Abstract
[Purpose]
Recent studies have shown that COVID-19 is often associated with altered gut microbiota composition and reflects disease severity. Furthermore, various reports suggest that the interaction between COVID-19 and host-microbiota homeostasis is mediated through the modulation of microRNAs (miRNAs). Thus, in this review, we aim to summarize the association between human microbiota and miRNAs in COVID-19 pathogenesis.
[Methods]
We searched for the existing literature using the keywords such “COVID-19 or microbiota,” “microbiota or microRNA,” and “COVID-19 or probiotics” in PubMed until March 31, 2021. Subsequently, we thoroughly reviewed the articles related to microbiota and miRNAs in COVID-19 to generate a comprehensive picture depicting the association between human microbiota and microRNAs in the pathogenesis of COVID-19.
[Results]
There exists strong experimental evidence suggesting that the composition and diversity of human microbiota are altered in COVID-19 patients, implicating a bidirectional association between the respiratory and gastrointestinal tracts. In addition, SARS-CoV-2 encoded miRNAs and host cellular microRNAs modulated by human microbiota can interfere with viral replication and regulate host gene expression involved in the initiation and progression of COVID-19. These findings suggest that the manipulation of human microbiota with probiotics may play a significant role against SARS-CoV-2 infection by enhancing the host immune system and lowering the inflammatory status.
[Conclusion]
The human microbiota-miRNA axis can be used as a therapeutic approach for COVID-19. Hence, further studies are needed to investigate the exact molecular mechanisms underlying the regulation of miRNA expression in human microbiota and how these miRNA profiles mediate viral infection through host-microbe interactions.
Keywords: SARS-CoV-2, COVID-19, human microbiota, dysbiosis, gut-lung axis, microRNAs
INTRODUCTION
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in a global health emergency, and its heterogeneity driven by mutations and recombinations has instigated constant and long-term health threats [1,2]. SARS-CoV- 2 belongs to the Coronaviridae family and is a single-stranded RNA virus. Its viral spike glycoprotein (S protein) specifically binds to the human angiotensin-converting enzyme 2 (ACE2) receptor on the surface of alveolar epithelial cells for entry into host cells [3]. Essentially, the host serine protease TMPRSS2 cleaves the S protein and mediates membrane fusion for virus entry into host cells [4,5]. Infection with SARS-CoV-2 triggers a cytokine storm characterized by an uncontrolled systemic hyper-inflammatory response and primarily affects the respiratory system with diverse clinical symptoms such as high fever, cough, and severe pulmonary conditions [6,7]. Diarrhea, nausea, and vomiting have also been observed in a substantial proportion of patients, indicating that the gastrointestinal (GI) tract is associated with COVID-19 [8]. As ACE2 receptors are expressed not only in the upper respiratory tract, but also in the GI tract, the virus has been detected in lung, blood, and fecal samples from COVID-19 patients [9,10]. In fact, the virus has been detected in fecal samples at a higher level than in nasopharyngeal specimens of young COVID-19 patients [11]. Therefore, SARS-CoV-2 may affect different people in different ways throughout various organ systems by directly and indirectly deregulating the host immune systems [7].
The human microbiome differs remarkably across age, sex, race, and ethnicity, suggesting that the specific and unique profiles of microbial population [12,13] perform many essential roles for human health [12]. In a healthy state, the human microbiota plays various roles, including energy recovery from primary and secondary metabolism, protection from pathogenic invasion, and modulation of inflammatory responses and the host immune system via interaction with host immune cells [14,15]. The importance of maintaining eubiotic conditions in microbial ecosystems is strongly associated with health and disease states, and it is known that multiple factors, such as lifestyle, diet, stress, and physical activity, can influence the status of the gut microbiome [14-17]. Growing evidence suggests that SARS-CoV-2 has been detected not only in the respiratory system (nasopharyngeal and mid-nasal specimens) but also in the GI tract (stool and rectal swabs) of COVID-19 patients. Infection of gut epithelial cells by SARS-CoV-2 can disrupt the gut microbiome, inducing dysbiosis, intestinal inflammation, and gastrointestinal symptoms [18,19]. Moreover, recent studies have shown that COVID-19 is often associated with altered gut microbiota composition, reflecting disease severity [20-23]. As growing evidence indicates the interaction between COVID-19 and host-microbiota homeostasis, researchers have been investigating the potential of manipulating gut microbiota with probiotics to enhance the host immune system [24,25]. The current review summarizes important findings on the direct association between SARS-CoV-2 infection and the gut-lung axis in addition to the pathological effects of dysbiosis on COVID-19 severity. We have also highlighted the potential roles of microRNAs (miRNAs) modulated by altered microbiota in COVID-19 pathogenesis. In conclusion, we suggest the implications of probiotics or miRNA-based therapies for the intervention and treatment of COVID-19.
