Abstract
Background
Candida albicans, the main human fungal pathogen, can cause fungal infection and seriously affect people's health and life. This study aimed to investigate the effects of ritonavir (RIT) on C. albicans and the correlation between SAP2 as well as ERG11 and drug resistance.
Results
Secreted aspartyl proteinases (Saps) activities and pathogenicity of C. albicans with different drug resistance were measured. M27‐A4 broth microdilution method was used to analyze the drug sensitivity of RIT combined with fluconazole (FCA) on C. albicans. After that, SAP2 and ERG11 mutations were examined by polymerase chain reaction (PCR) and sequencing, and quantitative real‐time PCR was utilized to determine the expression of the two genes. By analyzing pz values, the Saps activity of cross‐resistant strains was the highest, followed by voriconazole (VRC)‐resistant strains, FCA‐resistant strains, itraconazole (ITR)‐resistant strains, and sensitive strains. The pathogenicity of C. albicans in descending order was as follows: cross‐resistant strains, VRC‐resistant strains, ITR‐resistant strains, FCA‐resistant strains, and sensitive strains. With the increase of RIT concentrations, the Saps activity was gradually inhibited. Drug sensitivity results showed that there was no synergistic effect between RIT and FCA. Additionally, no gene mutation sites were found in SAP2 sequencing, and 17 synonymous mutations and 6 missense mutations occurred in ERG11 sequencing. Finally, the expression of SAP2 and ERG11 was significantly higher in the resistant strains compared with the sensitive strains, and there was a positive liner correlation between SAP2 and ERG11 messenger RNA expression (r = .6655, p < .001).
Conclusion
These findings may help to improve our understanding of azole‐resistant mechanisms of C. albicans and provide a novel direction for clinical therapeutics of C. albicans infection.
Keywords: Candida albicans, ERG11, SAP2, secreted aspartyl proteinases
These findings may help to improve our understanding of azole‐resistant mechanisms of C. albicans and provide a novel direction for clinical therapeutics of C. albicans infection.
Abbreviations
- C. albicans
Candida albicans
- FCA
fluconazole
- ITR
itraconazole
- RIT
ritonavir
- Saps
secreted aspartyl proteinases
- VRC
voriconazole
1. BACKGROUND
Candida albicans is a common opportunistic fungus that usually colonized in the gastrointestinal tract, reproductive tract, mouth and skin, and can cause a variety of diseases, from superficial mucosal diseases to life‐threatening systemic diseases. 1 Approximately 75% of women are likely to experience at least one vulvovaginal candidiasis event in their lifetime, and about 5% female population have recurrent infections. 2 Additionally, deep infection may remain existed or contribute to secondary candidiasis, which can seriously endanger people's health. 3 At present, the treatment of candidiasis mainly depends on antifungal drugs, such as polyenes, azoles and echinocandins. 4 However, with the widespread use of antibiotics and the changes in the living environment of C. albicans, 5 the morbidity and drug resistance of C. albicans are increasing year by year, which is a major problem in the clinical treatment of C. albicans infection. Therefore, it is vital to explore the pathogenic and drug‐resistant mechanisms of C. albicans, and to search for novel therapeutic strategies.
C. albicans has many pathogenic virulence properties, including the formation of biofilms, cell phenotypes transformation, adhesins on the cell surface and the expression of invasive proteases. 6 Secreted aspartyl proteinases (Saps), an invasive protease, are the primary virulence factor of C. albicans, and play important roles in the infection and pathogenesis of C. albicans. 7 Previous study had shown that Saps were closely related to the occurrence and development of vaginitis caused by C. albicans. 8 Therefore, it is possible to find drugs targeting Saps for the treatment of C. albicans infection. A study of Dos Santos 9 showed that a protease inhibitor ritonavir (RIT) significantly reduced the infectious rate of C. albicans in human immunodefeciency virus (HIV) patients, and indicated that RIT and Saps belonged to the aspartic protease family, which may be used as a new drug to inhibit Saps. Other studies have reported that 0.1 mg/ml RIT could significantly suppress adhesion of C. albicans to epithelial cells and acrylics, 10 as well as 8 mg/L of RIT inhibited 44% of growth in C. albicans. 11 When using RIT alone, due to the high concentration of RIT and the increase metabolic burden of human liver, it is necessary to adopt combination therapy against C. albicans.
SAP2, considered as a key virulence factor, can provide nutrients for its own growth by degrading macromolecular proteins on the mucosal surface, and simultaneously increase the ability of C. albicans to adhere and invade the host. Our previous research had found that the messenger RNA (mRNA) expression of SAP2 in the itraconazole (ITR)‐resistant strains was higher than that in the ITR‐sensitive strains, which indicated that SAP2 may be associated with the drug resistance of C. albicans. 12 Furthermore, lanosterol 14 ‐demethylase (14‐DM/ERG11p), an important component in ergosterol synthesis, is the target enzyme of azole drugs, and is encoded by Ergosterol 11 (EGR11). 13 Azole drugs, such as fluconazole (FCA) achieved antifungal effects by binding with ERG11 and inactivating the target enzymes. However, if ERG11 mutates, the molecular configuration of the target enzyme ERG11p will change, thus reducing the affinity between azole drugs and ERG11p and leading to drug resistant. Suchodolski et al. 14 found that the increased expression of ERG11 in C. albicans contributed to the increased production of ergosterol, and promoted drug resistance of strains. However, the expression of SAP2 and ERG11 in C. albicans with different drug resistance and the relationship between SAP2 as well as ERG11 and drug resistance remain unclear.
In this study, clinical C. albicans strains with different drug resistance were used, and the Saps activities in different drug resistant strains were measured in vitro and in vivo. After that, the effects of RIT on Saps activities in C. albicans were determined, and the synergistic effects of FCA combined with RIT on C. albicans were investigated. Further, SAP2 and ERG11 in different drug resistant strains were sequenced, and the relationship between SAP2 as well as ERG11 and drug resistance was explored. These findings will provide a theoretical basis for controlling the occurrence of drug resistance, and provide a new direction for clinical therapeutics of diseases caused by C. albicans.
2. METHODS
2.1. Experimental strains
A total of 50 clinical C. albicans strains isolated and identified by the Dermatovenereal Fungus Laboratory of The Second Hospital of Shanxi Medical University were used. Among them, there were 10 FCA‐resistant strains, 10 ITR‐resistant strains, 10 voriconazole (VRC)‐resistant strains, 10 FCA, ITR, or VRC‐sensitive strains, and 10 FCA, ITR and VRC cross‐resistant strains. Additionally, standard strain C. albicans ATCC1106, and quality control strains Candida krusei ATCC6258 and Candida parapsilosis ATCC22019 were purchased from the Fungus and Mycosi Research Center, Peking University Medical Science.
