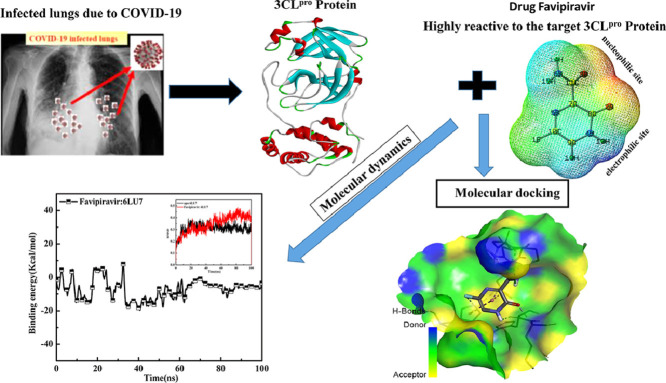
Abstract
As per date, around 20 million COVID-19 cases reported from across the globe due to a tiny 125 nm sized virus: SARS-CoV-2 which has created a pandemic and left an unforgettable impact on our world. Besides vaccine, medical community is in a race to identify an effective drug, which can fight against this disease effectively. Favipiravir (F) has recently attracted too much attention as an effective repurposed drug against COVID-19. In the present study, the pertinency of F has been tested as an antiviral option against viral protease (3CLpro) of SARS-CoV-2 with the help of density functional theory (DFT) and MD Simulation. Different electronic properties of F such as atomic charges, molecular electrostatic properties (MEP), chemical reactivity and absorption analysis have been studied by DFT. In order to understand the interaction and stability of inhibitor F against viral protease, molecular docking and MD simulation have been performed. Various output like interaction energies, number of intermolecular hydrogen bonding, binding energy etc. have established the elucidate role of F for the management of CoV-2 virus for which there is no approved therapies till now. Our findings highlighted the need to further evaluate F as a potential antiviral against SARS-CoV-2.
Keywords: Favipiravir, SARS-CoV-2, Density functional theory, Molecular docking, Electronic properties
Graphical abstract
1. Introduction
Our mother World is facing a global health catastrophe due to the pandemic situation created by the disease COVID-19 caused by a member of coronavirus family of viruses, which is exposed to the humanity from the last one and half years known as SARS-CoV2 [1]. COVID-19 infections normally evident with symptoms like high temperature, cough, myalgia, weakness, polypnea and severe respiratory problems. The disease can also progress to acute respiratory distress syndrome (ARDS) and death [2]. It is reported fact that the risk of serious symptoms and death in COVID-19 cases increases with age (> 60 years) and in people with other serious medical disorders like heart, lung, kidney, liver disease, diabetes, severe obesity [3]. From the beginning of COVID-19 spread, researchers and medical practitioners are working endlessly to find an efficient answer in the form of drug and vaccine against the disease. Studies are carried out on many repurposed and new drugs, interferon based treatments, cell based and plasma based therapies [4], [5]. Many already tested drugs/drug combinations like antiviral, antimalarial, immunosuppressant, Chinese medicines, 3CLpro inhibitors, JAK inhibitors are already being used on large number of COVID effected patients. Among them, some already used repurposed drugs are: Ivermectin, Doxycycline, Nitazoxanide/Azithromycin, Remdesivir, Oseltamivir, Hydroxychloroquine, Favipiravir (F), Lopinavir/ritonavir, Boceprevir, Telaprevir etc. [6], [7], [8], [9], [10], [11], [12], [13]. Similarly, phytoconstituents of some medicinal plants such as Tinosporacordifolia, Choline, Azadirachta Indica etc. are also used to treat the COVID-19 disease [14], [15]. One of the mostly used anti-viral drug: F which was originally designed for influenza has recently attracted too much attention as an effective repurposed drug against COVID-19 [16]. F (T-705) is a synthetic prodrug, discovered during assessing the antiviral activity of chemical agents active against the influenza virus in Japan [16]. F is already licensed as suitable drug for COVID-19 treatment in Wuhan, China, where the disease initially appeared in December 2019 [17]. After Chaina, as the pandemic spread in most places of Europe (Italy, Germany), Russia (Ukraine, Uzbekistan, Moldova, and Kazakhstan), Asia (Japan, Saudi Arabia, UAE, Egypt, Turkey, Bangladesh) and USA, F was used everywhere as an emergency medicine to treat most of the COVID patients [18], [19], [20]. F also has received emergency approval by the drug controller general of India (DGGI) for treatment of COVID-19 in June, 2020 [19]. Favipiravir comes into its active form F-RTP after it undergoes phosphoribosylation. It is a target specific drug which acts as a substrate for RNA-dependent RNA-polymerase (RdRp) enzyme, which is mistaken by the viral enzyme as a purine nucleotide and so the viral protein allows it to inhibit by forming viral protein-RdRp complex form exonuclease (ExoN) [21]. During inhibition into the viral RdRp enzyme, facile insertion of F into viral RNA happens sparing human DNA. After inhibition, it leads to termination of the viral protein synthesis as it gets incorporated in the viral RNA strand preventing protein's further extension.
The envelope surface of the coronavirus is covered with spike glycoproteins (S), membrane proteins (M) and envelope proteins (E) [22,23]. The main envelope of virus comprises a spiral nucleocapsid which is formed by genomic RNA and phosphorylated nucleocapsid (N) protein [24]. The S proteins of the virus initiate the attachment and entry to the host cells through the receptor binding domain (RBD) which is loosely attach to the virus surface [25]. To enter the host human cells, all coronavirus uses some key receptors. For SARS-CoV-2, the key receptor is angiotensin converting enzyme 2 (ACE2) [26]. After entering the host cell, airway trypsin-like protease (HAT), cathepsins and trans membrane protease serine 2 (TMPRSS2) split the S proteins of the virus and establish the penetration changes. So, development of targeted spike glycoprotein therapeutics against SARS-CoV-2 will definitely be a suitable option to combat COVID-19.
It is also reported that SARS CoV-2-RdRp complex is at least 10-fold more active than any other reported viral RdRp known [27]. For CoVs, the ExoN has been shown to remove certain nucleoside analogues (NAs) after insertion by RdRp into nascent RNA, which reduces their antiviral effects [28], [29], [30]. Despite this bad effect, several NAs like Favipiravir, Oseltamivir are currently being tested as anti CoV candidates. RdRp is one of the most intriguing and promising drug targets for SARS-CoV-2 drug development and F is one of such most effective RdRp inhibitor. The reason behind using an RdRp inhibitor like F to 3CLpro protein (6LU7) is that like all positive RNA viruses for CoV-2, RdRp (nsp12 protein) lies at the core of viral replication machinery. Due to its viral life cycle, lack of host homologous, high level of sequence and structural conservation nsp12 becomes an optimal target for therapeutics. But till now due to the lack of sufficient fundamental data, proper guide to design of an efficient antiviral therapeutics and their mechanism of action is not available. So RdRp inhibitors are chosen a promising target drugs since they are small sized NAs. It is reported that F can induces mutagenesis in vitro during influenza virus infection as it can inactivate the virus either by killing them or by changing their surface structure so the virus cannot be able to enter the host cells [31]. Though the proper interaction mechanism of mutagenesis is still unknown, we are expecting similar activity of F against SARS CoV-2 protein. We hope that F with its defined mode of action may well find a place as an anti RdRp component in combination therapies targeting corona viruses. Still the optimal dose of F is difficult to establish from the limited preclinical in vitro data. The above mentioned two way applicability of Favipiravir has motivated us to work on this specific antiviral drug against CoV-2 virus since it has recently received a lot of attention to treat Covid-19 patient [32] all over the World. However, there is a scarcity of information on the structural effectivity, chemical reactivity prediction on F. Therefore, in the present work for the first time, the structural effectivity of the F molecule has been investigated by using density functional theory (DFT). In the present Covid-19 situation, molecular dynamics (MD) simulations have endorsed researcher to create rational scientific advances. Advancement in this technology have made them an essential tool for scientists researching drug development [33]. Our in silico studies on structural analysis with the help of virtual screening, chemical analysis, molecular docking and MD on Favipiravir as ligand and 3CLpro main spike protease of CoV2 as receptor have established that COVID infections can be nullified by Favipiravir as it helps to shield the living host cell from the possible detrimental impact from CoV2 infection.