METHODS
We searched for existing scientific literature using the keywords “COVID-19 or microbiota,” “microbiota or microRNA,” “COVID-19 or probiotics” in PubMed until March 31, 2021. Next, we reviewed the articles related to the alteration of human microbiota by SARS-CoV infection in respiratory and GI tract to understand the involvement gut-lung axis in COVID-19 pathogenesis. In addition, we have summarized the interaction between human microbiota and miRNA expression in the progression of COVID-19.
The altered gut and lung microbiota in the pathogenesis and prognosis of COVID-19
Multiple studies have shown the involvement of human microbiota in innate and adaptive immunity and respiratory infections [26]. The healthy human lung has low density but harbors a high microbial diversity with the most prevalent bacterial compositions of Bacteroides, Firmicutes, and Proteobacteria [27]. However, in case of COVID-19, the lung microbiota in bronchoalveolar lavage fluid of COVID-19 patients shows significant differences in microbiota composition, with enrichment of pathogenic and commensal bacteria, indicating microbial dysbiosis in COVID-19 [28]. In addition, Fat et al. have evaluated the lung microbiota in lung biopsies from 20 patients with fatal COVID-19 and observed that Acinetobacter, Brevundimonas, Burkholderia, and Chryseobacterium were commonly present with mixed fungal infections in these patients [29]. To date, few studies have analyzed the nasopharyngeal microbiota of patients with COVID-19 patients [30-32]. Hoque et al. have investigated the profiles and genomic determinants of the microbiome in nasopharyngeal specimens with and without COVID-19. They found that the typical composition of bacterial phyla is represented by Proteobacteria, Tenericutes, Actinobacteria, and Cyanobacteria in the COVID-19 metagenome [30] (Table 1). Although the dominant phyla showed no statistically significant differences among groups in concordance with another study [31], COVID-19 still affected the prevalence of certain microbes such as Proteobacteria and Cyanobacteria, suggesting that microbiomes in the respiratory tract might have a significant impact on the pathogenesis and severity of COVID-19 [30]. Moreover, Budding et al. have uncovered evidence for an association between decreased pharyngeal microbial diversity and SARS-CoV-2 infection and suggested an age-dependency of pharyngeal microbiota dysbiosis [32].
Table 1.
Alteration of the predominant microbiome in healthy verses COVID-19 patients.
| Specimens | Non-COVID-19 | COVID-19 patients | Ref. |
|---|---|---|---|
| Nasopharyngeal specimens | Firmicutes (56.47%) | Proteobacteria (35.59%) | [30] |
| Bacteroidetes (14.59%) | Tenericutes (18.09%) | ||
| Actinobacteria (14.12%) | Actinobacteria (17.42%) | ||
| Fusobacteria (2.38%) | Cyanobacteria (11.23%) | ||
| Firmicutes (7.6%) | |||
| Bacteroidetes (6.2%) | |||
| Fecal specimens | Romboutsia | Streptococcus | [20] |
| Faecalibacterium | Rothia | ||
| Fusicatenibacter | Veillonella | ||
| Eubacterium hallii group | Erysipelatoclostridium | ||
| Actinomyces | |||
| Eubacterium | Depletion of commensal symbionts | [21] | |
| Faecalibacterium prausnitzii | Eubacterium ventriosum | ||
| Roseburia | Faecalibacterium prausnitzii | ||
| Lachnospiraceae taxa | Roseburia | ||
| Lachnospiraceae taxa | |||
| Enrichment of opportunistic pathogens | |||
| Clostridium hathewayi | |||
| Actinomyces viscosus | |||
| Bacteroides nordii |
Along with lung microbiota, the composition and diversity of gut microbiota are also altered in COVID-19 patients, suggesting a bidirectional association between the respiratory and GI tract microbiota [18,33]. The commensal microbiota ecosystem in the gut is complex and dynamic, and plays an important role in regulating the host immune system and metabolism [34]. During COVID-19 infection, the disrupted gut microbiome contributes to the severity of COVID-19 by promoting gut permeability and systemic inflammation, which leads to impaired ACE2 expression [19,35]. Studies have shown that the gut microbiota of COVID-19 patients is characterized by decreased levels of beneficial bacteria with a predominance of opportunistic bacteria [20,21] (Table 1). A cross-sectional study evaluating the intestinal