Abstract
Rare monogenic disorders often share molecular etiologies involved in the pathogenesis of common diseases. Congenital disorders of glycosylation (CDG) and deglycosylation (CDDG) are rare pediatric disorders with symptoms that range from mild to life threatening. A biological mechanism shared among CDG and CDDG as well as more common neurodegenerative diseases such as Alzheimer’s disease and amyotrophic lateral sclerosis, is endoplasmic reticulum (ER) stress. We developed isogenic human cellular models of two types of CDG and the only known CDDG to discover drugs that can alleviate ER stress. Systematic phenotyping confirmed ER stress and identified elevated autophagy among other phenotypes in each model. We screened 1049 compounds and scored their ability to correct aberrant morphology in each model using an agnostic cell-painting assay based on >300 cellular features. This primary screen identified multiple compounds able to correct morphological phenotypes. Independent validation shows they also correct cellular phenotypes and alleviate each of the ER stress markers identified in each model. Many of the active compounds are associated with microtubule dynamics, which points to new therapeutic opportunities for both rare and more common disorders presenting with ER stress, such as Alzheimer’s disease and amyotrophic lateral sclerosis.
Subject terms: Stress signalling, Cellular imaging, High-throughput screening, Neurodevelopmental disorders
Introduction
The study of rare monogenic disorders has yielded a number of insights into the molecular mechanisms underlying the pathobiology of more common diseases [1, 2]. Numerous diseases affecting both the central and peripheral nervous system involve elevated endoplasmic reticulum (ER) stress [3, 4]. In particular, ER stress has been implicated in diseases including Parkinson’s disease, Alzheimer’s disease, and amyotrophic lateral sclerosis (ALS) [5]. This suggests that therapeutic agents that ameliorate the effects of ER stress in monogenic disorders could have benefits across a broad range of disorders. Screens to identify such agents in the context of complex neurodegenerative diseases are challenging to implement; however, the etiology of a number of monogenic diseases is in large part attributed to ER stress including the congenital disorders of glycosylation (CDG) and deglycosylation (CDDG) [6–8]. Of particular interest, mutations in PMM2, the gene that encodes phosphomannomutase 2, result in the most common CDG [9]. Studies suggest that in PMM2-CDG, cells with weaker ER stress responses are more vulnerable to damage than cells with stronger ER stress responses [10]. Moreover, mutations in DPAGT1, which encodes the target of the well-known ER stress inducer tunicamycin [11], result in another CDG with systemic phenotypes [12, 13].
Here we utilize a morphological profiling and screening paradigm to identify agents that protect against the cellular stresses resulting from CDG and CDDG causal mutations. We focus specifically on mutations in PMM2 and DPAGT1, and in NGLY1, which causes the only reported CDDG [8]. We used genetic engineering to generate CDG and CDDG genotypes in a karyotypically normal human cell line in order to create cellular models amenable to mutation-specific phenotype identification. These CDG and CDDG cell lines were used in high-content small molecule screens to identify compounds that revert the imaging phenotypes caused by these mutations. Specifically, we screened 1049 annotated compounds representing a broad chemical space and multiple target classes. In order to validate the performance of the screen, we selected 16 compounds that were ranked amongst the best at phenotype reversion in the screen (protective compounds) and 10 compounds that did not affect aberrant phenotypes (non-active negative control compounds). We then evaluated these compounds in assays designed to test how well they revert mutational phenotypes in the three cellular models.
Materials and methods
CRISPR/Cas9 genome editing of hTERT RPE-1 cells
CDG and CDDG lines were generated by CRISPR/Cas9 genome editing of hTERT RPE1 (ATCC, CRL 4000TM) at the Columbia Stem Cell Core Facility. Promoter (U6) and gRNA scaffolds were synthesized by IDT and cloned into the pCR-Blunt II-TOPO plasmid (ThermoFisher Scientific, cat. K280002). Nucleofector (Lonza) was employed to introduce gRNA and Cas9-GFP plasmids into hTERT RPE-1 cells. After nucleofection, single colonies were manually picked into either 96-well plates or 10 cm dishes, incubated for ten days to reach confluency (96-well plate) or visible colonies (10 cm dish). For each colony, DNA was extracted by the KAPA Mouse Genotyping Kit (KAPA Biosystems) and genotyped by Sanger sequencing.
Guide RNA scaffold and termination signal: GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTT.
gRNA NGLY1: GGTGATTGCCAGAAGAACTAAGG, PMM2: GAATTCAATGAAAGTACCCCTGG, DPAGT1: CATGATCTTCCTGGGCTTTGCGG.
Chemicals
Tunicamycin (cat. 3516) and salubrinal (cat. 2347) were from Tocris. Rapamycin (cat. HY-10219) was from MedChemExpress. All screened compounds were provided by Janssen Pharmaceuticals.
Proliferation measurements
Cells were seeded in 96-well tissue culture plates and treated the next day as described in “Results”. At the indicated time points, the MTT assay was performed as described [14]. One-way ANOVA multiple comparisons and Dunnett test (GraphPad Prism software, v.8.2.0) were used to determine the equality of the means of different samples. The confidence level (p) was 0.05.
Quantitative RT-PCR
Total RNA was extracted by RNeasy Plus Mini kit (QIAGEN, cat. 74136) and reverse-transcribed with random primers using Superscript IV Reverse Transcriptase kit (ThermoFisher Scientific, cat. 18091200). One µL of cDNA was used in each qPCR reaction on a QuantStudio 5 (ThermoFisher Scientific) using SYBR Green PCR Master Mix (ThermoFisher Scientific, cat. 4364344). PCR primers detecting spliced and unspliced XBP1 expression were as described [15], and for human GAPDH were ACAGTCAGCCGCATCTTCTT and TTGATTTTGGAGGGATCTCG. The relative expression levels of target genes were normalized to that of the reference GAPDH gene by using the ΔΔCt method [16]. The fold change in expression for each sample is relative to parental hTERT RPE-1 cells treated with vehicle.
Immunoblot analysis
Cell lysates were prepared as described in [14], resolved in SDS-PAGE, transferred to PVDF membrane (Immobilon-P, Millipore, cat. IPVH00010), stained with appropriate antibodies (Supplementary Table 1) and developed as described [14]. Western blots were quantitatively analyzed via laser-scanning densitometry using NIH ImageJ v1.52k software.
Immunocytochemistry
hTERT RPE-1 and the isogenic mutant lines were seeded on glass coverslips, fixed with 4% paraformaldehyde (PFA), permeabilized with 1% Triton X-100/PBS, blocked and stained in 1% BSA/ 0.1% Triton X-100/PBS and mounted in Prolong Antifade DAPI (Invitrogen). Antibodies and dilutions are listed in Supplementary Table 1. Imaging was performed on an inverted Zeiss AxioObserver Z1 fluorescent microscope equipped with an AxioCam 503 mono camera and filters for 405, 488, and 568 nm. Images were acquired with Zen 2 software and post-processing was performed with AdobePhotoshop CC.
