Abstract
Pyrene Fluorescent Assay is established to monitor the dynamic actin nucleation, elongation, capping and disassembly in vitro. This technique provides an easy handle procedure and straightforward visual data analysis. By coupling actin purification and polymerization assays in this protocol, the readers could quickly get the affordable and straightforward assays to study actin dynamics.
Keywords: Actin Purification, Pyrene Assay, Actin Polymerization, Actin Nucleation, Formin
Background
Actin dynamics play essential roles in crucial aspects of cellular biological functions, such as cell motility, endosomal trafficking, scaffolding and responses to external stimuli (Pollard and Cooper, 2009; Weinberg and Drubin, 2012; Sun et al., 2018 ). Rearrangements of actin cytoskeleton including nucleation, elongation, stabilization, capping, crosslinking and depolymerization are tightly controlled by actin-binding-proteins (ABPs) (Pollard, 2016). In vitro studies of the biochemical functions of those ABPs in actin regulation provides insights in understanding the mechanism of actin network regulation. Here, we provide an easy understanding protocol which can be applied for actin cytoskeleton-related biochemistry experiments, including actin purification, pyrene actin assay. The protocol provides approaches to monitor the actin assembly with other actin-binding-proteins in vitro and help researchers to learn about the actin dynamics process under different conditions. This protocol provides a clear table template for users to prepare a reaction mixture. Complicated calculation steps are simplified. This protocol uses high throughput multi-well methods instead of cuvette measurements to provide higher throughput and good parallel comparison among samples.
Materials and Reagents
Pipette tips
SnakeSkinTM Dialysis Tubing, 10K MWCO, 22 mm (Thermo Fisher, catalog number: 68100)
Corning® Costar® 96-Well Cell Culture Plates (Sigma, catalog number: CLS3595)
Greiner 96-well plates, polypropylene, black polypropylene wells flat bottom (Sigma, catalog number: M9685)
HiPrepTM 16/60 Sephacryl S-300 HR (GE Healthcare, catalog number: 17116701)
Nalgene Oak Ridge High-Speed PPCO Centrifuge Tubes (Thermo Scientific, catalog number: 3119-0050)
Thinwall Polypropylene Tube (Beckman Coulter, catalog number: 326819)
Bottle Assembly, Polycarbonate, for Rotor Type 45Ti (Beckman Coulter, catalog number: 355622)
6-384-well plates
Rabbit muscle acetone powder (Pel-Freez, USA, catalog number: 41995) (storage temperature: -20 °C or below)
Pyrene-labeled actin (Cytoskeleton, lnc, catalog number: AP05-A) (storage temperature: -80 °C)
Tris, ULTRA PURE (MP Biomedicals, catalog number: 4819623)
ATP (MP Biomedicals, catalog number: 210000810)
DTT (Gold Biotechnology, catalog number: DTT50)
Calcium chloride (CaCl2) (Sigma, catalog number: 746495-100G)
Potassium chloride (KCl) (Sigma, catalog number: P9541-1KG)
Magnesium chloride (MgCl2) (Nacalai Tesque, catalog number: 20908-65)
EGTA (VWR, catalog number: VWRC0732-100G)
HCl (Sigma, catalog number: 320331)
100x G-buffer without CaCl2 (see Recipes)
1x G-buffer (see Recipes)
10x KME (see Recipes)
F-buffer (see Recipes)
Equipment
Pipettes
Beaker (Schott-Duran, catalog number: SCOT211063604(E))
Ultracentrifuge Optima XE (Beckman Coulter, model: OptimaTM XE)
JA25.50 rotor (Beckman Coulter, model: JA-25.50)
Ti50.2 rotor (Beckman Coulter, model: Type 50.2 Ti)
Ti55 rotor (Beckman Coulter, model: SW 55 Ti)
ÄKTApurifier (GE Healthcare, USA)
Fluorescence spectrophotometer Cytation 5 (BioTek, USA)
Mortar and pestle (Local market)
Homogenizer (VWR, catalog number: VWRI432-0208)
Spatula (Local Market)
Ultrasonic Processor for Small and Medium Volume Applications (Sonics & Materials, INC. catalog number: VCX 750)
Freezer
Software
Origin software (Originlab Corporation, USA, https://www.originlab.com/)
Procedure
-
Actin Purification
Day 1:
-
1
Prepare 1 L 1x G-buffer (see Recipes).
