Abstract
Tissue clearing techniques are useful for large-scale three-dimensional fluorescence imaging of thick tissues. However, high-resolution imaging deep inside tissues has been challenging, as it is extremely sensitive to light scattering and spherical aberrations. Here, we present a water-based optical clearing and mounting media, SeeDB2, which is designed for high numerical aperture (NA) objective lenses with oil or glycerol immersion. Using quick and simple soaking procedures, the refractive indices of samples can be matched either to that of immersion oil (1.52) or glycerol (1.46), thus minimizing light scattering and spherical aberrations. Fine morphology and various fluorescent proteins are highly preserved during the clearing and imaging process. Our method is useful for the three-dimensional fluorescence imaging of neuronal circuitry at synaptic resolution using confocal and super-resolution microscopy. SeeDB2 is also useful as a mounting media for the super-resolution imaging of fluorescent proteins.
Keywords: Tissue clearing, Fluorescence imaging, Confocal imaging, Super-resolution imaging, Connectome, SeeDB2
Background
Biological tissues are organized in 3D. In addition, many of important cellular machineries, e.g., synapses in neurons, are at sub-micron scale. Therefore, there have been increasing demands for a method for sub-micron-scale 3D imaging. Serial electron microscopy techniques (e.g., FIB-SEM or SBF-SEM) are promising, but they cannot make the best use of the genetic fluorescent labeling tools available in modern life science. To facilitate 3D imaging with fluorescence microscopy, a number of tissue clearing techniques have been developed in recent years (Richardson and Lichtman, 2015 and 2017). They are designed for large-scale 3D imaging, and some of them allow for whole-brain, and even whole-body-scale fluorescence imaging of fixed samples combined with confocal, two-photon, or light-sheet microscopy. However, many of them have not been fully optimized for high-resolution imaging.
In fluorescence microscopy, lateral resolution (d) is given as:d = 0.61λ/NA where λ is the wavelength of the light and NA represents the numerical aperture. The resolution improves as d decreases. Therefore, we need to use high NA objective lenses for high-resolution imaging.
NA is defined as:
NA = n sinα
where n is the refractive index (RI) of the immersion media, and α is the half angular aperture. Therefore, many of the high NA objective lenses are designed for oil (RI = 1.52) or glycerol (RI = 1.46) immersion for the best resolution. Previously, high NA objective lenses are intended to image thin sections or just the surface of samples. However, if we try to image deeper in the samples with these objective lenses, image quality will be easily impaired due to “spherical aberrations”. As RI of tissue samples are lower than that of immersion oil (RI = 1.52) and a glass coverslip (RI ~1.52), the excitation light will refract at the interface between the coverslip and samples, and it will no longer converge onto a small focal spot. This is known as spherical aberrations, reducing resolution and brightness in microscopy.
To minimize spherical aberrations, index matching of samples is crucial. However, many of the existing mounting media and clearing solutions have low RI, ranging from 1.33 (water) to 1.46 (glycerol-based mounting media). Even our previous clearing agent, SeeDB (RI = 1.49), did not reach RI 1.52 ( Ke et al., 2013 , 2014). 2,2'-thiodiethanol (TDE, RI = 1.52) has been previously proposed for index matching for oil-immersion objective lenses and has been widely used for synthetic fluorescent dyes ( Staudt et al., 2007 ). However, a major drawback of TDE is that most of fluorescent proteins are totally quenched in TDE. To overcome this limitation, we developed a new tissue clearing agent, SeeDB2S, that has a high refractive index (RI = 1.52), but also highly preserves fluorescent proteins ( Ke et al., 2016 ). We also formulated SeeDB2G (RI = 1.46) for glycerol-immersion objective lenses. As fluorescent proteins are better preserved than in commercialized mounting media, SeeDB2G/S are also useful as mounting media for high-resolution imaging. SeeDB2G/S is particularly powerful for high-resolution confocal microscopy and super-resolution microscopy of fluorescent proteins in tissues, sections, and cells.