2. Materials and methods
2.1. Potential inhibitor F and its Preparation
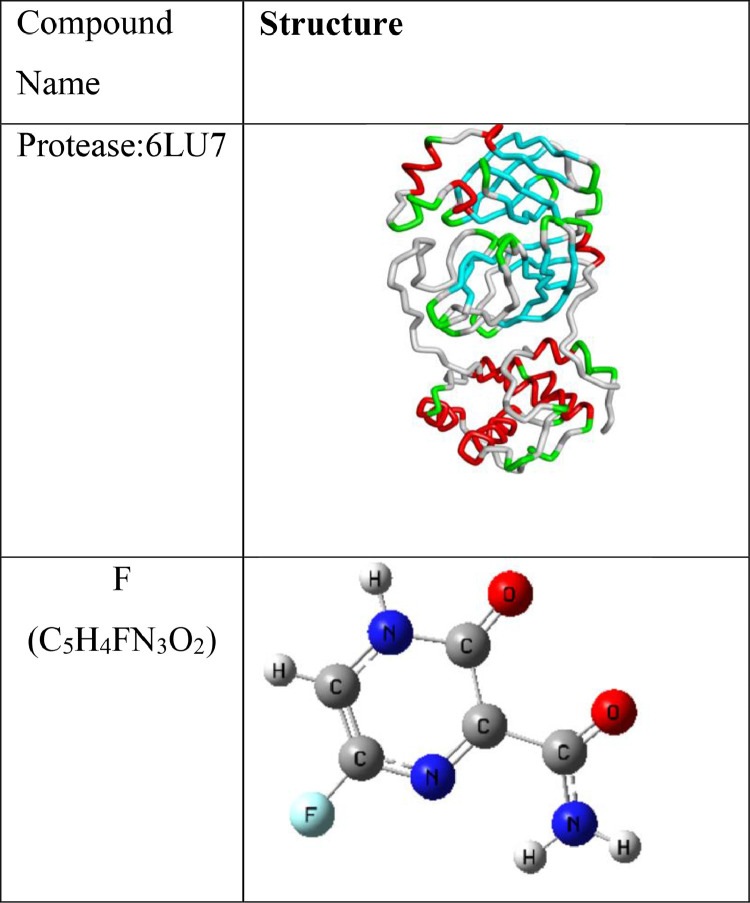
Favipiravir (T-705) is a synthetic prodrug (C5H4FN3O2), which is used in the antiviral activity of chemical agents against the influenza virus [31]. It has a good bioavailability (∼94%), 54% protein binding affinity. F-RTP binds to inhibits RdRp, which ultimately prevents viral transcription and replication [10,30]. Virtual screening of the drug has done before the checking of inhibition capability. SWISS ADME software (https://www.swissadme.ch) and ADMET (https://vnnadmet.bhsai.org/) software were used for the virtual screening [34]. Drug-likeness rules like Lipinski's rule of five (Ro5), Veber's rule, MDDR-like rule, Egan rule, Ghose filter, Muegge rule etc., were used for preliminary drug screening [35], [36], [37]. For Drug preparation, the ligand in ‘SDF’ format was obtained directly from the PubChem (National Library of Medicine) (https://pubchem.ncbi.nlm.nih.gov/) and converted to ‘PDB’ format with the help of Auto Dock tools [38]. Molecular structure of the ligand was optimized by using DFT with the basis set 6-311G (d,p) [39] using the Gaussian 09 program [40].
2.2. Potential target protein structure for SARS-CoV-2 and preparation
SARS-CoV-2 is a virus having positively sensed single stranded RNA. The protein structure of CoV-2 contains spike (S), membrane (M), envelope (E) and nucleocapsid (N) [41,42]. The structures of CoV-2 virus and already known CoV virus is very similar [43]. So the identification process of (3CLpro) of CoV-2 was appeared to be much faster [44,45]. 3CLpro is located at the 3 ends, which exhibits excessive variability and can be treated as a potential target or anti-coronaviruses inhibitors screening [46].
In the present study, we have used 3CLpro proteases (6LU7) as main target protein of drug molecules. The 3D structure of the 6LU7 was retrieved from the Protein Data Bank website (https://www.rcsb.org) [47] and existing water molecules available in the structure were removed (Fig. 1 ). Polar hydrogen were added in protein structure and inbuilt ligand was removed from the protein structure with the help of Discovery studio 2020 [48]. All of the above steps were performed in AutoDock [31], Molecular Graphics Laboratory (MGL) tools [49]. The output protein structure was saved in PDB format for further study.
Fig. 1.
Structure of receptor protein (6LU7) [from Protein data bank] and F as ligand molecule [from Gauss view].
2.3. Computational method for structural analysis
All the theoretical calculations, including the optimization of ground state geometries were carried out using the Gaussian 09 program [50,51]. The geometry of F was optimized, with density functional theory (DFT) using Becke3-Lee-Yang-Parr (B3) [52] exchange functional combined with the (LYP) [53] correlation functional with the standard 6-311G basis set. Excited state calculation of molecule was performed using time dependent DFT (TD-DFT). The frontier molecular orbital (FMOs) are simulated for the molecule using Koopman's theorem [54]. MEP surface mapped with electrostatic potential surface and atomic charges (Mulliken and natural) were derived by using optimized structure.
2.4. Molecular docking, molecular dynamics, binding energy
In computer-assisted drug designing, molecular docking is considered as the tool which helps in energy minimization and finding binding affinity between protein and ligand. In the present study protein-ligand interaction was studied by using the software used for Molecular docking is AutoDock 4.2 and AutoDock vina [38]. MGL tools were used for the preparation of protein and ligands for molecular docking. We have removed all the usual bound ligand present on the 3D structure PDB: 6LU7 protein. Reason for deleting the bound ligand present on the 3D structure of protein is that because of it ligand will not easily set in the pocket region and that will give incorrect results in docking. We have also removed all the Hetatoms present on the protein since they can block up binding sites and cause problems with protein-ligand interaction. The output structure of macromolecule is saved in pdbqt format. The removal of all the usual bound ligand and Hetatoms have been done with the help of software “Biovia Discovery Studio Visualizer”. While performing the docking we have set the following configuration parameters like exhaustiveness = 8, energy difference= 3 kcal/mol, Grid box center with coordinate x= -10.729204, y=12.417653, z=68.816122 of position of the target protein. The target protein is prepared for docking by saving in the pdbqt format. Similarly, drug is synthesized and saved in the pdbqt format using AutoDockVina. Docking is accomplished using CMD (Command Prompt) by issuing various commands. Result of docking provide us 9 different possible possess of interaction between drug and receptor. The most stable receptor-ligand complex should be chosen after all ligands have been docked. The decision of the stable receptor-ligand complex would be made based on the binding affinity (G) (Kcal\mol), dipole moment of ligand (in Debye), number of hydrogen bonds, Inhibition constant (µM) and drieding energy. The ligand which forms the highest number of bonds with the target protein mostly shows better complex formation. Different non-bonded hydrogen bonds for different output poses, dipole moment and drieding energy have been analyzed with the help of visualizing Discovery Studio visualizer 2020 version 20.1.0.19295 [48]. Inhibition constant have been computed by using the below mentioned equation:
| (1) |
where G is binding affinity, R is universal constant and T is the room temperature (298 K).
For reliability of the docking results, we have performed docking for same protein-ligand pair with the help of another docking software Pyrx [55]. We have performed MD simulation for calculating various thermodynamics parameters like potential energy (Epot), root mean square deviation (RMSD) for backbone, root mean square fluctuation (RMSF) for protein Cα Solvent accessible surface area (SASA), intermolecular hydrogen bonds, and binding energy of the ligand: protein complex structure with the help of Linux based platform ‘‘GROMACS 5.1 Packageˮ [56] with GROMOS43A2 force fields [57]. According to the procedure followed for MD simulation, TIP3P water model has been used and 4Na+ ions were added to maintain the neutrality of ligand: protein complex structure in a cubic box, with a buffer distance of 10 Å and volume as 893,000 A3. For energy minimization of complex, time varying (1–100,000 ps) steepest descent algorithm with for 50,000 steps was used.
To calculate the interaction free energies for the protein: ligand complex structure (ΔGbind), the MMPBSA (Molecular Mechanics Poisson-Boltzmann Surface Area) method [58] sourced from the Adaptive Poisson-Boltzmann Solver (APBS) and GROMACS packages have been used. To calculate ΔGbind, the snapshots at every 100 ps between 0 and 100,000 ps (100 ns) were collected. After completing the MD simulation of the complex using the single trajectory approach ΔGbind calculation generally initiates. For the bound protein: ligand complex ΔGbind can be given as:
| (1) |
| (2) |
| (3) |
| (4) |
Where,-TΔS is the conformational energy change due to binding, is solvation free energy change, is the molecular mechanical energy changes in gas phase, is the covalent energy, is electrostatic energy, is Van der Waals energy changes. is the sum of , , and changes. While covalent energy is the combination of bond, angle and torsion and is the sum of polar and nonpolar contributions.To calculate the binding energy the data was collected between 0 ps and 100,000 ps (100 ns). For RMSD and RMSF, we have run multiple simulations independently.
2.5. Computational details
MD simulations and corresponding energy calculations have been computed in a single system using HP Intel Core i5 - 1035G1 CPU and 8 GB of RAM with Intel UHD Graphics and a 512 GB SSD.
3. Results and discussion
3.1. Optimized structure
The most stable structure of F in the ground state was optimized using B3LYP/6-311G*(d,p) level of theory (Fig. 1). Different bond lengths (Å) and bond angles (°) of F are shown in the Table 1 . The ground state electronic energy of the optimized structure is -3813.169 Kcal/mol. F possess a very high value of dipole moment of 5.23 Debye due to its C1 point group symmetry. The high value of dipole moment may increase the bioactivity of the probe system [59]. Bioactivity is a significant property of a drug that promotes bonding or complex formation of drug with the target protein [60].
Table 1.
Optimized geometrical parameters (bond length (Å) and bond angle (°) of F by using using B3LYP/6-311G*(d,p) method.
| Bond | Bond length (Å) | Bond | Bond angle (°) |
|---|---|---|---|
| 1F-10C | 1.35193 | 1F-10C-5N | 116.389 |
| 10C-5N | 1.32542 | 1F-10C-9C | 120.115 |
| 10C-9C | 1.36362 | 10C-9C-13H | 124.772 |
| 9C-13H | 1.08705 | 9C-10C-5N | 123.496 |
| 9C-4N | 1.35277 | 13H-9C-4N | 118.588 |
| 4N-12H | 1.03458 | 9C-4N-12H | 118.770 |
| 4N-8C | 1.39439 | 9C-4N-8C | 125.190 |
| 8C-2O | 1.22978 | 4N-8C-7C | 111.958 |
| 8C-7C | 1.46753 | 2O-8C-7C | 127.300 |
| 7C-11C | 1.51046 | 5N-7C-11C | 117.310 |
| 11C-3O | 1.23114 | 5N -7C- 8C | 122.588 |
| 11C-6N | 1.34506 | 8C-7C-11C | 120.093 |
| 6N-15H | 1.01544 | 3O-11C-6N | 123.785 |
| 6N-14H | 1.01788 | 11C-6N-15H | 121.908 |
| 7C-5N | 1.31311 | 11C-6N-14H | 119.153 |
3.2. Frontier molecular orbitals analysis
The highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) plays an important role in the investigation of chemical stability/reactivity, and related optical properties of the molecules [61]. The energy gap (Eg) can be easily calculated by taking the difference between energy of HOMO and LUMO orbitals of the probe molecule. Table 2 shows the computed theoretical energies of HOMO (EHOMO) and the LUMO (ELUMO) orbitals. The value of Eg for F was computed as 4.029 eV (Table 2). The ionization potential (IP) can be computed as -EHOMO and the electron affinity (EA) is computed as -ELUMO.
