Abstract
Despite the frequent detection of KRAS driver mutations in patients with colorectal cancer (CRC), no effective treatments that target mutant KRAS proteins have been introduced into clinical practice. In this study, we identified potential effector molecules, based on differences in gene expression between CRC patients carrying wild-type KRAS (n = 390) and those carrying KRAS mutations in codon 12 (n = 240). CRC patients with wild-type KRAS harboring mutations in HRAS, NRAS, PIK3CA, PIK3CD, PIK3CG, RALGDS, BRAF, or ARAF were excluded from the analysis. At least 11 promising candidate molecules showed greater than two-fold change between the KRAS G12 mutant and wild-type and had a Benjamini-Hochberg-adjusted P value of less than 1E-08, evidence of significantly differential expression between these two groups. Among these 11 genes examined in cell lines transfected with KRAS G12 mutants, BMP4, PHLDA1, and GJB5 showed significantly higher expression level in KRAS G12A, G12D, and G12V transfected cells than in the wild-type transfected cells. We expect that this study will lead to the development of novel treatments that target signaling molecules functioning with KRAS G12-driven CRC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11010-021-04172-8.
Keywords: BMP4, PHLDA1, Colorectal cancer, KRAS mutation, Therapeutic targets
Introduction
The incidence and mortality rates of colorectal cancer (CRC) have recently been increasing in Japan [1]. Surgical resections can cure CRC in the early stage, and advances in pharmacotherapy have also improved the treatment outcomes in patients with unresectable and advanced/recurrent-stage CRC. However, the five-year survival rate in patients with advanced stage IV CRC is quite low at approximately 18% [2]. Therefore, new therapeutic drugs, particularly molecular targeted agents with fewer adverse drug reactions, need to be developed for improving the prognosis in CRC patients [3]. Advanced CRC is typically treated with monoclonal antibodies targeting epidermal growth factor receptor (EGFR), such as cetuximab or panitumumab, used alone or in combination with standard chemotherapy, but CRC patients harboring KRAS mutations do not respond to the antibody-based anti-EGFR treatment [4].
RAS proteins, including KRAS as one of the molecules that play a central role in intracellular signaling pathways, appear to be involved in a wide range of processes including cell proliferation, differentiation, metabolism, and cell death [5–7]. Therefore, drugs that directly target RAS proteins that are ubiquitously expressed as house-keeping genes are more likely to have unanticipated reactions with other proteins in the body [8]. Wild-type KRAS has been shown to act as a tumor suppressor gene during the differentiation of myeloid cells [9] and inhibit lung carcinogenesis in murine teratomas [10]. Literature surveys suggest that the wild-type KRAS could play an onco-suppressor role [11–13]. KRAS mutations are observed in approximately 40% of patients with CRC and occur frequently in codon 12 or 13 and less frequently in codons 146 or 61. A study focusing on immortalized human bronchial epithelial cells reported differences in the degree of constitutive activation of the KRAS protein, rates of increase in tumor cell proliferation, and the degree of activation of proliferative signals downstream of KRAS, depending on the mutation sites in the KRAS gene [14]. In addition, downstream effector molecules of KRAS signaling pathways were shown to differ according to tumor type [15]. These observations raised the possibility that the mechanism by which activated KRAS binds preferentially to its downstream partners’ genes, and how these interactions after cell determination, may differ among humans.
KRAS mutations are considered to occur during initiation or early event in colorectal carcinogenesis [16, 17], but not in the malignant progression of CRC because it has been found in dysplastic lesions and adenomatous polyps, and such mutations alone are insufficient for the sustained growth of cancer. Once the KRAS mutations occur, the KRAS activation signaling will be sustained for over 10 years in the somatic evolution of adult cancers. More specifically, the presence of KRAS mutations alone is considered to be insufficient for malignant transformations unless they function in cooperation with a particular set of other cancer-related genes in vivo. If this is true, identification of signaling molecules functioning in cooperation with KRAS may allow for the development of a new strategy for suppressing cancer without the use of KRAS inhibitors. MEK inhibitors are being evaluated for their clinical efficacy in targeting CRC with KRAS mutations and have a greater dependence on MAPK pathway signaling [18]; however, it seems that MAPK pathway inhibition during the treatment of CRC with KRAS mutation remains elusive [6, 19, 20]. Furthermore, studies have shown that MEK inhibitors did not improve overall survival in patients with advanced non-small cell lung cancer (NSCLC) [21] or pancreatic cancer [22] harboring KRAS mutations. An effective combination therapy using TBK1 and MEK or BET inhibitors has also been reported in aggressive murine KRAS-driven lung cancer [23]. In addition to MEK inhibitors, a recent study revealed that a covalent KRAS inhibitor could inhibit tumor cell growth in NSCLC with KRAS G12C mutation [24, 25], but not in CRC [26].
Although many KRAS-associated molecules play an important role in regulating KRAS transcription [27], the regulatory mechanisms underlying its activation in vivo have not been fully elucidated. In this study, we first comprehensively analyzed the mutations and expressions of known genes involved in the KRAS signaling pathway in patients with CRC. The KRAS G12 mutation is found at a characteristically high frequency and is associated with worse overall survival in patients with CRC [28]. Therefore, next, we explored the potential effector molecules whose gene expression levels differed between CRC patients with wild-type KRAS and those with a KRAS mutation in codon 12. We then validated these candidate genes by transfecting KRAS mutants into human cells. Effective therapies targeting KRAS signaling pathway have not yet been introduced in clinical practice. Moreover, RAS proteins have been dismissed as undruggable targets for many years (5, 6). We hope that this study paves the way for the development of novel treatments that target signaling molecules functioning in the KRAS G12-driven CRC.
Materials and methods
Subjects
We performed the Whole Exome Sequencing (WES) and Comprehensive Cancer Panel (CCP) using blood samples and fresh surgical specimens. We then conducted Gene Expression Profiling (GEP) using matched tumor and adjacent normal tissues from each patient. Tumor-specific single nucleotide variants (SNVs) were determined by comparing tumor tissue with blood cell data from the same patient. Between January 2014 and January 2017, the samples were obtained from 906 patients with CRC treated with surgery at the Shizuoka Cancer Center Hospital, Shizuoka, Japan (Table 1).
Table 1.