Materials and Reagents
1.5 ml Eppendorf tube (Eppendorf, for 1 ml solution)
5 ml Eppendorf tube (Eppendorf, catalog number: 0030119401; optional for 3 ml solution, for thick brain slices or whole-mount samples)
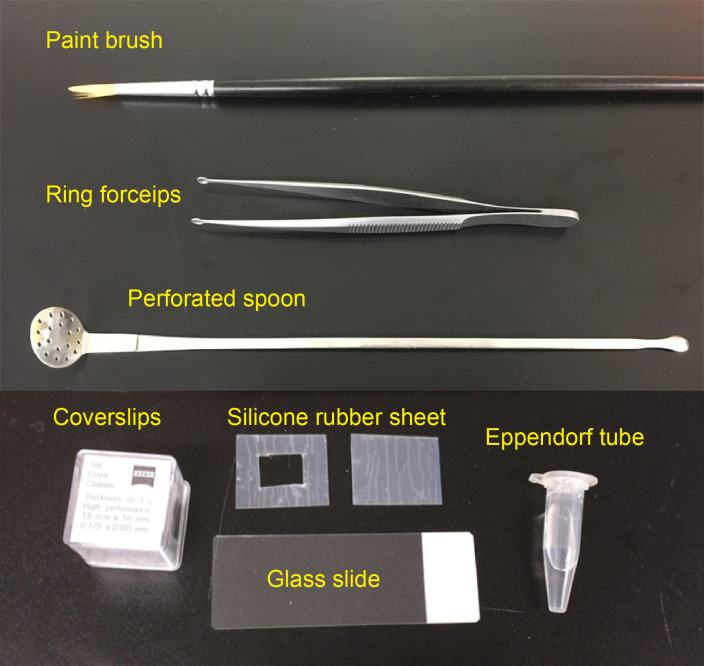
Paint brush (for thin brain slices, No. 1-2, see Figure 1)
-
Silicone rubber sheet (translucent, 0.2 mm thick; e.g., AS ONE, catalog number: 6-9085-13; see Figure 1)
Note: Various thickness of silicone rubber sheets are available from Togawa Rubber (AS ONE), Professional Plastics, CS Hyde, etc., ranging from 0.1 mm to 8.0 mm. The thickness should match that of brain slices.
Glass slide (76 mm x 26 mm; MATSUNAMI Glass; Figure 1)
-
Coverslips (18 x 18 mm, No 1.5H; e.g., Paul Marienfeld, catalog number: 0107032 or ZEISS, catalog number: 474030-9000-000; Figure 1)
Note: No 1.5H (170 ± 5 μm thick) is highly recommended for super-resolution imaging.
Sodium chloride (NaCl)
Sodium hydrogen phosphate dodecahydrate (Na2HPO4·12H2O)
Potassium chloride (KCl)
Potassium dihydrogen phosphate (KH2PO4)
4% paraformaldehyde (PFA; e.g., NACALAI TESQUE, catalog number: 26126-25) in PBS
-
20% Saponin (NACALAI TESQUE, catalog number: 30502-42) in ddH2O with filter sterilization
Note: Different vendors prepare saponin from different species of plants. We strongly recommend NACALAI TESQUE. Brownish lots (often found in Sigma-Aldrich) should be avoided.