2.2. Detection of Saps activity in vitro
Base on the methods of Li et al. 15 and Barros et al., 16 the Saps activity was measured in vitro using bovine serum albumin (BSA) agar medium. Briefly, 50 clinical strains were inoculated in Sabouraud dextrose agar (SDA) Medium (Beijing Luqiao Technology Co. Ltd), and cultured at 25°C for 48 h. After that, a fresh single colony was selected, and the fungal suspension was adjusted to 1 × 107 CFU/ml with sterile saline. Afterwards, fungal suspension (5 μl) was inoculated to the center of the BSA agar medium (Beijing Luqiao Technology Co. Ltd), or the BSA agar medium containing 32, 8, 2, and 0 μg/ml RIT. After cultured at 37°C for 72 h, the size of colony ring and surrounding transparent ring were examined. The Saps activity was evaluated by the ratio of colony diameter to diameter of the dense white zone of precipitation around phospholipase (PL) positive colonies (pz value). 17 The formula of pz value was shown as follows: pz value = (colony diameter)/(colony diameter + the diameter of the transparent ring). The experiment was repeated three times for each strain.
2.3. In vivo pathogenicity of C. albicans with different drug resistance
A total of 60 SPF female mice with 6 weeks old were obtained from the Animal Center of Shanxi Medical University. All mice were free to drink and eat during the experiment. The mice were fed under controlled temperature (24 ± 2°C) and humidity (50 ± 5%) conditions, with a 12 h light/dark cycle. After 3 days of adaptive feeding, the mice were randomly divided into six groups (n = 10): cross‐resistant group, FCA‐resistant group, ITR‐resistant group, VRC‐resistant group, sensitive group, and control group. The clinical strains with different drug resistance were inoculated in SDA medium, and cultured at 25°C for 48 h. Sterile saline was used to adjust fungal suspension to 1 × 107 CFU/ml. The mice in the cross‐resistant group, FCA‐resistant group, ITR‐resistant group, VRC‐resistant group, and sensitive group were injected with 0.2 ml cross‐resistant fungal suspension, FCA‐resistant fungal suspension, ITR‐resistant fungal suspension, VRC‐resistant fungal suspension, and sensitive fungal suspension, respectively, via tail vein. 12 The mice in the control group were injected with 0.2 ml saline into caudal vein. The appetite and mental state of the mice were observed three times a day. After observed for 30 days, the mortality and survival rates were counted.
After the experiment, all the mice were killed by cervical dislocation. The protocol was approved by the Ethics Committee of the Second Hospital of Shanxi Medical University (approval number: [2019]YX[24D]).
2.4. Drug sensitivity assay under different conditions
The purified C. albicans was resuspended in the RPMI1640 medium (Beijing Luqiao Technology Co. Ltd), and the fungal suspension was adjusted to 5 × 103 CFU/ml by the RPMI1640 medium. The C. albicans suspension in a free state was prepared. The preparation of C. albicans suspension under biofilm condition was shown as follows. The fungal suspension was adjusted to 5 × 106 CFU/ml by the RPMI1640 medium. The suspension (100 μl) was added to each well of a 96‐well plate (except the negative control), and cultured at 37°C for 1 h. After washing with phosphate‐buffered saline (PBS) twice, 100 μl RPMI1640 medium was added, and then cultured at 37°C for 24 h. After washing with PBS three times, the medium was discarded, and the fungal suspension was cultured for another 24 h to form mature biofilm.
The sensitivity of RIT combined with FCA on C. albicans was determined using the Clinical and Laboratory Standards Institute (CLSI) standard M27‐A4 broth microdilution method as previously described. 18 The concentrations of FCA were prepared in double distilled water from 0.125 to 128 μg/ml by a two‐fold dilution method; while the concentrations of RIT were prepared in dimethyl sulfoxide from 2 to 256 μg/ml. Under free conditions, each concentration of FCA (100 μl) was added to a 96‐well plate, and then each concentration of RIT (100 μl) was added. Afterwards, the fungal suspension was added to each well of the 96‐well plate. The control groups were added 100 μl PRMI1640 medium. Under biofilm conditions, the biofilm of C. albicans was firstly established in a 96‐well plate, and different concentrations of FCA or RIT (100 μl) was added. RPMI1640 medium was used as the control. After cultured at 37°C for 48 h, the minimal inhibitory concentration (MIC) values were read based on the CLSI. 19 When MIC ≤ 2, the strains were identified as sensitive to FCA (s); when MIC = 4, the strains were dose‐dependent sensitive to FCA; when MIC ≥ 8, the strains were resistant to FCA (R).
The interaction between RIT and FCA on C. ablicans was analyzed based on the values of FICI.20, 21 The formula of FICI was shown as follows: FICI = (MICFCA in combination/MICFCA alone) + (MICRIT in combination/MICRIT alone). If FICI ≤ 0.5, FCA and RIT have synergistic effects; if 0.5 < FICI ≤ 1, FCA and RIT have additive effects; if 1 < FICI < 4, there is no interaction between FCA and RIT; if FICI ≥ 4, RIT and FCA have antagonistic effects.
2.5. DNA extraction, and SAP2 and ERG11 sequencing
DNA was extracted from C. albicans using OMEGA D3370 yeast DNA extraction kit (Guangzhou Feyou Biotechnology Co., Ltd) following the manufacturer's instructions. The primer sequences of ERG11 and SAP2 were shown in Table 1, and synthesized by Sangon Biotech (Shanghai) Co., Ltd. The volume of polymerase chain reaction (PCR) system was 25 μl, including 2X TaqPCR Mix 12.5 μl, forward primer 1 μl, reverse primer 1 μl, DNA 2.5 μl, and double distilled water 8 μl. The PCR reaction was initiated at 94°C for 10 min, followed by 94°C for 45 s, 52°C/48.5°C (SAP2/ERG11) for 45 s, and 72°C for 2 min, for a total of 30 cycles, and finally ended at 72°C for 10 min.
Table 1.
The primer sequences of ERG11 and SAP2
| Process | Primer | Sequence (5′‐3′) | Gene code | Length (bp) |
|---|---|---|---|---|
| PCR | SAP2 | F: ATGTTTTTAAAGAATATTTTCATTGCTCTTGC | NC032096 | 1197 |
| R: TTAGGTCAAGGCAGAAATACTGGA | ||||
| ERG11 | F: CAAGAAGATCATAACTCAAT | X13296 | 1641 | |
| R: CAGAACACTGAATCGAAAGA | ||||
| RT‐qPCR | SAP2 | F: TGGTATTCTTATGGGTGGTC | / | 357 |
| R: TTAGCAGCAGCAGTATCC | ||||
| ERG11 | F: CAAGTGGTTCATCAGCTTCAC | / | 272 | |
| R: TTATTTGTCCCGTGGCAG |
Abbreviations: PCR, polymerase chain reaction; RT‐qPCR, quantitative real‐time PCR.
The PCR products were purified and two directional sequenced by Sangon Biotech (Shanghai) Co., Ltd. The gene sequences of the PCR products were analyzed by Blast software, and then were compared with the known sequences in GenBank database (NC032096/X13296). Additionally, the sequences are translated into amino acid sequences to identify gene mutation sites by chromas and secentral softwares.