-
2
Rehydrate and grind the 2.5 g lyophilized rabbit skeletal muscle acetone powder in 50 ml G-buffer.
-
3
Clear the solution by centrifugation at 27,000 × g for 1 h at 4 °C in the JA25.50 rotor (Beckman Coulter).
-
4
Collect the solubilized actin in the supernatant.
-
5
Polymerize actin in a 250 ml beaker by adding 50 mM KCl and 2 mM MgCl2 for 1 h followed by addition of 0.8 M KCl for 30 min at 4 °C.
-
6
Pellet filamentous actin (F-actin) by centrifugation at 150,000 × g for 3 h at 4 °C using Ti50.2 rotor (Beckman Coulter).
-
7
Pour off the supernatant and gently wash the surface of the pellet twice with 1 ml G-buffer.
-
8
Transfer the pellet to a 10 ml homogenizer using a spatula (Do not let the pellet dry) and add
1 ml G-buffer to the homogenizer.
-
9
Wash the centrifuge tube with G-buffer several times (use a total of 7 ml G-buffer). (This step allows the maximum recovery of actin)
-
10
Homogenize the pellet by a homogenizer.
-
11
Brief sonicate the pellet using the Ultrasonic Processors on ice using 1 s on, 1 s off cycles at 30% amplitude for 4 s.
-
12
Take out the homogenized solution and dialysis against G-buffer for 36 h at 4 °C using snakeskinTM dialysis tubing. Change the old G-buffer with a fresh one every 12 h (this step will allow the F-actin depolymerize).
Day 2:
-
13
Change the old G-buffer with the fresh buffer every 12 h.
Day 3:
-
14
Clean the monomeric actin by spinning at 200,000 × g for 2.5 h at 4 °C using a Ti55 rotor and collect the supernatant.
-
15
Equilibrate the Sephacryl S-300 HR column with 150 ml 1x G-buffer.
-
16
Load the supernatant collected from step A14 to the column using 5 ml sample loop.
-
17
Perform size exclusion chromatography using 1x G-buffer.
-
18
Collect the monomeric actin peak after gel filtration chromatography (elution volume is around 50-60 ml) (Figure 1).
-
19
Determine the concentration of individual fractions from gel filtration and store the G-actin solution at 4 °C (The G-actin could be stored at 4 °C without significant activity lost).
-
1
-
Actin Polymerization Assay
Turn on the Fluorescence spectrophotometer Cytation 5 and start the software.
-
Set up the reaction program:
Parameter: excitation/emission: 365 nm/407 nm
Time interval: 15 s
Cycle number: 299
Temperature: Room temperature
Prepare 1 ml 10x KME buffer and 10 ml 1x G-buffer (see Recipes).
Calculate the polymerization reaction materials needed based on Table 1. For one reaction, actin mix volume is 25 μl; maximum additional protein volume is 10 μl, 10x KME buffer volume is 12 μl and tops up with 1x G-buffer to total volume is 120 μl.
Prepare target protein stock according to Table 1.
Prepare actin mix based on Table 2 and keep on ice for 5 min.
Prepare reaction mix (without 10x KME) in Corning® Costar® 96-well cell culture plates according to Table 1.
Add 12 μl 10x KME to each well together using a multichannel pipette and gently mix the reaction solution.
Transfer 110 μl reaction solution into a 96-well black plate and initialize the Fluorescence spectrophotometer for data acquisition.
-
Plot the data using Origin software (Figure 1 is a typical example for actin polymerization assay).
*G-buffer is used to top up the reaction volume to 120 μl.
**Maximum total volume of protein and protein buffer is 10 μl.
*Use the value of the actual G-actin and pyrene-labeled actin stocks employed for the experiments.