Low-melting point agarose (e.g., Thermo Fisher Scientific, catalog number: 16520100)
Omnipaque 350 (e.g., DAIICHI SANKYO, Omnipaque 350 Injection, 50 ml in 1 bottle; also available from GE healthcare)
Histodenz (e.g., Sigma-Aldrich, catalog number: D2158)
Sodium azide (e.g., Sigma-Aldrich, catalog number: 13412)
1 M stock of Tris-HCl, pH 7.6 (e.g., NACALAI TESQUE, catalog number: 35436-01), used to prepare Tris-EDTA buffer
0.5 M stock of EDTA, pH 8.0 (e.g., Dojindo, catalog number: 347-07481), used to prepare Tris-EDTA buffer
-
Immersion media:
Glycerol (e.g., Type G Immersion Liquid, Leica Microsystems, catalog number: 11513910; Glycerine solution, Leica Microsystems, catalog number: 11513872) for SeeDB2G
Oil (Type F, Olympus, MOIL-30; also available from Leica, Zeiss, etc.) for SeeDB2S
(Optional) SeeDB2 Trial Kit (Wako Pure Chemical Industries, catalog number: 294-80701)
Phosphate-buffered saline (PBS) (see Recipes)
Tris-EDTA buffer, pH 7.6 (see Recipes)
Permeabilization solution (see Recipes)
Solution 1 (see Recipes)
Solution 2 (see Recipes)
-
SeeDB2 solutions (see Recipes)
SeeDB2G with saponin (clearing)
SeeDB2G (mounting)
SeeDB2S with saponin (clearing)
SeeDB2S (mounting)
Figure 1. Materials required for preparing imaging chamber and slice mounting.
Equipment
Perforated spoon (optional for handling thick brain slices, custom-ordered, flat head, head diameter = 15 mm, see Figure 1 and Video 1)
Ring forceps (optional for whole-mount samples, e.g., Natsume Seisakusho, NAPOX, catalog number: A-26, see Figure 1)
Vibratome (e.g., LinearSlicer, DOSAKA, model: PRO7N)
Seesaw shaker (e.g., Bio Craft, model: BC-700)
Rotator (e.g., TAITEC, model: RT-30mini, catalog number: 0057154-000)
-
Fluorescence microscope
Confocal microscope (e.g., Olympus, model: FV3000; Leica Microsystems, model: Leica TCS SP8; Nikon, model: A1+)
Super-resolution microscope (e.g., Zeiss, model: LSM 880 with Airyscan; Leica Microsystems, model: Leica TCS SP8 with HyVolution 2; Leica Microsystems, model: Leica TCS SP8 STED)
-
Objective lenses
Examples: 63x oil-immersion (NA = 1.4, WD = 0.14 mm) (Leica Microsystems, model: HC PL APO 63x/1.4 Oil CS2, catalog number: 15506350); 100x oil-immersion (NA = 1.4, WD = 0.13 mm) (Leica Microsystems, model: HC PL APO 100x/1.4 Oil CS2, catalog number: 15506325); 63x glycerol-immersion (NA = 1.3, WD = 0.28 mm) (Leica, model: HCX PL APO 63x/1.30 GLYC CORR, catalog number: 11506193); 63x oil-immersion (NA = 1.4, WD = 0.19 mm) (Carl Zeiss, model: Plan-Apochroamt 63x/1.40 Oil DIC, catalog number: 440762-9904-000)
Video 1. Imaging chamber preparation and sample mounting.

Cut a silicone rubber sheet (1 mm-thick and 0.2 mm-thick are shown in this video) into an appropriate size with scissors or a utility knife. A silicone rubber sheet will adhere to a glass slide, by removing air between the rubber sheet and the glass slide. Mount cleared samples (cleared samples are often difficult to find) using a perforated spoon and a paint brush. Use No. 1.5H coverslips to seal the samples.
Procedure
-
Choice of optical clearing protocol
SeeDB2G (RI = 1.46): SeeDB2G is optimized for glycerol-immersion lenses to achieve the highest resolution, but water- and multi-immersion objective lenses are also useful with reasonable resolution. SeeDB2G is useful for larger tissues due to lower viscosity.
SeeDB2S (RI = 1.52): SeeDB2S is optimized for high-NA oil-immersion objective lenses. Since SeeDB2S is viscous and oil-immersion lenses typically have a short W.D. (0.1-0.2 mm), we recommend using relatively thin samples (< 300 μm thick slices) for SeeDB2S.
