Abstract
The use of genomic testing is rapidly emerging as an important clinical tool both for cancer diagnosis and for guiding treatment decisions in a wide range of malignancies, including gastrointestinal (GI) cancers such as colorectal cancer (CRC). Advances in technologies such as polymerase chain reaction and next‐generation sequencing methods have made it possible to noninvasively screen for CRC through, for example, the use of blood‐ or stool‐based testing, with high specificity. Tests are also available that can provide prognostic information beyond traditional clinicopathologic factors such as tumor size, grade, and nodal status, which can enable clinicians to more accurately risk stratify patients for recurrence. Lastly, in the setting of resected CRC, tests are now available that can detect circulating tumor DNA as a means for noninvasive minimal/molecular residual disease monitoring, thereby potentially guiding the use of adjuvant chemotherapy and/or escalating or de‐escalating therapy. The Gastrointestinal Cancer Therapy Expert Group (GICTEG) recently convened a virtual meeting to discuss current issues related to genomic testing in GI cancer, with the goal of providing guidance on the use of these tests for the practicing community oncologist, for whom GI cancer may be only one of many tumor types encountered. This article provides a summary of the discussion and highlights the key opinions of the GICTEG on this topic.
Implications for Practice
The Gastrointestinal Cancer Therapy Expert Group seeks to provide practical guidance and opinion on the treatment of gastrointestinal malignancies, including colorectal cancer (CRC), for the practicing community oncologist in situations for which guidelines from established bodies, such as the National Comprehensive Cancer Network and the American Society of Clinical Oncology, may be less clear. In the present report, clinical guidance on the use of molecular assays for a range of clinical indications in CRC is presented, including the use of circulating tumor DNA to detect minimal/molecular residual disease in patients with successfully resected early‐stage CRC.
Keywords: Genomic testing, Gastrointestinal cancers, Colorectal cancer, Gastrointestinal Cancer Therapy Expert Group, Circulating tumor DNA, Minimal residual disease
Short abstract
The Gastrointestinal Cancer Therapy Expert Group (GICTEG) recently convened a virtual meeting to discuss current issues related to genomic testing in gastrointestinal cancer, with the goal of providing guidance on the use of these tests for the practicing community oncologist. This article provides a summary of the discussion and highlights the key opinions of the GICTEG on this topic.
Introduction
Despite improvements in prevention, screening, as well as advances in potentially curative surgical and adjuvant therapies, colorectal cancer (CRC) remains one of the most commonly diagnosed cancers and is a leading cause of cancer death worldwide. Molecular assays are increasingly being used to prognosticate patients, guide treatment decisions, and monitor for disease recurrence in CRC. Several tests are now available and/or are under development that offer the promise of simpler, more specific, less costly, and less invasive methods for screening and early detection, stratification of patients for recurrence risk, guidance regarding the utility of adjuvant treatment, and monitoring for disease recurrence. Treatment guidelines have not yet been established on the use of molecular testing and/or other emergent technologies in CRC.
About the GICTEG and Role of Funding Sources
The Gastrointestinal Cancer Therapy Expert Group (GICTEG) comprises expert physicians and clinical researchers who have dedicated their careers to the treatment of patients with gastrointestinal (GI) malignancies. The purpose of the group is to meet periodically to discuss important developments related to management of GI cancers, with a particular emphasis on new findings and/or areas for which guidance from established bodies, such as the National Comprehensive Cancer Network (NCCN) and the American Society for Clinical Oncology (ASCO), may be unresolved and/or less well established. The goal is to elicit the group's collective opinions on a given topic as it relates to their own clinical practices and, more importantly, how this might impact oncologists in the community setting who may not be as extensively versed in the treatment of GI cancers. Importantly, this article is not intended to replace any existing guidance or guidelines, nor to be an exhaustive review of the topic(s) in question. Rather, it is intended to present a concise synopsis of the most important data in the area and summarize the opinion of the expert group, as gleaned from the meeting discussion.
In May of 2020, members of the GICTEG convened a virtual meeting to discuss current and emergent issues surrounding the use of molecular testing in colorectal and other GI cancers. The faculty members of the GICTEG were selected by Total Health Information Services, a medical information company, based on their expert experience on this topic. An unrestricted educational grant for this activity was provided by Natera Oncology. The faculty and Total Health jointly selected the main topics and general outline for the discussion. It is recognized that the panelists may or may not have relationships with corporate entities both related and/or unrelated to the topic in question. Content of the discussions, and any expert opinions presented herein, is intended to be based on the panelists’ own expert clinical experience and insight, in alignment with current guidelines, and is understood not to be influenced by any corporate relationship or interest.