Table 2.
The calculated electronic properties of the F (all values are in eV).
| S. No. | Molecular properties | Values |
|---|---|---|
| 1. | HOMO | -6.756 |
| 2. | LUMO | -2.727 |
| 3. | Energy gap (Eg) | 4.029 |
| 4. | Ionization potential (IP) | 6.756 |
| 5. | Electron affinity (EA) | 2.727 |
| 6. | Electrophilicity Index (ω) | 5.58 |
| 7. | Chemical Potential (μ) | -4.741 |
| 8. | Electronegativity (χ) | 4.741 |
| 9. | Softness (S) | 0.496 |
| 10. | Hardness (η) | 2.014 |
Different FMO related molecular parameters (global hardness (η), electronegativity (χ), chemical potential (μ) and electrophilicity (ω), and chemical softness (S)) of the Fare calculated by using the following formulae in the framework of Koopmans’ theorem [54]:
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
We have observed a moderate value of Eg (4.029 eV) which suggest the chemical reactiveness and optically polarizable nature of F molecule. Using the values of IP and EA, one can calculate χ, η and ω parameters, which are helpful in analyzing the reactivity of the molecule. The calculated value of IP as 6.756 eV and EA as 2.727 eV indicate high reactivity of F [62]. Higher value of χ (4.741 eV) also confirms the higher reactivity of F. Organic molecules can be classified on the basis of ω values [63]. An organic molecule will be of minimal electrophiles if its ω value is less than 0.8 eV, moderate electrophiles with 0.8 < ω < 1.5 eV and strong electrophiles if its ω value is greater than 1.5 eV [64]. In the present case our probe molecule F has ω value as 5.58 eV, which represent its strong electrophile nature. Chemical potential of -4.741 eV also indicates the electron-withdrawing character of F [64]. Chemical η value of a system measures the resistance to alter the electron distribution, which is associated with the reactivity of the system. All the FMO related parameters suggested the higher reactivity of F towards the target protein.
3.3. Charge analysis
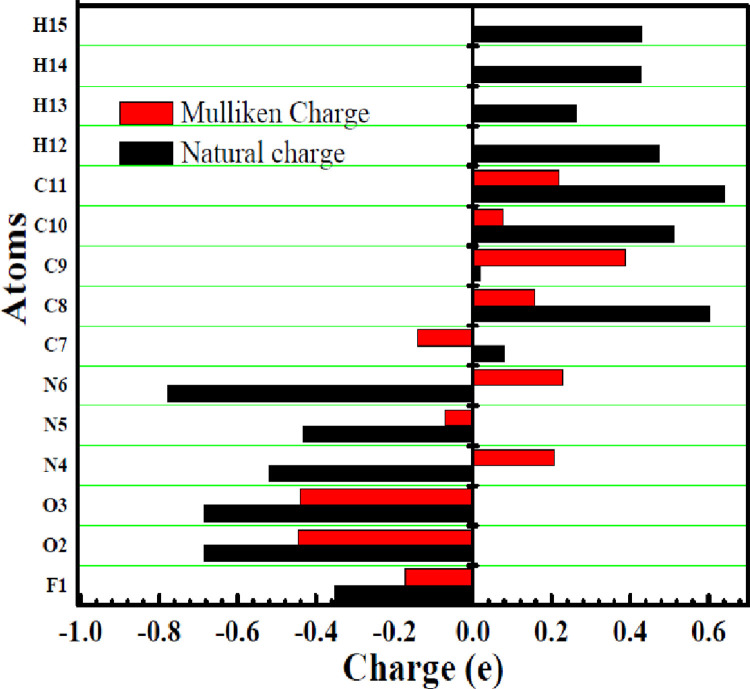
Mulliken and natural charges play a significant role, for predicting the nucleophilic, molecular polarizability and electrophilic reactive regions of a certain molecule [65]. Different computed Mulliken and natural charges of the F drug are shown Fig. 2 and supporting document (SD1). Observed charge analysis shows that all oxygen atoms possess negative charges. O2 atom has the highest negative charge (NBO: -0.684e and Mulliken: -0.44648e) rather than O3 atom. N6 atom has the highest negative charge (NBO: -0.779 e and Mulliken: 0.22745 e) rather than N5 atom. All the carbon atoms have positive charges. C9 atom possess the maximum positive Mulliken charge of 0.38868e. Fluorine atoms (F1) display negative charge (NBO: - 0.353 e and Mulliken: - 0.17395 e). The H12, H14, H15 atoms have the highest positive charge about rather than other H13 hydrogen atom due to the attachment to electron withdrawing nitrogen group. Variation of charge in the probe molecule represent the tendency of the molecule to accept/donate electrons and hence the higher reactiveness of the system. Presence of electron donor and acceptor groups in the F displays the capability of forming complex with external target molecule.
Fig. 2.
Mulliken and natural charges distribution of the F.
3.4. MEP analysis
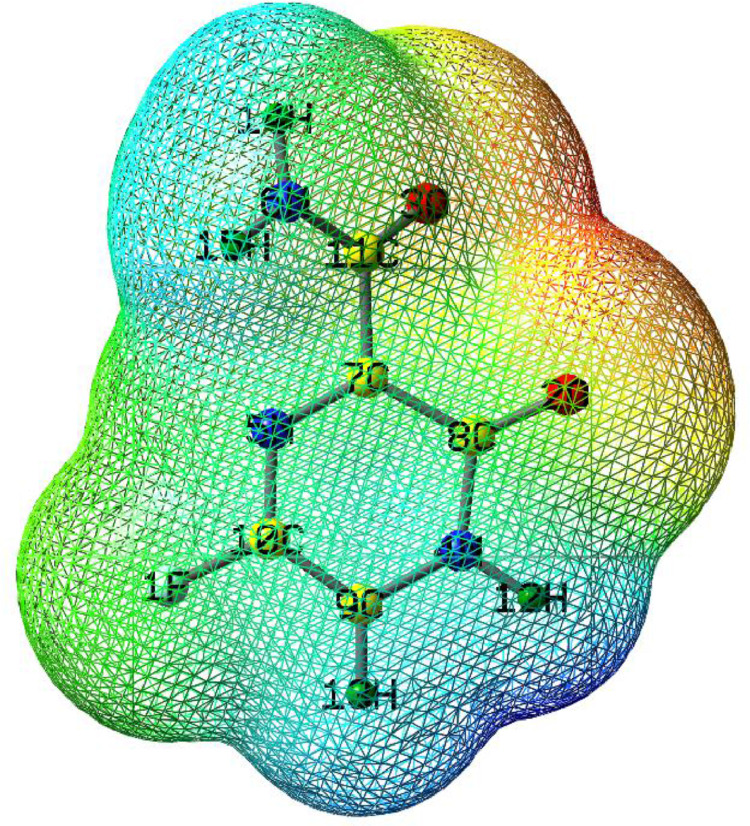
The MEP surface of the optimized molecule F is shown in the Fig. 3 . The MEP surface of the molecule represent the electrophilic and nucleophilic reactive site of the molecules [66]. Different colors in the MEP shows the different electrostatic potential regions. Red colour in the MEP represent electron-rich sites, orange color partially negative charge, yellow colors lightly electron-rich regions, blue color positive charge and green color neutral sites. In the MEP surface of F, red color display electron rich regions over the O2 and O3 oxygen atoms. This indicates the lone pair electrons around the oxygen atoms. The hydrogen atoms such as H12, H14, and H15 specifies with blue color show the electrophilic sites (electron-poor). Remaining regions of green color display the zero potential (neutral regions). The existence of electrophilic and nucleophilic regions of F clearly demonstrate the possibilities of favourable chemical reactions of F with target protein system.
Fig. 3.
Molecular electrostatic potential map of the molecule F (Red colour; represent electron-rich sites, orange color; partially negative charge, yellow color; lightly electron-rich regions, blue; positive charge and green color; neutral sites).
3.5. Absorption analysis
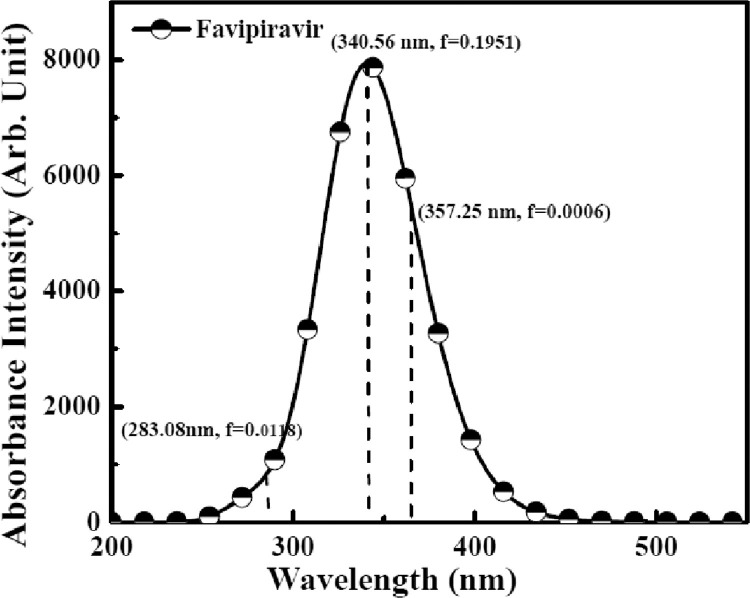
The theoretical UV-Vis spectrum of F is shown in the Fig. 4 . The computed absorption excitation energies (E), and oscillator strength (f) are reported in Table 3 . According to results, the λmax appears at 357.25 nm and f is 0.0006 for S0 →S1 transition. The main contribution for the formation of the absorption band at 340.56 nm and the f is 0.1951 for S0 →S2 transition. S0 →S3 transition correspond to the 283.08 nm wavelength with 0.0118 value of f. The strong band in the absorption spectrum is observed at 340.56 nm which is due to n→π* transition and a weak band at 283.08 nm due to π→π* transition. These electronic transitions correspond to the π→π* and n→π*, makes the probe system highly unstable, which confirms its ability of binding to the target protein.