In this protocol, we describe the protocol for high-resolution/super-resolution imaging of mouse brain slices. However, SeeDB2 is also useful for high-resolution imaging of other organisms (e.g., Drosophila), other organs and sample types (mouse oocytes), cultured cells, and frozen sections. See the original paper ( Ke et al., 2016 ) and SeeDB Resources for details.
Note: SeeDB Resources (https://sites.google.com/site/seedbresources/) provide updated information from the authors, see Ke et al., 2014 .
-
Choice of objective lenses and slice thickness
Because of the working distance limitations in high-NA lenses, only one-fourth to half of the depth accessible by 20x lens can be reached by 63x or 100x lenses. Be sure to select neurons located close to the surface. See Table 1 showing examples of objective lenses used for confocal imaging.
+, optimal; ±, non-optimal; -, not suitable. Data based on Leica and Zeiss objective lenses.
-
SeeDB2G (Figure 2)
Dissect the mouse brain after an intracardiac perfusion of 4% PFA in PBS. Post-fix the brain sample in 4% PFA at 4 °C with gentle shaking overnight.
Wash the sample in PBS three times with gentle shaking (10 min each).
Embed the sample in 4% low-melting point agarose/PBS with desired orientation and then cut the brain with a vibratome.
-
Transfer the sample into the permeabilization solution and incubate with shaking overnight (> 16 h) at 4 °C. Antibody staining and counterstaining should be performed prior to the clearing process.
Optional: Perform antibody staining in 3 ml using 5 ml Eppendorf tubes on a rotator. After blocking with blocking buffer (0.5% skim milk, 0.25% fish gelatin, 2% saponin in PBS) for 24 h at 4 °C, incubate samples with primary antibodies in washing buffer (2% saponin in PBS) for 24 h. After three washes with washing buffer, incubate samples with secondary antibodies and DAPI for 12-16 h. Wash samples three times (2 h each) in washing buffer.
Note: We recommend 2% saponin for thick mouse brain samples; however, for the thinner slices, lower concentrations of Triton X-100 are also workable. Antibodies are able to penetrate up to 200-300 μm in depth.
Transfer the sample into a new tube filled with solution 1 and place it on a rotator. Incubate for 6-10 h for whole-mount samples, 2-4 h for slice samples (200-500 μm).
Transfer the sample into a new tube filled with solution 2 and place it on a rotator. Incubate for 6-10 h for whole-mount samples, 2-4 h for slice samples (200-500 μm).
-
Transfer the sample into a new tube filled with SeeDB2G with saponin and place it on a rotator. Incubate for 6-10 h for whole-mount samples, 2-4 h for slice samples (200-500 μm). An example of transmission images is shown in Figure 4.
Notes:
Do not store samples in solutions containing saponin. For thin slice or neonate samples, prolonged incubation (> 24 h) in solutions containing saponin may cause damage to the sample.
Store the sample in SeeDB2G (without saponin) till mounting and imaging.SeeDB2-cleared samples can be stored in Eppendorf at 4 °C for up to 6 months. Sodium azide (0.05%) should be added for the long-term storage.
-
SeeDB2S (Figure 2)
Dissect the mouse brain after intracardiac perfusion of 4% PFA in PBS. Post-fix the brain sample in 4% PFA at 4 °C with gentle shaking overnight.
Wash the sample in PBS three times with gentle shaking (10 min each).
Embed the sample in 4% low-melting point agarose/PBS with desired orientation and then cut the brain with a vibratome.
-
Transfer the sample into permeabilization solution and incubate with shaking overnight (> 16 h) at 4 °C. Antibody staining and counterstaining should be performed prior to the clearing process.
Note: We recommend 2% saponin for thick mouse brain samples; however, for the thinner slices, lower concentrations of Triton X-100 are also workable. Antibodies penetrate up to 200-300 μm depth.