Senescence detection
Cells were seeded in 6 well plates, and stained using Senescence β-Galactosidase Staining Kit (Cell Signaling Technology, cat. 9860) according to manufacturer’s instruction. The images were acquired, and the number of stained cells was counted using Zeiss Primovert inverted brightfield/phase contrast microscope equipped with AxioCam ERc5s camera.
Apoptosis detection
Cells were stained using APC-labeled Annexin-V (BD Biosciences, cat. 550474) and propidium iodide (PI) according to the manufacturer’s instructions and analyzed immediately on FACSCelesta (BD Biosystems). Data were processed using FlowJo v. 10.5.3 and Prism8 v8.2.0 software.
Autophagy detection
Cells were collected by trypsinization, fixed with 4% PFA/PBS for 15 min at RT. Fixed cells were permeabilized using Intracellular Staining Permeabilization Wash buffer (BioLegend, cat.421002) according to the manufacturer instructions, stained with appropriate primary and secondary antibodies (Supplementary Table 1) and analyzed on a FACSCelesta cytometer (BD Biosystems). The data were processed using FlowJo v. 10.5.3 and Prism8 v8.2.0 software.
Cellular morphology assessment by immunostaining
Cells were seeded in 96-well plates, treated with tested compounds, vehicle (DMSO) or positive controls. After 24 h, cells were fixed with 4% PFA/PBS, blocked with 1% BSA/PBS and stained with phalloidin-568 (ThermoFisher Scientific, cat. A12380). After two washes with PBS, cells were stained with 300 nM DAPI (BD Pharmingen, cat. 564907). Imaging was performed with an inverted Zeiss AxioObserver Z1 epifluorescent microscope equipped with an AxioCam 503 mono camera, and images acquired with the Zen 2 software. Post-processing was performed AdobePhotoshop CC software.
High-content imaging and compound screening
The hTERT RPE-1 cells and NGLY1−/−, PMM2F119L/−, and DPAGT1+/− mutant lines were plated in 384 well plates at a density of 3000 cells per well. The next day compounds were added to the cells at a final concentration of 10 µM and incubated for 24 h. Cells were stained with MitoTracker Red (Molecular Probes, cat. M7512) mitochondrial stain for 30 min according to the manufacturer’s protocol, then media was removed, cells were fixed in 4% PFA/PBS, permeabilized with 0.1% NP-40, and blocked with 3% BSA/ PBS overnight. For staining, ConcanavalinA-488 (Molecular Probes, cat. C11252), phalloidin-547 (Molecular Probes, cat. A22283), and DAPI were added to the wells, then washed before imaging. Images were acquired on a Molecular Devices Image Express microscope at 4 fields per well. Feature extraction from images was done with Perkin-Elmer Columbus Image Analysis software, and feature analysis and hit determination was performed using TIBCO Spotfire analysis package.
Secondary validation of selected compounds
Candidate and control compounds from the high-throughput screen were validated using two assays, MTT and RT-qPCR for sXBP1 expression as described above. Dose–response curves on hTERT RPE-1 cells identified the lowest non-toxic concentration for each of the candidate compounds. Cells were treated for 24 h with Group I (5.0 nM), Group II (1.0 µM), or non-active control (10 µM) compounds. Post treatment, total RNA was extracted by RNeasy Plus Mini kit for RT-qPCR. Alternatively, cells were subjected to MTT assay at days 0, 1, 3, and 5 post-treatment.
Statistical analyses
Results are expressed as mean ± SEM for a minimum of three independent experiments. Sample size and statistical tests are detailed in the figure legends. Statistical analysis was performed using one-way ANOVA followed by Dunnett multiple comparisons post-test to compare each condition to vehicle-treated controls. P values ≤ 0.05 were considered significant.
Results
Establishment of precision human cellular models of CDG and CDDG
Genome editing was used to generate hTERT RPE-1 cell lines that mimic genotypes associated with CDG and CDDG. All known CDDG patients possess complete loss of function of NGLY1 [8]. We designed gRNAs to generate the recurrent NGLY1 R401X missense variant, but after repeated attempts were unable to obtain the homozygous R401X genotype. Therefore, we screened clones for knock-out of NGLY1 resulting from biallelic indel formation (NGLY1−/−). PMM2-CDG often results from compound heterozygous mutations that reduce enzymatic activity [6, 17, 18]. Compound heterozygous PMM2 lines were generated by monoallelic knock-in of the second most recurrent and very severe mutation (F119L) [9, 19], and then screening for an indel on the second allele (PMM2F119L/−). We generated DPAGT1+/− lines by monoallelic knockout via indel formation. All genotypes were confirmed by Sanger sequencing (Fig. 1A, Supplementary Fig. 1A–C, E).
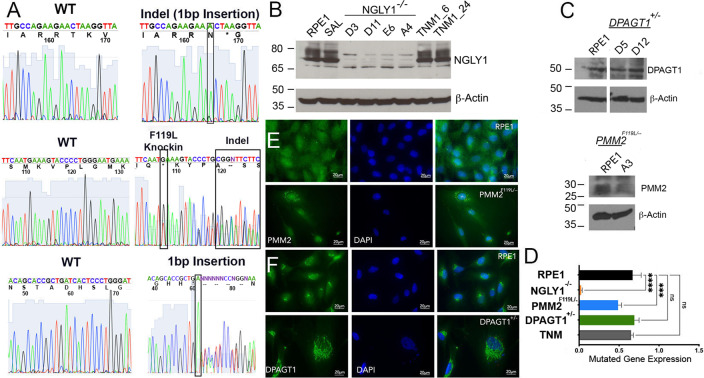
Fig. 1. CDG and CDDG cellular models were validated by Sanger sequencing, immunoblot and immunofluorescence staining for target proteins.
A Electropherogram traces for parental RPE-1 cells, CDDG—NGLY1−/− D11 (top), and CDG—PMM2F119L/−A3 (middle) and DPAGT1+/− D5 (bottom) lines. B, C Representative immunoblot images for target proteins in RPE-1 and isogenic CDDG (B) and CDG (C) lines. RPE-1 cells treated with TNM (1 µM for either 6 h or 24 h) were used as a positive control. D Quantification of the target protein levels in the edited RPE-1 cells (n ≥ 3 per genotype). Expression relative to levels in parental RPE-1 cells. ***P < 0.001, ****P < 0.0001, one-way ANOVA followed by Dunnett multiple comparisons post-test. E, F Representative immunofluorescence images for staining for PMM2 and DPAGT1 in parental RPE-1, CDG—PMM2 F119L/− A3, and CDG DPAGT1+/− D5 lines. Scale bar: 20 µm.
