pellets
Top with MilliQ dH2O to 100 ml
Store at room temperature
-
Resuspension buffer (50 mM sodium citrate, pH 4.7)
Dilute 50 ml of 20x citrate buffer into 1 L with MilliQ dH2O
Adjust to pH 4.7 if necessary
Store at room temperature
-
Equilibration buffer (50 mM sodium citrate, pH 4.7 with 15% (v/v) 1-propanol)
Dilute 25 ml of 20x citrate buffer into 425 ml with MilliQ dH2O
Adjust to pH 4.7 if necessary
Add 75 ml of 1-propanol
Store at room temperature
-
Elution buffer [50 mM sodium citrate, pH 4.7 with 65% (v/v) 1-propanol]
Dilute 25 ml of 20x citrate buffer into 175 ml with MilliQ dH2O
Adjust to pH 4.7 if necessary
Add 325 ml of 1-propanol
Store at room temperature
-
Ashing solution (2 M H2SO4, 0.44 M HClO4)
Make this solution fresh
Measure 8.5 ml of MilliQ dH2O
Add 1.11 ml of sulfuric acid (MW = 98.08 g/mol)
Add 0.376 ml of 70% perchloric acid (MW = 100.46 g/mol)
-
Reducing solution (3 mM ammonium molybdate, 0.25 M sodium acetate, 1% ascorbic acid)
Make this solution fresh
-
Weigh out the following:
185.38 mg of ammonium molybdate tetrahydrate (MW = 1235.86)
1.701 g of sodium acetate trihydrate (MW = 136.08)
0.5 g of L-ascorbic acid sodium salt (MW = 198.11)
Dissolve in 50 ml of MilliQ dH2O
-
0.1 M phosphate buffer
Weigh out 1.56 g of sodium phosphate monobasic dihydrate (MW = 156.01 g/mol)
Dissolve in 100 ml of MilliQ dH2O
-
Phosphate standards (6 concentrations; 8 mM to 0.25 mM)
Add 80 μl of 0.1 M phosphate buffer to 920 μl of MilliQ dH2O for 8 mM phosphate
Serially dilute 2-fold in MilliQ dH2O until 0.25 mM phosphate
Note: Other inorganic phosphate can be used as a standard.
1H NMR and ESI-MS analysis of teichoic acids
-
Saturated sodium carbonate solution
Gradually stir sodium carbonate (MW = 105.99) into 100 ml of MilliQ dH2O until it is saturated and stops dissolving
-
1 M ammonium bicarbonate
Dissolve 1.58 g of ammonium bicarbonate (MW = 79.06) in 20 ml of MilliQ dH2O
-
5 mM ammonium bicarbonate
Dilute 250 μl of 1 M ammonium bicarbonate into 50 ml with MilliQ dH2O
-
Dilute ammonium hydroxide
Dilute 100 μl of ammonium hydroxide solution into 1 ml with MilliQ dH2O
Acknowledgments
The protocol for crude WTA extraction for PAGE was adapted from Meredith et al. (2008) and Kho and Meredith (2018). The protocol for crude lipoteichoic acid extraction for PAGE was adapted from Kho and Meredith (2018). The protocol for TA-PAGE and staining was adapted from Wolters et al. (1990), Pollack and Neuhaus (1994), Meredith et al. (2008) and Kho and Meredith (2018). The protocol for WTA precipitation for NMR was adapted from Bernal et al. (2009). The LTA hydrophobic interaction chromatography protocol was adapted from Morath et al. (2001), Draing et al. (2006), Gründling and Schneewind (2014) and Kho and Meredith (2018). The phosphate content analysis protocol was adapted from Draing et al. (2006) and Kho and Meredith (2018). NMR analysis of S. aureus LTA was adapted from Morath et al. (2001) and Kho and Meredith (2018). The protocol for TA dephosphorylation and ESI-MS was adapted from Eugster et al. (2011), Shen et al. (2017) and Kho and Meredith (2018).
Funding source: Funding was provided by the Eberly College of Science at the Pennsylvania State University.