Transfer the sample into a new tube filled with solution 1 and place it on a rotator. Incubate for 2-4 h for slice samples (200-500 μm).
Transfer the sample into a new tube filled with solution 2 and place it on a rotator. Incubate for 2-4 h for slice samples (200-500 μm).
Transfer the sample into a new tube filled with SeeDB2G with saponin and place it on a rotator. Incubate for 2-4 h for slice samples (200-500 μm).
Transfer the sample into a new tube filled with SeeDB2S with saponin and place it on a rotator. Incubate for 2-4 h for slice samples (200-500 μm).
-
Transfer the sample into a new tube filled with SeeDB2S (without saponin, 0.01% sodium azide can be added for long-term storage) for mounting. An example of transmission images are shown in Figure 4.
Note: Do not store samples in solutions containing saponin. For thin slice or neonate samples, prolonged incubation (> 24 h) in solutions containing saponin may cause damages to the sample.
-
Imaging chamber preparation and sample mounting (Video 1 and Figure 3)
-
(Optional) Stand the sample in SeeDB2G/S for 2-4 h to remove the air bubbles.
Note: If air bubbles are heavily accumulated in the tube, transfer the sample to a new tube filled with SeeDB2G/S and incubate it for 2-4 h.
Cut the silicone rubber sheet into appropriate size using scissors or a utility knife.
Remove the protection sheets (stuck to both sides, only found in 0.2 mm silicone sheet) and press the sheet onto the glass slide. Push the rubber sheet to remove air bubbles between the glass slide and the rubber sheet.
-
Leave a drop of SeeDB2G/S (from the storage tube, not a fresh one) into the chamber, and pick up the brain slice with a perforated spoon and a paint brush to mount the sample.
Note: Do not place the brain slices onto a dry surface. Do not expose SeeDB2S solution or SeeDB2S-cleared samples to the air for long time (> 5 min).
(Optional) Remove the air bubbles in the imaging chamber with a paint brush.
-
Gently press the cover glass onto the imaging chamber and use the spillover of SeeDB2G/S to seal the cover glass. See Figure 3 for mounted samples on a glass slide.
Note: Do not use nail polish to seal the sample. The excess amount of SeeDB2G/S will form a tight film after air drying.
Use an appropriate immersion media for high-resolution fluorescence imaging. Use glycerol (Refractive index = 1.46) for SeeDB2G, and oil (Refractive index = 1.52; Type F) for SeeDB2S. SeeDB2G/S cannot be used as an immersion media. Figure 5 shows an example of confocal images. Figure 6 shows super-resolution images (Airyscan and STED).
-
Anticipated Results (Figure 4)
Table 1. Examples of objective lenses.
| Objective lens | 10x air | 20x multi | 40x oil | 63x glycerol | 63x oil | 100x oil |
| N.A. | 0.4 | 0.75 | 1.3-1.4 | 1.3 | 1.4 | 1.4-1.46 |
| W.D. (mm) | 2.2-3.1 | 0.68 | 0.21-0.24 | 0.3 | 0.14-0.19 | 0.11-0.13 |
| Immersion | - | Water/glycerol/oil | Oil | Glycerol | Oil | Oil |
| Lateral resolution | 2 μm | 0.5 μm | 0.3 μm | 0.25 μm | 0.21 μm | 0.2 μm |
| Confocal | + | + | + | + | + | + |
| Super-resolution | - | - | ± | ± | + | + |
| SeeDB2G | ± | + | ± | + | ± | ± |
| SeeDB2S | ± | ± | + | ± | + | + |
| Suitable sample thickness | < 2 mm | < 1 mm | < 500 μm | < 500 μm | < 300 μm | < 200 μm |
Figure 2. Graphic protocol for SeeDB2.