Genomic Testing in CRC Screening and Ongoing Clinical Trials
Although not intended to be a comprehensive list of all CRC and/or pan‐cancer screening tests that are currently under investigation, Table 1 provides an overview of some of the genomic tests and technologies that have been used to screen for CRC, including commercially available and investigational tests [1, 2, 3, 4, 5, 6, 7, 8, 9]. These noninvasive tests aim to assess for genomic and epigenomic changes in bodily fluids (i.e., “liquid biopsies”), which can detect the presence of cancer. Exact Sciences’ Cologuard, one of the more widely known noninvasive screening methods, is a stool‐based assay that evaluates fecal hemoglobin as well as several DNA markers, including KRAS mutations, and epigenomic markers like aberrant methylation of the NDRG4 and BMP3 genes. The performance characteristics of the test were assessed in a prospective, multisite, point‐in‐time study of adults at average risk for CRC, ages 50–84, who were scheduled to undergo colonoscopy. The test has a reported sensitivity of 92.3% for detecting CRC as compared with 73.8% for fecal immunohistochemical testing (p = .002); sensitivity of the test was also better for detection of advanced precancerous lesions (42.4% vs. 23.8%; p < .001), polyps with high‐grade dysplasia (69.2% vs. 46.2% p = .004), and serrated sessile polyps of ≥1cm (42.4% vs. 5.1%; p < .001); however, the specificity of the test was lower (i.e., more false positives) [7, 8]. Several investigational blood‐based assays for CRC screening are also highlighted in Table 1.
Table 1.
Selected genomic tests for CRC screening commercially available and/or under investigation
| Company | Test description and uses | Relevant clinical trials |
|---|---|---|
| Guardant [1, 2] |
ctDNA‐based blood test (LUNAR‐2) currently under investigation for use in detecting CRC in an average risk population |
ECLIPSE (NCT04136002): CRC screening study initiated in 2019; goal to enroll ~10,000 patients ages 45–84 planning to have a colonoscopy |
| Grail [3, 4] | Pan‐cancer early detection blood test using cf nucleic acids (DNA and RNA) |
PATHFINDER (NCT04241796): prospective, multicenter study enrolling ~6,200 participants; primary goal is to guide appropriate diagnostic workup for different cancer types |
| Freenome [5, 6] |
Multiomics platform using cfDNA, cfRNA, proteomics, epigenetics, as well as computational biology and machine learning methods to generate early tumor and immune cancer signatures |
PREEMPT CRC (NCT04369053): clinical study enrolling ~14,000 average risk participants undergoing colonoscopy; primary goal is to validate blood‐based test for CRC screening |
| Exact Sciences [7, 8, 9] |
ColoGuard Stool–based hemoglobin and DNA test evaluates KRAS mutations, β‐actin, and aberrant methylation of NDRG4 and BMP3 |
NCT01397747 (DeeP‐C) |
|
Liquid biopsy blood test (CancerSEEK) currently under development for early detection of CRC and other cancers; not available for patients at present |
Initial evaluation of CancerSEEK test published as part of the DETECT‐A study; study evaluated >10,000 women aged 65 to 75 with no prior cancer history |
Abbreviations: cf, cell‐free; CRC, colorectal cancer; ctDNA, circulating tumor DNA.
Discussion
The group considered several questions related to the use of these tests as a means for screening, including where and how these tests are currently being used, the target population, and current guidance on their use. Overall, most medical oncologists generally become involved in the care of cancer patients once the diagnosis is made, and for that reason these noninvasive screening tests are usually ordered by primary care physicians and/or gastroenterologists as part of health maintenance visits. Because of this, most in the group had not used any of the available assays for early CRC detection (Table 1) outside of the setting of a clinical trial. The group recognizes that patients frequently have knowledge of these tests (e.g., through media, internet, and/or social media sources) and often have questions about using them. With regards to the utility of blood‐based genomic tests for CRC, the recent work of Lennon and colleagues was noted [9]. In this prospective, interventional study of over 10,000 women not known to have cancer, those with a positive blood test were followed up with positron emission tomography–computed tomography scanning to confirm and localize the cancer. The group thought that studies of this type could be a useful model for other population screening methodologies as a means to improve the level of evidence and provide additional confidence before this type of testing may be routinely used in practice. Several important ongoing efforts including the GRAIL programs and ECLIPSE will help to refine the role of cell‐free approaches in early CRC detection [2, 4]. It was agreed that the current gold standard for screening for early CRC remains colonoscopy.