As expected, NGLY1−/− lines do not express NGLY1 protein and the levels of PMM2 in PMM2F119L/− were decreased by ~50% (Fig. 1B–D). The expression level of DPAGT1 was different between the two DPAGT1+/− clones analyzed despite confirmation of monoallelic disruption of DPAGT1 (Fig. 1C and Supplementary Fig. 1C). Clone D5, however, consistently expressed ~50% of DPAGT1 relative to parental cells. This clone was used in the high-content screens discussed below.
Consistent with published studies [20, 21], we found DPAGT1 localized to the perinuclear space, and PMM2 was diffuse throughout the cytosol and nucleus (Fig. 1E, F, Supplementary Fig. 1D). Interestingly, PMM2F119L was found in cytosolic puncta suggestive of protein aggregation (Fig. 1E). We failed to detect NGLY1 by immunocytochemistry using multiple NGLY1 antibodies (not shown).
CDG and CDDG lines exhibit elevated ER stress and autophagy responses
Although a common molecular feature of CDG is elevated levels of ER stress [22], systematic examination of ER stress in CDDG has not been performed. In order to establish the ER stress profiles of the cellular models, we first established a baseline in isogenic RPE-1 cells using a moderate concentration of the N-linked glycosylation inhibitor and ER stress inducer tunicamycin [23, 24] and the ER stress inhibitor salubrinal [25]. We examined ER stress using markers from each of the three recognized pathways of ER stress (Fig. 2A): (1) detection of eIF2α Ser51 phosphorylation (p-eIF2α) and nuclear translocation of ATF4 and/or CHOP, (2) presence of spliced XBP1 mRNA (sXBP1), and (3) cleavage of ATF6 [24, 26]. As expected, tunicamycin treatment resulted in strong induction of p-eIF2α, elevated expression of sXBP1, and reduced levels of ATF6(90) and corresponding increases of ATF6(60) due to cleavage (Fig. 2B–E, TNM).
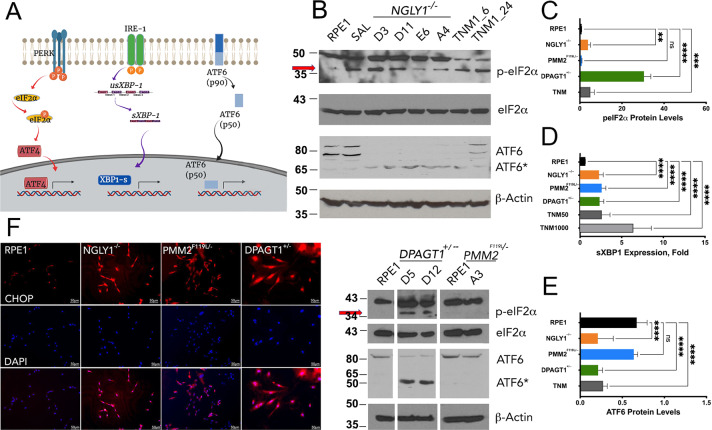
Fig. 2. Cellular models of CDG and CDDG exhibit elevated ER stress responses.
A Schematic of the three major ER stress pathways (analyzed markers are framed). B Representative immunoblot images for ER stress markers in RPE-1 and different clones of isogenic CDDG (top) and CDG (bottom) lines. RPE-1 cells treated with TNM (1 µM for either 6 h or 24 h) and salubrinal (SAL, 50 µM for 24 h) were used as negative and positive controls, respectively. C Quantification of p-eIF2α levels relative to total eIF2α protein (n ≥ 3 per each genotype or treatment). D Expression of spliced XBP1 transcript (n ≥ 10 per each genotype or treatment). E Quantification of ATF6(90) levels in parental and edited RPE-1 cells. (n ≥ 3 per each genotype or treatment). Expression data in C–E are relative to levels in parental RPE-1 cells. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA followed by Dunnett multiple comparisons post-test. F Nuclear localization of CHOP/ATF4 in parental and edited RPE-1 cells (scale bars are 50 µm).
All CDG and CDDG lines exhibited increased ER stress responses relative to untreated RPE-1 (Fig. 2B–F), but the distinct genotypes showed differential activation of the key ER stress response pathways. For example, NGLY1−/− and DPAGT1+/− lines exhibited activation of all three established ER stress pathways, whereas PMM2F119L/− had only significant increases in XBP1 splicing (Fig. 2B–E). In fact, the only pathway induced across all CDG and CDDG lines was splicing of XBP1.
Low, but significant, levels of apoptosis were detected in PMM2 F119L/− and NGLY1−/− but not in DPAGT1+/− lines (Fig. 3A, B). Apoptosis levels in mutant cell lines were comparable to low levels of induced ER stress (Fig. 3B, TNM50) and significantly lower than would be expected from high ER stress conditions (Fig. 3B, TNM1000) further suggesting that CDG and CDDG lines exhibit lower chronic ER stress responses.
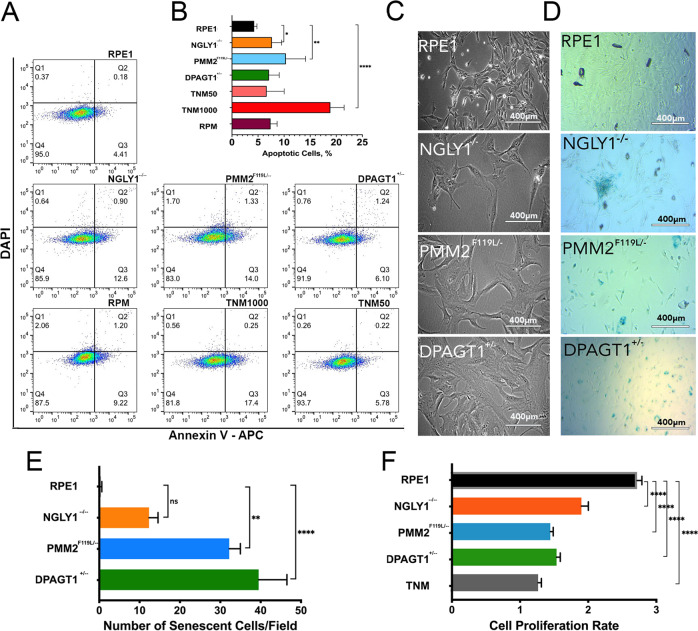
Fig. 3. CDG and CDDG cellular models exhibit low levels of apoptosis, line-dependent levels of senescence and reduced proliferation.