Fig. 4.
UV-visible spectra of F molecule computed by TD-DFT/6-311G*(d,p) method.
Table 3.
UV-visible absorption data of F molecule with transition level and oscillator strength.
|
Theoretical |
||||
|---|---|---|---|---|
| Transition | λ (nm) | E (eV) | (f) | |
| Absorption | S0 →S1 | 357.25 | 3.4705 | 0.0006 |
| S0 → S2 | 340.56 | 3.6406 | 0.1951 | |
| S0 → S3 | 283.08 | 4.3798 | 0.0118 | |
3.6. Analysis of drug likeness properties of favipiravir
Among all of Drug likeness Ro5 rule or ‘‘a rule of thumbˮ is the most important rule. The filters of Ro5 are followed by: Molecular weight less than equals to 500, H-bond donors less than equals to 5, H-bond acceptor less than equals to 10, MLOGP less than equals to 4.15 and molar refractivity between 30 and 140. Those drugs which follows the Ro5 with some required pharmacological properties can be used as potential candidate for vocally active drug in humans [34]. F follows the Ro5 so it can be proposed as chemical compound with compulsory pharmacological properties as a potential candidate for orally active drug in humans [34]. From the ADMET analysis, we can conclude that F has no cyto-toxicity effect and no hERG (human Ether-a-go-go Related-Gene) Blocker can have the maximum suggested dose as 170 mg/day (SD2).
3.7. Analysis of molecular docking results
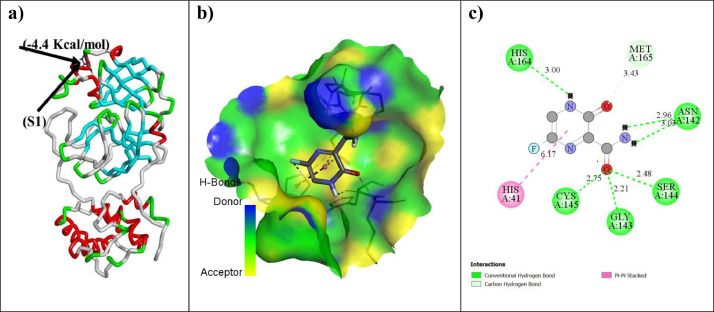
Different favorable poses for F and protein obtained from molecular docking with binding energy, dreiding energy and inhibition constant are shown in SD 3. For F among all poses best binding complex (F: 6LU7) structure was obtained as pose 3. The basis for the selection of best pose is the maximum number of hydrogen bonded interactions (7), hydrophobic interaction (1), binding energy (-4.4 kcal/mol) and lowest value of dreiding energy (53.04) and inhibition constant (5.9 × 10−4M) of the complex at 300 K (room temperature).
According to the available crystal structure, SARS-CoV-2 CLpro has four binding pockets named as S1–S4 within its protease that function as active sites. The active site is made up of the base protein's backbone and side chain residues. The S1 binding site is created by Phe-140, Asn-142, Ser-144, Cys145, His-163, His-172, and Glu-166 side chain residues and Leu-141, Gly-143, His-164, and Met-165 backbone. S1’ active site is formed by some side chains and backbone as His-41, Val 42, Asn-119, Thr-25, Cys-145, Gly-143, Thr-26. The other subsites are S2 (Tyr-54, Asp-187, Met-49, His-41 Arg-188) and S4 (Met-165, Leu-167, Pro-168, Ala-191, Gln-192, Glu-166, Arg-188, Thr-190). F is normally bound to the S1 chain residues of HIS164, MET165, ASN142, SER144, GLY143 and CYS145 of 6LU7. Weak interactions like “hydrogen bonded interactions and hydrophobic interactions” always stabilize the ligands at the target protein site by varying the binding affinity [67]. Due to their weak nature, hydrogen and hydrophobic interaction have significant roles in stabilizing the energetically favored ligands in a suitable pocket of the environment of protein structures. In defining the stability of any protein: ligand complex, larger the number of these interactions better is the possibility of binding affinity between ligand: protein complexation and so the possibility of stable complex structure [67]. Maximum number of hydrogen bonded interactions (both conventional and carbon hydrogen bonds) were observed for pose 3 of docked F: 6LU7 structure between 6LU7 (Residues: HIS164, MET165, ASN142, SER144, GLY143, CYS145) and F (atoms: O, H) (Fig. 5 , Table 4 ). For the same pose structure one hydrophobic interaction was observed for docked structure between 6LU7 (Residue: HIS 41,) and F (Fig. 5, Table 4). Docking score for F and protein by Pyrx is almost similar to the obtained results by AutoDock vina, which verifies our molecular docking results (SD 4). To validate the possibility of strong interaction and stability between F and the receptor protein 6LU7, inhibition constant, dipole moment and dreiding energy were considered as important parameters. Lowest value of dreiding energy and lower value of inhibition constant (ki) for pose 3 structure as comparison to other available poses validate the strong interaction of F ligand as inhibitor towards the receptor protein 6LU7 [68, 69]. Maximum value of dipole moment for a complex structure always represent the better stability of the formed complex structure between receptor protein and ligand (Table 4). So molecular docking results indicate that F can be easily inhibited inside the favorable pocket of receptor 6LU7 protein and can form a stable F:6LU7 complex by hydrogen and hydrophobic bonding between them.
Fig. 5.
(a) Binding energies F, (b) 3D view of Donor:acceptor surface for best pose in terms of H-bond interaction (c) 2D view of possible types of interaction in pose for F:6LU7 (Green: Conventional Hydrogen bond, Dark pink:π-π bond, sky blue: carbon hydrogen bond).
Table 4.
Various interaction parameters for the docked structure of Favipiravir (as FAV) with receptor protein 6LU7.
| Ligand |
Binding affinity (kcal/mol) |
Hydrogen bonded interaction (donor::acceptor, distance in Å)[Type of bond) | Hydrophobic bonded interaction(donor::acceptor, distance in Å) | Dreiding energy(ligand) | Dipole moment of ligand(Debye) | InhibitionConstant(M)Ki= |
|---|---|---|---|---|---|---|
| Favipiravir | -4.4 | (A:GLY143:HN-:: FAV:O, 2.21) [Conventional Hydrogen Bond] (A:SER144:HN -:: FAV:O, 2.48) [Conventional Hydrogen Bond] (A:CYS145:HN -: FAV:O, 2.75) [Conventional Hydrogen Bond] (:FAV:H -: A:HIS164:O, 3.00) [Conventional Hydrogen Bond] (:FAV:H -: A:ASN142:OD1, 2.96) [Conventional Hydrogen Bond] (:FAV:H -: A:ASN142:OD1, 3.03) [Conventional Hydrogen Bond] (A:MET165:CA -:: FAV:O, 3.43) [Carbon Hydrogen Bond] |
(A:HIS41-:: FAV, 6.17) |
53.04 |
2.770 |
5.9 × 10−4 |
3.8. Analysis of molecular dynamics (MD) simulation results
The comparison of minimum potential energy (Epot) of the stabilized structures of protein in its apo state with individually docked ligand compound have been done simultaneously (Table 5 , SD 5a). Protein in its apo state has an average Epot of -0.3 × 106 8.8 Kcal/mol. The average Epot for individual F, in presence of 6LU7 was obtained as at the order of ∼ -0.1 × 106 4.9 (Table 5, SD 5a). Under the equilibrium condition (NVT and NPT) we have verified the stability of F: 6LU7 complex structure. The temperature (T), density (D), pressure (P) and volume (V) of complex structure with varying time trajectory from 0 to 100 ps have been calculated SD 5b–d). After the implementation of NVT and NPT conditions MD simulations were operated for complete time trajectory 0–100 ns. Output for all thermodynamic parameters for protein in its apo state and with all complex formations with possible ligands are shown in Table 5, SD 5b–d.
Table 5.