**Volume is based on real reaction volume.
Figure 2. Pyrene actin polymerization assay example.
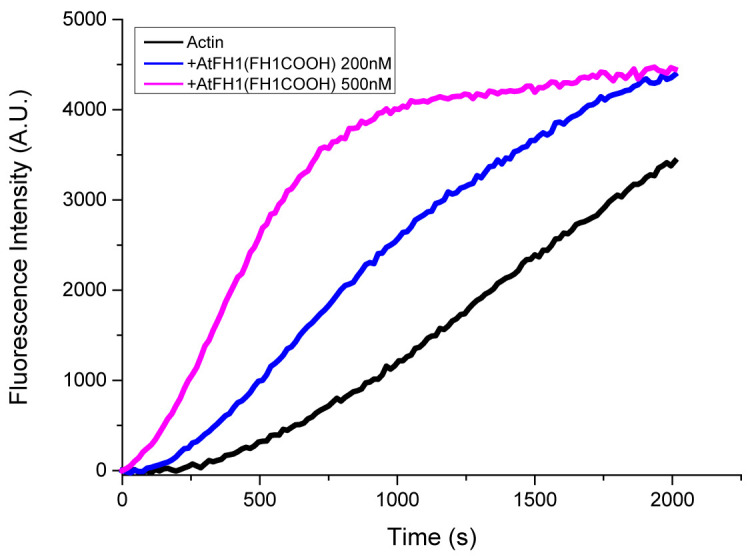 Two micromolar actin mixed with 5% pyrene-labeled actin was polymerized with indicated concentrations of Arabidopsis formin AtFH1(FH1COOH). Black, actin only; Blue, 200 nM AtFH1(FH1COOH); Purple, 500 nM AtFH1(FH1COOH). These three curves are the controls for the typical behavior of actin and formin.
Two micromolar actin mixed with 5% pyrene-labeled actin was polymerized with indicated concentrations of Arabidopsis formin AtFH1(FH1COOH). Black, actin only; Blue, 200 nM AtFH1(FH1COOH); Purple, 500 nM AtFH1(FH1COOH). These three curves are the controls for the typical behavior of actin and formin.
Figure 1. Typical gel filtration profile of G-actin purification.
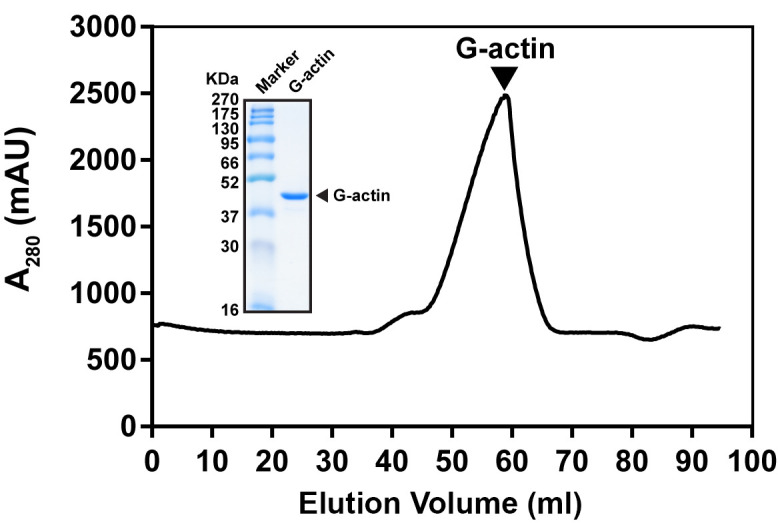
Actin was purified from Sephacryl S-300 HR column. G-actin was eluted at 50-60 ml. Inset showed SDS-PAGE analysis of G-actin eluted peak fraction.