2.6. RNA extraction and real‐time quantification PCR
According to the manufacturer's protocols, column yeast total RNA extraction and purification kit (Sangon Biotech (Shanghai) Co., Ltd) utilized to isolate total RNA from C. albicans. Afterwards, the concentration and quality of total RNA were measured using a microplate reader. Subsequently, RNA was reverse‐transcribed to cDNA using a cDNA synthesis kit (Roche Diagnostics Products Co., Ltd.) following the manufacturer's recommendations. The primer sequences of ERG11 and SAP2 were shown in Table 1. The quantitative real‐time PCR (RT‐qPCR) reaction was begun at 95°C for 10 min, followed by a total of 45 cycles at 95°C for 10 s, 55°C for 15 s and 72°C for 20 s. ACT1 was served as a housekeeping gene, and the relative expression levels of ERG11 and SAP2 was calculated using the method. 22
2.7. Statistical analysis
Each experiment repeated three times (n = 3). SPSS 22.0 software (SPSS, Inc.) was utilized for statistical analyses. Data were expressed as mean ± SD) The Log‐rank test in Kaplan–Meier was used to compare the survival time among the groups. One‐way analysis of variance was applied for the comparison of multiple groups. p < .05 was considered as statistical significance. The correlation between SAP2 expression and ERG11 expression was analyzed using Pearson linear correlation coefficient.
3. RESULTS
3.1. Saps activity analysis of C. albicans in vitro
The Saps activity of all experimental strains with different drug resistance was determined. The pz values of cross‐resistant strains, FCA‐resistant strains, ITR‐resistant strains, VRC‐resistant strains and sensitive strains were 0.59 ± 0.023, 0.631 ± 0.01, 0.701 ± 0.028, 0.623 ± 0.011, and 0.795 ± 0.016, respectively. By the analysis of variance, there were significant differences of pz values among the cross‐resistant strains, FCA‐resistant strains, ITR‐resistant strains, VRC‐resistant strains and sensitive strains (p < .05, Figure 1A). The results indicated that the Saps activity of cross‐resistant strains was the highest, followed by the VRC‐resistant strains, FCA‐resistant strains, ITR‐resistant strains, and sensitive strains.
Figure 1.
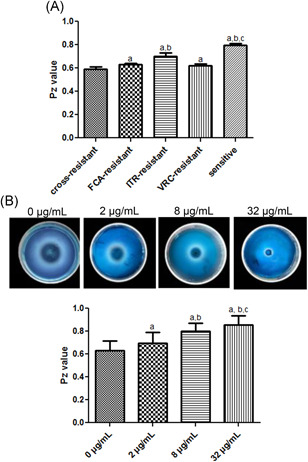
(A) pz values of Candida albicans with different drug resistance. a: p < .05, compared with the cross‐resistant strains. b: p < .05, compared with the FCA‐resistant strains. c: p < .05, compared with the ITR‐resistant strains. (B) Secreted aspartyl proteinases (Saps) activity of C. albicans treated with different concentrations of ritonavir (RIT). Upper: The formation of colony ring and transparent ring in C. albicans with different concentrations of RIT. Lower: pz value of C. albicans with different concentrations of RIT. a: p < .05, compared with the strains treated with 0 μg/ml RIT. b: p < .05, compared with the strains treated with 2 μg/ml RIT. c: p < .05, compared with the strains treated with 8 μg/ml RIT. FCA, fluconazole; ITR, itraconazole; VRC, voriconazole
Subsequently, different concentrations of RIT were used to treat C. albicans, and the effects of RIT on Saps activity were evaluated. It was obvious that all C. albicans formed transparent rings, that is, all these strains produced Saps (Figure 1B). Compared with the control group (0 μg/ml), the pz values in the strains treated with RIT were gradually increased with the increase of RIT concentrations (p < .05, Figure 1B). This suggested that with the increase of RIT concentrations, the Saps activity of C. albicans could be suppressed.
3.2. Pathogenicity analysis of C. albicans with different drug resistance in mice
The mice blood was cultured in SDA medium after the mice treated with C. albicnas, including cross‐resistant, FCA‐resistant, ITR‐resistant, VRC‐resistant and sensitive strains. After cultured at 37°C for 48 h, smooth milky white cheese‐like colonies were found (Figure 2A), which indicated that the fungemia mice model was successfully established. 23 There was no death in the control group during a 30‐day observation period. The median survival time of the cross‐resistant group, FCA‐resistant group, ITR‐resistant group, VRC‐resistant group, and sensitive group was 12, 22, 18, 13.5, and 20 days, respectively (Table 2). The survival rates at 10, 20, and 30 days in different groups were shown in Table 2. In addition, the survival curves showed that the survival rate in escalating order was as follows: cross‐resistant group, VRC‐resistant group, ITR‐resistant group, FCA resistant group and sensitive group (Figure 2B). The results indicated that the pathogenicity of cross‐resistant strains was the highest, followed by VRC‐resistant strains, ITR‐resistant strains, FCA‐resistant strains, and sensitive strains.
Figure 2.
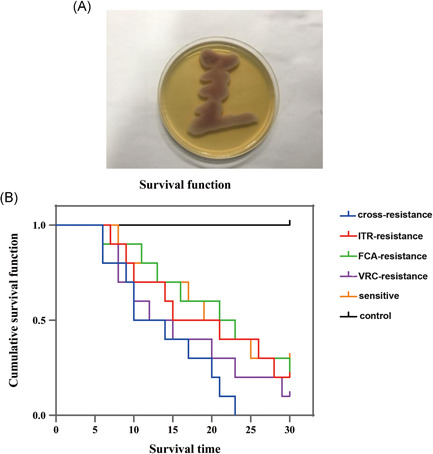
(A) The blood culture result of mice treated with C. albicans on Sabouraud dextrose agar medium at 37°C for 48 h. (B) Survival curves of mice treated with C. albicans with different drug resistance. FCA, fluconazole; ITR, itraconazole; VRC, voriconazole
Table 2.
Median survival time and survival rate of mice in each group
| Groups | Median survival time (day) | Survival rate (%) | Log‐rank test | |||
|---|---|---|---|---|---|---|
| 10 Day | 20 Day | 30 Day | χ 2 | P | ||
| Cross‐resistant | 12 | 50 | 20 | 0 | 25.62 | 0.00 |
| FCA‐resistant | 22 | 90 | 60 | 20 | ||
| ITR‐resistant | 18 | 70 | 50 | 20 | ||
| VRC‐resistant | 13.5 | 60 | 30 | 10 | ||
| Sensitive | 20 | 80 | 50 | 30 | ||
| Control | 30 | 100 | 100 | 100 | ||
Abbreviations: FCA, fluconazole; ITR, itraconazole; VRC, voriconazole.
3.3. Drug sensitivity under free and biofilm conditions
M27‐A4 broth microdilution method was carried out to determine the drug sensitivity of FCA combined with RIT on C. albicans under free and biofilm conditions. The MIC50 values of FCA combined with RIT under free and biofilm conditions were presented in Table 3. By analyzing FICI, it was found that under free conditions, additive effects of FCA and RIT were observed in 8 strains with 0.5 < FICI ≤ 1, and no interaction between FCA and RIT was shown in 42 strains with 1 < FICI ≤ 4 (Table 4). However, under biofilm conditions, additive effects of FCA and RIT were shown in 12 strains, and no interaction was in 38 strains (Table 4). These results showed that RIT and FCA had no synergistic effects on C. albicans with different drug resistance.
Table 3.