Genomic Testing in Early‐Stage CRC
Prognostic Versus Predictive
In discussing genomic testing options that inform recurrence risk in early‐stage CRC, the group first noted the importance of highlighting the difference between tests that provide prognostic versus predictive information. Tests that provide prognostic information inform on the overall risk of disease recurrence in patients with surgical resection (i.e., disease biology), whereas tests that provide predictive information may inform decisions regarding the role of adjuvant or postoperative therapies to decrease recurrence risk (i.e., response to therapy).
Oncotype and Immunoscore
Molecular assays that may inform recurrence risk assessment in early‐stage CRC, including Oncotype Dx and Immunoscore, are highlighted in Table 2. Oncotype DX Colon (12‐gene assay) has been used clinically as a means to assess risk of CRC recurrence in patients with stage II disease based on its recurrence scoring system; however, it has not been validated as a means for predicting response to adjuvant chemotherapy. The prognostic performance of the test has also been validated across several major CRC trials, including QUASAR, CALGB 9581, NSABP C‐07, and SUNRISE [10, 11, 12, 13]. Immunoscore is an image‐based risk assessment tool that evaluates the host immune response (CD3+ and CD8+ cells) in the tumor microenvironment; it is intended to be used adjunctively with standard tumor, lymph node, metastasis (TNM) classification, measuring the density of CD3+ and CD8+ T lymphocyte populations in the center and at the periphery of the tumor (Table 2) [14, 15, 16, 17, 18, 19, 20]. The test has been validated across several studies as a powerful prognostic tool in the treatment of patients with stage II and III CRC (Table 2).
Table 2.
Selected genomic tests for risk assessment in CRC
| Test | Test description and uses | Relevant clinical trials |
|---|---|---|
| Oncotype DX Colon [10, 11, 12, 13] |
12‐gene genomic test (7 cancer‐related, 5 reference); validated for use in stage II MMR‐P and stage III A/B colon cancers; used to estimate recurrence risk (low, intermediate, high) and help guide treatment decisions; not predictive of adjuvant chemotherapy benefit |
Validation studies confirm Recurrence Score (0–100); result predicts risk for local recurrence (QUASAR, CALGB 9581, NSABP C‐07, SUNRISE) |
| Immunoscore Colon [14, 15, 16, 17, 18, 19, 20] |
Image analysis–based risk assessment tool targeting CD3+ and CD8+ cells and examining host immune response at the tumor site; intended for use adjunctively with TNM classification |
In patients with stage II disease Immunoscore predicted a high‐risk subgroup (Immunoscore Low) with increased recurrence at 5 years (23% vs. 8%); suggested 7 of 10 high‐risk patients with stage II disease might be spared chemotherapy Patients with stage III disease with Immunoscore High status had significantly better DFS (HR = 0.59; p = .0013), whereas 1 of 2 low‐risk (T1‐3/N1) patients with Immunoscore Low status had significantly worse 3‐year DFS (78% vs. 92%) |
Abbreviations: CRC, colorectal cancer; DFS, disease‐free survival; MMR‐P, mismatch repair proficient; TNM, tumor, node, metastasis.
Discussion
Although well‐accepted clinical and pathologic factors, including lymphatic/vascular invasion, perineural invasion, T4 staging, and clinical obstruction or perforation, remain essential in risk assessment and adjuvant chemotherapy decision‐making for early‐stage CRC, the group weighed in on the prognostic‐versus‐predictive value of the growing number of molecular assays for risk assessment. The panelists agreed that tests such as Oncotype DX Colon and Immunoscore represent prognostic tests, and they have not been validated as predictive of chemotherapy benefit. Although these tests may inform whether patients are at particularly high risk of recurrence (i.e., prognosis), it is unclear whether or to what degree adjuvant chemotherapy may reduce that risk. The group therefore agreed that these tests cannot inform adjuvant chemotherapy decisions at the present time, given the lack of validated predictive information. This is in accordance with current NCCN guidelines, which note insufficient data to recommend the use of Oncotype or other available multigene assays (e.g., ColDx, ColoPrint) as a means to guide adjuvant chemotherapy treatment [21]. Of note, the prognostic, but not predictive, value of these tests in CRC stands in contrast to other cancer types, such as breast cancer, for which genomic tests (i.e., Oncotype Dx, MammaPrint, Breast Cancer Index) have been validated to inform the benefit of adjuvant therapies.