Data obtained from MD simulations of time resolved trajectory for receptor protein 6LU7 in its apo state and for the complex state with the ligand F.
| S. No | Parameter |
Apfo protease (6LU7) |
F:6LU7 |
||
|---|---|---|---|---|---|
| Mean | Range | Mean | Range | ||
| MD Simulation Result | |||||
| 1. | SR Columbic Interaction Energy (Kcal/mol) | NA | NA | -9.21 | 0–20 |
| 2. | SR LJ Interaction Energy (Kcal/ mol) | NA | NA | -19.30.7 | 0- -30 |
| 3. | RMSD (nm) | 0.30 | 0.13–0.36 | 0.34 | 0.10-0.45 |
| 4. | Inter H-Bonds | NA | NA | 5 | 0-9 |
| 5. | Radius of gyration(nm) | 2.18 | 2.13–2.24 | 2.210.1 | 2.20-2.21 |
| 6. | SASA (nm2) | 33 | 30–35 | 8 | 6-10 |
| MM/PBSA Results | |||||
| 7. | Potential Energy (Kcal /mol) | -0.3 × 1068.8 | -7.0 × 105 - -1.3 × 106 | -0.1 × 1064.9 | -3.4 × 104 - -2.8 × 105 |
| 10. | Binding energy(∆G)(Kcal/mol) | NA | NA | -7.20.08 | -20 - 10 |
| 11. | Van der Waal Energy(∆Evdw) (Kcal/mol) | NA | NA | -0.650.03 | -1 - 1 |
| 12. | Electrostatic Energy(∆Eelec)(Kcal/mol) | NA | NA | -0.170.01 | -0.5 - 0.5 |
| 13. | Polar solvation energy(∆Epolar) (Kcal/mol) | NA | NA | -6.340.08 | 0 - -8 |
| 14. | Nonpolar energy (∆Enonpolar) (Kcal/mol) | NA | NA | -0.060.007 | 0 - -1 |
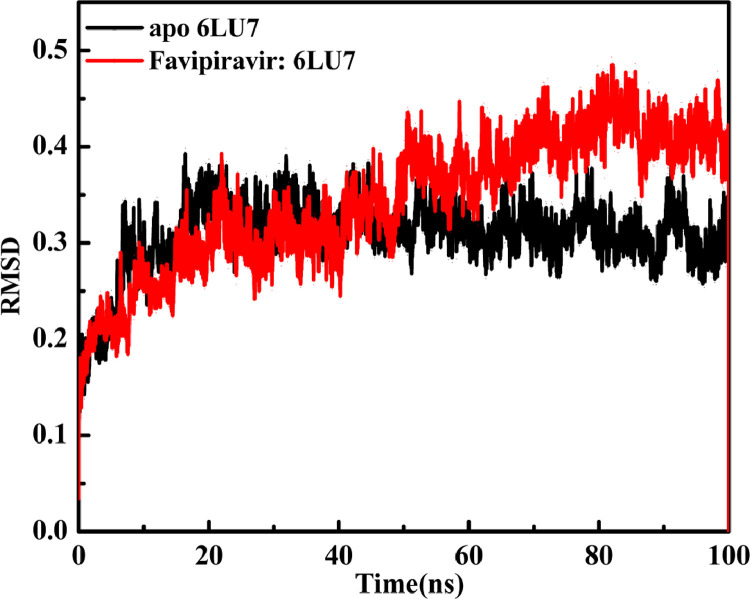
During simulation, the average distance between the atoms of the protein is calculated using RMSD analysis. This analysis reveals information about protein/complex structure, stability, and equilibrium. To identify the conformation stability and convergence of 6LU7 bound with F, Simulation result was obtained by calculating RMSD of backbone atoms. RMSD of both apo protein and complex backbone against the time of the simulation were presented as the trajectories. In RMSD for F:6LU7 a variation between 0.10 and 0.45 compared to receptor protein 6LU7 variation 0.13–0.36 again indicates that the less fluctuation in F:6LU7 complex structure during binding with receptor protein. The average RMSD value for the F:6LU7 (0.34 nm) and host protein (0.30 nm) is almost same which confirms the possibility of stable F:6LU7 complex structure (Fig. 6 ).
Fig. 6.
Root mean square deviation (RMSD) graphs for apo state of receptor protein 6LU7 and in complex (Favipiravir:6LU7) with receptor protein 6LU7 up to 100 ns.
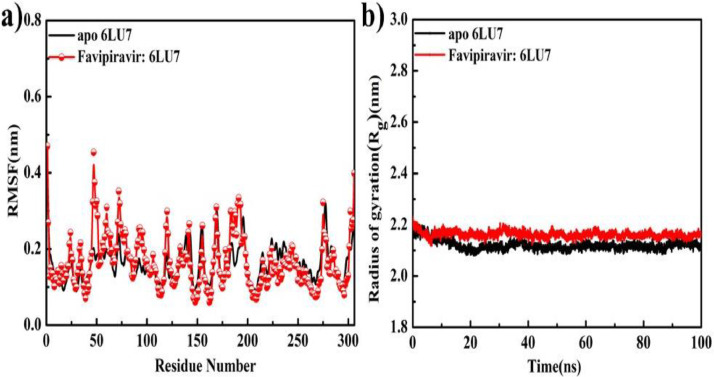
Similarly for RMSF of F:6LU7, less fluctuation between the receptor and inhibitor was observed. Perfect similarity in RMSF values confirmed that F:6LU7 complex structure does not affected the protein backbone (Fig. 7 a). The radius of gyration (Rg) tells us about the compressed nature of a complex structure or backbone receptor protein [70]. Variation of Rg value throughout the total-time trajectory (0 ps to 100 ns) showed that F: 6LU7 has quite stable and compressed structure. Rg of F: 6LU7 and apo 6LU7 show a perfect match having an average value of 2.18 nm with a fluctuation between 2.13 and 2.24 nm (Fig. 7b, Table 5).
Fig. 7.
(a) RMSF for apo state 6LU7 and of complex F:6LU7 structure, (b) Rg for apo state 6LU7 and of complex F:6LU7, structure in time trajectory (0–100 ns).
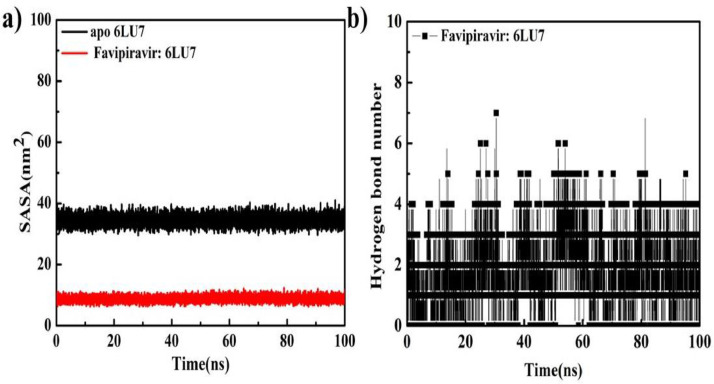
Solvent accessible surface area (SASA) tells about the area of receptor contact to the solvents. The greater value of SASA means that more of the drug is inserted into the water. And lower the value of SASA means that more of the drug is covered by the protein means better complexation. The SASA value for apo protein was calculated between 30 and 35 nm2 with a 33 nm2 mean value however, for F:6LU7 complex structure the SASA value was observed between 6 and 10 nm2 which satisfied the possibility of better complexation of F drug with receptor 6LU7 (Fig. 8 a, Table 5). Stability of ligand: receptor protein complex structure is always dependent on the contribution of nonbonded interactions.
Fig. 8.
(a): SASA area for protein 6LU7 in its apo state and for complex F:6LU7 structure and (b) Intermolecular hydrogen bond numbers for complex F:6LU7 structure for the total time trajectory, 0–100,000 ps.
Intermolecular nonbonded hydrogen bonded interaction between ligand and receptor protein plays a dominant role to define the stability of complex structure. For present simulation a 3.5 Å cut-off condition is used to identify the proper nonbonded hydrogen bonded interaction. The number of hydrogen bonded interactions for F: 6LU7 were observed to be varying between 0 and 7 (Fig. 8b). The obtained numbers of intermolecular hydrogen bonded interactions between inhibitor: receptor through MD simulations were perfectly matched with the obtained molecular docking result.
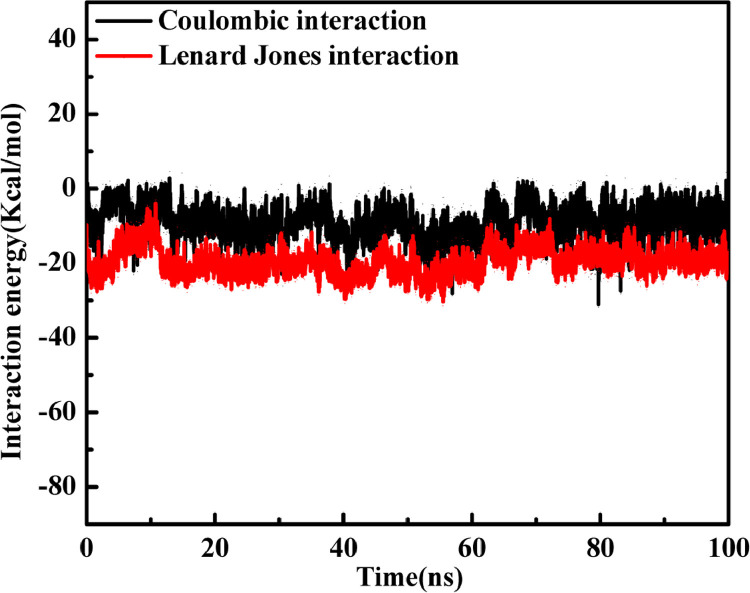
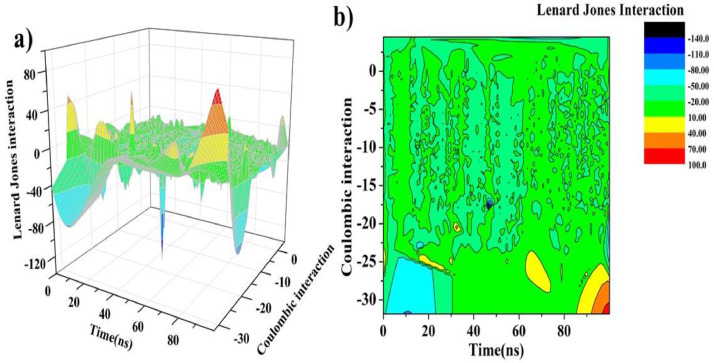
To validate the strength of interaction between a ligand and receptor nonbonded interaction energy also plays a very important role. In the present study, for complex structure F:6LU7, we have observed a variation of short-range Coulombic interaction (Coul-SR) energy and Lennard Jones (LJ-SR) energy (Table 5) over the full-time trajectory (Fig. 9 ). For complex structure LJ-SR has shown greater effect on the binding affinity than the Coul-SR. For F:6LU7 complexation higher effect of LJ-SR energy (-19.30.7 Kcal/mol) compared to Coul-SR (-9.21 Kcal/mol) is represented by 3D view and with color contour representation of LJ-SR energy and Coul-SR energy with respect to the time trajectory 0 ps to 100,000 ps (Fig. 10 ). The 3D view of LJ-SR and Coul-SR shows that the complex structure is completely stable in full time trajectory which shows the best interaction between protein and ligand.