The minimal inhibitory concentration (MIC50) of fluconazole (FCA) combined with ritonavir (RIT) under free and biofilm conditions (unit: g/ml)
| Number | Free conditions (MIC50) | Biofilm conditions (MIC50) | ||||||
|---|---|---|---|---|---|---|---|---|
| FCA | FCA/RIT | RIT | FICI | FCA | FCA/RIT | RIT | FICI | |
| CA‐1 | 0.5 | 0.5 | >128 | 2 | 2 | 2 | >256 | 2 |
| CA‐4 | 2 | 2 | >128 | 2 | 8 | 4 | >256 | 1 |
| CA‐6 | 64 | 16 | >128 | 0.75 | 128 | 128 | >256 | 2 |
| CA‐7 | 32 | 32 | >128 | 2 | 128 | 128 | >256 | 2 |
| CA‐9 | 64 | 64 | >128 | 2 | 128 | 64 | >256 | 1 |
| CA‐10 | 4 | 4 | >128 | 2 | 16 | 8 | >256 | 1.5 |
| CA‐11 | 2 | 2 | >128 | 2 | 8 | 8 | >256 | 2 |
| CA‐12 | 2 | 0.5 | >128 | 0.75 | 8 | 8 | >256 | 2 |
| CA‐14 | 2 | 2 | >128 | 2 | 16 | 16 | >256 | 2 |
| CA‐15 | 2 | 2 | >128 | 2 | 8 | 2 | >256 | 0.75 |
| CA‐18 | 64 | 64 | >128 | 2 | 128 | 128 | >256 | 2 |
| CA‐20 | 0.12 | 0.12 | >128 | 2 | 0.5 | 0.5 | >256 | 2 |
| CA‐21 | 2 | 2 | >128 | 2 | 8 | 8 | >256 | 2 |
| CA‐22 | 1 | 1 | >128 | 2 | 4 | 4 | >256 | 2 |
| CA‐23 | 4 | 1 | >128 | 1.25 | 64 | 64 | >256 | 2 |
| CA‐24 | 16 | 16 | >128 | 2 | 64 | 64 | >256 | 2 |
| CA‐25 | 2 | 2 | >128 | 2 | 4 | 4 | >256 | 2 |
| CA‐26 | 64 | 64 | >128 | 2 | 128 | 128 | >256 | 2 |
| CA‐27 | 0.5 | 0.25 | >128 | 1.5 | 4 | 1 | >256 | 0.75 |
| CA‐28 | 1 | 1 | >128 | 2 | 4 | 4 | >256 | 2 |
| CA‐29 | 4 | 4 | >128 | 2 | 64 | 64 | >256 | 2 |
| CA‐30 | 0.25 | 0.25 | >128 | 2 | 0.5 | 0.5 | >256 | 2 |
| CA‐31 | 2 | 0.5 | >128 | 0.75 | 8 | 2 | >256 | 0.75 |
| CA‐32 | 32 | 16 | >128 | 1.5 | 128 | 128 | >256 | 2 |
| CA‐33 | 4 | 2 | >128 | 1 | 64 | 64 | >256 | 2 |
| CA‐35 | 0.5 | 0.5 | >128 | 2 | 2 | 1 | >256 | 1 |
| CA‐37 | 8 | 8 | >128 | 2 | 64 | 32 | >256 | 1 |
| CA‐38 | 2 | 1 | >128 | 1.5 | 8 | 8 | >256 | 2 |
| CA‐40 | 16 | 16 | >128 | 2 | 64 | 64 | >256 | 2 |
| CA‐41 | 8 | 8 | >128 | 2 | 64 | 64 | >256 | 2 |
| CA‐42 | 4 | 2 | >128 | 1 | 64 | 16 | >256 | 0.75 |
| CA‐45 | 8 | 8 | >128 | 2 | 32 | 32 | >256 | 2 |
| CA‐46 | 64 | 32 | >128 | 1 | 128 | 128 | >256 | 2 |
| CA‐48 | 16 | 16 | >128 | 2 | 64 | 64 | >256 | 2 |
| CA‐49 | 2 | 2 | >128 | 2 | 64 | 64 | >256 | 2 |
| CA‐52 | 32 | 32 | >128 | 2 | 128 | 128 | >256 | 2 |
| CA‐54 | 64 | 64 | >128 | 2 | 128 | 128 | >256 | 2 |
| CA‐56 | 64 | 32 | >128 | 1 | 128 | 128 | >256 | 2 |
| CA‐58 | 0.5 | 0.25 | >128 | 1.5 | 64 | 16 | >256 | 0.75 |
| CA‐59 | 64 | 32 | >128 | 1 | 128 | 128 | >256 | 2 |
| CA‐60 | 8 | 8 | >128 | 2 | 16 | 4 | >256 | 0.75 |
| CA‐61 | 2 | 1 | >128 | 1 | 16 | 16 | >256 | 2 |
| CA‐63 | 8 | 8 | >128 | 2 | 64 | 16 | >256 | 0.75 |
| CA‐66 | 0.5 | 0.25 | >128 | 1.5 | 1 | 64 | >256 | 1 |
| CA‐72 | 64 | 64 | >128 | 2 | 128 | 128 | >256 | 2 |
| CA‐77 | 1 | 1 | >128 | 2 | 8 | 8 | >256 | 2 |
| CA‐89 | 0.25 | 0.25 | >128 | 2 | 1 | 1 | >256 | 2 |
| CA‐90 | 4 | 2 | >128 | 1.5 | 32 | 16 | >256 | 1.2 |
| CA‐94 | 32 | 32 | >128 | 2 | 128 | 128 | >256 | 2 |
| CA‐99 | 0.25 | 0.25 | >128 | 2 | 0.5 | 0.5 | >256 | 2 |
Table 4.
The sensitization rate of FCA combined with RIT under free and biofilm conditions
| FCA combined with RIT | ||
|---|---|---|
| Free condition (rate) | Biofilm condition (rate) | |
| Synergistic (FICI ≤ 0.5) | 0 | 0 |
| Additive (0.5 < FICI ≤ 1) | 8 (16%) | 12 (24%) |
| No interaction (1 < FICI ≤ 4) | 42 (84%) | 38 (76%) |
| Antagonistic (FICI > 4) | 0 | 0 |
Abbreviations: FCA, fluconazole; FICI, fractional inhibitory concentration index; RIT, ritonavir.
3.4. SAP2 and ERG11 sequencing analyses
SAP2 gene sequencing results showed that among 50 strains of C. albicans, 45 strains were successfully sequenced, and no gene mutation sites were found in all sequencing strains.
For ERG11 gene sequencing, 43 strains of C. albicans were successfully sequenced, and 35 strains had base mutations, including 17 kinds of synonymous mutations (C363T, T462C, A504G, C558T, T696C, C805T, T1143C, A1167G, A1173G, A1230G, C1257T, T1350C, T1431C, C1443T, T1449C, A1587G, T1617C) and 6 kinds of missense mutations (T495A, C515T, A530C, T541C, G1609A, G1693C). As shown in Table 5, these mutations corresponded to the mutations of six different amino acids (D116E, T123I, K128T, Y132H, V488I, A516P). There were no mutations for standard strain ATCC11006.
Table 5.