Table 1. Actin polymerization calculation example table.
| Reaction number | Reaction condition |
G-buffer (73-83 μl)* |
10x KME | 10 μM Actin |
Protein buffer (0-10 μl)** |
Calculated protein stock (0-10 μl)** |
|---|---|---|---|---|---|---|
| 1 | Actin Ctrl | 81 | 12 | 25 | 2 | 0 |
| 2 | Actin + Protein A | 81 | 12 | 25 | 0 | 2 |
| … | … | 81 | 12 | 25 | 0 | 2 |
Table 2. Actin mix calculation example table.
| Actin mix | Stock Concentration | Final Concentration | Volume (200 μl) |
|---|---|---|---|
| Globular Actin (G-Actin) | 30 μM* | 9.5 μM | 63.3 μl |
| Pyrene-labeled Actin | 15 μM * | 0.5 μM | 6.67 μl |
| EGTA | 5 mM | 200 μM | 8 μl |
| MgCl2 | 2.5 mM | 109.7 μM | 8.78 μl |
| G-Buffer | 113.22 μl** (Top up to 200 μl) |
Data analysis
All the in vitro pyrene actin assays were analyzed in Microsoft Excel or Origin software. Pyrene fluorescence intensity (Y-axis) is plotted as a function of time (X-axis). The final data could be plotted using the time window for the different purpose. To monitor the initial nucleation activity, usually < 1,000-1,500 s is sufficient to reflect the initial nucleation rate. The protein activities for different steps of actin polymerization may vary for some proteins depends on the protein stability during storage. Appropriate controls should be included in the same experiment (e.g., Actin only control should be included all the time to know whether actin is active). For non-stable proteins, the use of freshly prepared protein is recommended for biological replications.
Notes
Pyrene actin assay results could be varied because of protein activity and practical reason. Replicate experiments are needed.
Timing is important for the experiments. In order to get better reproducible results, time control of sample handling is necessary. For example, some actin-binding protein (such as formin) will rapidly accelerate actin polymerization in seconds.
Try to avoid creating any bubbles during pipetting.
Use cut tips to pipet filamentous actin.
Prepare fresh G-buffer and F-buffer for each day experiments.
-
Fluorescence spectrophotometer Cytation 5 selection option:
Monochromator-based Detection Modes (6-384-well plates):
UV-Vis absorbance (230-999 nm, 1 nm increment)
Fluorescence intensity (250-700 nm (850 nm option))
Time resolved fluorescence (secondary)
Luminescence
Recipes
-
100x G-buffer without CaCl2 (10 ml)
2 ml of 1 M Tris-HCl, pH 8
1 ml of 0.2 M ATP, pH 7
0.5 ml of 1 M DTT, pH 7
Add ddH2O to 10 ml
Make 110 μl aliquots and store at -20 °C
-
1x G-buffer (10 ml)
100 μl 100x G-buffer
9.9 ml cold ddH2O
10 μl 0.1 M CaCl2
Prepare fresh 1x G-buffer for one day’s experiment and store on ice
-
10x KME
500 mM KCl
10 mM MgCl2
10 mM EGTA
Store at room temperature
-
F-buffer (10 ml)
9 ml G-buffer
1 ml 10x KME
Prepare fresh F-buffer for one day’s experiment and store on ice
Acknowledgments
This study was supported by was NTU startup grant (M4081533), NIMBELS (NIM/01/2016), MOE Tier 2 (MOE2016-T2-1-005S), and MOE Tier 1 (RG38/17-S) to Y. Miao in Singapore. The protocol was adapted from Sun et al. (2018).
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Pollard T. D.(2016). Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 8(8) pii: a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pollard T. D. and Cooper J. A.(2009). Actin, a central player in cell shape and movement. Science 326(5957): 1208-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun H., Qiao Z., Chua K. P., Tursic A., Liu X., Gao Y. G., Mu Y., Hou X. and Miao Y.(2018). Profilin negatively regulates formin-mediated actin assembly to modulate PAMP-triggered plant immunity. Curr Biol 28(12): 1882-1895 e1887. [DOI] [PubMed] [Google Scholar]
- 4. Weinberg J. and Drubin D. G.(2012). Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol 22(1): 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