Characteristics of the colorectal cancer patients
| KRAS wild-type | KRAS mutated | aP value | |
|---|---|---|---|
| Total number | 534 | 372 | |
| Tumor type | |||
| Colon | 302 | 194 | |
| Rectum | 232 | 178 | 0.20 |
| Location of the primary tumor | |||
| Anal | 3 | 1 | |
| Ascending | 69 | 76 | |
| Cecum | 18 | 37 | |
| Descending | 21 | 8 | |
| Sigmoid | 132 | 57 | |
| Transverse | 59 | 15 | |
| Rectum | 232 | 178 | |
| Clinical stage | |||
| Stage I | 39 | 52 | |
| Stage II | 123 | 89 | |
| Stage III | 303 | 187 | |
| Stage IV | 63 | 42 | |
| Unknown | 6 | 2 | |
| Age, y | |||
| <45 | 39 | 13 | |
| 46–55 | 58 | 24 | |
| 56–65 | 128 | 56 | |
| ≥66 | 309 | 85 | |
| Gender | |||
| Male | 337 | 194 | |
| Female | 197 | 178 | 0.001 |
| Smoking status | |||
| Nonsmokers | 188 | 165 | |
| Smokers | 346 | 207 | 0.006 |
| Unknown | 0 | 0 | |
| Pack-yearsb | |||
| 0 | 188 | 165 | |
| Light smokers (>0 to <20) | 90 | 69 | |
| Heavy smokers (≥20) | 237 | 129 | 0.079 |
| Smokers but pack-years unknown | 19 | 9 | |
| Unknown | 0 | 0 | |
| Drinking status | |||
| Nondrinkers | 118 | 98 | |
| Drinkers | 334 | 209 | 0.09 |
| Unknown | 82 | 65 | |
aP value by Fisher exact test
bPack-years defined as number of packs of cigarettes smoked per day times of years of smoking
WES/CCP and GEP were performed using the Ion Proton system and Agilent system, respectively. Details of the experimental procedures have been described in previous reports [29–32].
Ethical statement
All experimental protocols were approved by the Institutional Review Board at the Shizuoka Cancer Center (Authorization Number: 25–33). Written informed consent was obtained from all patients for the participation in this study. All experiments using clinical samples were carried out in accordance with the approved guidelines [33].
Cell lines
The human 293 embryonic kidney cell line and human CRC cell line, Caco-2, were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum at 37 °C in 5% CO2. Both 293 and Caco-2 cells have wild-type KRAS as well as BRAF and PIK3CA, which are direct downstream effectors of RAS signaling.
Construction of KRAS expression vector
To construct the KRAS cDNA expression vectors to transduce the entire KRAS coding exons representing either the mutant or wild-type forms, the respective cDNA was synthesized using a 1 μg of total RNA isolated from normal breast tissue. The cDNA was amplified using the primers for the KRAS sequence including a Kozak translation initiation sequence containing an ATG initiation codon for proper initiation of translation. The polymerase chain reaction (PCR) products were cloned into the pcDNA3.1 D/V5-His vector (Thermo Fisher Scientific) downstream to the human cytomegalovirus promoter to express the KRAS protein fused with a V5-epitope tag at its C-terminus. Site-directed mutagenesis was performed according to the manufacturer’s protocol (In-Fusion HD Cloning Kit, TaKaRa, Japan). The resulting pcDNA3.1D/V5-His/KRAS vectors were designated as pKRAS-WT, pKRAS-A, pKRAS-C, pKRAS-D, pKRAS-R, pKRAS-S, and pKRAS-V, and they harbored wild-type, G12A, G12C, G12D, G12R, G12S, and G12V mutants at codon12 of the KRAS cDNA, respectively. A pcDNA3.1 D/V5-His/LacZ (named pLacZ) served as a negative control.
Transfection of KRAS expression vectors into cells
The 293 cells had a high transfection efficiency (90% or more), and the Caco-2 cells were transfected using TransIT-293 transfection reagent (Mirus Bio LLC, Madison) or Lipofectamine 3000 (Invitrogen) and Opti-MEM, as previously described [34]. The cells were seeded at 3–5 × 105 cells/well in 6-well plates; 24–48 h later, when the cells reached 70–80% confluence, they were transfected with pKRAS-WT, pKRAS-A, pKRAS-C, pKRAS-D, pKRAS-R, pKRAS-S, pKRAS-V, or pLacZ expression vector. After 4–5 h, the medium was replaced with DMEM, and the cells were incubated for 24 or 48 h.
Western blot analyses of transfected cells
Western blot analyses of the cells transfected with either of the vectors indicated above were performed essentially as described [34]. The protein samples were size fractionated using a gradient 12% SDS polyacrylamide gel, and a commercially available antibodies were used for the detection of the V5 peptide tag (Thermo Fisher Scientific) and β-actin protein (Sigma Chemical Co, St. Louis, MO).
Validation of candidate genes using real-time quantitative RT-PCR analysis
A total RNA from cells transfected with pKRAS expression vectors as described above was isolated using Isogen reagent (Nippon Gene, Japan), and the cDNA was synthesized. The cDNA was subjected to the real-time quantitative RT-PCR (qPCR) using the Universal Master Mix according to the manufacturer’s specifications. Primers and TaqMan probes for candidate genes were used along with commercially available online (Thermo Fisher Scientific). The qPCR signal obtained with the optimal cycling parameters for each gene was normalized to β-actin.
Statistical analysis
A significant difference in gene expression between the KRAS wild-type and KRAS-mutated CRC was calculated using Welch’s t-test, and the significance level was set to 1E-08 by Benjamini-Hochberg (BH) correction for multiple testing. In the comparative analysis of candidate genes, Welch’s t test was applied to compare gene expression levels among the vector-transfected cells. Fisher’s exact test was used to compare the subjects between the groups.
Results
Whole exome sequencing and deep sequencing of the custom cancer panel in CRC
We used WES to analyse 1074 cancer-related genes from 27 databases [29] in paired tumor tissue and blood samples to detect genetic changes in CRC. Simultaneously, we used the CCP comprising 409 target genes to conduct deep sequencing of tumor tissue samples to validate the WES data. The mean depth of coverage of the target regions was 115-fold for WES and 1027-fold for CCP. KRAS mutations were detected in 41.0% of all cases (372/906), which was consistent with the frequencies for these mutations observed in previous studies [35]. The concordance rate between the WES and CCP for KRAS mutations was 91.4% (340/372). The non-coincident was composed of WES-negative (15/372) and CCP-negative (17/372) for KRAS mutations.
Among KRAS-mutated CRC samples, the frequencies KRAS mutations were as follows: G12, 64.5% (240/372); G13, 20.2% (75/372); A146, 8.1% (30/372); Q61, 2.7% (10/372); K117, 2.7% (10/372); Q22, 0.5% (2/372); A59, 0.5% (2/372), and 0.3% (1/372) for A14, G77 and Y64 mutations. Within the KRAS G12 mutations, the frequencies of the various types of substitutions were 47.9% for KRAS G12D (115/240), 26.3% for G12V (63/240), 9.2% for G12A (22/240), 8.8% for G12C (21/240), 5.8% for G12S (14/240), and 2.1% for G12R (5/240).