Base mutation sites and amino acid substitution in ERG11 gene of C. albicans
| Strains | Resistance to FCA/ITR/VRC | Base mutation sits | Amino acid substitution |
|---|---|---|---|
| CA‐1 | VRC | T1431C | No |
| CA‐6 | FCA | C363T/T462C/C558T/T696C/C805T/A1587G | No |
| CA‐7 | FCA | T462C/T495A/C558T/T1143C/A1230G/C1257T/T1431C/A1587G/T1617C | D116E |
| CA‐9 | FCA/ITR/VRC | T462C/T495A/A530C/A1587G/T1617C | D116E/K128T |
| CA‐10 | VRC | T1431C | No |
| CA‐11 | ITR | T462C/T495A/A1167G/A1587G | D116E |
| CA‐14 | S | T462C/T495A/A1587G | D116E |
| CA‐15 | VRC | C363T/T462C/C558T/T696C/C805T/T1143C/A1173G/C1257T/T1350C/C1443T/T1449C/G1609A | V488I |
| CA‐18 | FCA/ITR/VRC | T462C/T495A/C515T/C1257T/A1587G | D116E/T123I |
| CA‐20 | S | C363T/C558T/C805T/A1167G/A1587G | No |
| CA‐23 | VRC | A1167G/C1443T/A1587G | No |
| CA‐26 | FCA/ITR/VRC | T462C/T495A/C558T/C1257T/T1350C/T1431C/A1587G/T1617C/G1693C | D116E/A516P |
| CA‐29 | ITR | T462C/T495A/A530C/C558T/C1257T | D116E/K128T |
| CA‐30 | VRC | T462C/T495A/A1230G/C1257T/T1350C/T1431C/A1587G/T1617C | D116E |
| CA‐31 | ITR | C363T/T462C/C558T/C805T/A1167G/A1587G | No |
| CA‐32 | FCA | T462C/T495A/A530C/A1587G | D116E/K128T |
| CA‐33 | ITR | T462C/T495A/A1587G | D116E |
| CA‐35 | ITR | C363T/T462C/C558T/T696C/C805T/T1143C/C1257T/T1350C/C1443T/T1449C/G1609A | V488I |
| CA‐37 | FCA/ITR/VRC | T462C/T495A/C558T/C805T/T1143C/A1173G/C1257T/T1350C/C1443T/T1449C | D116E |
| CA‐38 | VRC | T462C/T495A/A1587G | D116E |
| CA‐40 | FCA/ITR/VRC | T462C/T495A/A530C/A1587G | D116E/K128T |
| CA‐41 | FCA | T462C/T495A/A1167G/A1587G | D116E |
| CA‐42 | ITR | A1167G/A1587G | No |
| CA‐45 | FCA/ITR/VRC | C363T/T462C/A504G/C558T/T696C/C805T/A1587G | No |
| CA‐48 | FCA | A1167G/C1257T/A1587G | No |
| CA‐49 | ITR | A1587G | No |
| CA‐52 | FCA/ITR/VRC | C363T/T462C/C558T/C805T/T1143C/A1173G/T1350C/C1443T/T1449C | No |
| CA‐54 | FCA/ITR/VRC | C363T/T462C/C558T/T696C/C805T/A1173G/T1350C/T1449C/G1609A | V488I |
| CA‐56 | FCA/ITR/VRC | T462C/T495A/A504G/A530C/T541C/C558T | D116E/K128T/Y132H |
| CA‐59 | VRC | T462C/T495A/A1587G | D116E |
| CA‐60 | FCA | C363T/T462C/C558T/C805T/T1143C | No |
| CA‐61 | ITR | A1230G/T1431C | No |
| CA‐63 | FCA | A1587G | No |
| CA‐66 | ITR | T1431C | No |
| CA‐94 | FCA | C363T | No |
Abbreviations: A, alanine; D, aspartic acid; E, glutamic acid; FCA, fluconazole; FICI, fractional inhibitory concentration index; H, histidine; I, isoleucine; K, lysine; P, proline; RIT, ritonavir; S, sensitive to FCA, ITR and VRC; T, threonine; V, valine; Y, tyrosine.
In the sensitive strains, two strains showed synonymous mutations and one strains had missense mutations (T495A→D116E). In the drug resistant strains (cross‐resistant, FCA‐resistant, ITR‐resistant and VRC‐resistant strains), there were 17 synonymous mutations and 6 missense mutations. Among them, in the single drug resistant strains (FCA‐resistant, ITR‐resistant and VRC‐resistant strains), 11 of 24 strains showed 3 missense mutations (D116E, K128T, V488I). In the cross‐resistant strains, 7 of 9 strains displayed 6 missense mutations, including D116E, K128T, V488I, T123I, Y132H and A516P. The missense mutation rates of each group were presented in Table 6. It is clear that the missense mutation rate of cross‐resistant strains (77.8%) was higher than that in the sensitive strains (10%), FCA‐resistant strains (37.5%), ITR‐resistant strains (44.4%), and VRC‐resistant strains (57.1%). By Fisher's exact probability analysis, there was a statistically significant difference in the missense mutation rate between the cross‐resistant strains and sensitive strains (p = .0452).
Table 6.
Missense mutation rates of each group in ERG11 gene sequencing
| Group | Number of strains | Number of strains with missense mutation | Missense mutation rates |
|---|---|---|---|
| Sensitive | 10 | 1 | 10% |
| Cross‐resistant | 9 | 7 | 77.8% |
| FCA‐resistant | 8 | 3 | 37.5% |
| ITR‐resistant | 9 | 4 | 44.4% |
| VRC‐resistant | 7 | 4 | 57.1% |
Abbreviations: FCA, fluconazole; ITR, itraconazole; VRC, voriconazole.
3.5. Correlation between the expression of SAP2 and as well as ERG11 and drug resistance
The expression of SAP2 in the sensitive, cross‐resistant, FCA‐resistant, ITR‐resistant, and VRC‐resistant strains was 1.715 ± 0.576, 5.380 ± 1.39, 3.879 ± 1.125, 4.385 ± 1.02, and 3.534 ± 1.162, respectively (Figure 3A). Compared with the sensitive strains, SAP2 expression was significantly upregulated in the cross‐resistant strains and single drug resistant strains (p < .05). However, SAP2 expression in the FCA‐resistant strains and VRC‐resistant strains was significantly lower than that in the cross‐resistant strains (p < .05). For ERG11, its expression was markedly higher in the cross‐resistant strains and single drug resistant strains compared to the sensitive strains (p < .05, Figure 3B). Additionally, there was no significant difference in the ERG11 expression among cross‐resistant, FCA‐resistant, ITR‐resistant and VRC‐resistant strains (p > .05).
Figure 3.
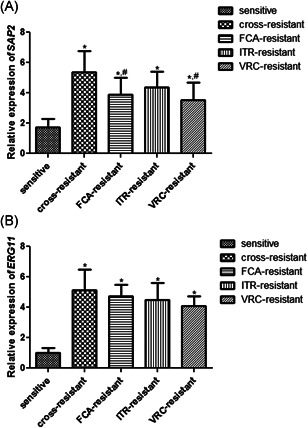
The mRNA expression of SAP2 (A) and ERG11 (B) in the sensitive, cross‐resistant, FCA‐resistant, ITR‐resistant and VRC‐resistant strains. *p < .05, compared with the sensitive strains; # p < .05, compared with the cross‐resistant strains. FCA, fluconazole; ITR, itraconazole; mRNA, messenger RNA; VRC, voriconazole
The results of Pearson linear correlation coefficient analysis showed that there was a positive liner correlation between SAP2 and ERG11 mRNA expression (r = .6655, p < .001).