The group discussed recent data from the IDEA France collaborative group highlighting the potential role of Immunoscore in adjuvant chemotherapy decision‐making for stage III colon cancer [22]. In this study, which investigated the efficacy of 3 versus 6 months of oxaliplatin‐based therapy in stage III colon cancer, Immunoscore was a significant and independent predictor of disease‐free survival (DFS; p = .003) after adjustment for sex, histological grade, stage, and microsatellite instability (MSI) status in multivariate analysis. Those with intermediate‐ or high‐risk Immunoscore results had a benefit of 6 months of adjuvant mFOLFOX6 (HR = 0.53; p = .0004), whereas those with low‐risk Immunoscore did not have a significant benefit of the 6‐month regimen relative to the 3‐month regimen. Although these data are compelling, the group agreed that the results would need to be prospectively validated before Immunoscore testing would be used to more definitively inform treatment decisions [22]. In addition, it is unclear mechanistically why a low Immunoscore signature would be associated with lack of benefit from chemotherapy. Most in the group agreed that, at the current time, recurrence risk in early CRC is best assessed on the basis of clinical and pathologic characteristics, pending further validation of additional assays in prospective clinical trials.
Other Molecular Markers
The group agreed with current NCCN guidance recommending universal MSI or mismatch repair (MMR) testing for all newly diagnosed patients with colon cancer, which may be performed using either immunohistochemistry or polymerase chain reaction (PCR) platforms [21]. In addition, the group noted that BRAF mutation testing should be reflexed in patients found to be MMR deficient as a means to identify patients who may benefit from genetic counseling. The presence of BRAF mutation would exclude conditions such as Lynch syndrome (hereditary nonpolyposis colorectal cancer) and, as such, germline testing would not be necessary in these individuals. The group also noted the prognostic impact of BRAF and KRAS mutations in CRC. In a study investigating patients with stage III CRC treated with adjuvant FOLFOX4 with or without cetuximab (n = 2,559), Taieb et al. found that, among patients with microsatellite‐stable tumors, KRAS and BRAF V600E mutations were associated with shorter DFS and overall survival [23]. These results suggest that KRAS and BRAF mutational testing may help to better stratify patients for adjuvant trials, though newer technologies may further refine stratification as discussed below [23]. Results from an earlier trial also suggested an adverse impact of KRAS (HR = 1.44; p < .001) and BRAF V600E mutation (HR = 1.37; p = .009) on DFS in patients with stage III CRC treated with FOLFOX [24].
Next‐Generation Sequencing and Molecular Residual Disease in Resectable CRC: What to Test and When?
The group considered the question of whether, in a patient with a clinically high risk for recurrence, they would do extended gene panel testing (e.g., next‐generation sequencing [NGS] analysis for BRAF, HER2, etc.) on the primary tumor or wait until a recurrence occurs and do genomic testing on a recurrence sample. Most in the group were in agreement that there is a high degree of concordance between the primary tumor and the metastatic site of recurrence for oncogenic driver alterations. Therefore, when selecting a tissue sample for genomic analysis, either primary tumor or metastatic site would be appropriate for extended panel testing, depending on adequacy of tissue specimen available. The group did, however, recommend that, among patients treated with molecularly selected biologic agents such as BRAF−, HER2−, and/or EGFR‐directed therapies, a new tissue biopsy or blood‐based assay as described below should be strongly considered upon disease progression, as this may identify resistance mechanisms and inform subsequent treatment options, ideally in the setting of a clinical trial. For patients with metastatic disease, the group believes that essential testing includes MMR/MSI, KRAS, NRAS, and BRAF (HER2 for wild‐type RAS tumors) determinations, and if NGS has been performed, evidence of NTRK gene fusion and tumor mutational burden should be assessed.
Circulating Tumor DNA Testing for CRC Recurrence
Although a large portion of patients with CRC may be cured by surgery alone, a group of patients benefit from the addition of adjuvant chemotherapy as a means to eradicate micrometastatic disease [25, 26]. Chemotherapy, however, may lead to significant, and in some cases potentially irreversible, toxicities such as oxaliplatin‐associated peripheral neuropathy. A risk stratification strategy in these patients may therefore help avoid unnecessary chemotherapy treatment and its associated toxicities. Circulating tumor DNA (ctDNA) is released into the plasma in significant quantities as a result of tumor cell turnover and multiple other mechanisms and, as such, detection of molecular/minimal residual disease (MRD) via ctDNA assessment has emerged as a promising tool to guide risk stratification in adjuvant treatment. Morris and Kopetz, using pooled data from several cohorts, noted an increasing rate of postoperative ctDNA detection, as may be expected, with increasing pathologic stage, with 12%, 27%, and 48% of patients with stage II, III, and IV disease, respectively, having detectable ctDNA after resection (p < 1 x 10‐5) [27].