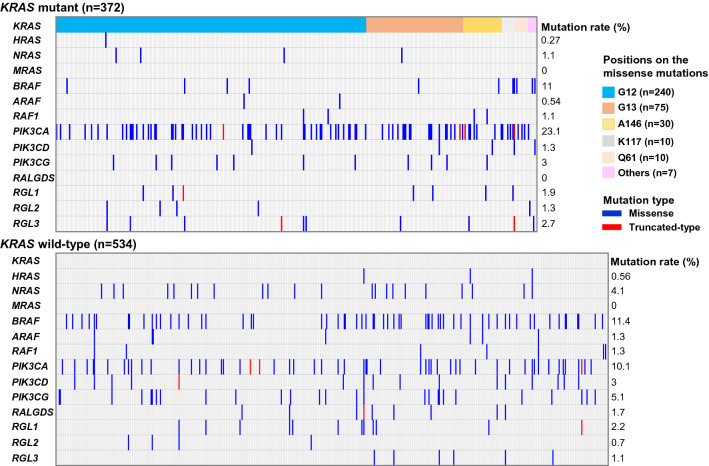
In KRAS mutated CRC samples, somatic mutations in PIK3CA (86/372, 23.1%) were the most frequently detected among the genes known to mediate RAS-associated responses. On the other hand, somatic mutations in BRAF (61/534, 11.4%), PIK3CG (27/534, 5.1%), PIK3CD (16/534, 3.0%), and NRAS (22/534, 4.1%) were frequently detected in KRAS wild-type CRC compared to KRAS-mutated CRC. The median tumor mutational burden (TMB) in KRAS wild-type CRC (n = 534) and KRAS mutated CRC (n = 372) were 8.27 mutations/Mb, and 13.27 mutations/Mb, respectively. Notably, somatic mutations in RALGDS were detected in KRAS wild-type CRC, but not in KRAS mutated CRC. It is intriguing that our WES analysis revealed that the RAS-associated genes were frequently mutated at high levels in patients with KRAS wild-type CRC compared to KRAS-mutated CRC (Fig. 1).
Fig. 1.
Genomic alterations in the KRAS-related genes in CRC. RAS-related genes were obtained from the NCI RAS Initiative [6]: a Mutation frequencies of genes that directly regulate RAS proteins in 906 colorectal cancer patients with (n = 372) or without (n = 534) KRAS mutations. Each column denotes an individual tumor. Left: percentage of samples with mutations in a given gene. Others (Pink Square) in the positions on KRAS mutations indicated Q22 (n = 2), A59 (n = 2), A14 (n = 1), G77 (n = 1), and Y64 (n = 1)
Comprehensive gene expression analysis of KRAS pathway-associated genes using DNA microarray
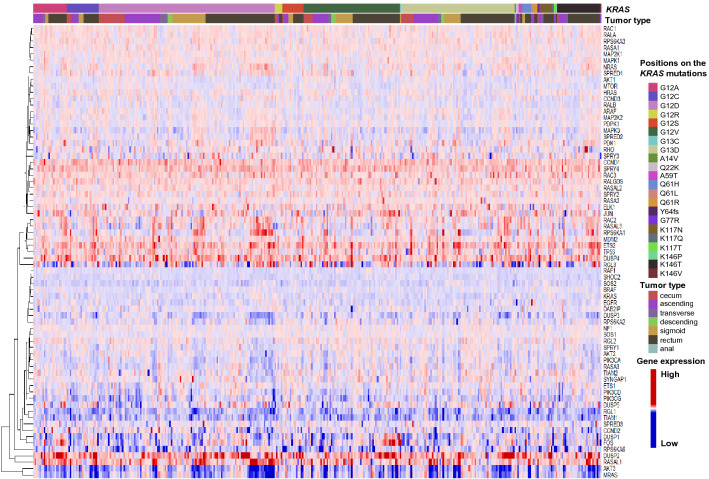
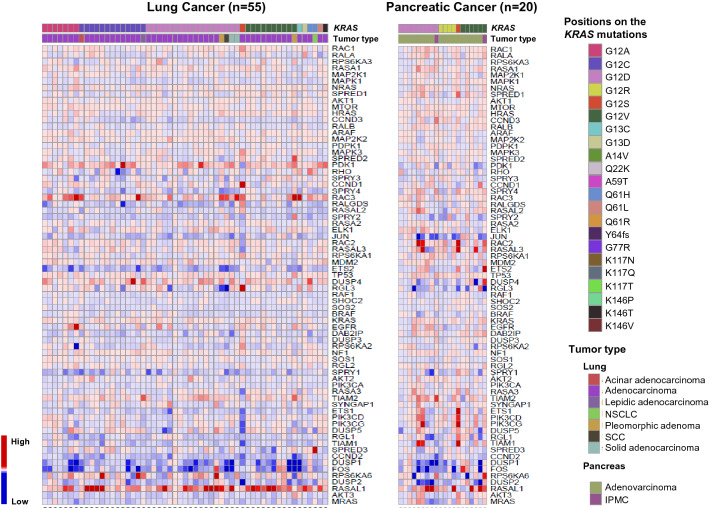
Of the known downstream genes in the KRAS pathway, increased expression was observed for CCND1, DUSP2, DUSP4, ETS2, JUN, RAC2, RAC3, SPRY4, ELK1, RALGDS, and RASAL1 in KRAS mutated CRC (Fig. 2). Conversely, the expression levels of CCND1, DUSP2, ETS2, JUN, and RALGDS were decreased in lung and pancreatic adenocarcinomas with KRAS mutations (Fig. 3). The signaling cascades downstream of the KRAS protein leading to the following pathways involving RAF/MAPK/ERK, PI3K/AKT, and RAL GDS/RAL have been well elucidated and are considered to differ according to the tumor type. It is noteworthy that transcription factors, such as ETS2, JUN, and ELK1, were upregulated in the KRAS mutated CRC, but not in lung and pancreatic cancers. Thus, the genes corresponding to these transcription factors may be promising targets for treating KRAS mutated CRC. However, the differences in expression levels of ETS2, JUN, and ELK1 between the KRAS mutant and the wild-type were not statistically significant (BH-adjusted P value, > 0.26).
Fig. 2.
A clustered heat map showing 65 of the KRAS pathway-associated genes that are differentially expressed in tumor tissues relative to adjacent normal tissues in 374 CRC with KRAS mutations. The tumor type in CRC indicates the location of the primary tumor (right upper panel). The expression levels (log2) are normalized for each gene and shown by the graded color scale
Fig. 3.