Competing interests
The authors declare no conflicts of interest or competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Bernal P., Zloh M. and Taylor P. W.(2009). Disruption of D-alanyl esterification of Staphylococcus aureus cell wall teichoic acid by the β-lactam resistance modifier(-)-epicatechin gallate . J Antimicrob Chemother 63(6): 1156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown S., Santa Maria J. P. Jr.and Walker S.(2013). Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67: 313-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Draing C., Pfitzenmaier M., Zummo S., Mancuso G., Geyer A., Hartung T. and von Aulock S.(2006). Comparison of lipoteichoic acid from different serotypes of Streptococcus pneumoniae . J Biol Chem 281(45): 33849-33859. [DOI] [PubMed] [Google Scholar]
- 4. Eugster M. R. and Loessner M. J.(2011). Rapid analysis of Listeria monocytogenes cell wall teichoic acid carbohydrates by ESI-MS/MS . PLoS One 6(6): e21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer W., Behr T., Hartmann R., Peter-Katalinic J. and Egge H.(1993). Teichoic acid and lipoteichoic acid of Streptococcus pneumoniae possess identical chain structures. A reinvestigation of teichoid acid(C polysaccharide) . Eur J Biochem 215(3): 851-857. [DOI] [PubMed] [Google Scholar]
- 6. Gründling A. and Schneewind O.(2007). Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus . J Bacteriol 189(6): 2521-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kho K. and Meredith T. C.(2018). Salt-induced stress stimulates a lipoteichoic acid-specific three component glycosylation system in Staphylococcus aureus . J Bacteriol. DOI: 10.1128/JB.00017-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim J. H., Seo H., Han S. H., Lin J., Park M. K., Sorensen U. B. and Nahm M. H.(2005). Monoacyl lipoteichoic acid from pneumococci stimulates human cells but not mouse cells. Infect Immun 73(2): 834-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kovács M., Halfmann A., Fedtke I., Heintz M., Peschel A., Vollmer W., Hakenbeck R. and Bruckner R.(2006). A functional dlt operon, encoding proteins required for incorporation of D-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol 188(16): 5797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mechler L., Bonetti E. J., Reichert S., Flotenmeyer M., Schrenzel J., Bertram R., Francois P. and Gotz F.(2016). Daptomycin tolerance in the Staphylococcus aureus pitA6 mutant is due to upregulation of the dlt operon . Antimicrob Agents Chemother 60(5): 2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meredith T. C., Swoboda J. G. and Walker S.(2008). Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus . J Bacteriol 190(8): 3046-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mistou M. Y., Sutcliffe I. C. and van Sorge N. M.(2016). Bacterial glycobiology: rhamnose-containing cell wall polysaccharides in Gram-positive bacteria. FEMS Microbiol Rev 40(4): 464-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morath S., Geyer A. and Hartung T.(2001). Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus . J Exp Med 193(3): 393-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Percy M. G. and Gründling A.(2014). Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68: 81-100. [DOI] [PubMed] [Google Scholar]
- 15. Pollack J. H. and Neuhaus F. C.(1994). Changes in wall teichoic acid during the rod-sphere transition of Bacillus subtilis 168 . J Bacteriol 176(23): 7252-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rismondo J., Percy M. G. and Grundling A.(2018). Discovery of genes required for lipoteichoic acid glycosylation predicts two distinct mechanisms for wall teichoic acid glycosylation. J Biol Chem 293(9): 3293-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen Y., Boulos S., Sumrall E., Gerber B., Julian-Rodero A., Eugster M. R., Fieseler L., Nystrom L., Ebert M. O. and Loessner M. J.(2017). Structural and functional diversity in Listeria cell wall teichoic acids. J Biol Chem 292(43): 17832-17844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silhavy T. J., Kahne D. and Walker S.(2010). The bacterial cell envelope. Cold Spring Harb Perspect Biol 2(5): a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swoboda J. G., Campbell J., Meredith T. C. and Walker S.(2010). Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 11(1): 35-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weidenmaier C. and Peschel A.(2008). Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6(4): 276-287. [DOI] [PubMed] [Google Scholar]
- 21. Winstel V., Xia G. and Peschel A.(2014). Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus . Int J Med Microbiol 304(3-4): 215-221. [DOI] [PubMed] [Google Scholar]
- 22. Wolters P. J., Hildebrandt K. M., Dickie J. P. and Anderson J. S.(1990). Polymer length of teichuronic acid released from cell walls of Micrococcus luteus . J Bacteriol 172(9): 5154-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wörmann M. E., Corrigan R. M., Simpson P. J., Matthews S. J. and Gründling A.(2011). Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes . Mol Microbiol 79(3): 566-583. [DOI] [PMC free article] [PubMed] [Google Scholar]