microbiota in fecal samples from COVID-19 patients and healthy controls has reported that the gut microbiota of COVID-19 patients is significantly dominated by opportunistic pathogens, such as Streptococcus, Rothia, Veillonella, and Actinomyces, with reduced bacterial diversity and beneficial symbionts compared with healthy controls [20]. Similarly, the gut microbiota of COVID-19 patients showed significant alterations during hospitalization, and intestinal dysbiosis was positively correlated with COVID-19 severity [21]. The baseline fecal abundance of opportunistic pathogens, such as Coprobacillus, Clostridium ramosum, and Clostridium hathewayi showed a positive correlation with COVID-19 severity. However, the anti-inflammatory bacterium, Faecalibacterium prausnitzii, was inversely correlated with the severity of COVID-19 in this study. In addition, Yeoh et al. have demonstrated that the dysbiotic gut microbiota composition is concordant with COVID-19 severity, which is associated with plasma concentrations of several cytokines, chemokines, and inflammation markers [22]. Even after recovery from COVID-19 and resolution of respiratory symptoms, gut microbiota dysbiosis was still detectable and persistent, suggesting implications for future immune-related health problems beyond COVID-19 [21,22]. Moreover, Gou et al. have suggested that the disrupted composition of gut microbiota may underlie the susceptibility of normal individuals to severe COVID-19, associated with an abnormal inflammatory status [36]. Prominently, these studies demonstrate a direct interaction between SARS-CoV-2 infection and microbiota dysbiosis of the respiratory and GI tracts. Therefore, severe COVID-19 patients may have disrupted the gut epithelial barrier, which may allow the virus to reach not only the gut-lung axis but also internal organs by entering the bloodstream [23].
The interaction of human microbiota and miRNAs in COVID-19 pathogenesis
The potential mechanism by which gut microbiota affects host pathophysiology is by precisely modulating the gene expression through miRNAs at the post-transcriptional level [37,38]. Nakata et al. have demonstrated that increased miR- 21-5p expression by commensal bacteria affects the intestinal epithelial permeability by regulating ADP ribosylation factor 4 [39]. Intestinal probiotics, Lactobacillus fermentum, and Lactobacillus salivarius can increase miR-155 and miR- 233 expression and enhance intestinal barrier function [40]. The exact mechanisms by which gut microbiota regulate miRNA expression remain largely unknown; however, different metabolites produced by gut microbiota may regulate the miRNA profiles of host cells. Notably, microbiota-derived extracellular vesicles harbor biologically active components, such as mRNAs and miRNAs, which may affect host gene expression [41]. Host miRNAs, such as fecal miRNAs mainly produced by intestinal epithelial cells, can be delivered to the gut microbiota and regulate the transcription and expression of microbial genes, shaping gut microbiota composition [42]. Collectively, these results indicate the implication of host miRNA expression for gut microbiota profiles and their bidirectional interaction in host homeostasis.
Previous studies have demonstrated that several viruses encode miRNAs that regulate host gene expression involved in promoting apoptosis in order to enhance their replication [43]. Aydemir et al. have identified 20 pre-miRNA candidates and 40 mature miRNAs encoded by SARS-CoV-2, which can regulate host gene expression by targeting NFKB, JAK/STAT, and TGFB signaling pathways [44] (Table 2). Therefore, viral miRNAs, such as SARS-CoV-mir-D8- 5p and SARS-CoV-mir-R1-5p, target many human genes involved in the transcription, metabolism, and immune systems, implicating the roles of miRNAs during viral infection [44,45]. Likewise, predicted 26 mature miRNAs from the SARS-CoV-2 genome might target human genes involved in innate antiviral immunity [46]. In SARS-CoV-2, small viral RNAs contribute to lung pathology by inducing pro-inflammatory cytokines, whereas their antagomirs specifically reduced the inflammatory lung pathology, highlighting the potential role of the small viral RNA antagomiRs in eliciting direct antiviral effects [47].
Table 2.