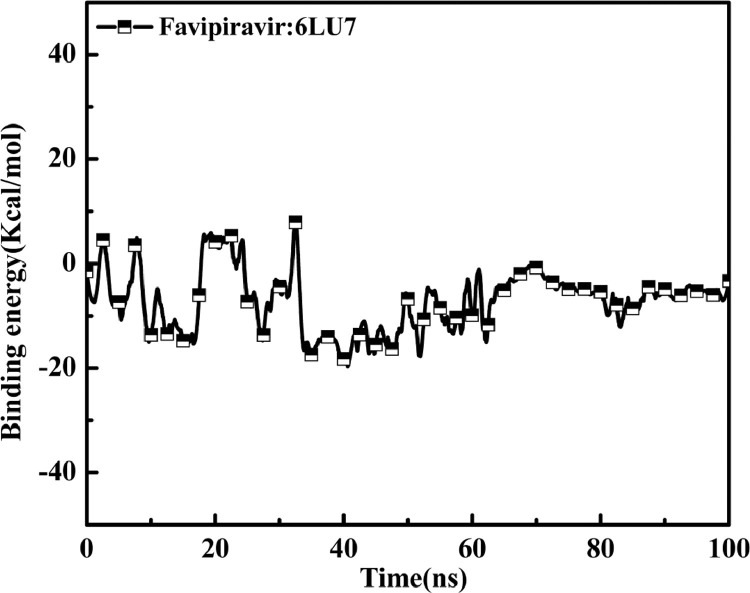
Fig. 11.
Variation of total binding energy for F with receptor protein in time trajectory 0–100 ns.
Fig. 9.
2D graph for F:6LU7complex:Variation of Coulombic interaction energy and Lenard Jones interaction energy with respect to whole time trajectory (0–100 ns).
Fig. 10.
For F:6LU7 complex: (a) Variation of Coulombic interaction energy and Lenard Jones interaction energy with respect to time trajectory 0 –100 ns (b) attached with color contour representation with specific color coding.
To find the ligand binding affinities towards receptor protein, we have applied the MM/PBSA method to compute the binding energy of the complex structure formation. According to the applied method Van der Waal energy (Evdw) and electrostatic energy (Eelectrostatic) have been computed and are shown in Table 5. For inhibitor F, the best binding affinity was observed for with the appearance of values of Evdw (-0.650.03 Kcal/mol) and Eelectrostatic (-0.170.01 Kcal/mol) (Table 5). All the energies needed for ΔG are polar and nonpolar solvation energies which are shown in the Table 5. The average values polar and nonpolar solvation energy are -6.340.08 Kcal/mol and -0.060.007 Kcal/mol, respectively. The nonbonding interaction energies of the binding region for the complex formation is generally indicated by the free binding energy (ΔG) values. The average ΔGbind for F:6LU7 complex structure was obtained as -7.20.08 Kcal/mol. From the results of ΔGbind we can conclude that drug F strongly binds with receptor 6LU7 protein and shows that the complex structure was completely stable after 70 ns. Though F shows a good binding affinity towards CoV-2 protease, there are other members of repurposed drugs also who are uqually effective towards the same protease in terms of their binding affinity ranges from -6 to -40 Kcal/mol. They are Chloroquine [71], Hydroxychloroquine [10], Nelfinavir [72], Azithromycin [6], Lopinavir [73], Ritonavir [73], Remdesivir [74] etc. Some of these drugs show better binding affinity and some also show less affinity towards 3CLpro protease- 6LU7.
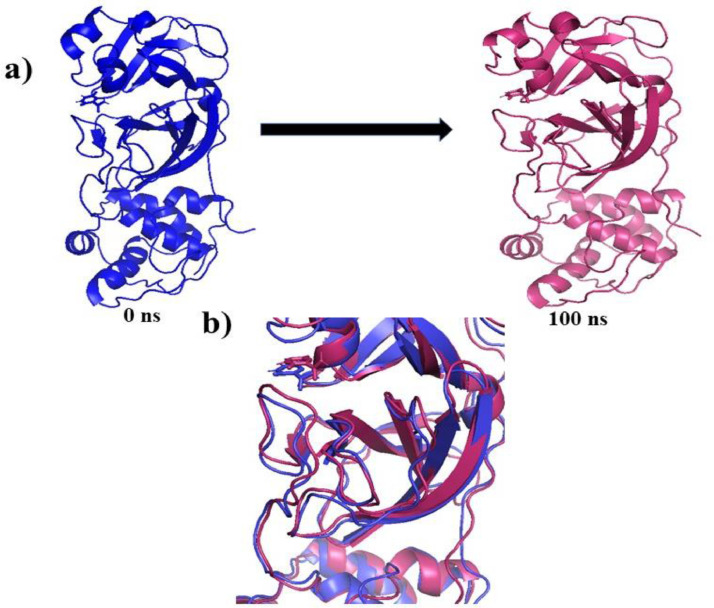
The check the possibility of better binding affinity of F towards 6LU7, superposition of conformational variation in the F:6LU7 complex structures of initial (0 ns) and final (100 ns) frames for variable time trajectories have been studied. At the beginning of the simulation, the ligand Favipiravir was placed outward of the binding site (blue, 0 ns), whereas at final stage it has moved more inside the binding cavity with a small rotation and change in the position of functional group (pink, 100 ns). The structural superimposition of the lignad bound proteins's initial and final frames revealed very small alterations at the binding location of ligand (Fig. 12 ). It revealed that the ligand orientation changes upon simulation for maximum convergence. Moreover, during the course of simulation small tilt of the ligand position indicates that F was docked well at the binding site of the 3CLpro (Fig. 12a,b). The small change is position through the time scale was also verified by the resulting RMSD after structural superposition which was observed as 1.467 Å.
Fig. 12.
(a) Structural position of first frame (0 ns) and last frame (100 ns), (b) Superposition of initial and final of F bound 3CLpro complex after MD simulation.
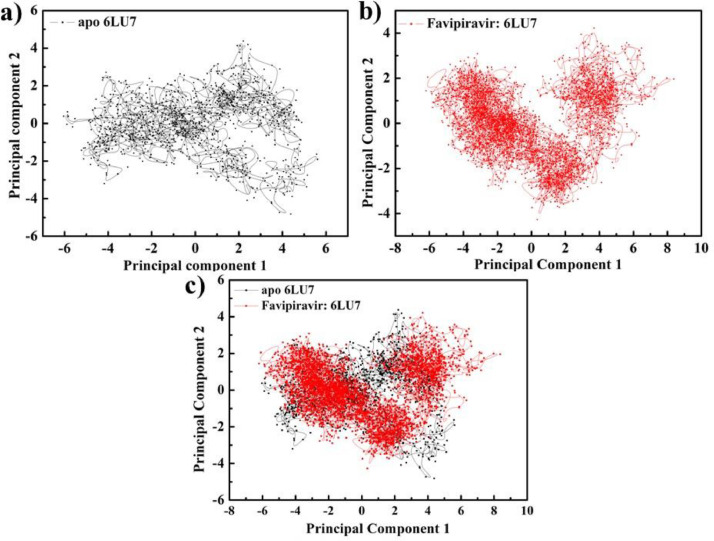
3.9. Principal component analysis and free energy landscape
Using molecular dynamics trajectories, principal component analysis (PCA) is normally used to determine the collective motion of the apo protein and bound ligand with protein. The PCA study is established on the eigenvectors (overall direction of motion of the atoms) and eigenvalues of the protein's Cα atom (atomic contribution of motion) [75]. To gain a better understanding of the structural and conformational changes in protein caused by binding of ligand, the MD trajectories of apo protein, along with F bound protein were examined with the PCA. To evaluate the conformational sampling of systems, tracing the covariance matrix for backbone atom positions is also used. The covariance trace value for the apo protein was detected to be 21.8 nm2 and a lower covariance trace value of 17.9 nm2 was found for the complex (F: 6LU7). Overall PCA analysis revealed that binding of F with apo 6LU7 results in a substantial change in the overall motion of 6LU7 with a compressed conformational space. (Fig. 13 (a–c)). The result also validate that F bound 3CLpro protein is more stable than the unbound protein.
Fig. 13.
Projection of protein atoms in phase space along the first two principal eigenvectors. (a) protein 6LU7 (b) Protein complexed with standard protease inhibitor F(c) Superimposed plot showing Protein 6LU7 unbound protein and protein complexed with inhibitor F.
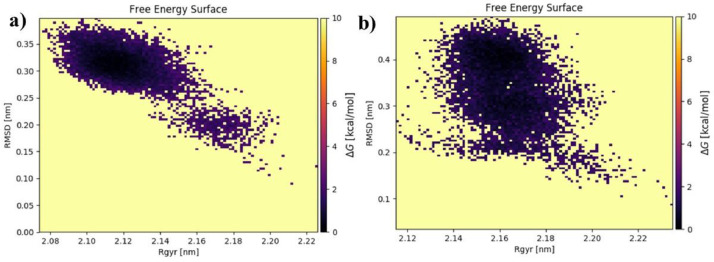
We have also examined the free energy landscape (FEL) against the first principal components PC (RMSD) to show the energy minima landscape of unbound protein and F bound 3CLpro protein (Fig. 14 (a,b)). The value of ∆G for both cases is 0–10 kcal/mol. The minimal energy area and size (shown in black) indicate the apo protein (6LU7) and F:6LU7 complex's stability. The stability of the protein and its related complex is indicated by smaller and more centred black patches.