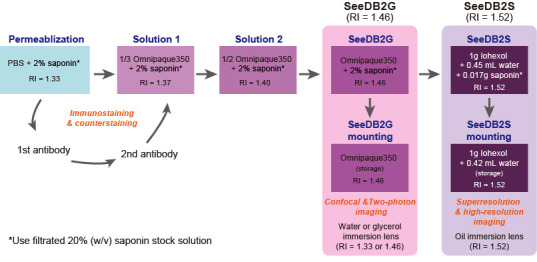
Figure 4. Transmission images of 500 μm adult mouse brain slices after optical clearing with SeeDB2G and SeeDB2S.

( Ke et al., 2016 ). Grids are 2.6 x 3.2 mm.
Figure 3. Slice sample ready for imaging.
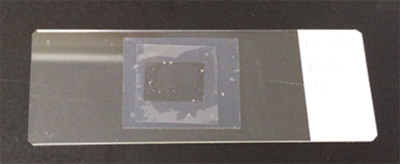
A 220 μm brain slice was mounted within the imaging chamber. A silicone rubber sheet (0.2 mm thick) was used for the spacer, and a coverslip is placed to seal the sample. The silicone rubber sheet will adhere to the glass slide and coverslip without adhesive. The spill-over of the SeeDB2G/S will form a tight film after air drying.
Figure 5. Confocal imaging of adult brain slices (Thy1-YFP-H mouse).
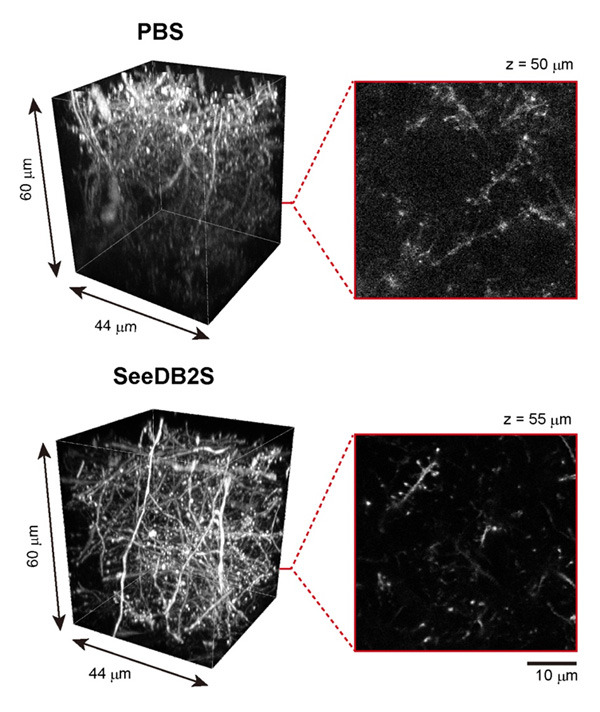
Adult brain slices (220 μm thick) were imaged with 63x oil-immersion lens under confocal microscopy (Leica TCS SP8). In the PBS sample (uncleared), due to light scattering and spherical aberrations, the resolution and fluorescence signals intensity were dampened. The highest resolution was maintained throughout all depths when cleared with SeeDB2S.
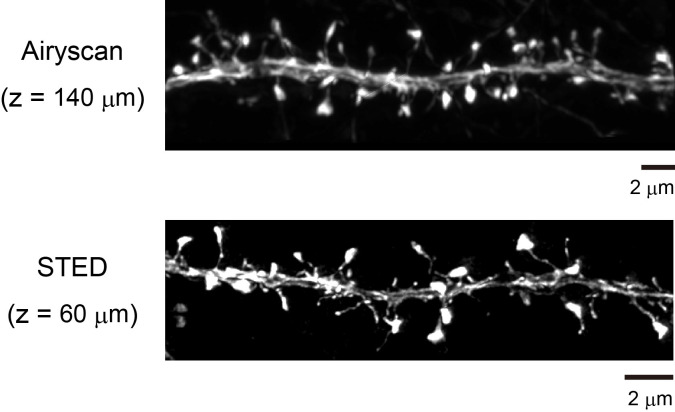
Figure 6. Super-resolution imaging of adult brain slices (Thy1-YFP-H mouse).