A Representative dot-blots of apoptosis detection (Annexin V staining) in RPE-1, CDG and CDDG lines. Tunicamycin was used as a positive control for apoptosis (1.0 µM, 24 h) and chronic ER stress (0.05 µM, 24 h). Rapamycin treatment (RPM, 500 nM) was used as a control for autophagy induction. B Quantification of apoptosis by flow cytometry (N = 3 experiments). C Representative phase-contrast images of cellular morphology. Scale bars, 400 µm. D Representative images of β-galactosidase senescence staining of parental RPE-1 and CDDG, NGLY1−/− D11, and CDG PMM2 F119L/− A3 and DPAGT1+/− D5 lines. E Quantification of senescence levels as indicated by β-galactosidase staining (10 fields were counted per each genotype). F Quantification of cellular proliferation rates for CDG and CDDG lines relative to parental RPE-1 (n ≥ 10 per each genotype or treatment). To define cell proliferation rate, ratio of OD590 at 72 h to OD590 at 24 h post seeding was calculated. *P < 0.05, **P < 0,01, ****P < 0.0001, one-way ANOVA followed by Dunnett multiple comparisons post-test.
Autophagy is known to play an important role in the response to ER stress and is seen as a marker of chronic ER stress [27]. Significant upregulation of autophagy was detected in all mutant cell lines using markers of early (p62/SQSTM1) and late (LAMP1) stages of autophagy followed by fluorescent microscopy or flow cytometry (Fig. 4 and Supplementary Fig. 2).
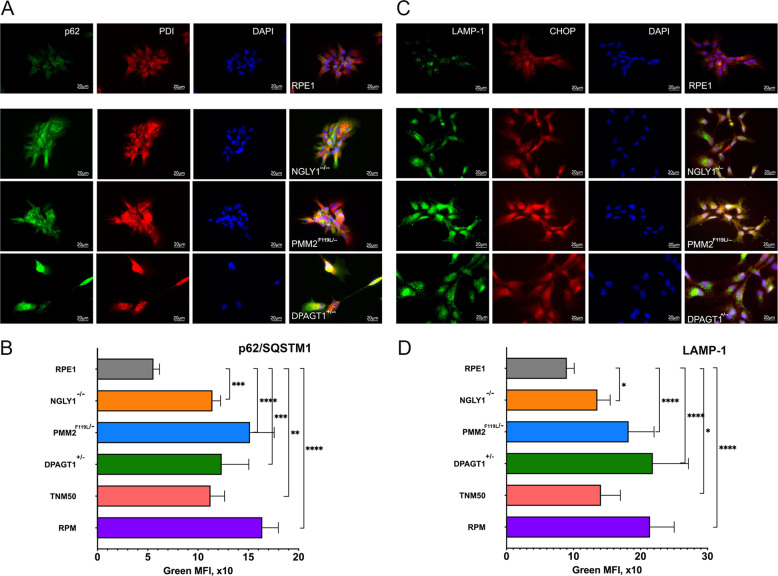
Fig. 4. CDG and CDDG lines exhibit elevated autophagy levels.
Representative immunofluorescence images of parental RPE-1 cells and CDDG and CDG cell lines stained with antibodies against A p62/SQSTM1 or C LAMP1 in combination with anti-CHOP or anti-PDI antibodies, respectively. Scale bar, 20 µm. Quantification of p62/SQSTM1 (B) and LAMP1 (D) staining by flow cytometry (n ≥ 3 per each genotype or treatment). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA followed by Dunnett multiple comparisons post-test.
CDG and CDDG lines exhibit distinctive morphological phenotypes and proliferation defects
CDG and CDDG lines were characterized for phenotypes useful for high-content imaging screens. All mutant cell lines exhibited a flat, extended morphology (Fig. 3C) reminiscent of cellular senescence that was not seen in the isogenic parental line. Indeed, β-galactosidase staining confirmed various levels of senescence among the mutant cell lines (Fig. 3D, E). All lines demonstrated slower proliferation compared to the isogenic RPE-1 line (Fig. 3F). DPAGT1+/− lines exhibited the slowest proliferation rates and were comparable to those observed in the parental line when subjected to chronic ER stress from low concentration tunicamycin exposure.
Primary drug screen identifies compounds able to reverse CDG and CDDG cellular morphology phenotypes
Our drug screening platform takes advantage of the distinctive cellular phenotypes that result from the CDG and CDDG mutations. In order to identify compounds able to correct aberrant morphological phenotypes in the mutant lines, we utilized a “cell painting” phenotypic assay [28, 29] based on stains for mitochondria, the actin cytoskeleton, endoplasmic reticulum, and nuclei (Fig. 5A). Machine learning algorithms were trained on acquired images of RPE-1 cells, and more than 300 cellular features, such as fluorescence intensity, presence and numbers of puncta, texture, and cellular shape and geometry were extracted and analyzed. Functional testing and validation of the cell painting assay was performed on CDG and CDDG cell lines (Fig. 5B). Importantly, hierarchical clustering and principal component analyses clearly distinguished mutant cells from each other and from parental RPE-1 cells (Fig. 5C, D). This demonstrates that there are distinct phenotypic, morphological changes that occur as a consequence of the CDG or CDDG mutation in each of the clones.
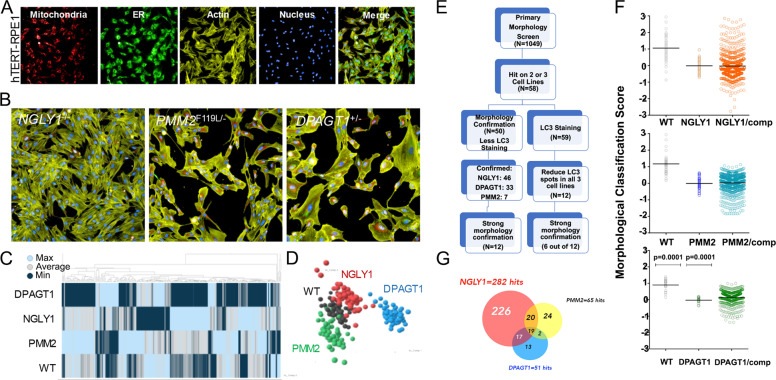
Fig. 5. High-throughput drug screen against cellular morphology via cell painting identifies compounds able to revert aberrant line-specific morphological characteristics.
A Cell painting images of parental RPE-1 cells. Cells were plated in 384-well tissue culture plate and stained with MitoTracker Red (mitochondrial stain), ConcanavalinA-488 (ER stain), phalloidin-547 (actin stain), and DAPI (nuclei) as described in the “Materials and methods” section. B Representative cell painting images of CDDG and CDG lines. C, D Hierarchical clustering and principal component analyses of extracted morphological features distinguishes CDG and CDDG cell lines from parental RPE-1 cells. E Screening workflow for selection of candidate compounds. F Representative scatter plots of primary screen results for each genotype. Each dot represents one well. For RPE-1 and vehicle treated isogenic CDG and CDDG cell lines, N = 56 replicate wells. For compound treatments, each compound was tested in N = 1 well. G Venn diagram comparing the number and overlap of compounds affecting each CDG/CDDG phenotype.