Heat map showing 65 of the KRAS pathway-associated genes that are differentially expressed in tumor tissues compared to adjacent normal tissues in 55 lung and 20 pancreatic cancers with KRAS mutations. The order of the KRAS-related genes is the same as in CRC samples (Fig. 2). Samples for lung (left) and pancreatic (right) cancers with KRAS mutations were obtained from our previous study [29]
Exploring of the drug-targetable oncogenes functioning with the KRAS-G12 mutant
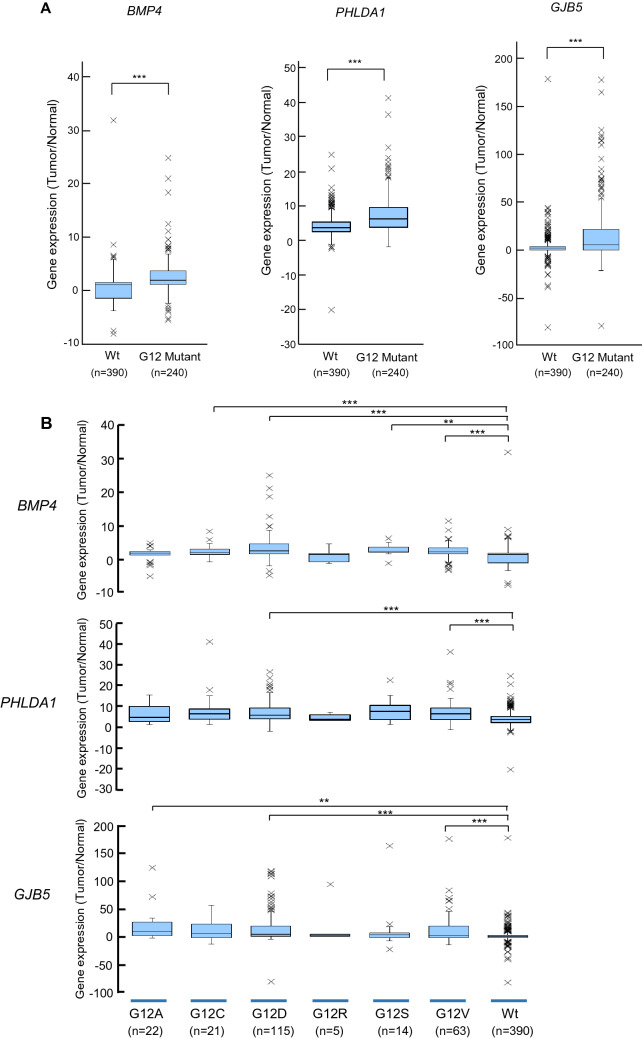
To exploit the novel KRAS G12 mutant targets, GEP was assessed in KRAS G12 mutated CRC (n = 240) and KRAS wild-type (n = 390). KRAS wild-type CRC harboring mutations in HRAS, NRAS, PIK3CA, PIK3CD, PIK3CG, RALGDS, RGL1-3, BRAF, ARAF, or RAF1 were excluded from the analysis because mutations in these genes directly affect KRAS-mediated signaling. The difference in the normalized signal intensities (fold change, FC) between the tumor and adjacent normal tissues was then calculated. The KRAS G12 mutated CRC (n = 240) and the selected KRAS wild-type CRC (n = 390) harbored APC mutation at 79.6% (191/240) and 74.9% (292/390), respectively; however, this difference that was not statistically significant (P = 0.21). On the other hand, the incidence of TP53 mutations showed a statistically significant difference (P < 0.01) between KRAS G12 mutated CRC (64.5%, 143/240) and KRAS wild-type CRC (83.1%, 324/390). There were number 13,222 genes that showed a positive FC value (mutant/wild-type) in KRAS G12 mutated CRC compared to the KRAS wild-type CRC. It was also noted that at least 11 promising candidate molecules showed greater than two-FC between KRAS G12 mutant and wild-type and had a BH-adjusted P value of less than 1E-08 and showed significant differential expression between these two groups (Table 2).
Table 2.
List of promising candidate genes that showed significant differences between KRAS G12 mutated and wild-type CRC
| Gene | Description | FCa | P -value | ||
|---|---|---|---|---|---|
| Welch’s t test | BH-adjusted | ||||
| 1 | HOXB6 | Homeobox B6 | 2.72 | 3.42E-16 | 4.50E-13 |
| 2 | PHLDA1 | Pleckstrin homology like domain family A member 1 | 3.26 | 6.02E-15 | 3.79E-12 |
| 3 | BMP4 | Bone morphogenetic Protein 4 | 2.05 | 1.22E-14 | 6.83E-12 |
| 4 | OTUB2 | OTU deubiquitinase, ubiquitin aldehydebinding 2 | 2.81 | 1.99E-14 | 1.05E-11 |
| 5 | TGFBI | Transforming growth factor beta 1 | 2.43 | 2.92E-14 | 1.36E-11 |
| 6 | SLC28A3 | Solute carrier family 28 member 3 | 6.78 | 6.76E-13 | 1.94E-10 |
| 7 | TMEM211 | Transmembrane protein 211 | 9.95 | 7.89E-12 | 1.42E-09 |
| 8 | DNAH2 | Dynein axonemal heavy chain 2 | 4.30 | 1.33E-11 | 2.27E-09 |
| 9 | FAM169A | Family with sequence similarity 169 member A | 3.36 | 2.15E-11 | 3.31E-09 |
| 10 | GJB5 | Gap junction protein beta 5 | 14.06 | 2.29E-11 | 3.46E-09 |
| 11 | C2orf70 (FAM166C) | Family with sequence similarity 166 member C | 2.66 | 2.39E-11 | 3.57E-09 |
aFC (Fold Change) in the normalized signal intensities between KRAS G12 mutated CRC and KRAS wild-type CRC
Validation of promising candidate genes in KRAS-mediated signaling
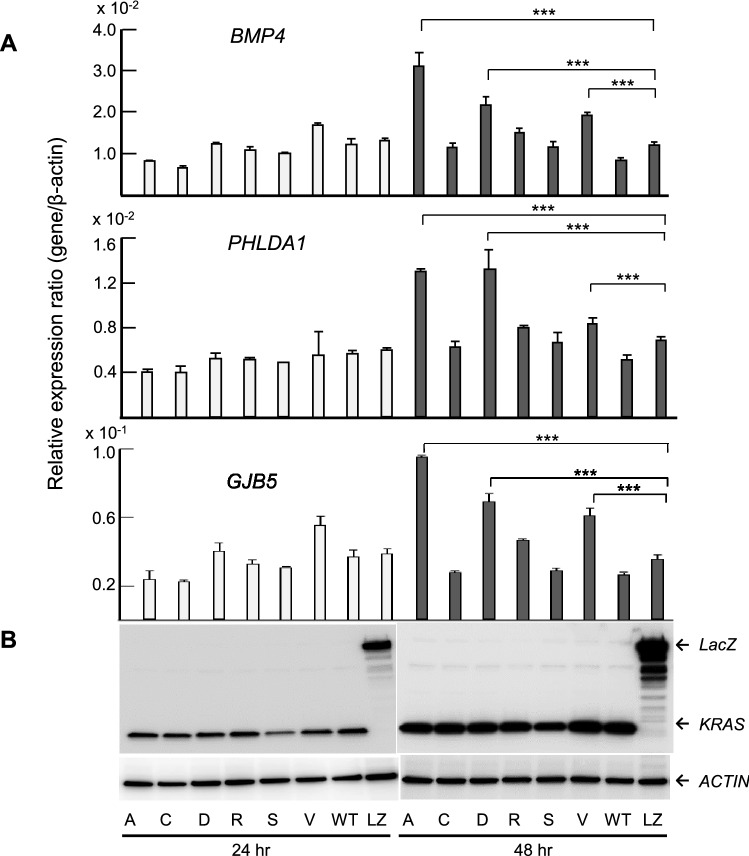
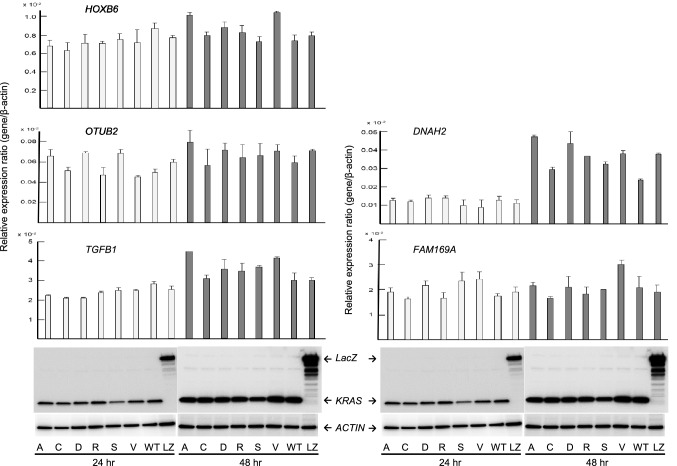
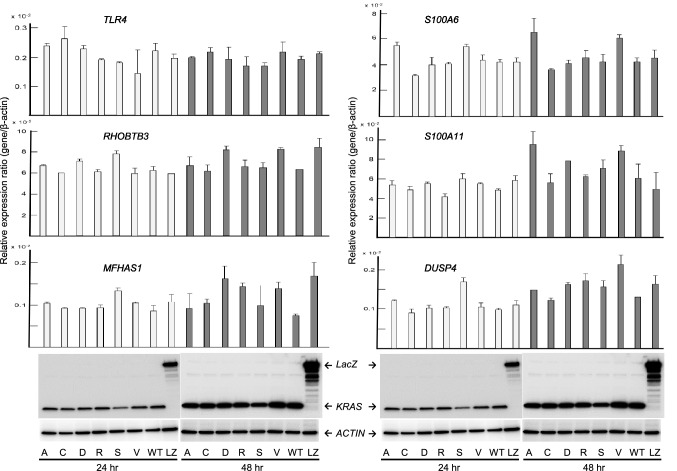
To verify the expression levels of the candidate genes in KRAS G12 mutated CRC, expression plasmids of KRAS variants, designated pKRAS-WT (wild-type), pKRAS-A (G12A), pKRAS-C (G12C), pKRAS-D (G12D), pKRAS-R (G12R), pKRAS-S (G12S), pKRAS-V (G12V), and pLacZ (control vector), were transfected into the human 293 embryonic kidney cells harboring KRAS wild-type. The level of gene expression in the transfected cells was analyzed using qPCR. The expression levels of the 11 candidate genes varied depending on the type of KRAS mutant, but the expression was effectively induced in G12A, G12D, and G12V mutants. Remarkably, as shown in Fig. 4, BMP4, PHLDA1, and GJB5 expression levels were significantly upregulated in the G12A-, G12D-, G12V- transfected cells, compared those in the WT-transfected cells, suggesting