Due to the minimized spherical aberrations, SeeDB2S is also useful for various kinds of super-resolution microscopy, such as STED (Leica), Airyscan (Zeiss), HyVolution2 (Leica), and SpinSR (Olympus). For large-scale super-resolution imaging, we first imaged with a 20x objective lens. Then, we focused on a particular neuron and obtained super-resolution images along the target neuron to save imaging time and data volume. We used Neurolucida (MBF Bioscience) for the quantification and reconstruction of neuronal morphology. Images adapted from Ke et al. (2016) .
Data analysis
Data analysis procedures have been described in the original publication ( Ke et al., 2016 ). No statistical analysis was performed in this protocol paper. We used Neurolucida (MBF Bioscience) for the 3D rendering in Figure 5.
Notes
-
The saponin solution is brownish
We strongly recommend saponin from NACALAI TESQUE. Different vendors prepare saponin from different species of plants. Brownish lots should be avoided. Alternatively, a low concentration of Triton X-100 can be used.
-
Histodenz does not dissolve completely
Prepare ~10 ml SeeDB2S solution in a 50 ml conical tube to prevent insufficient mixing of iohexol power and Tris buffer. Use rotator for gentle mixing.
-
White or yellow aggregations in SeeDB2 solutions
To avoid bacterial contamination, filter and sterilize the 20% saponin stock solution. Add 0.01%-0.1% of sodium azide to each clearing solution and store the solutions containing saponin at 4 °C.
-
Water evaporation
The surface of SeeDB2 forms a film as the water evaporates from the surface. SeeDB2 should be sealed in the imaging chamber during imaging.
-
Sealing samples in the imaging chambers/glass slides
Nail polish should not be used to seal the samples. For thick samples, we routinely use a silicone rubber. To seal samples on glass slides, we use an excess amount of SeeDB2, which then forms a tight film after water evaporation.
-
Samples are floating within SeeDB2
To avoid movement artifacts during imaging, we prepared brain slices that are slightly thicker than the thickness of the spacer (translucent silicone rubber sheet). For example, we prepared 220 μm brain slices when we used 200 μm-thick rubber sheets. For small samples, it is also useful to prepare SeeDB2 with 0.1% low-melting agarose to immobilize the sample; we used low-melting agarose to image mouse oocyte samples. Embedding small samples in agarose may also be useful to avoid the loss of samples during clearing, as it is often difficult to find invisible samples from clearing solution.
-
Movement artifacts
Because SeeDB2 allows for imaging up to the limit of the working distance (W.D.) of the objective lens, glass slides are often pushed up by the objective lenses as you get closer to the upper limit, which leads to movement artifacts. We recommend using a clamp or placing a weight on the glass slide in order to avoid the movement of samples during imaging.
-
Detecting sample surface
Because the refractive index of SeeDB2 is the same as that of coverslips, it is difficult to determine the surface of the sample when you perform the fluorescence imaging. Make sure to attach the sample to the coverslip (see Note 6).
-
Photo-bleaching
Various fluorescent proteins (e.g., TagBFP2, ECFP, mTurquoise2, EGFP, EYFP, CyOFP, tdTomato, tdKatushka2) are very stable and resistant to photo-bleaching in SeeDB2 solution. Indeed, SeeDB2 performs much better than commercialized mounting media for fluorescent proteins. However, some Alexa dyes and DAPI are easily photo-bleached in SeeDB2. In these situations, we suggest 2,2’-Thiodiethanol (TDE, refractive index up to 1.52) as an alternative for chemical dyes ( Staudt et al., 2007 ), however, fluorescent proteins are quenched in TDE.