We screened 1049 annotated compounds representing a broad chemical space and multiple target classes on CDG and CDDG cell lines (Fig. 5E). The compound library was assembled with publicly available compounds that have known biological activities as well as Janssen proprietary compounds that have evidence of bioactivity compiled from multiple internal data sets. NGLY1−/−, PMM2F119L/− and DPAGT1+/− lines were treated for 24 h with 10 µM of each compound. Parental RPE-1 cells treated with vehicle (DMSO) served as a positive control while vehicle-treated CDG and CDDG lines served as negative controls (Fig. 5F). Post-treatment, the cell-painting assay was performed and a morphology score was computed for each compound’s ability to revert morphology of mutant cell lines toward that of parental cells. Results of the primary morphology screen identified 58 compounds that had positive effects in two or three cell lines (Fig. 5F, G). Because CDG/CDDG cell lines demonstrate elevated autophagy levels, primary screening hits were subsequently assessed for their ability to modulate autophagy by immunocytochemistry with LC3 as the marker. Twelve candidate compounds reduced autophagy in all three cell lines (Group I, Supplementary Table 2), and 10 additional candidate compounds (confirmed in at least 2 cell lines) that had a minimal, or no effect on autophagy (Group II, Supplementary Table 2).
Evaluation of compounds for amelioration of ER stress and proliferation defects
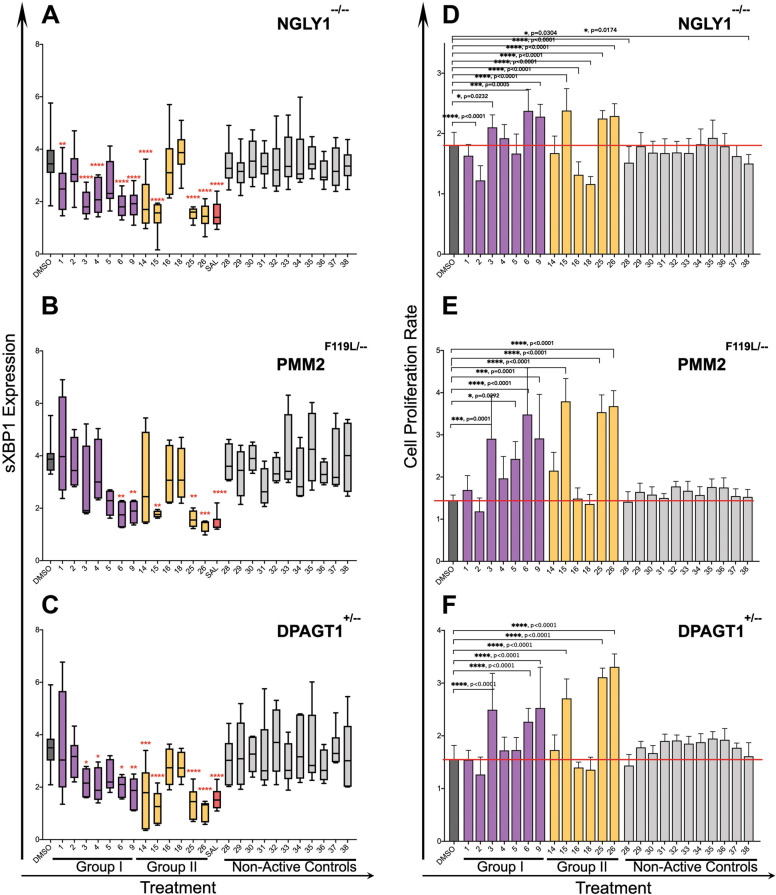
The top six candidates from Group I and Group II were further evaluated their ability to alleviate ER stress in the CDG and CDDG lines. Compounds that improved phenotypes in all three genetic lines were also included for further testing. In order to validate the efficiency and potency of the primary screen, we selected 10 non-active compounds in the cell-painting assay for comparison to candidate compounds. In total, we tested 16 active compounds and 10 non-active control compounds for their effects on cell proliferation and sXBP1 expression. We focused on sXBP1 expression because it was the only ER stress marker dysregulated across all three models, and the only significant ER stress marker in PMM2F119L/− (Fig. 2C–E).
All active compounds affected one or both assays in at least one of the cell lines (Fig. 6). Compound effects appeared to be mechanism and cell line-dependent in the proliferation and sXBP1 assays. For example, Group I compounds showed effects on all lines (Fig. 6, purple bars) while the autophagy inducing compounds (Group II) were generally more efficient in the NGLY1−/− and DPAGT1+/− lines (Fig. 6, yellow bars). Compounds 1, 3, 4, 5, and 14 decreased sXBP1 expression in CDDG lines compared to DMSO treated controls (dark gray bar) but did not affect proliferation (Fig. 6A, D). None of the ten non-active control compounds had an effect on sXBP1 expression (Fig. 6A–C, gray) or proliferation (Fig. 6D–F, gray bars). It is interesting to note that salubrinal—a highly selective inhibitor of eIF2α phosphorylation—was as effective as some active compounds at reducing sXBP1 expression. This is particularly interestingly for PMM2F119L/− cells, which do not show induction of p-eIF2α.
Fig. 6. Validation that active compounds correct proliferation defects and an ER stress phenotype shared across the CDG and CDDG lines.
CDG and CDDG cell lines were treated with optimized concentrations of candidate and non-active control compounds and expression of sXBP1 and cell proliferation were assessed. A–C Expression levels of sXBP1 in NGLY1−/− (A), PMM2F119L/− (B), and DPAGT1+/− (C) clones treated with candidates or non-active compounds. Results are presented as a ratio of sXBP1 levels in compound-treated cells to vehicle treated cells (n ≥ 10 per each genotype or treatment). D–F Modulation of proliferation rate of NGLY1−/− (D), PMM2F119L/− (E), and DPAGT1+/− (F) lines treated with candidates or non-active compounds (n ≥ 10 per each genotype or treatment). To define cell proliferation rate, ratio of OD590 at 72 h to OD590 at 24 h post seeding (before treatment) was calculated. Data presented are an average for all clones available for a specific mutation. **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA followed by Dunnett multiple comparisons post-test.
We found no correlation between reduction of sXBP1 expression and repair of proliferation for compounds 3, 4, 5, and 14 in CDG cell lines. Group I candidates 6 and 9 and Group II candidates 15, 25, and 26 all effectively reduced sXBP1 expression (Fig. 6A–C), and showed restoration of proliferation in CDG and CDDG lines, (Fig. 6D–F). Active compounds, but not non-active controls, were able to revert aberrant cellular morphology similar to that of vehicle treated controls (Supplementary Fig. 3). Together, these data validate the effectiveness of the screen, and identify sets of compounds that are able to correct aberrant cellular phenotypes associated with CDG and CDDG genotypes.