4. DISCUSSION
C. albicans, the main human fungal pathogen, can cause fungal infection and seriously affect people's health and life. In this study, we found that all the 50 clinical strains formed transparent rings, and by analyzing pz values, the Saps activity of cross‐resistant strains was the highest, followed by the VRC‐resistant strains, FCA‐resistant strains, ITR‐resistant strains, and sensitive strains. The results of in vivo mice model showed that the pathogenicity of C. albicans in descending order was as follows: cross‐resistant strains, VRC‐resistant strains, ITR‐resistant strains, FCA‐resistant strains, and sensitive strains. When strains were treated with RIT, the Saps activity was inhibited with the increase of RIT concentrations. Drug sensitivity results showed that there was no synergistic effect between RIT and FCA on C. albicans with different drug resistance. SAP2 sequencing showed that no gene mutation sites were found in all sequencing strains. For ERG11 sequencing, 35 strains had base mutations, including 17 kinds of synonymous mutations and 6 kinds of missense mutations (T495A, C515T, A530C, T541C, G1609A, G1693C). Additionally, there was a positive liner correlation between SAP2 and ERG11 mRNA expression (r = .6655, p < .001).
In the previous study, the proteases of 37 C. albicans, 7 C. glabrata, 5 C. parapsilosis, and 12 C. tropicalis were determined, and it was found that 74.56% of the strains detected Saps, 44.73% of the strains detected PL, 24 which indicated that the production of Saps and PL may be the important virulence factor of Candida. Our research showed that 50 clinical strains of C. albicans cultured on BSA medium formed obvious transparent rings, which proved that each strains produced Saps. Price et al. 17 defined the pz value, and proposed that the smaller pz value was, the higher Saps activity was; pz value equalled to 1 meant the strains did not produce Saps. Sanitá et al. 25 isolated C. albicans from the healthy subjects (HS), diabetics (DOC) and non‐diabetics with oral candidiasis (NDOC), and measured the activity of Saps, as well as found that C. albicans from NDOC showed the lower Saps activity compared to the C. albicans from HS and DOC. Our results showed that Saps activity of cross‐resistant strains was significantly higher than that in the sensitive strains. It was speculated that the Saps activity of C. albicans may be associated with the source of strains and their drug resistance. However, the specific reasons should be further explored.
To evaluate the pathogenicity of C. albicans with different drug resistance, an intravenous infection model of systemic candidiasis was established. The mice were injected with fungal suspension by tail vein, and blood culture results proved the model was successfully constructed. In the current study, the median survival time of the cross‐resistant group, FCA‐resistant group, ITR‐resistant group, VRC‐resistant group, and sensitive group was 12, 22, 18, 13.5, and 20 days, respectively. This implied that the pathogenicity of cross‐resistant strains was the highest, followed by VRC‐resistant strains, ITR‐resistant strains, FCA‐resistant strains, and sensitive strains. Schaller et al. 26 reported that C. albicans lacking SAP1 or SAP2 could reduce tissue damage and decrease the expression of proinflammatory cytokines, which indicated that the potential for Saps to cause tissue damage may be related to epithelial‐induced proinflammatory cytokine responses. Another study showed that some Saps, particularly Sap2, could mediate the acute inflammatory response of inflammatory corpus‐dependent vaginal epithelial cells to C. albicans. 8 Combined with our results, it was further proved that Saps activity may be closely related to the pathogenicity of C. albicans, and it was speculated that higher Saps activity in drug‐resistant strains may enhance the adhesion of C. albicans, promote tissue damage and inflammatory response, resulting in high mortality of mice. However, more studies are needed to be performed to investigate the potential mechanisms.
RIT has been reported to be a kind of HIV protease inhibitors, and to directly inhibit Saps of C. albicans. 27 A study of Tsang et al. 28 showed that when C. albicans exposed to 100 μmol/L RIT, its adhesion decreased by 50%. In our study, different concentrations of RIT were used to treat with C. albicans, and the pz values were determined. It was obvious that the Saps activity of C. albicans was gradually inhibited with the increase concentrations of RIT. Therefore, we hypothesized that RIT could suppress toxicity of C. albicans by inhibiting Saps activity, and RIT combined with azole drugs (FCA) may have synergistic effects on the inhibition of C. albicans. To verify the hypothesis, drug sensitivity experiments were carried out. The results showed that under free and biofilm conditions, RIT and FCA had no synergistic effects on C. albicans; the additive rates of RIT combined with FCA were respectively 16% and 24%; others displayed no interaction between FCA and RIT. Previous study has indicated that RIT decreased the growth of C. albicans and the activity of Saps in a nitrogen‐limited medium, the yeast carbon base and BSA (YCB‐BSA). 11 After comparing the components of RPMI1640 medium and YCB‐BSA medium, we speculated that the reasons may be the different growing environment of C. albicans and the activity of Saps. Therefore, the drug sensitivity assays of RIT combined with FCA are needed to be further performed on the YCB‐BSA medium, or the synergistic effects between FCA and RIT should be confirmed through an animal model.
Saps are the primary virulence factor of C. albicans, and Sap2 is an important extracellular protease in the Saps family. Whether SAP2 mutates in nature and its relationship with candida albicans resistance to azole drugs are unknown. In this study, SAP2 gene was sequenced, and it was found that there were no gene mutation sites in all sequencing strains. The possible reasons can be that SAP2 gene sequence is relatively conservative and rarely mutates, which may have nothing to do with the drug resistance of C. albicans. However, some scholars believed that the high expression of SAP2 in C. albicans was associated with drug resistance. Copping et al. 29 found that the expression of SAP2 in the FCA‐resistant strains was higher than that in the sensitive strains, which indicated that SAP2 may be related to the drug resistance of C. albicans. Another study demonstrated that upregulation of SAP2 in C. albicans exposed to FCA may be an adaptive mechanism. 30 Our results showed that the mRNA expression of SAP2 was significantly upregulated in the resistant strains compared with the sensitive strains, and the SAP2 expression was the highest in the cross‐resistant strains. Therefore, we speculated that high expression of SAP2 may be related to the drug resistance of C. albicans.