Recipes
-
Phosphate-buffered saline (PBS)
80 g NaCl
29 g Na2HPO4·12H2O
2 g KCl
2 g KH2PO4
Add ddH2O to prepare 1 L PBS
-
Tris-EDTA buffer, pH 7.6
10 mM Tris-HCl
0.25 mM EDTA
-
Permeabilization solution
100 μl 20% saponin solution + 900 μl PBS
-
Solution 1
100 μl 20% saponin solution + 333 μl Omnipaque350 + 567 μl ddH2O
-
Solution 2
100 μl 20% saponin solution + 500 μl Omnipaque350 + 400 μl ddH2O
-
SeeDB2 solutions
Notes:
Tris-EDTA buffer pH 7.6 (Recipe 2) is used to prepare SeeDB2S from Histodenz powder. Do not use PBS. Phosphate buffer will generate white precipitates after long-term storage and impair imaging quality.
The solution of 0.01% sodium azide can be included in the buffer for long-term preservation. Do not store clearing solution for long time.
-
Pre-made SeeDB2 solutions are also commercialized from FUJIFILM Wako Pure Chemical.
Solutions Composition SeeDB2G with saponin (clearing) 2% saponin in Omnipaque350 SeeDB2G (mounting) Use Omnipaque350 directlyOr 7,500 μl Tris-EDTA (pH 7.6) + 10 g HistodenzSeeDB2S with saponin (clearing) 3,780 μl Tris-EDTA (pH 7.6) + 420 μl 20% saponin solution + 10 g Histodenz SeeDB2S (mounting) 4,200 μl Tris-EDTA (pH 7.6) + 10 g HistodenzAdd 4,200 μl Tris-EDTA to a 50 ml conical tube, and then add 10 g Histodenz. Use a rotator to fully dissolve the Histodenz in the conical tube
Acknowledgments
This protocol was adapted from our original publication of the protocol ( Ke et al., 2016 ). We thank J.R. Sanes for providing the Thy1-YFP-H mouse line, R. Sakaguchi for help with video, and M. Leiwe for proofreading. This work was supported by grants from the PRESTO program of the Japan Science and Technology Agency (JST), the Brain/MINDS project of the Japan Agency for Medical Research and Development (AMED), the Mitsubishi Foundation, the Strategic Programs for R&D (President’s Discretionary Fund) of RIKEN, the JSPS KAKENHI (grant numbers 23680038, 15K14336, 16K14568, 16H06456, 17H06261), and RIKEN CDB intramural grant. M.-T.K. was supported by RIKEN Foreign Postdoctoral Researcher program. The imaging experiments were supported by the RIKEN Kobe Light Microscopy Facility. Animal experiments were supported by Laboratory for Animal Resources and Genetic Engineering (LARGE) at the RIKEN Center for Life Science Technologies.
Competing interests
M.-T.K. and T.I. have filed a patent application on SeeDB2, assigned to RIKEN.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Ke M. T., Fujimoto S. and Imai T.(2013). SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16(8): 1154-1161. [DOI] [PubMed] [Google Scholar]
- 2. Ke M. T., Fujimoto S. and Imai T.(2014). Optical clearing using SeeDB. Bio-protocol 4(3): e1042. [Google Scholar]
- 3. Ke M. T., Nakai Y., Fujimoto S., Takayama R., Yoshida S., Kitajima T. S., Sato M. and Imai T.(2016). Super-resolution mapping of neuronal circuitry with an index-optimized clearing agent. Cell Rep 14(11): 2718-2732. [DOI] [PubMed] [Google Scholar]
- 4. Richardson D. S. and Lichtman J. W.(2015). Clarifying tissue clearing. Cell 162(2):246-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Richardson D. S. and Lichtman J. W.(2017). SnapShot: tissue clearing. Cell 171(2): 496–496..e1. [DOI] [PubMed] [Google Scholar]
- 6. Staudt T., Lang M. C., Medda R., Engelhardt J., Hell S. W.(2007). 2,2'-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc Res Tech. 70(1):1-9. [DOI] [PubMed] [Google Scholar]