Fig. 14.
Free energy landscape of the first principal components for (a) protein 6LU7 (b) Protein complexed with standard protease inhibitor F.
4. Conclusion
Favipiravir, which was originally designed for influenza has recently attracted too much attention as an effective repurposed drug against COVID-19. Our present work represents the computational work on the applicability of F as an emerging antiviral option against SARS-CoV-2 pandemic. Different geometrical optimized bond lengths, bond angles, dipole moment, point group symmetry, electronic energy were computed theoretically. Variation of charge, C1 point group symmetry, high value of dipole moment (5.23 Debye) represent the reactiveness of the F towards the target protein. Computed FMO parameters of F validates chemical reactiveness. F shows the possibility of HOMO to LUMO charge transfer with the involvement of lone pairs and pi (π) bonds. UV-vis spectra confirms the contribution of lone pairs and π bonds in the charge transfer. All these parameters confirms the reactivity and hence the ability of F to bind with the target protein. Molecular docking results shows strong binding affinity (-4.4 Kcal/mol), low inhibition constant (5.9 × 10−4M) and maximum number of interactions (both conventional and carbon hydrogen bonds) and hydrophobic interaction for F:6LU7 complex structure. Several thermodynamic parameters (Epot, T, V, D, interaction energies, ΔGbind, Rg, SASA energy) obtained by MD simulation results validated the stability of F:6LU7 complex structure. For F: 6LU7 complex structure, the number of hydrogen bonded interactions were observed to be in between 0 and 7 which was perfectly matched with the docking results. We have observed similar RMSD and RMSF for the backbone of the complex and free 6LU7 protein, which again validate the possibility of strong binding of anti-viral drug F and 6LU7. The perfect closeness of average RMSD between F:6LU7 (0.34 nm) and host protein (0.30 nm) confirmed the total inheritance of proposed drug inside host protein. More negative value of Lennard-Jones interaction energy (-19.30.7 Kcal/mol) in compare to Coulombic interaction (-9.21 Kcal/mol) represent its higher effect in the F:6LU7 complex formation. Lower value of ΔGbind (-7.20.08 Kcal/mol) computed by MD simulation confirmed the stable complexation between F drug and 6LU7 protein. Our simulated data for Favipiravir as an inhibitor for the 3CLpro of COVID-19 allows us to characterize the behavior of Favipiravir in the binding site of target 3CLpro protease as well as to elucidate the fundamental biochemical processes for inhibitor the target receptor protein. We are very much confinent that our in silico data obtained from DFT and MD Simulations on repurposed inhibitor Favipiravir against target 3CLpro protease 6LU7 will be useful for drug discovery and further research and development to fight against the deadly disease COVID-19.
CRediT authorship contribution statement
Pooja Yadav: Writing – original draft, Data curation, Visualization, Investigation. Meenakshi Rana: Visualization, Investigation, Software, Validation, Writing – review & editing. Papia Chowdhury: Conceptualization, Software, Methodology, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.131253.
Appendix. Supplementary materials
References
- 1.Gorbalenya A.E., Baker S.C., Ralph S.B., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Leontovich A.M., Neuman B.W., Penzar D., Poon L.L.M., Samborskiy D.V., Sidorov I., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aytür Y.K., Köseoğlu B.F., Taşkıran Ö.Ö., Ordu-Gökkaya N.K., Delialioğlu S.Ü., Tur B.S, Sarıkaya S., Şirzai H., Tiftik T.T., Alemdaroğlu E., Ayhan F.F. Pulmonary rehabilitation principles in SARS-COV-2 infection (COVID-19): a guideline for the acute and subacute rehabilitation. Turk. J. Phys. Med. Rehabil. 2020;66(2):104. doi: 10.5606/tftrd.2020.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L., He W., Yu X., Hu D., Bao M., Liu H., Zhou J., Jiang H. Coronavirus disease 2019 in elderly patients: characteristics and prognostic factors based on 4-week follow-up. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skariyachan S., Gopal D., Chakrabarti S., Kempanna P., Uttarkar A., Muddebihalkar A.G., Niranjan V. Structural and molecular basis of the interaction mechanism of selected drugs towards multiple targets of SARS-CoV-2 by molecular docking and dynamic simulation studies-deciphering the scope of repurposed drugs. Comput. Biol. Med. 2020;1(126) doi: 10.1016/j.compbiomed.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang B., Guo H., Ling L., Ji J., Niu J., Gu Y. The chronic adverse effect of chloroquine on kidney in rats through an autophagy dependent and independent pathways. Nephron. 2020;144(1):53–64. doi: 10.1159/000503882. [DOI] [PubMed] [Google Scholar]
- 6.Kelleni M. Nitazoxanide/Azithromycin combination for COVID-19: a suggested new protocol for COVID-19 early management. Pharmacol. Res. 2020;157:104874. doi: 10.1016/j.phrs.2020.104874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rana M., Chowdhury P. arXiv; 2020. Ivermectin and Doxycycline Combination as a Promising Drug Candidate Against SARS-CoV-2 Infection: A Computational Study. preprint arXiv:2012.00653. [Google Scholar]
- 8.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B. 2017:93. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pooja P.C. arXiv; 2020. Repurposing the Combination Drug of Favipiravir, Hydroxychloroquine and Oseltamivir as a Potential Inhibitor against SARS-CoV-2: A Computational Study. preprint arXiv:2012.00652. [Google Scholar]
- 10.Zhang R., Mylonakis E. In inpatients with COVID-19, none of remdesivir, hydroxychloroquine, lopinavir, or interferon β-1a differed from standard care for in-hospital mortality. Ann. Intern. Med. 2021;174(2):JC17. doi: 10.7326/ACPJ202102160-017. [DOI] [PubMed] [Google Scholar]
- 11.Uzunova K., Filipova E., Pavlova V., Vekov T. Insights into antiviral mechanisms of remdesivir, lopinavir/ritonavir and chloroquine/hydroxychloroquine affecting the new SARS-CoV-2. Biomed. Pharmacother. 2020;24 doi: 10.1016/j.biopha.2020.110668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar Y., Singh H., Patel C.N. In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. J. Infect. Public Health. 2020;13(9):1210–1223. doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padhi A.K., Tripathi T. Targeted design of drug binding sites in the main protease of SARS-CoV-2 reveals potential signatures of adaptation. Biochem. Biophys. Res. Commun. 2021;28(555):147–153. doi: 10.1016/j.bbrc.2021.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhury P. In silico investigation of phytoconstituents from Indian medicinal herb ‘Tinosporacordifolia (giloy)’against SARS-CoV-2 (COVID-19) by molecular dynamics approach. J. Biomol. Struct. Dyn. 2020:1–18. doi: 10.1080/07391102.2020.1803968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury P., Pathak P. Neuroprotective immunity by essential nutrient “Choline” for the prevention of SARS CoV2 infections: an in silico study by molecular dynamics approach. Chem. Phys. Lett. 2020;761 doi: 10.1016/j.cplett.2020.138057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arab-Zozani M., Hassanipour S., Ghoddoosi-Nejad D. Favipiravir for treating patients with novel coronavirus (COVID-19): protocol for a systematic review and meta-analysis of randomised clinical trials. BMJ Open. 2020;10(7) doi: 10.1136/bmjopen-2020-039730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Islam M.S., Sobur M.A., Akter M., Nazir K.N.H., Rahman M.T., Toniolo A. Coronavirus disease 2019 (COVID-19) pandemic, lessons to be learned! J. Adv. Vet. Anim. Res. 2020;7(2):260. doi: 10.5455/javar.2020.g418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon A., Selisko B., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., Coutard B., Peersen O. Rapid incorporation of favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11(1):1–9. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal U., Reyma R., UdwadiaZarir F. Favipiravir: a new and emerging antiviral option in COVID-19. Med. J. Armed Forces India. 2020;76:372–376. doi: 10.1016/j.mjafi.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhagavathula A.S., Rahmani J., Aldhaleei W.A., Kumar P., Rovetta A. Global, regional and national incidence and case-fatality rates of novel coronavirus (covid-19) across 154 countries and territories: a systematic assessment of cases reported from January to March 16, 2020. MedRxiv. 2020 doi: 10.1101/2020.03.26.20044743. Submitted for publication. [DOI] [Google Scholar]
- 21.Shannon A., Selisko B., Le N.T.T., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., Coutard B. Favipiravir strikes the SARS-CoV-2 at its Achilles heel, the RNA polymerase. BioRxiv. 2020 doi: 10.1101/2020.05.15.098731. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padhi A.K., Tripathi T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol. Transl. Sci. 2020;3:1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorden D.E., Gwendolyn M.J., Bouhaddou M. A SARS-CoV-2- human protein-protein interaction map reveals drug targets and potential drug-repurposing. BioRxiv. 2020 doi: 10.1101/2020.03.22.002386. [DOI] [Google Scholar]
- 24.Priyadarsini S., Panda S., Singh R., Behera A., Biswal P., Mech P., Ramaiyan K. In silico structural delineation of nucleocapsid protein of SARS-CoV-2. J. Entomol. Zool. Stud. 2020;8(2):06–10. [Google Scholar]