that these genes can be added to the list of candidates of KRAS G12A, G12D, or G12V target genes in CRC. To re-verify the expression data of BMP4, PHLDA1, and GJB5 were validated in the KRAS mutants-transfected 293 cells, and real-time RT-PCR analysis was performed for the KRAS mutant transfected Caco-2 CRC cells. Although the measured gene expression level was different between the 293 and Caco-2 cells, the effect of KRAS mutant transduction, that is, G12D, G12A, and G12V, was confirmed in Caco-2 cells (Fig. 5). This inconsistency in induced gene expression between the 293 and Caco-2 cells may be attributed to differences in transfection efficiency, susceptibility, and cellular differentiation, the nature of which should be explored further. The up-regulation of these genes was re-verified in an independent experiment (data not shown). BMP4, PHLDA1, and GJB expression levels in pairs of tumors and adjacent normal tissues from the patients with CRC obtained using GEP were significantly higher (P < 0.001) in the KRAS G12 mutant compared with those in the wild-type (Fig. 6a). The KRAS G12D and G12V mutants also showed increased expression levels (P < 0.001) in comparison with the wild-type (Fig. 6b) Western blot analysis using the V5-tagged antibody showed no difference in the KRAS protein levels between the pKRAS-WT and pKRAS mutated cells. The entire transfection experiment was repeated twice, showing the same KRAS protein level in the transfected cells. The other eight genes (genes shown in Table 2) were not verified by qPCR (Fig. 7). In addition to the 11 candidate genes, we analyzed the TLR4, RHOBTB3, MFHAS1, S100A6, S100A11, and DUSP4 genes that had a BH-adjusted P value of less than 1E-09 between KRAS G12 mutant and wild-type, but less than two-fold, which have been implicated in the oncogenic functions (Supplementary Table). None of these genes showed a significantly different expression levels in KRAS G12 mutant transfected cells from those in the wild-type or LacZ transfected control cells. (Fig. 8).
Fig. 4.
Promising candidate genes are validated using qPCR in the KRAS G12 mutant transfected 293 cells: a Relative expression ratio is defined as the ratio between the expression level of a gene to that of the internal reference gene, β-actin. White and black columns indicate the expression levels at 24 and 48 h after transfection, respectively. The assays are carried out in triplicates and means ± standard deviation are plotted, b KRAS protein expression in the 293 cells transfected with KRAS mutants, wild-type, or LacZ control vector analyzed using Western blot with V5 and β-actin antibodies. The β-actin is used as a loading control. A, C, D, R, S, V, WT, and LZ indicate G12A-, G12C-, G12D-, G12R-, G12S-, G12V-, wild-type-, and LacZ transfected cells, respectively. The asterisk indicates ***P value < 0.001
Fig. 5.
Expression of BMP4, PHLDA1, and GJB5 are validated using qPCR in the KRAS G12 mutant transfected Caco-2 cells: a Relative expression ratio is defined as the ratio between the expression level of a gene to that of the internal reference gene, β-actin. b KRAS protein expression in the Caco-2 cells are analyzed using Western blot with V5 and β-actin antibodies. The assays are carried out the same as that show in Fig. 4. The asterisk indicates ***P value < 0.001
Fig. 6.
BMP4, PHLDA1, and GJB5 expression levels in CRC with KRAS G12 mutant and wild-type (a) or KRAS-G12A, -G12C, -G12D, -G12R, -G12S, -G12V mutants, and wild-type (b). The expression level (log2) was normalized for each gene. *** indicates P < 0.001; ** indicates P < 0.01
Fig. 7.
Five genes, excluding BMP4, PHLDA1, and GJB5 shown in Table 2 are validated using qPCR in the KRAS G12 mutant transfected cell. All genes show a difference in up-regulation but this difference is not significant compared to KRAS wild-type or LacZ transfected cells. SLC28A3, TMEM211, and C2orf70 genes shown in Table 2 are not detected by qPCR. The assays are carried out in triplicate, and means ± standard deviations are plotted
Fig. 8.