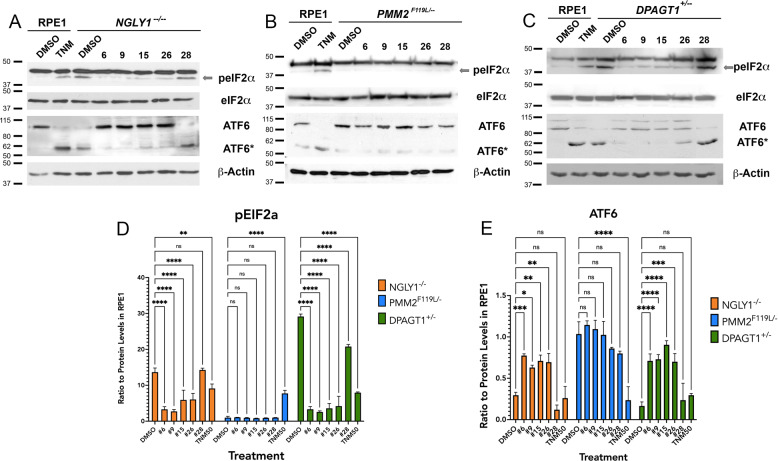
We then tested whether lead active compounds could alleviate markers of the other ER stress pathways dysregulated in our models (Fig. 2). We treated the CDG and CDDG lines with optimized concentrations of the top two Group I (6, 9) and Group II (15, 26) compounds as well as a non-active control compound (28), and then assessed levels of p-eIF2α and ATF6 (Fig. 7). As expected, all candidate compounds effectively decreased levels of p-eIF2α and decreased ATF6(90) cleavage relative to vehicle-treated control NGLY1−/− cells (Fig. 7A, D, E). The PMM2F119L/− line did not exhibit abnormal levels of p-eIF2α or ATF6(90) and none of the candidate compounds adversely affected expression of these markers (Fig. 7B, D, E). Similar to NGLY1−/−, all candidate compounds were able to effectively alleviate markers of ER stress in the DPAGT1+/− line (Fig. 7C–E). We note that the non-active control compound tested (#28) was able to reduce p-eIF2α levels in DPAGT1+/− cells; however, it was not as effective as candidate compounds (Fig. 7E). We also note that, despite an indication of autophagy induction by Group II compounds 15 and 26 in the primary screen, we did not see a significant increase (or reduction) of SQSTM1/p62 protein levels following treatment with any of the tested compounds in any of the cellular models (Supplementary Fig. 4).
Fig. 7. Select candidate compounds are able to alleviate the multiple ER stress phenotypes identified in each of the CDG and CDDG lines.
Following treatment of CDG and CDDG cell lines with optimized concentrations of candidate compounds (6, 9, 15, 26) and a non-active control compound (28), p-eIF2α and ATF6 levels were assessed by immunoblot. A–C Representative images of immunoblots. TNM, tunicamycin (50 nM). (D, E) Quantification of p-eIF2α levels in levels in CDG/CDDG treated cells. (n = 3 per each genotype and treatment). Expression data in (D, E) are relative to expression levels in parental RPE-1 cells. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.001 by one-way ANOVA followed by Dunnett multiple comparisons post-test.
Table 1 provides a summary of correction for proliferation and ER stress markers for each compound tested in each of the CDG and CDDG lines. Taken together, our screen identified several highly-effective lead compounds that are able to correct proliferation defects as well as alleviate molecular markers of multiple ER stress pathways dysregulated in cellular models of CDG and CDDG.
Table 1.
Correlation between the proliferation, sXBP1 and ATF6 expression and peIF2α levels in CDG and CDDG cell lines treated with candidate and non-active control compounds.
| Compounds | NGLY1−/− | PMM2F119L/− | DPAGT1+/− | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proliferation | sXBP1 | peIF2α | ATF6 | Proliferation | sXBP1 | peIF2αa | ATF6a | Proliferation | sXBP1 | peIF2α | ATF6 | |
| Candidates Group I | ||||||||||||
| 1 | ** | ns | ns | ns | ns | ns | ||||||
| 2 | **** | ns | ns | ns | ns | ns | ||||||
| 3 | ns | **** | *** | ns | **** | * | ||||||
| 4 | ns | **** | ns | ns | ns | ** | ||||||
| 5 | ns | * | * | ns | ns | ns | ||||||
| 6 | **** | **** | **** | *** | **** | ** | ns | ns | *** | ** | **** | **** |
| 9 | *** | **** | **** | * | *** | ** | ns | ns | **** | *** | **** | **** |
| Candidates Group II | ||||||||||||
| 14 | ns | **** | ns | ns | ns | *** | ||||||
| 15 | **** | **** | **** | ** | **** | ** | ns | ns | **** | **** | **** | **** |
| 16 | *** | ns | ns | ns | ns | ns | ||||||
| 18 | **** | ns | ns | ns | ns | ns | ||||||
| 25 | ns | **** | **** | ** | **** | **** | ||||||
| 26 | ns | **** | **** | ** | **** | *** | ns | ns | **** | **** | **** | *** |
| Non-active controls | ||||||||||||
| 28 | ns | ns | ns | ns | ns | ns | ns | ns | ns | ns | **** | ns |
| 29 | ns | ns | ns | ns | ns | ns | ||||||
| 30 | ns | ns | ns | ns | ns | ns | ||||||
| 31 | ns | ns | ns | ns | ns | ns | ||||||
| 32 | ns | ns | ns | ns | ns | ns | ||||||
| 33 | ns | ns | ns | ns | ns | ns | ||||||
| 34 | ns | ns | ns | ns | ns | ns | ||||||
| 35 | ns | ns | ns | ns | ns | ns | ||||||
| 36 | ns | ns | ns | ns | ns | ns | ||||||
| 37 | ns | ns | ns | ns | ns | ns | ||||||
| 38 | ns | ns | ns | ns | ns | ns | ||||||
aNo phenotype for these ER stress markers.
*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
A central challenge to the development of novel therapies is the availability of screenable models that focus on disease-relevant phenotypes. Screens based on mutation-induced phenotypes, such as morphological differences, allows one to establish a screening assay without a full understanding of the molecular mechanisms that drive disease pathology. This creates an opportunity for the identification of new therapeutic targets as well as uncovering new insights related to etiology.
The objective of the high-content, phenotypic screen described here was to rapidly identify small molecules capable of alleviating ER stress in cellular models of monogenic disease. The rationale for our screen is that ER stress responses should be applicable across a variety of cell types, and drugs capable of alleviating ER stress will help treat symptoms of disease. Backed by the growing body of evidence linking ER stress to multiple neurological conditions and to CDG and CDDG, we reasoned that using ER stress markers as a functional readout combined with cellular phenotypes can serve as a proxy for overall cellular health on a disease background. It was important to develop the CDDG and CDG models in a cell type with uniform morphology that permits rapid and easily quantifiable morphology changes. We note that a screen in a more disease-relevant cell type such as hiPSC-derived neurons may be more applicable; however, such approaches have a number of drawbacks that our approach addresses. For instance, common neuronal differentiation methods yield a heterogeneous population of cells with differing levels of maturity and morphology that renders potential molecular or morphological phenotypes difficult to identify or interpret. Moreover, the labor intensiveness and cost of differentiation methods often makes large-scale screens prohibitive. Rather, a multi-tiered strategy whereby large screens are performed on genetic cellular models with high confidence phenotypes and lead compounds are then validated in more relevant cellular and/or animal models is more efficacious.