- 25.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang H. Structure of Mpro from COVID- 19 virus and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 26.Velavan T.P., Meyer C.G. The COVID-19 epidemic. Trop. Med. Int. Health. 2020;25(3):278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdelnabi R., et al. Understanding the mechanism of the broad-spectrum antiviral activity of favipiravir (T-705): key role of the F1 motif of the viral polymerase. J. Virol. 2017;91:e00487. doi: 10.1128/JVI.00487-17. -17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferron F. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuta Y., Takahashi K., Fukuda Y., Kuno M., Kamiyama T., Kozaki K., Nomura N., Egawa H., Minami S., Watanabe Y., Narita H., Shiraki K. in vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 2002;46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baranovich T., Wong S.S., Armstrong J., et al. 705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J. Virol. 2013;87:3741e3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dabbous M.H, Sherief A.E., Manal H.E., Ahmed F.S., Fatma F.S.E., Ghafar M.S.A.E., Soliman S., Elbahnasawy M., Badawi R., Tageldin M.A. Efficacy of favipiravir in COVID-19 treatment: a multi-center randomized study. Arch. Virol. 2021;166:949–954. doi: 10.1007/s00705-021-04956-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Padhi A.K., Rath S.L., Tripathi T. Accelerating COVID-19 research using molecular dynamics simulation. J. Phys. Chem. B. 2021 Jul 28 doi: 10.1021/acs.jpcb.1c04556. [DOI] [PubMed] [Google Scholar]
- 34.Daina A., Michielin O., Zoete V. Swiss ADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benson N.C., Daggett V. A comparison of multiscale methods for the analysis of molecular dynamics simulations. J. Phys. Chem. B. 2012;116(29):8722–8731. doi: 10.1021/jp302103t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipinski A.C. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Veber D.F., Johnson S.R., Cheng H.Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 38.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and auto dock Tools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becke A. A new inhomogeneity parameter in density-functional theory. J. Chem. Phys. 1997;10(20):8554–8560. doi: 10.1063/1.475007. [DOI] [Google Scholar]
- 40.Frisch M.J. Gaussian Inc; 2004. Gaussian 09, Revision D.01. [Google Scholar]
- 41.Woo P.C.Y., Lau S.K.P., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.K., Poon L.L.M., Wong S.S.Y., Guan Y., Peiris J.S.M., Yuen K.Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorden D.E., Gwendolyn M.J., Bouhaddou M. A SARS-CoV-2- human protein-protein interaction map reveals drug targets and potential drug-repurposing. bioRxiv. 2020 doi: 10.1101/2020.03.22.002386. [DOI] [Google Scholar]
- 43.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Chang S. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang F., Deng L., Zhang L., Cai Y., Cheung C.W., Xia Z. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19) J. Gen. Intern. Med. 2020;35(5):1545–1549. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1751298. 1-0. [DOI] [PubMed] [Google Scholar]
- 46.Deng X., StJohn S.E., Osswald H.L., O'Brien A., Banach B.S., Sleeman K., Ghosh A.K., Mesecar A.D., Bake S.C. Coronaviruses resistant to a 3C-like protease inhibitor are attenuated for replication and pathogenesis, revealing a low genetic barrier but high fitness cost of resistance. J. Virol. 2014;88:11886–11898. doi: 10.1128/JVI.01528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S., Feng Z., Ghosh S., Goodsell D.S., Green R.K., Guranovi V., Guzenko D., B.P. H., Kalro T., Liang Y., Zardecki C. RCSB protein data bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DassaultSyst`emes BIOVIA . DassaultSyst`emes; San Diego: 2017. Discovery Studio Modeling Environment, Release. [Google Scholar]
- 49.A. Tools, 1.5. 6 (ADT)/MGL tools 1.5. 6 The Scripps Research Institute, CA, USA (2012). Available at http://mgltools.scripps.edu.
- 50.Frisch M.J., et al. Gaussian Inc.; Wallingford CT: 2004. Gaussian 09, Revision D.01. [Google Scholar]
- 51.Smitha M., Sheena Mary Y., Mary Y.S., Serdaroglu G., Chowdhury P., Rana M., Umamahesvari H., Sarojini B.K., Mohan B.J., Pavithran R. Modeling the DFT structural and reactivity studies of a pyrimidine-6-carboxylate derivative with reference to its wavefunction-dependent, MD simulations and evaluation for potential antimicrobial activity. J. Mol. Struct. 2021;1237 [Google Scholar]
- 52.(a) Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]; (b) Becke A.D. Density-functional thermochemistry. V. Systematic optimization of exchange-correlation functional. J. Chem. Phys. 1997;107:8554–8560. [Google Scholar]
- 53.Lee C., Yang W., Parr R.G. Development of the colle-salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 54.Koopmans T. By assigning wave functions and eigenvalues for the individual electrons in an atom. Physica. 1934;1:104–113. [Google Scholar]
- 55.Dallakyan S., Olson A.J. Chemical Biology. Humana Press; New York, NY: 2015. Small-molecule library screening by docking with PyRx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 56.Berendsen H.J.C., Van der Spoel D., Van Drunen R. GROMACS: a message passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 57.V.Gunsteren W.F., Billeter S.R., Eising A.A., Hünenberger P.H., Krüger P., Mark A.E., Scott W.R.P., Tironi I.G. Biomolecular Simulation: The GROMOS96 Manual and User Guide. 1996 [Google Scholar]
- 58.Rashmi K., Rajendra K., Andrew L. g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 59.Mary Y.S., Thomas R., Narayana B., Samshuddin S., Sarojini B.K., Armaković S., Armaković S.J., Pillai G.G. Theoretical studies on the structure and various physico-chemical and biological properties of a terphenyl derivative with immense anti-protozoan activity. Polycycl. Aromat. Compd. 2019:1–16. [Google Scholar]
- 60.Tripathi M.K., Singh P., Sharma S., Singh T.P., Ethayathulla A.S., Kaur P. Identification of bioactive molecule from withaniasomnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1790425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rana M., Singla N., Chatterjee A., Shukla A., Chowdhury P. Investigation of nonlinear optical (NLO) properties by charge transfer contributions of amine functionalized tetraphenylethylene. Opt. Mater. 2016;62:80–89. [Google Scholar]
- 62.Jasmine G.F., Amalanathan M., Roy S.D.D. Molecular structure and charge transfer contributions to nonlinear optical property of 2-methyl-4-nitroaniline: a DFT study. J. Mol. Struct. 2016;1112:63–70. [Google Scholar]
- 63.Parr R.G., Yang W. Oxford University Press; New York: 1989. Functional Theory of Atoms and Molecules. [Google Scholar]
- 64.Domingo L.R., Aurell M.J., Perez P., Contreras R. Quantitative characteri- zation of the global electrophilicity power of common diene/dienophile pairs in Diels-Alder reactions. Tetrahedron. 2002;58:417–423. [Google Scholar]
- 65.Sagaama A., Noureddine O., Brandán S.A., Jarczyk-Jędryka A., Flakus H.T., Ghalla H., Issaoui N. Molecular docking studies, structural and spectroscopic properties of monomeric and dimeric species of benzofuran-carboxylic acids derivatives: DFT calculations and biological activities. Comput. Biol. Chem. 2020;87 doi: 10.1016/j.compbiolchem.2020.107311. [DOI] [PubMed] [Google Scholar]
- 66.Sheikhi M., Shahab S., Filippovich L., Yahyaei H., Dikusar E., Khaleghian M. New derivatives of (E,E)-azomethines: design, quantum chemical modeling, spectroscopic (FT-IR, UV/Vis, polarization) studies, synthesis and their applications: experimental and theoretical investigations. J. Mol. Struct. 2018;1152:368–385. [Google Scholar]
- 67.Patil R., Das S., Stanley A., Yadav L., Sudhakar A., Varma A.K. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS One , 2010;5(8):e12029. doi: 10.1371/journal.pone.0012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stephen L.M., et al. DREIDING: a generic force field for molecular simulations. J. Phys. Chem. 1990;94(26):8897–8909. doi: 10.1021/j100389a010. [DOI] [Google Scholar]
- 69.Padhi A.K., Dandapat J., Uversky V.N., Tripathi T. Structural proteomics-driven targeted design of favipiravir-binding site in the RdRp of SARS-CoV-2 unravels susceptible hotspots and resistance mutations. Preprints. 2021 doi: 10.20944/preprints202105.0492.v1. [DOI] [Google Scholar]
- 70.Dharmendra K.Y., et al. Molecular insights into the interaction of Rons and Thieno [3,2-c]pyran analogs with SIRT6/COX-2: a molecular dynamics study. Sci. Rep. 2018;8:4777. doi: 10.1038/s41598-018-22972-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Braz H.L., Moraes Silveira de J.A., Marinho A.D.M.E., De M., Moraes Filho de M.O., Monteiro H.S., Jorge R.J. In silico study of azithromycin, chloroquine and hydroxychloroquine and their potential mechanisms of action against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;56(3) doi: 10.1016/j.ijantimicag.2020.106119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Musarrat F., Chouljenko V., Dahal A., Nabi R., Chouljenko T., Jois S.D., Kousoulas K.G. The anti-HIV drug nelfinavir mesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARSCoV-2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID-19 infections. J. Med. Virol. 2020;92(10):2087–2095. doi: 10.1002/jmv.25985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nutho B., Mahalapbutr P., Hengphasatporn K., Pattaranggoon N.C., Simanon N., Shigeta Y., Hannongbua S., Rungrotmongkol T. Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry. 2020;59(18):1769–1779. doi: 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- 74.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. Iscience. 2021;24(1) doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gupta S., Singh A.K., Kushwaha P.P., Prajapat K.S, Shuaib M., Senapati S., Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J. Biomol. Struct. Dyn. 2020;9:1–2. doi: 10.1080/07391102.2020.1776157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.