List of miRNAs related to COVID-19 pathogenesis.
| Description | Ref. | ||
|---|---|---|---|
| Viral miRNAs | SARS-CoV-2-mir-D8-5p, SARS-CoV-2-mir-D10-3p, SARS-CoV-2-mir-D9-5p etc.: Regulate NFκB, JAK/STAT and TGFβ signaling pathways | [44] | |
| Host miRNAs | 7 key miRNAs linked to viral pathogenicity and host responses: miR-8066, miR-5197, miR-3611, miR-3934-3p, miR-1307-3p, miR-3691-3p, and miR-1468-5p | [51] | |
| Differential miRNA expression in the peripheral blood from COVID-19 patients as compared to healthy controls | [52] | ||
| - Up-regulation: miR-16-2-3p, miR-6501-5p, miR-618, miR-61-3p | |||
| - Down-regulation: miR-183-5p, miR-627-5p, miR-144-3p, miR-21-5p | |||
| Target genes and their targeting miRNAs related to viral entry | [54-58] | ||
| - ACE2: miR-1246, miR-200c-3p, miR-125a-5p | |||
| - TMPRSS2: let-7-5p, let-7d-5p | |||
Host miRNAs can interfere with viral replication both directly and indirectly by inducing antiviral reactions in the progression of viral infection [48] (Table 2). It has been demonstrated that the abundance and profile of miRNAs are associated with the severity and mortality of COVID-19 in aged patients, indicating the essential roles of host cellular miRNAs in the pathogenesis of COVID-19 [49]. Likewise, the low expression and lack of differential expression of miRNAs have been predicted to promote susceptibility of lung epithelial cells to SARS-CoV-2 infection [50]. Arisan et al. have identified 7 key miRNAs (miR-8066, miR-5197, miR- 3611, miR-3934-3p, miR-1307-3p, miR-3691-3p, and miR- 1468-5p) with significant links to viral pathogenicity and host responses [51]. When evaluating the miRNA expression pattern in peripheral blood, miR-16-2-3p, miR-6501-5p, and miR-618 were the most upregulated miRNAs in COVID-19 patients compared to the control group. In contrast, the expression of other miRNAs, such as miR-183-5p, miR-627- 5p, and miR-144-3p, was significantly reduced in patients compared to healthy donors, and were associated with the dysregulation of immune function through differential miRNA expression profile [52]. Recently, the potential functions of 13 host miRNAs in SARS-CoV-2 infection have been systematically reviewed, suggesting their potential roles in the interaction between miRNAs and viral activity [53]. Moreover, several studies have investigated host miRNAs that directly target the expression of ACE2 and TMPRSS2, which are critical for viral entry and insertion. For example, miR-1246, miR-200c-3p, and miR-125a-5p were predicted to regulate ACE2 expression levels and their associated pathways [54-56], while let-7a-5p and let-7d-5p were reported to negatively correlate with TMPRSS2 expression [56-58]. These results demonstrate the impact of host cellular miRNAs on SARS-CoV-2 infection and COVID-19 pathogenesis.
DISCUSSION
In this review, we summarize the current literature to date that provides evidence for the interaction of lung and gut microbiota in the pathogenesis and prognosis of COVID-19, with implications for modulating miRNA expression by altered microbiome signatures (Figure 1).
Figure 1.
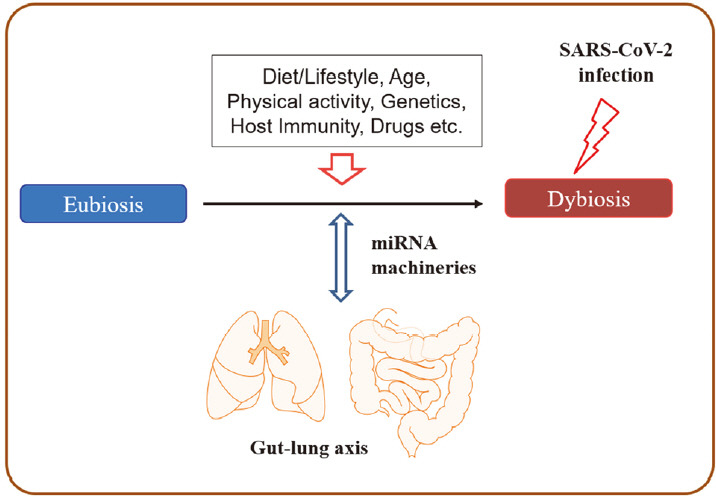
Interplays between human microbiota and miRNAs in COVID-19 pathogenesis.