A majority of the active compounds in our screen, #s 3, 4, 6, 9, 15, 25, are reported to affect microtubules (Supplementary Table 3) either through direct effects on microtubules themselves or by targeting proteins (e.g. kinases) that regulate microtubule dynamics [30–36]. Compounds structurally similar to compounds 6, 9, 15, 25 and 26 (Supplementary Table 4) identified here are reported to prevent ER stress in multiple cellular systems through modulation of JAK/STAT and growth factor signaling among others (Supplementary Table 3) [37–41]. Multiple compounds converging on a biological process (the regulation of microtubules) suggests this is a legitimate therapeutic avenue.
Our study describes, to our knowledge, the first example of a high-throughput screen on genetically modified human cells for three monogenic diseases with a shared endogenous molecular phenotype. Here we focused on a biological process, ER stress, thought to unite a number of rare and more common diseases, and successfully identified bioactive ER stress diminishing compounds through unbiased morphological screening. This work has shown it is possible to develop cellular models that possess screenable phenotypes able to identify compounds that alleviate molecular and morphological phenotypes caused by the underlying genetic mutation, thereby establishing a platform to identify targeted and common treatments for monogenic disorders. Due to the genetic heterogeneity of CDGs, it will be important to determine whether the compounds identified here also alleviate ER stress-related phenotypes in other genetic causes of CDG.
Beyond establishing a paradigm for identifying therapeutic compounds for rare monogenic diseases, this work suggests a direction for identifying compounds able to alleviate the symptoms related to ER stress in more common diseases characterized by ER stress including neurodegenerative diseases. Loss of microtubule mass or altered microtubule dynamics in axons and dendrites are major contributors to neurodegenerative diseases such as ALS, Parkinson’s disease, Alzheimer’s disease, and several tauopathies [42]. Future studies will determine whether compounds that affect microtubule dynamics are able to prevent disease-relevant phenotypes in cellular models of neurodegenerative diseases.
Supplementary information
Acknowledgements
We thank Guy Ludwig and other past and present members of the Institute for Genomic Medicine for insights and helpful discussions. We also thank Barbara Corneo and Alejandro Garcia Diaz of the Columbia Stem Cell Core Facility for assistance with CRISPR/Cas9 editing to generate the CDG and CDDG models.
Author contributions
Conceptualization: D.B.G., M.J.B., Y.-F.L., M.V.W. and J.W. Data curation, formal analysis, investigation, and methodology: I.V.L., M.V.W., S.S. and Y.-F.L. Project Administration: I.V.L., M.V.W., A.A-O., J.W. and M.J.B. Supervision: M.J.B., J.W. and D.B.G. Validation: I.V.L., M.V.W., S.S. and M.J.B. Visualization: I.V.L. and M.J.B. Writing—original draft: I.V.L., M.J.B. and D.B.G. Writing—review & editing: I.V.L., M.V.W., M.B.H., M.J.B. and D.B.G.
Competing interests
DBG is a founder of and holds equity in Praxis, serves as a consultant to AstraZeneca, and has received research support from Janssen, Gilead, Biogen, AstraZeneca and UCB. MBH serves as a consultant to the Muscular Dystrophy Association, and receives research support from Biogen. All other authors declare no competing interests.
Footnotes
Edited by C. Munoz-Pinedo
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Michael J. Boland, Email: mb4129@columbia.edu
David B. Goldstein, Email: dg2875@columbia.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-04045-4.
References
- 1.Peltonen L, Perola M, Naukkarinen J, Palotie A. Lessons from studying monogenic disease for common disease. Hum Mol Genet. 2006;15:R67–R74. doi: 10.1093/hmg/ddl060. [DOI] [PubMed] [Google Scholar]
- 2.Allen AS, Bellows ST, Berkovic SF, Bridgers J, Burgess R, Cavalleri G, et al. Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol. 2017;16:135–43. doi: 10.1016/S1474-4422(16)30359-3. [DOI] [PubMed] [Google Scholar]
- 3.Roussel BD, Kruppa AJ, Miranda E, Crowther DC, Lomas DA, Marciniak SJ. Endoplasmic reticulum dysfunction in neurological disease. Lancet Neurol. 2013;12:105–18. doi: 10.1016/S1474-4422(12)70238-7. [DOI] [PubMed] [Google Scholar]
- 4.Jung J, Michalak M, Agellon LB. Endoplasmic reticulum malfunction in the nervous system. Front Neurosci. 2017;11:220. doi: 10.3389/fnins.2017.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hetz C, Saxena S. ER stress and the unfolded protein response in neurodegeneration. Nat Rev Neurol. 2017;13:477–91. doi: 10.1038/nrneurol.2017.99. [DOI] [PubMed] [Google Scholar]
- 6.Jaeken J. Congenital disorders of glycosylation. Ann N Y Acad Sci. 2010;1214:190–8. doi: 10.1111/j.1749-6632.2010.05840.x. [DOI] [PubMed] [Google Scholar]
- 7.Freeze HH. Understanding human glycosylation disorders: biochemistry leads the charge. J Biol Chem. 2013;288:6936–45. doi: 10.1074/jbc.R112.429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enns GM, Shashi V, Bainbridge M, Gambello MJ, Zahir FR, Bast T, et al. Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genet Med. 2014;16:751–8. doi: 10.1038/gim.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthijs G, Schollen E, Pardon E, Veiga-Da-Cunha M, Jaeken J, Cassiman JJ, et al. Mutations in PMM2, a phosphomannomutase gene on chromosome 16p13, in carbohydrate-deficient glycoprotein type I syndrome (Jaeken syndrome) Nat Genet. 1997;16:88–92. doi: 10.1038/ng0597-88. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Zhao Y, Zhou K, Freeze HH, Zhang YW, Xu H. Insufficient ER-stress response causes selective mouse cerebellar granule cell degeneration resembling that seen in congenital disorders of glycosylation. Mol Brain. 2013;6:52. doi: 10.1186/1756-6606-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heifetz A, Keenan RW, Elbein AD. Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry. 1979;18:2186–92. doi: 10.1021/bi00578a008. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Rush JS, Karaoglu D, Krasnewich D, Lubinsky MS, Waechter CJ, et al. Deficiency of UDP-GlcNAc:Dolichol Phosphate N-Acetylglucosamine-1 Phosphate Transferase (DPAGT1) causes a novel congenital disorder of Glycosylation Type Ij. Hum Mutat. 2003;22:144–50. doi: 10.1002/humu.10239. [DOI] [PubMed] [Google Scholar]