Previous study has implied that the drug resistance of C. albicans is a complex process involving multiple mechanisms.18, 31 ERG11 has been proved to play an important role in drug resistance. We sequenced ERG11 gene in the all clinical strains, and found 17 kinds of synonymous mutations and 6 kinds of missense mutations. These missense mutations corresponded to six different amino acids substitutions, including D116E, T123I, K128T, Y132H, V488I, A516P. D116E represented that after T495A missense mutation, the encoded amino acid was changed from aspartic acid to glutamic acid at 116th position. Our study showed that D116E was occurred not only in the sensitive strains, but also in single drug‐resistant strains and cross‐resistant strains, which indicated that D116E may not be connected with drug resistance. The results were in accordance with previous study. 32 K128T and V488I substitutions were observed in the single drug resistant strains and the cross‐resistant strains. Wang et al. 33 reported that the point mutation of K128T could not affect C. albicans resistance to FCA, which was inconsistent with our results. Thus, the relationship between K128T mutation and resistance to azoles should be further explored. Manastir et al. 34 demonstrated that V488I mutation in ERG11 gene were determined in the FCA‐resistant isolates, and may be associated with the azoles resistance of C. albicans. In addition, a study of Wu et al. 35 indicated that substitution T123I and Y132H in ERG11 gene could confer resistance to FCA and VRC. Our results showed that T123I and Y132H substitutions were only found in cross‐resistant strains, which hinted that T123I and Y132H mutations in ERG11 may be lead to cross‐resistance of C. albicans to azoles. A516P appeared in the cross‐resistant strains, and was a new mutation point that was not found in the previous papers. Consequently, further studies are needed to be carried out on the resistance of C. albicans to azoles caused by A516P.
In addition, the upregulation of ERG11 in C. albicans can promote ergosterol synthesis, thus increasing the drug resistance. However, an in vitro study had found this resistance is reversible, suggesting that the high expression of ERG11 may be an adaptive mechanism in the resistant strains, and not directly related to the resistance of C. albicans to azoles. 36 In this study, RT‐qPCR showed that the expression of ERG11 was markedly higher in the cross‐resistant strains and single drug resistant strains compared to the sensitive strains. The results were consistent with the previous reports. 37 Because of the complexity of drug resistance mechanisms, we further analyzed the relationship between SAP2 as well as ERG11 expression and drug resistance. Pearson linear correlation coefficient analysis indicated that there was a positive liner correlation between SAP2 and ERG11 mRNA expression. This implied that ERG11 and SAP2 may have a certain relationship in C. albicans, which jointly lead to the emergence of resistance to azole drugs.
5. CONCLUSION
In conclusion, the Saps activity and pathogenicity of C. albicans in the cross‐resistant strains were the highest, and RIT could inhibit the Saps activity. However, there was no synergistic effect between RIT and FCA on C. albicans with different drug resistance. Additionally, the expression of SAP2 and ERG11 was higher expressed in the resistant strains, and there was a positive liner correlation between SAP2 and ERG11 mRNA expression. All these results may help to improve our understanding of azole‐resistant mechanisms of C. albicans and provide a novel direction for clinical therapeutics of diseases caused by C. albicans.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Wenli Feng and Jing Yang designed the research. Wenli Feng, Jing Yang, Yan Ma, Zhiqin Xi, and Xiaoqin Zhao did the experiment and obtained the data. Xiaoxia Zhao and Min Zhao analyzed and explained the data. Wenli Feng and Jing Yang drafted the manuscript, and Wenli Feng revised. All authors have read and approved the final version.
ACKNOWLEDGMENTS
This study was supported by Research and Development Key Projects of Shanxi Province (Project number: 201903D321123 and 201603D321063), Basic Research Project supported by Shanxi Province (Project number: 201701D121171), Research Project Supported by Health Commission of Shanxi Province (Project number: 201601050, and 2018048), the General Project of the National Natural Science Foundation of China (Project number: 82072262) and the 5th of emerging industry leading talents projects in Shanxi Province. The funders had roles in design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Feng W, Yang J, Ma Y, et al. The effects of secreted aspartyl proteinase inhibitor ritonavir on azoles‐resistant strains of Candida albicans as well as regulatory role of SAP2 and ERG11 . Immun Inflamm Dis. 2021;9:667‐680. 10.1002/iid3.415
Contributor Information
Wenli Feng, Email: wenlifeng2010@163.com.
Jing Yang, Email: yangjing7962@126.com.
DATA AVAILABILITY STATEMENT
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Wang Y. Looking into Candida albicans infection, host response, and antifungal strategies. Virulence. 2015;6(4):307‐308. 10.1080/21505594.2014.1000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sobel JD. Vulvovaginal candidosis. Lancet. 2007;369(9577):1961‐1971. 10.1016/S0140-6736(07)60917-9 [DOI] [PubMed] [Google Scholar]
- 3. Vila T, Romo JA, Pierce CG, McHardy SF, Saville SP, Lopez‐Ribot JL. Targeting Candida albicans filamentation for antifungal drug development. Virulence. 2017;8(2):150‐158. 10.1080/21505594.2016.1197444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Odds FC, Brown AJ, Gow NA. Antifungal agents: mechanisms of action. TIM. 2003;11(6):272‐279. 10.1016/s0966-842x(03)00117-3 [DOI] [PubMed] [Google Scholar]
- 5. Xie JL, Polvi EJ, Shekhar‐Guturja T, Cowen LE. Elucidating drug resistance in human fungal pathogens. Future Microbiol. 2014;9(4):523‐542. 10.2217/fmb.14.18 [DOI] [PubMed] [Google Scholar]
- 6. Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119‐128. 10.4161/viru.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Staniszewska M, Bondaryk M, Pilat J, Siennicka K, Magda U, Kurzatkowski W. Virulence factors of Candida albicans Przegl Epidemiol. 2012;66(4):629‐633. [PubMed] [Google Scholar]
- 8. Pericolini E, Gabrielli E, Amacker M, et al. Secretory aspartyl proteinases cause vaginitis and can mediate vaginitis caused by Candida albicans in mice. mBio. 2015;6(3):e00724. 10.1128/mBio.00724-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dos Santos AL. HIV aspartyl protease inhibitors as promising compounds against Candida albicans Andre Luis Souza dos Santos. World J Biol Chem. 2010;1(2):21‐30. 10.4331/wjbc.v1.i2.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gruber A, Berlit J, Speth C, et al. Dissimilar attenuation of Candida albicans virulence properties by human immunodeficiency virus type 1 protease inhibitors. Immunobiology. 1999;201(1):133‐144. 10.1016/S0171-2985(99)80052-7 [DOI] [PubMed] [Google Scholar]