The diversity and abundance of microbiota signatures among individuals strongly affect human health and disease- related inflammatory and metabolic conditions [12-16]. In a healthy state, the human microbiota plays various roles, including energy recovery from metabolism, protection from pathogenic invasion, and modulation of the host immune system [14,15]. Several factors can modify human microbiota, including host genetics, lifestyle, diet, age, stress, physical activity, and antibiotics, and they continuously affect the healthy microbiota throughout the life of the host [14-17]. In particular, the frequency and duration of physical activity have strong evidence for a positive association with gut microbiome diversity [15]. In addition, host cells affect the composition and productivity of gut microbiota by releasing nonspecific factors (e.g., antimicrobial peptides, secreted immunoglobulin A and mucins, and epithelial barrier) and specific factors, such as miRNAs [17,59]. For example, hsamiR- 515-5p and hsa-miR-1226-5p can enter bacterial cells and have a specific effect on gut bacterial growth [17]. In addition, the gut microbiota interacts with the host through various ligands (e.g., pathogen-associated molecular patterns) and produces bioactive metabolites, including shortchain fatty acids, anti-inflammatory and anti-proliferative lipids, essential vitamins (vitamins B and K), hormones, and serotonin [14]. Consequently, gut microbiota can influence host pathophysiology, and host factors reciprocally shape the bacterial ecosystem across individuals.
Growing evidence on host-microbe interactions indicates that microbiota is an essential mediator in communication between the gut and other organs and maintaining human health via the gut microbiota-miRNA interactions [60,61]. Interplays between microbiota and miRNAs have been reported in several lung diseases [62], implicating the beneficial effects of probiotics on the modulation of miRNAs. Thus, the administration of probiotics may function through several signaling pathways, resulting in the prevention and treatment of various pathological conditions [63]. As microbiome dysbiosis is linked with the severity of COVID-19, the implication of probiotics in modulating the severity has been suggested as a promising weapon against COVID-19 [24,25,64]. Several studies focusing on the gut microbiome and probiotics in COVID-19 have been conducted, and several registered clinical trials have focused on the usage of probiotics in COVID-19 [24].
MicroRNAs produced by all living organisms and viruses are well-conserved and regulate the expression of their target genes [65]. Although the exact mechanisms of host and viral miRNAs in SARS-CoV-2 infection are not well-defined, it is evident that virus-encoded miRNAs and host cellular miRNAs are associated with the initiation and severity of COVID-19. SARS-CoV-2 encoded miRNAs alter host gene expression to create a favorable environment for viral infection. Interestingly, host miRNAs can play dual roles as both antiviral and proviral factors [45]. The number of host miRNAs targeting the SARS-CoV-2 genome sequence inhibits viral replication by regulating innate antiviral immunity [53]. Other host microRNAs can increase the stability of the viral genome and inhibit decay, which is beneficial for viral replication and propagation [66]. All these results indicate the potential impact of miRNAs on SARS-CoV-2 and host interplay, suggesting the development of therapeutic approaches for miRNAs against COVID-19. Therefore, miRNA-based antiviral therapies, such as miRNA mimics or inhibitors, can be used for the intervention and treatment of COVID-19 [67,68].
The United States Centers for Disease Control and Prevention (CDC) has reported that 45% of COVID-19 patients requiring hospitalization are ≥ 65 years old, and the highest mortality and morbidity against COVID-19 have been reported in older patients with underlying chronic diseases associated with inflammation (https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/index.html). Elderly patients with certain medical conditions may be highly vulnerable to the infection with SARS-CoV-2 because they are associated with altered gut microbiota and the integrity of epithelial barriers [69]. In addition, the expression of ACE2, which is critical for viral interaction with host cells, has been shown to increase with age, accounting for increased susceptibility to older patients [70]. Moreover, the abundance and profile of miRNAs have also been associated with the severity and mortality of COVID-19 in aged patients, indicating the essential roles of host cellular miRNAs in the pathogenesis of COVID-19 [49].
In conclusion, the human microbiota-miRNA axis can be used as a promising therapeutic approach for the management of COVID-19. Therefore, further studies are essential to investigate the exact molecular mechanisms of how altered human microbiota regulate the miRNA expression profile and how these deregulated miRNA profiles mediate viral infection through host-microbe interactions.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2020R1F1A1049665).