- 13.Würde AE, Reunert J, Rust S, Hertzberg C, Haverkämper S, Nürnberg G, et al. Congenital disorder of glycosylation type Ij (CDG-Ij, DPAGT1-CDG): extending the clinical and molecular spectrum of a rare disease. Mol Genet Metab. 2012;105:634–41. doi: 10.1016/j.ymgme.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Lebedeva IV, Su ZZ, Vozhilla N, Chatman L, Sarkar D, Dent P, et al. Mechanism of in vitro pancreatic cancer cell growth inhibition by melanoma differentiation-associated gene-7/interleukin-24 and perillyl alcohol. Cancer Res. 2008;68:7439–47. doi: 10.1158/0008-5472.CAN-08-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fonseca SG, Ishigaki S, Oslowski CM, Lu S, Lipson KL, Ghosh R, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest. 2010;120:744–55. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Van Schaftingen E, Jaeken J. Phosphomannomutase deficiency is a cause of carbohydrate-deficient glycoprotein syndrome type I. FEBS Lett. 1995;377:318–20. doi: 10.1016/0014-5793(95)01357-1. [DOI] [PubMed] [Google Scholar]
- 18.van de Kamp JM, Lefeber DJ, Ruijter GJ, Steggerda SJ, den Hollander NS, Willems SM, et al. Congenital disorder of glycosylation type Ia presenting with hydrops fetalis. J Med Genet. 2007;44:277–80. doi: 10.1136/jmg.2006.044735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kjaergaard S, Schwartz M, Skovby F. Congenital disorder of glycosylation type Ia (CDG-Ia): phenotypic spectrum of the R141H/F119L genotype. Arch Dis Child. 2001;85:236–9. doi: 10.1136/adc.85.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 21.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. A subcellular map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 22.Lecca MR, Wagner U, Patrignani A, Berger EG, Hennet T. Genome-wide analysis of the unfolded protein response in fibroblasts from congenital disorders of glycosylation type-I patients. FASEB J. 2005;19:240–2. doi: 10.1096/fj.04-2397fje. [DOI] [PubMed] [Google Scholar]
- 23.Cóppola-Segovia V, Cavarsan C, Maia FG, Ferraz AC, Nakao LS, Lima MM, et al. ER stress induced by tunicamycin triggers alpha-synuclein oligomerization, dopaminergic neurons death and locomotor impairment: a new model of Parkinson’s disease. Mol Neurobiol. 2017;54:5798–806. doi: 10.1007/s12035-016-0114-x. [DOI] [PubMed] [Google Scholar]
- 24.Shang J. Quantitative measurement of events in the mammalian unfolded protein response. Methods Enzymol. 2011;491:293–308. doi: 10.1016/B978-0-12-385928-0.00016-X. [DOI] [PubMed] [Google Scholar]
- 25.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–9. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 26.Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nat Rev Microbiol. 2015;13:71–82. doi: 10.1038/nrmicro3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rashid HO, Yadav RK, Kim HR, Chae HJ. ER stress: Autophagy induction, inhibition and selection. Autophagy. 2015;11:1956–77. doi: 10.1080/15548627.2015.1091141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gustafsdottir SM, Ljosa V, Sokolnicki KL, Anthony Wilson J, Walpita D, Kemp MM, et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS ONE. 2013;8:e80999. doi: 10.1371/journal.pone.0080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coxon CR, Anscombe E, Harnor SJ, Martin MP, Carbain B, Golding BT, et al. Cyclin-dependent kinase (CDK) inhibitors: structure-activity relationships and insights into the CDK-2 selectivity of 6-substituted 2-arylaminopurines. J. Med Chem. 2017;60:1746–67. doi: 10.1021/acs.jmedchem.6b01254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis S, Lamb ML, Wang T, Almeida L, Block MH, Davies AM, et al. Discovery of 5-chloro-N2-[(1S)-1-(5-fluoropyrimidin-2-yl)ethyl]-N4-(5-methyl-1H-pyrazol-3-yl)pyrimidine-2,4-diamine (AZD1480) as a novel inhibitor of the Jak/Stat pathway. J Med Chem. 2011;54:262–76. doi: 10.1021/jm1011319. [DOI] [PubMed] [Google Scholar]
- 32.Jordan MA. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr. Med Chem. Anticancer Agents. 2002;2:1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 33.Kadir S, Astin JW, Tahtamouni L, Martin P, Nobes CD. Microtubule remodelling is required for the front-rear polarity switch during contact inhibition of locomotion. J. Cell Sci. 2011;124:2642–53. doi: 10.1242/jcs.087965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rochlin MW, Wickline KM, Bridgman PC. Microtubule stability decreases axon elongation but not axoplasm production. J Neurosci. 1996;16:3236–46. doi: 10.1523/JNEUROSCI.16-10-03236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahenk Z, Brady ST. Axonal tubulin and microtubules: morphologic evidence for stable regions on axonal microtubules. Cell Motil Cytoskeleton. 1987;8:155–64. doi: 10.1002/cm.970080207. [DOI] [PubMed] [Google Scholar]
- 36.Sengottuvel V, Fischer D. Facilitating axon regeneration in the injured CNS by microtubules stabilization. Commun Integr Biol. 2011;4:391–3. doi: 10.4161/cib.15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurel PS, Hatch AL, Higgs HN. Connecting the cytoskeleton to the endoplasmic reticulum and Golgi. Curr Biol. 2014;24:R660–R672. doi: 10.1016/j.cub.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meares GP, Liu Y, Rajbhandari R, Qin H, Nozell SE, Mobley JA, et al. PERK-dependent activation of JAK1 and STAT3 contributes to endoplasmic reticulum stress-induced inflammation. Mol Cell Biol. 2014;34:3911–25. doi: 10.1128/MCB.00980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen MD, Mushynski WE, Julien JP. Cycling at the interface between neurodevelopment and neurodegeneration. Cell Death Differ. 2002;9:1294–306. doi: 10.1038/sj.cdd.4401108. [DOI] [PubMed] [Google Scholar]
- 40.Qin H, Buckley JA, Li X, Liu Y, Fox TH, 3rd, Meares GP, et al. Inhibition of the JAK/STAT pathway protects against alpha-synuclein-induced neuroinflammation and dopaminergic neurodegeneration. J. Neurosci. 2016;36:5144–59. doi: 10.1523/JNEUROSCI.4658-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun QA, Hess DT, Wang B, Miyagi M, Stamler JS. Off-target thiol alkylation by the NADPH oxidase inhibitor 3-benzyl-7-(2-benzoxazolyl)thio-1,2,3-triazolo[4,5-d]pyrimidine (VAS2870) Free Radic Biol Med. 2012;52:1897–902. doi: 10.1016/j.freeradbiomed.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matamoros AJ, Baas PW. Microtubules in health and degenerative disease of the nervous system. Brain Res. Bull. 2016;126:217–25. doi: 10.1016/j.brainresbull.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.