Validation of TLR4, RHOBTB3, MFHAS1, S100A6, S100A11, and DUSP4 genes that had a BH-adjusted P value less than 1.00E-10 between KRAS G12 mutated and wild-type CRC, which have been implicated in the oncogenic function. All genes were not validated by qPCR. The Western blot analysis of transfected cell is the same as that show in Fig. 6. The assays are carried out in triplicate and means ± standard deviation were plotted
Discussion
In this study, we identified BMP4, PHLDA1, and GJB5 as the most likely genes that are activated downstream of the KRAS G12-driver mutation in CRC, especially the G12A, G12D, and G12V mutations. On the other hand, transfection of the G12C, G12R, and G12S mutants showed lower expression of BMP4, PHLDA1, and GJB5, but higher than those of the wild-type, compared with the G12A, G12D, and G12V mutants. Presently, the detailed mechanism for these differential expression profiles is not clear; however, specific KRAS mutations have unique biological and clinical behaviors. Hunter et al. [36] have systemically examined the biochemical and biophysical properties of common KRAS mutants and showed that a cell line harboring the G12A mutation, which had high affinity for RAF kinase and low intrinsic GTPase activity, showed the highest sensitivity to MEK inhibitor, suggesting that G12A mutation intensely affects the downstream signal of KRAS. In our present study, the highest induction was caused by G12A mutant in several genes (Figs.4 and 5). Additionally, the G12D mutation, which is predicted to be a low RAF activator, is associated with PI3K, but not RAF kinase and does not induce ERK phosphorylation in NIH3T3 cells. G12V, which is predicted to be a moderate RAF activator [36], is associated with both RAF kinase and PI3K in NIH3T3 cells [37]. Therefore, it is suggested that the signals of KRAS mutation have different biological properties depending on mutation type and differentially affect the final gene expression process in the signal transduction cascade. The genes identified in our study may be involved in CRC development ant progression by directly or indirectly regulating the expression of these genes, depending on the type of KRAS mutation. To clarify the detailed mechanisms of KRAS mutation-induced differential gene expression patterns, further investigations are necessary. Furthermore, in CRC, G12A, G12D, and G12V mutations account for 85% of all KRAS G12 mutations. Therefore, it may also contribute to the acceleration of personalized medicine for CRC patients with these mutations. Our study has added these genes to the list of those that are possibly involved in colorectal carcinogenesis.
BMP4 belongs to the TGFβ superfamily and has been reported to be involved in the regulation of various biological processes such as tissue organization of colonic epithelial cells, interaction between epithelial cells and stromal cells, epithelial-mesenchymal transition (EMT) induction, and metastasis [38, 39]. Additionally, BMP4 has been reported to promote colon cancer cell invasiveness and tumor formation [40]. Therefore, it is suggested that genes induced by the activation of BMP4-dependent signaling may be involved in the carcinogenesis and progression of CRC. In contrast, another study showed that BMP4 was involved in the suppression of colon cancer cell growth and that the activated KRAS down-regulated BMP4 via the ERK pathway [41]. A possible explanation for this apparent controversy could be that these differential roles accounted to the differences in cell lines used among those studies. Aberrant activation of the Wnt/β-catenin pathway enhances BMP4 signaling in colorectal cancer cells [42]. Therefore, although there was a possibility that BMP4 expression was increased by inactivation of APC in CRC, no difference was observed in the frequency of APC mutation depending on the presence or absence of KRAS mutations in this study. PHLDA1 may be a transcriptional activator that is induced by various external stimuli and acts as a mediator of apoptosis, proliferation, differentiation, and cell migration depending on the cell type and physiological context [43]. It has also been suggested that PHLDA1 is a putative epithelial stem cell marker in the small and large human intestine and contributes to the migration and proliferation of colon cancer cells [44], and it may contribute to the understanding of the oncogenic mechanism of colorectal carcinogenesis. However, the mechanistic basis for KRAS activation and/or PHLDA1 in CRC has not been fully elucidated, and it should be determined by further investigation. GJB5 is a member of the connexin family that regulates cell adhesion, proteolysis, and motility. Connexins have been shown to function as tumor suppressors in cancer [45, 46] and have been reported to regulate EMT, tumor cell differentiation, and angiogenesis [47]. Among different members of the connexin family, GJB5 has not been described in association with colorectal cancer or RAS signaling, and the role of GJB5 in colorectal carcinogenesis remains largely unknown. Therefore, it is prudent to exclude this gene as a drug-targetable candidate in CRC at this time.
In recent years, various combinations of existing molecular targets [48], synthetic lethal partners [49], and immune checkpoint inhibitors [50] for RAS-activating signals have been extensively developed, and tumor suppressive effects have been shown in animal models. The genes identified in this study may be effective targets when used in combination with existing inhibitors of the MAPK pathway, such as MEK or BRAF inhibitors. The role of the genes identified in this study in the carcinogenesis and progression of CRC with KRAS G12 mutations may be a modulation of the cancer phenotype, the nature of which should be elucidated in future studies. We believe that our study will lead to further functional characterization of genes in the context of KRAS-based individualized therapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by a KAKENHI Grant-in-Aid for Scientific Research (18K07957) from the Japan Society for the Promotion of Science (JSPS).
Declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hashiguchi Y, Muro K, Saito Y, Ito Y, Ajioka Y, Hamaguchi T, Hasegawa K, Hotta K, Ishida H, Ishiguro M, Ishihara S, Kanemitsu Y, Kinugasa Y, Murofushi K, Nakajima T, Oka S, Tanaka T, Taniguchi H, Tsuji A, Uehara K, Ueno H, Yamanaka T, Yamazaki K, Yoshida M, Yoshino T, Itabashi M, Sakamaki K, Sano K, Shimada Y, Tanaka S, Uetake H, Yamaguchi S, Yamaguchi N, Kobayashi H, Matsuda K, Kotake K, Sugihara K. Japanese society for cancer of the colon and rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol. 2020;25:1–42. doi: 10.1007/s10147-019-01485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oki E, Ando K, Nakanishi R, Sugiyama M, Nakashima Y, Kubo N, Kudou K, Saeki H, Nozoe T, Emi Y, Maehara Y. Recent advances in treatment for colorectal liver metastasis. Ann Gastroenteral Surg. 2018;2:167–175. doi: 10.1002/ags3.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE, Lieu C, Lindor NM, Minsky BD, Monzon FA, Sargent DJ, Singh VM, Willis J, Clark J, Colasacco C, Rumble RB, Temple-Smolkin R, Ventura CB, Nowak JA. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American society for clinical pathology, college of American pathologists, association for molecular pathology, and the American society of clinical oncology. J Clin Oncol. 2017;35:1453–1486. doi: 10.1200/JCO.2016.71.9807. [DOI] [PubMed] [Google Scholar]
- 4.Roock WD, Claes B, Bernasconi D, Schutter JD, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, Dosso SD, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Fiore FD, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Cutsem EV, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 5.Ledford H. The ras renaissance. Nature. 2015;520:278–280. doi: 10.1038/520278a. [DOI] [PubMed] [Google Scholar]
- 6.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, Settleman J, Giovannini M, Jacks T. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoyama N, Kim Y-J, Hirabayashi Y, Tabe Y, Takamori K, Ogawa H, Iwabuchi K. Kras promotes myeloid differentiation through Wnt/β -catenin signaling. FASEB BioAdv. 2019;1:435–449. doi: 10.1096/fba.2019-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Wang Y, Vikis HG, Johnson L, Liu G, Li J, Anderson MW, Sills RC, Hong HL, Devereux TR, Jacks T, Guan K-L, You M. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]
- 11.James RM, Arends MJ, Plowman SJ, Brooks DG, Miles CG, West JD, Patek CE. K-ras proto-oncogene exhibits tumor suppressor activity as its absence promotes tumorigenesis in murine teratomas. Mol Cancer Res. 2003;1:820–825. [PubMed] [Google Scholar]
- 12.Spandidos DA, Sourvinos G, Tsatsanis C, Zafiropoulos A. Normal ras genes: their onco-suppressor and pro-apoptotic functions (review) Int J Oncol. 2002;21:237–241. doi: 10.3892/ijo.21.2.237. [DOI] [PubMed] [Google Scholar]
- 13.Singh A, Sowjanya AP, Ramakrishna G. The wild-type Ras: road ahead. FASEB J. 2005;19:161–169. doi: 10.1096/fj.04-258hyp. [DOI] [PubMed] [Google Scholar]
- 14.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, Tsao A, Liu S, Larsen JE, Wang J, Diao L, Coombes KR, Chen L, Zhang S, Abdelmelek MF, Tang X, Papadimitrakopoulou V, Minna JD, Lippman SM, Hong WK, Herbst RS, Wistuba II, Heymach JV, Powis G. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–239. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan TL, Amzallag A, Bagni R, Yi M, Afghani S, Burgan W, Fer N, Strathern LA, Powell K, Smith B, Waters AM, Drubin D, Thomson T, Liao R, Greninger P, Stein GT, Murchie E, Cortez E, Egan RK, Procter L, Bess M, Cheng KT, Lee C-S, Lee LC, Fellmann C, Stephens R, Luo J, Lowe SW, Benes CH, McCormick F. Differential effector engagement by oncogenic KRAS. Cell Rep. 2018;22:1889–1902. doi: 10.1016/j.celrep.2018.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margetis N, Kouloukoussa M, Pavlou K, Vrakas S, Mariolis-Sapsakos T. K-ras mutations as the earliest driving force in a subset of colorectal carcinomas. Vivo. 2017;31(4):527–542. doi: 10.21873/invivo.11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Smits AMM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 18.Bahrami A, Hassanian SM, ShahidSales S, Farjami Z, Hasanzadeh M, Anvari K, Aledavood A, Maftouh M, Ferns GA, Khazaei M, Avan A. Targeting RAS signaling pathway as a potential therapeutic target in the treatment of colorectal cancer. J Cell Physiol. 2018;233:2058–2066. doi: 10.1002/jcp.25890. [DOI] [PubMed] [Google Scholar]
- 19.Wilson CY, Tolias P. Recent advances in cancer drug discovery targeting RAS. Drug Discov Today. 2016;21:1915–1919. doi: 10.1016/j.drudis.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Brand TM, Wheeler DL. KRAS mutant colorectal tumors Small GTPase. 2012;3:34–39. doi: 10.1038/onc.2010.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janne PA, van den Heuvel MM, Barlesi F, Cobo M, Mazieres J, Crino L, Orlov S, Blackhall F, Wolf J, Garrido P, Poltoratskiy A, Mariani G, Ghiorghiu D, Kilgour E, Smith P, Kohlmann A, Carlile DJ, Lawrence D, Bowen K, Vansteenkiste J. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer, the select-1 randomized clinical trial. JAMA. 2017;317:1844–1853. doi: 10.1001/jama.2017.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brauswetter D, Gurbi B, Varga A, Varkondi E, Schwab R, Banhegyi G, Fabian O, Keri G, Valyi-Nagy I, Petak I. Molecular subtype specific efficacy of MEK inhibitors in pancreatic cancers. PLoS ONE. 2017;12:e0185687. doi: 10.1371/journal.pone.0185687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitajima S, Asahina H, Chen T, Guo S, Quiceno LG, Cavanaugh JD, Merlino AA, Tange S, Terai H, Kim JW, Wang X, Zhou S, Xu M, Wang S, Zhu Z, Thai TC, Takahashi C, Wang Y, Neve R, Stinson S, Tamayo P, Watanabe H, Kirschmeier PT, Wong K-K, Barbie DA. Overcoming resistance to dual innate immune and MEK inhibition downstream of KRAS. Cancer Cell. 2018;34:439–452. doi: 10.1016/j.ccell.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janes MR, Zhang J, Li L-S, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, Feng J, Chen JH, Li S, Li S, Long YO, Thach C, Liu Y, Zarieh A, Ely T, Kucharski JM, Kessler LV, Wu T, Yu K, Wang Y, Yao Y, Deng X, Zarrinkar PP, Brehmer D, Dhanak D, Lorenzi MV, Hu-Lowe D, Patricelli MP, Ren P, Liu Y. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. 2018;172:578–589. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 26.McCormick F. Sticking it to KRAS: covalent inhibitors enter the clinic. Cancer Cell. 2020;37:3–4. doi: 10.1016/j.ccell.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malumbres M, Barbacid M. RAS oncogens: the first 30 years. Nat Rev. 2003;3:7–13. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 28.Jones RP, Sutton PA, Evans JP, Clifford R, McAvoy A, Lewis J, Rousseau A, Mountford R, McWhirter D, Malik HZ. Specific mutations in KRAS codon 12 are associated with worse overall survival in patients with advanced and recurrent colorectal cancer. Br J Cancer. 