- 11. Blanco MT, Hurtado C, Perez‐Giraldo C, Moran FJ, Gonzalez‐Velasco C, Gomez‐Garcia AC. Effect of ritonavir and saquinavir on Candida albicans growth rate and in vitro activity of aspartyl proteinases. Med Mycol. 2003;41(2):167‐170. 10.1080/mmy.41.2.167.170 [DOI] [PubMed] [Google Scholar]
- 12. Feng W, Yang J, Pan Y, Xi Z, Qiao Z, Ma Y. The correlation of virulence, pathogenicity, and itraconazole resistance with SAP activity in Candida albicans strains. Can J Microbiol. 2016;62(2):173‐178. 10.1139/cjm-2015-0457 [DOI] [PubMed] [Google Scholar]
- 13. Kushima H, Tokimatsu I, Ishii H, Kawano R, Watanabe K, Kadota JI. A new amino acid substitution at G150S in lanosterol 14‐alpha demethylase (Erg11 protein) inmulti‐azole‐resistant trichosporonasahii. Medical mycology journal. 2017;58(1):E23‐E28. 10.3314/mmj.16-00027 [DOI] [PubMed] [Google Scholar]
- 14. Suchodolski J, Muraszko J, Korba A, Bernat P, Krasowska A. Lipid composition and cell surface hydrophobicity of Candida albicans influence the efficacy of fluconazole‐gentamicin treatment. Yeast. 2020;37(1):117‐129. 10.1002/yea.3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li W, Yu D, Gao S, Lin J, Chen Z, Zhao W. Role of Candida albicans‐secreted aspartyl proteinases (Saps) in severe early childhood caries. Int J Mol Sci. 2014;15(6):10766‐10779. 10.3390/ijms150610766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barros LM, Boriollo MF, Alves AC, Klein MI, Goncalves RB, Hofling JF. Genetic diversity and exoenzyme activities of Candida albicans and Candida dubliniensis isolated from the oral cavity of Brazilian periodontal patients. Arch Oral Biol. 2008;53(12):1172‐1178. 10.1016/j.archoralbio.2008.06.003 [DOI] [PubMed] [Google Scholar]
- 17. Price MF, Wilkinson ID, Gentry LO. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia. 1982;20(1):7‐14. 10.1080/00362178285380031 [DOI] [PubMed] [Google Scholar]
- 18. Feng W, Yang J, Yang L, et al. Research of Mrr1, Cap1 and MDR1 in Candida albicans resistant to azole medications. Exp Ther Med. 2018;15(2):1217‐1224. 10.3892/etm.2017.5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng W, Yang J, Ji Y, et al. Mrr2 mutations and upregulation are associated with increased fluconazole resistance in Candida albicans isolates from patients with vulvovaginal candidiasis. Lett Appl Microbiol. 2020;70(2):95‐101. 10.1111/lam.13248 [DOI] [PubMed] [Google Scholar]
- 20. Santos DA, Hamdan JS. In vitro antifungal oral drug and drug‐combination activity against onychomycosis causative dermatophytes. Med Mycol. 2006;44(4):357‐362. 10.1080/13693780500536893 [DOI] [PubMed] [Google Scholar]
- 21. Tamura T, Asahara M, Yamamoto M, et al. In vitro susceptibility of dermatomycoses agents to six antifungal drugs and evaluation by fractional inhibitory concentration index of combined effects of amorolfine and itraconazole in dermatophytes. Microbiol Immunol. 2014;58(1):1‐8. 10.1111/1348-0421.12109 [DOI] [PubMed] [Google Scholar]
- 22. Xu G, Ao R, Zhi Z, Jia J, Yu B. miR‐21 and miR‐19b delivered by hMSC‐derived EVs regulate the apoptosis and differentiation of neurons in patients with spinal cord injury. J Cell Physiol. 2019;234(7):10205‐10217. 10.1002/jcp.27690 [DOI] [PubMed] [Google Scholar]
- 23. Kovacs R, Czudar A, Horvath L, Szakacs L, Majoros L, Konya J. Serum interleukin‐6 levels in murine models of Candida albicans infection. Acta Microbiol Immunol Hung. 2014;61(1):61‐69. 10.1556/AMicr.61.2014.1.6 [DOI] [PubMed] [Google Scholar]
- 24. Das VM, Ballal M. Proteinase and phospholipase activity as virulence factors in Candida species isolated from blood. Revista Iberoamericana de Micologia. 2008;25(4):208‐210. [DOI] [PubMed] [Google Scholar]
- 25. Sanita PV, Zago CE, Pavarina AC, Jorge JH, Machado AL, Vergani CE. Enzymatic activity profile of a Brazilian culture collection of Candida albicans isolated from diabetics and non‐diabetics with oral candidiasis. Mycoses. 2014;57(6):351‐357. 10.1111/myc.12162 [DOI] [PubMed] [Google Scholar]
- 26. Schaller M, Korting HC, Borelli C, Hamm G, Hube B. Candida albicans‐secreted aspartic proteinases modify the epithelial cytokine response in an in vitro model of vaginal candidiasis. Infect Immun. 2005;73(5):2758‐2765. 10.1128/IAI.73.5.2758-2765.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pichova I, Pavlickova L, Dostal J, et al. Secreted aspartic proteases of Candida albicans, Candida tropicalis, Candida parapsilosis and Candida lusitaniae. Inhibition with peptidomimetic inhibitors. Eur J Biochem. 2001;268(9):2669‐2677. 10.1046/j.1432-1327.2001.02152.x [DOI] [PubMed] [Google Scholar]
- 28. Tsang CS, Hong I. HIV protease inhibitors differentially inhibit adhesion of Candida albicans to acrylic surfaces. Mycoses. 2010;53(6):488‐494. 10.1111/j.1439-0507.2009.01743.x [DOI] [PubMed] [Google Scholar]
- 29. Copping VM, Barelle CJ, Hube B, Gow NA, Brown AJ, Odds FC. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J Antimicrob Chemother. 2005;55(5):645‐654. 10.1093/jac/dki088 [DOI] [PubMed] [Google Scholar]
- 30. do Rosario Esteves Guimaraes C, de Freitas HF, Barros TF. Upregulation of secreted aspartyl proteinase genes of fluconazole‐sensitive Candida albicans isolates. Mol Biol Rep. 2019;46(6):6147‐6154. 10.1007/s11033-019-05049-2 [DOI] [PubMed] [Google Scholar]
- 31. Feng W, Yang J, Xi Z, et al. Regulatory role of ERG3 and Efg1 in azoles‐resistant strains of Candida albicans isolated from patients diagnosed with vulvovaginal Candidiasis. Indian J Microbiol. 2019;59(4):514‐524. 10.1007/s12088-019-00833-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sardari A, Zarrinfar H, Mohammadi R. Detection of ERG11 point mutations in Iranian fluconazole‐resistant Candida albicans isolates. Curr Med Mycol. 2019;5(1):7‐14. 10.18502/cmm.5.1.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang YB, Wang H, Guo HY, Zhao YZ, Luo SQ. Analysis of ERG11 gene mutation in Candida albicans. Di 1 Jun Yi Da Xue Xue Bao. 2005;25(11):1390‐1393. [PubMed] [Google Scholar]
- 34. Manastir L, Ergon MC, Yucesoy M. Investigation of mutations in Erg11 gene of fluconazole resistant Candida albicans isolates from Turkish hospitals. Mycoses. 2011;54(2):99‐104. 10.1111/j.1439-0507.2009.01766.x [DOI] [PubMed] [Google Scholar]
- 35. Wu Y, Gao N, Li C, Gao J, Ying C. A newly identified amino acid substitution T123I in the 14alpha‐demethylase (Erg11p) of Candida albicans confers azole resistance. FEMS Yeast Res. 2017;17(3), 10.1093/femsyr/fox012 [DOI] [PubMed] [Google Scholar]
- 36. Ribeiro MA, Paula CR. Up‐regulation of ERG11 gene among fluconazole‐resistant Candida albicans generated in vitro: is there any clinical implication? Diagn Microbiol Infect Dis. 2007;57(1):71‐75. 10.1016/j.diagmicrobio.2006.04.019 [DOI] [PubMed] [Google Scholar]
- 37. Feng W, Yang J, Xi Z, et al. Mutations and/or overexpressions of ERG4 and ERG11 genes in clinical azoles‐resistant isolates of Candida albicans. Microb Drug Resist. 2017;23(5):563‐570. 10.1089/mdr.2016.0095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