REFERENCES
- 1.Islam MR, Hoque MN, Rahman MS, Alam A, Akther M, Puspo JA, Akter S, Sultana M, Crandall KA, Hossain MA. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci Rep. 2020;10:14004. doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paules CI, Marston HD, Fauci AS. Coronavirus infections-more than just the common cold. JAMA. 2020;323:707–8. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 3.Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–15. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, Bikdeli B, Ahluwalia N, Ausiello JC, Wan EY, Freedberg DE, Kirtane AJ, Parikh S, Maurer MS, Nordvig AS, Accili D, Bathon JM, Mohan S, Bauer KA, Leon MB, Krumholz HM, Uriel N, Mehra MR, Elkind MSV, Stone GW, Schwartz A, Ho DD, Pilezikian JPB, Landry DW. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–32. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redd WD, Zhou JC, Hathorn KE, McCarty TR, Bazarbashi AN, Thompson CC, Shen L, Chan WW. Prevalence and characteristics of gastrointestinal symptoms in patients with severe acute respiratory syndrome coronavirus 2 infection in the United States: a multicenter cohort study. Gastroenterology. 2020;159:765–7. doi: 10.1053/j.gastro.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–4. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, Yang Y, Liu B, Wang W, Wei C, Yang J, Ye G, Cheng Z. The presence of SARSCoV- 2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92:833–40. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 11.Han MS, Seong MW, Kim N, Shin S, Cho SI, Park H, Kim TS, Park SS, Choi EH. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg Infect Dis. 2020;26:2497–9. doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brooks AW, Priya S, Blekhman R, Bordenstein SR. Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 2018;16:e2006842. doi: 10.1371/journal.pbio.2006842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216:20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manor O, Dai CL, Kornilov SA, Smith B, Price ND, Lovejoy JC, Gibbons SM, Magis AT. Health and disease markers correlate with gut microbiome composition across thousands of people. Nat Commun. 2020;11:5206. doi: 10.1038/s41467-020-18871-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burchill E, Lymberopoulos E, Menozzi E, Budhdeo S, McIlroy JR, Macnaughtan J, Sharma N. The unique impact of COVID-19 on human gut microbiome research. Front Med (Lausanne) 2021;8:652464. doi: 10.3389/fmed.2021.652464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP, Schertzer JD, Larche MJ, Davidson DJ, Verdu EF, Surette MG, Bowdish DM. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21:455–66. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, Guo F, Zhang X, Luo R, Huang C, Lu H, Zheng B, Zhang J, Yan R, Zhang H, Jiang H, Xu Q, Guo J, Gong Y, Tng L, Li L. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis. 2020;71:2669–78. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, Wan Y, Chung ACK, Cheung CP, Chen N, Lai CKC, Chen Z, Tso EYK, Fung KSC, Chan V, Ling L, Joynt G, Hui DSC, Chan FKL, Chan PKS, Na SC. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–55. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, Chung AC, Cheung CP, Tso EY, Fung KS, Chan V, Ling L, Joynt G, Hui DSC, Chow KM, Ng SSS, Li TCM, Ng RW, Yip TC, Wong GLH, Chan FK, Wong CK, Chan PK, Ng SC. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HS. Do an altered gut microbiota and an associated leaky gut affect COVID-19 severity? mBio. 2021;12:e03022–20. doi: 10.1128/mBio.03022-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Din AU, Mazhar M, Waseem M, Ahmad W, Bibi A, Hassan A, Ali N, Gang W, Qian G, Ullah R, Shah T, Ullah M, Khan I, Nisar MF, Wu J. SARS-CoV-2 microbiome dysbiosis linked disorders and possible probiotics role. Biomed Pharmacother. 2021;133:110947. doi: 10.1016/j.biopha.2020.110947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chhibber-Goel J, Gopinathan S, Sharma A. Interplay between severities of COVID-19 and the gut microbiome: implications of bacterial co-infections? Gut Pathog. 2021;13:14. doi: 10.1186/s13099-021-00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He Y, Wen Q, Yao F, Xu D, Huang Y. Gut-lung axis: the microbial contributions and clinical implications. Crit Rev Microbiol. 2017;43:81–95. doi: 10.1080/1040841X.2016.1176988. [DOI] [PubMed] [Google Scholar]
- 27.Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019;20:1279–90. doi: 10.1038/s41590-019-0451-9. [DOI] [PubMed] [Google Scholar]
- 28.Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, Zhou Z, Yang J, Zhong J, Yang D, Guo L, Zhang G, Li H, Xu Y, Chen M, Gao Z, Wang J, Ren L, Li M. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with Coronavirus disease 2019. Clin Infect Dis. 2020;71:713–20. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan J, Li X, Gao Y, Zhou J, Wang S, Huang B, Wu J, Cao Q, Chen Y, Wang Z, Luo D, Zhou T, Li R, Shang Y, Nie X. The lung tissue microbiota features of 20 deceased patients with COVID-19. J Infect. 2020;81:e64–7. doi: 10.1016/j.jinf.2020.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoque MN, Rahman MS, Ahmed R, Hossain MS, Islam MS, Islam T, Hossain MA, Siddiki AZ. Diversity and genomic determinants of the microbiomes associated with COVID-19 and non-COVID respiratory diseases. Gene Rep. 2021;23:101200. doi: 10.1016/j.genrep.2021.101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Maio F, Posteraro B, Ponziani FR, Cattani P, Gasbarrini A, Sanguinetti M. Nasopharyngeal microbiota profiling of SARSCoV- 2 infected patients. Biol Proced Online. 2020;22:18. doi: 10.1186/s12575-020-00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Budding AE, Sieswerda E, Wintermans BB, Bos MP. An age dependent pharyngeal microbiota signature associated with SARSCoV-2 infection (4/21/2020) Available at SSRN: https://ssrn.com/abstract=3582780or http://dx.doi.org/10.2139/ssrn.3582780.