2017;116:923–929. doi: 10.1038/bjc.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagashima T, Yamaguchi K, Urakami K, Shimoda Y, Ohnami S, Ohshima K, Tanabe T, Naruoka A, Kamada F, Serizawa M, Hatakeyama K, Matsumura K, Ohnami S, Maruyama K, Mochizuki T, Kusuhara M, Shiomi A, Ohde Y, Terashima M, Uesaka K, Onitsuka T, Nishimura S, Hirashima Y, Hayashi N, Kiyohara Y, Tsubosa Y, Katagiri H, Niwakawa M, Takahashi K, Kashiwagi H, Nakagawa M, Ishida Y, Sugino T, Takahashi M, Akiyama Y. Japanese version of The cancer genome atlas, JCGA, established using fresh frozen tumors obtained from 5143 cancer patients. Cancer Sci. 2020;111:687–699. doi: 10.1111/cas.14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohshima K, Hatakeyama K, Nagashima T, Watanabe Y, Kanto K, Doi Y, Ide T, Shimoda Y, Tanabe T, Ohnami Su, Ohnami S, Serizawa M, Maruyama K, Akiyama Y, Urakami K, Kusuhara M, Mochizuki T, Yamaguchi K. Integrated analysis of gene expression and copy number identified potential cancer driver genes with amplification-dependent overexpression in 1454 solid tumors. Sci Rep. 2017;7:641. doi: 10.1038/s41598-017-00219-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnami S, Ohshima K, Nagashima T, Urakami K, Shimoda Y, Saito J, Naruoka A, Hatakeyama K, Mochizuki T, Serizawa M, Ohnami Su, Kusuhara M, Yamaguchi K. Comprehensive characterization of genes associated with the TP53 signal transduction pathway in various tumors. Mol Cell Biochem. 2017;431:75–85. doi: 10.1007/s11010-017-2977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatakeyama K, Nagashima T, Ohshima K, Ohnami Su, Ohnami S, Shimoda Y, Serizawa M, Maruyama K, Naruoka A, Akiyama Y, Urakami K, Kusuhara M, Mochizuki T, Yamaguchi K. Mutational burden and signatures in 4000 Japanese cancers provide insights into tumorigenesis and response to therapy. Cancer Sci. 2019;110:2620–2628. doi: 10.1111/cas.14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ministry of Health, Labour and Welfare. Japanese ethical guidelines for human genome/gene analysis research. 2017. https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/hokabunya/kenkyujigyou/i-kenkyu/index.html. Accessed 17 Sept 2019
- 34.Ohnami S, Sato Y, Yoshimura K, Ohnami Su, Sakamoto H, Aoki K, Ueno H, Ikeda M, Morizane C, Shimada K, Sakamoto Y, Esaki M, Saito I, Hirose H, Saito D, Sugimura H, Kosuge T, Okusaka T, Yoshida T. His595Tyr polymorphism in the methionine synthase reductase (MTRR) gene is associated with pancreatic cancer risk. Gastroenterology. 2008;135:477–488. doi: 10.1053/j.gastro.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 35.Serebriiskii IG, Connelly C, Frampton G, Newberg J, Cooke M, Miller V, Ali S, Ross JS, Handorf E, Arora S, Lieu C, Golemis EA, Meyer JE. Comprehensive characterization of RAS mutations in colon and rectal cancers in old and young patients. Nat Commun. 2019;10:3722. doi: 10.1038/s41467-019-11530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 37.Céspendes MV, Sancho FJ, Guerrero S, Parreño M, Casanova I, Pavón MA, Marcuello E, Trias M, Cascante M, Capellà G, Mangues R. K-ras Asp12 mutant neither interacts with Raf, nor signals through Erk and is less tumorigenic than K-ras Val12. Carcinogenesis. 2006;27:2190–2200. doi: 10.1093/carcin/bg1063. [DOI] [PubMed] [Google Scholar]
- 38.Ma J, Zeng S, Zhang Y, Deng G, Qu Y, Guo C, Yin L, Han Y, Cai C, Li Y, Wang G, Bonkovsky HL, Shen H. BMP4 promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett. 2017;411:117–129. doi: 10.1016/j.canlet.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 39.Theriault BL, Shepherd TG, Mujoomdar ML, Nachtigal MW. BMP4 induces EMT and Rho GTPase activation in human ovarian cancer cells. Carcinogenesis. 2007;28:1153–1162. doi: 10.1093/carcin/bgm015. [DOI] [PubMed] [Google Scholar]
- 40.Yokoyama Y, Watanabe T, Tamura Y, Hashizume Y, Miyazono K, Ehata S. Autocrine BMP-4 signaling is a therapeutic target in colorectal cancer. Cancer Res. 2017;77:4026–4038. doi: 10.1158/0008-5472.CAN-17-0112. [DOI] [PubMed] [Google Scholar]
- 41.Duerr E-M, Mizukami Y, Moriichi K, Gala M, Jo W-S, Kikuchi H, Xavier RJ, Chung DC. Oncogenic KRAS regulates BMP4 expression in colon cancer cell lines. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1223–G1230. doi: 10.1152/ajpgi.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J-S, Crooks H, Dracheva T, Nishanian TG, Singh B, Jen J, Waldman T. Oncogenic β-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 2002;62:2744–2748. [PubMed] [Google Scholar]
- 43.Nagai MA. Pleckstrin homology-like domain, family A, member 1 (PHLDA1) and cancer (Review) Biomed Rep. 2016;4:275–281. doi: 10.3892/br.2016.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakthianandeswaren A, Christie M, D’Andreti C, Tsui C, Jorissen RN, Li S, Fleming NI, Gibbs P, Lipton L, Malaterre J, Ramsay RG, Phesse TJ, Ernst M, Jeffery RE, Poulsom R, Leedham SJ, Segditsas S, Tomlinson IPM, Bernhard OK, Simpson RJ, Walker F, Faux MC, Church N, Catimel B, Flanagan DJ, Vincan E, Sieber OM. PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res. 2011;71:3709–3719. doi: 10.1158/0008-5472.CAN-10-2342. [DOI] [PubMed] [Google Scholar]
- 45.Shettar A, Damineni S, Mukherjee G, Kondaiah P. Gap junction β-2 expression is negatively associated with the estrogen receptor status in breast cancer tissues and is a regulator of breast tumorigenesis. Oncol Rep. 2018;40:3645–3653. doi: 10.3892/or.2018.6764. [DOI] [PubMed] [Google Scholar]
- 46.Zhang D, Chen C, Li Y, Fu X, Xie Y, Li Y, Huang Y. Cx31.1 acts as a tumor suppressor in non-small cell lung cancer (NSCLC) cell lines through inhibition of cell proliferation and metastasis. J Cell Mol Med. 2012;16:1047–1059. doi: 10.1111/j.1582-4934.2011.01389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aasen T, Mesnil M, Naus CC, Lampe PD, Laird DW. Gap junctions and cancer: communicating for 50 years. Nat Rev Cancer. 2016;16:775–788. doi: 10.1038/nrc.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manchado E, Weissmueller S, Morris JP, IV, Chen C-C, Wullenkord R, Lujambio A, Stanchina E, Poirier JT, Gainor JF, Corcoran RB, Engelman JA, Rudin CM, Rosen N, Lowe SW. A combinatorial strategy for treating KRAS mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Downward J. RAS synthetic lethal screens revisited: still seeking the elusive prize? Clin Cancer Res. 2015;21:1802–1809. doi: 10.1158/1078-0432.CCR-14-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dietlein F, Kalb B, Jokic M, Noll EM, Strong A, Tharun L, Ozretic L, Kunstlinger H, Kambartel K, Randerath WJ, Jungst C, Schmitt A, Torgovnick A, Richters A, Rauh D, Siedek F, Persigehl T, Mauch C, Bartkova J, Bradley A, Sprick MR, Trumpp A, Rad R, Saur D, Bartek J, Wolf J, Buttner R, Thomas RK, Reinhardt HC. A synergistic interaction between Chk1- and MK2 inhibitors in KRAS-mutant cancer. Cell. 2015;162:146–159. doi: 10.1016/j.cell.2015.05.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.