- 33.de Oliveira GLV, Oliveira CNS, Pinzan CF, de Salis LVV, Cardoso CRB. Microbiota modulation of the gut-lung Axis in COVID-19. Front Immunol. 2021;12:635471. doi: 10.3389/fimmu.2021.635471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodriguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gou W, Fu Y, Yue L, Chen G-D, Cai X, Shuai M, Xu F, Yi X, Chen H, Zhu YJ, Xiao ML, Jiang Z, Miao Z, Xiao C, Shen B, Wu X, Zhao H, Ling W, Wang J, Chen YM, Guo T, Zheng JS. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. MedRxiv. 2020 [Google Scholar]
- 37.Zhao Y, Zeng Y, Zeng D, Wang H, Zhou M, Sun N, Xin J, Khalique A, Rajput DS, Pan K, Shu G, Jing B, Ni X. Probiotics and microRNA: their roles in the host-microbe interactions. Front Microbiol. 2020;11:604462. doi: 10.3389/fmicb.2020.604462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behrouzi A, Ashrafian F, Mazaheri H, Lari A, Nouri M, Riazi Rad F, Hoseini Tavassol Z, Siadat SD. The importance of interaction between MicroRNAs and gut microbiota in several pathways. Microb Pathog. 2020;144:104200. doi: 10.1016/j.micpath.2020.104200. [DOI] [PubMed] [Google Scholar]
- 39.Nakata K, Sugi Y, Narabayashi H, Kobayakawa T, Nakanishi Y, Tsuda M, Hosono A, Kaminogawa S, Hanazawa S, Takahashi K. Commensal microbiota-induced microRNA modulates intestinal epithelial permeability through the small GTPase ARF4. J Biol Chem. 2017;292:15426–33. doi: 10.1074/jbc.M117.788596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, Garcia F, Olivares M, Rodriguez-Cabezas ME, Galvez J. Differential intestinal anti-inflammatory effects of lactobacillus fermentum and lactobacillus salivarius in DSS mouse colitis: impact on microRNAs expression and microbiota composition. Mol Nutr Food Res. 2017;61:1700144. doi: 10.1002/mnfr.201700144. [DOI] [PubMed] [Google Scholar]
- 41.Macia L, Nanan R, Hosseini-Beheshti E, Grau GE. Host- and microbiota-derived extracellular vesicles, immune function, and disease development. Int J Mol Sci. 2019;21:107. doi: 10.3390/ijms21010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Chen WD, Wang YD. The roles of the gut microbiota-miRNA interaction in the host pathophysiology. Mol Med. 2020;26:101. doi: 10.1186/s10020-020-00234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cullen BR. MicroRNAs as mediators of viral evasion of the immune system. Nat Immunol. 2013;14:205–10. doi: 10.1038/ni.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aydemir MN, Aydemir HB, Korkmaz EM, Budak M, Cekin N, Pinarbasi E. Computationally predicted SARS-COV-2 encoded microRNAs target NFKB, JAK/STAT and TGFB signaling pathways. Gene Rep. 2021;22:101012. doi: 10.1016/j.genrep.2020.101012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farshbaf A, Mohtasham N, Zare R, Mohajertehran F, Rezaee SA. Potential therapeutic approaches of microRNAs for COVID-19: challenges and opportunities. J Oral Biol Craniofac Res. 2020;11:132–7. doi: 10.1016/j.jobcr.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saini S, Saini A, Thakur CJ, Kumar V, Gupta RD, Sharma JK. Genome-wide computational prediction of miRNAs in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed target genes involved in pulmonary vasculature and antiviral innate immunity. Mol Biol Res Commun. 2020;9:83–91. doi: 10.22099/mbrc.2020.36507.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morales L, Oliveros JC, Fernandez-Delgado R, tenOever BR, Enjuanes L, Sola I. SARS-CoV-encoded small RNAs contribute to infection-associated lung pathology. Cell Host Microbe. 2017;21:344–55. doi: 10.1016/j.chom.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trobaugh DW, Klimstra WB. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fulzele S, Sahay B, Yusufu I, Lee TJ, Sharma A, Kolhe R, Isales CM. COVID-19 virulence in aged patients might be impacted by the host cellular microRNAs abundance/profile. Aging Dis. 2020;11:509–22. doi: 10.14336/AD.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chow JT, Salmena L. Prediction and Analysis of SARS-CoV-2- targeting microRNA in human lung epithelium. Genes (Basel) 2020;11:1002. doi: 10.3390/genes11091002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arisan ED, Dart A, Grant GH, Arisan S, Cuhadaroglu S, Lange S, Uysal-Onganer P. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12:614. doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C, Hu X, Li L, Li JH. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J Clin Lab Anal. 2020;34:e23590. doi: 10.1002/jcla.23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marchi R, Sugita B, Centa A, Fonseca AS, Bortoletto S, Fiorentin K, Ferreira S, Cavalli LR. The role of microRNAs in modulating SARS-CoV-2 infection in human cells: a systematic review. Infect Genet Evol. 2021;91:104832. doi: 10.1016/j.meegid.2021.104832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan AT, Khalid Z, Zahid H, Yousaf MA, Shakoori AR. A computational and bioinformatic analysis of ACE2: an elucidation of its dual role in COVID-19 pathology and finding its associated partners as potential therapeutic targets. J Biomol Struct Dyn. 2020;1-17 doi: 10.1080/07391102.2020.1833760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu D, Chatterjee S, Xiao K, Riedel I, Wang Y, Foo R, Bar C, Thum T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J Mol Cell Cardiol. 2020;148:46–9. doi: 10.1016/j.yjmcc.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nersisyan S, Shkurnikov M, Turchinovich A, Knyazev E, Tonevitsky A. Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. PLoS One. 2020;15:e0235987. doi: 10.1371/journal.pone.0235987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukhopadhyay D, Mussa BM. Identification of Novel Hypothalamic MicroRNAs as promising therapeutics for SARS-CoV-2 by regulating ACE2 and TMPRSS2 expression: an in silico analysis. Brain Sci. 2020;10:666. doi: 10.3390/brainsci10100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pierce JB, Simion V, Icli B, Perez-Cremades D, Cheng HS, Feinberg MW. Computational analysis of targeting SARS-CoV-2, viral entry proteins ACE2 and TMPRSS2, and interferon genes by host microRNAs. Genes (Basel) 2020;11:1354. doi: 10.3390/genes11111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang CS, Kao CY. Current understanding of the gut microbiota shaping mechanisms. J Biomed Sci. 2019;26:59. doi: 10.1186/s12929-019-0554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masotti A. Interplays between gut microbiota and gene expression regulation by miRNAs. Front Cell Infect Microbiol. 2012;2:137. doi: 10.3389/fcimb.2012.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng Q, Chen WD, Wang YD. Gut Microbiota: an integral moderator in health and disease. Front Microbiol. 2018;9:151. doi: 10.3389/fmicb.2018.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casciaro M, Di Salvo E, Pioggia G, Gangemi S. Microbiota and microRNAs in lung diseases: mutual influence and role insights. Eur Rev Med Pharmacol Sci. 2020;24:13000–8. doi: 10.26355/eurrev_202012_24205. [DOI] [PubMed] [Google Scholar]
- 63.Davoodvandi A, Marzban H, Goleij P, Sahebkar A, Morshedi K, Rezaei S, Mahjoubin-Tehran M, Tarrahimofrad H, Hamblin MR, Mirzaei H. Effects of therapeutic probiotics on modulation of microRNAs. Cell Commun Signal. 2021;19:4. doi: 10.1186/s12964-020-00668-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stavropoulou E, Bezirtzoglou E. Probiotics as a weapon in the fight against COVID-19. Front Nutr. 2020;7:614986. doi: 10.3389/fnut.2020.614986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 66.Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2- miR-122 complex. Proc Natl Acad Sci U S A. 2012;109:941–6. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hum C, Loiselle J, Ahmed N, Shaw TA, Toudic C, Pezacki JP. MicroRNA mimics or inhibitors as antiviral therapeutic approaches against COVID-19. Drugs. 2021;81:517–31. doi: 10.1007/s40265-021-01474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sevgin O, Sevgin K. Systematic review of microRNAs in the SARS-CoV-2 infection: are microRNAs potential therapy for COVID-19? J Genet Genome Res. 2021;8:053. [Google Scholar]
- 69.Bana B, Cabreiro F. The microbiome and aging. Annu Rev Genet. 2019;53:239–61. doi: 10.1146/annurev-genet-112618-043650. [DOI] [PubMed] [Google Scholar]
- 70.Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]