In discussing genomic ctDNA testing for MRD, the group noted important differences between the tumor‐informed versus tumor‐uninformed (or agnostic) assays (Fig. 1). Tumor‐agnostic assays evaluate large panels of genes, methylation patterns, and fragmentomic ctDNA signatures that are common drivers and/or are frequently mutated in CRC as a means to detect residual disease. Tumor‐informed assays, by comparison, create a tissue‐based, specific signature of the patient's tumor that can be tested over time in the plasma. There are advantages and limitations to both approaches [28]. A tumor‐agnostic approach may detect a wide range of potentially druggable mutations in the tumor (if annotated and reported) and is therefore well suited to study heterogeneous tumors and/or the adaptive evolution of tumors over time in response to treatment. By comparison, the tumor‐informed approach uses a specific genomic fingerprint for the patient's tumor that is not expected to change over time and/or with treatment (i.e., generally more truncal genomic changes). Although this does not enable, nor is it intended to identify, potentially actionable targets over time, it has high sensitivity and specificity to detect recurrent disease that is present as ctDNA in the blood. An example of a tumor‐agnostic assay is LUNAR‐1 (Guardant Health, Redwood City, CA), and an example of a tumor‐informed assay is the Signatera ctDNA test developed by Natera (Fig. 1).
Figure 1.
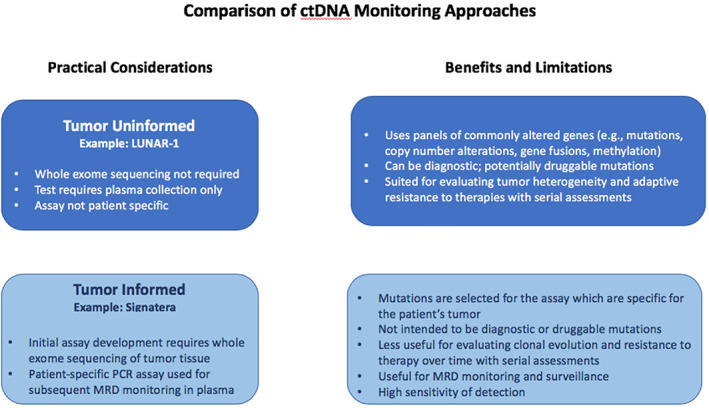
A comparison of ctDNA monitoring approaches. ctDNA assessments in CRC can be either tumor uninformed (top) or tumor informed (bottom); practical considerations and the benefits and limitations of each approach are shown. Whereas the former method is more appropriately used to assess tumor heterogeneity, clonal evolution, and/or resistance to therapies in CRC, the latter approach is more patient specific and may be better suited for assessment of MRD and ongoing surveillance for patients with initially resected CRC.Abbreviations: CRC, colorectal cancer; ctDNA, circulating tumor DNA; MRD, molecular/minimal residual disease; PCR, polymerase chain reaction.
The group noted findings from Parikh and colleagues (ASCO 2019) using a tumor‐uninformed approach that incorporates both genetic alterations and epigenetic markers to identify patients at high risk for recurrence (LUNAR‐1 Assay) [29]. In this study, 72 patients with CRC underwent standard of care (SOC) that included surgery with or without neoadjuvant therapy (n = 42) or surgery with adjuvant therapy with or without neoadjuvant therapy (n = 30). Plasma samples were collected after a median of 31 and 37 days, respectively, following surgery or completion of adjuvant therapy, respectively, and median follow‐up was 515 days. The study comprised approximately one third of patients with stage 0–II disease, one third with stage III disease, and one third with stage IV disease at time of resection. Detection of ctDNA following SOC therapy had a recurrence positive predictive value (PPV) of 100% and an negative predictive value (NPV) of 76%, yielding a hazard ratio for recurrence of 9.22 (p < .0001). Similar findings were observed in the surgery (HR = 8.7) and adjuvant therapy (HR = 9.3) cohorts (for both, PPV, 100%; NPV, 76%, both p < .0001). These findings suggest that ctDNA detection post resection using a tumor‐uninformed approach may help identify patients at high risk for recurrence who may potentially benefit from adjuvant therapy as well as a modified therapy approach.
The group reviewed additional evidence supporting the use of ctDNA to detect MRD. Tie and colleagues found that in a population‐based cohort study of 96 patients with stage III CRC, ctDNA was detectable in 21% of postsurgical samples and 17% of samples post chemotherapy, and ctDNA positivity was associated with a significantly increased risk of recurrence (Table 3). In multivariate analysis, postsurgical ctDNA status (HR = 7.5; p < .001) demonstrated the strongest association with relapse‐free interval, followed by clinical risk factors (HR = 2.5; p = .008) [30]. Similar findings were also observed by the same group in a study examining ctDNA in patients with stage II CRC [31]. Reinert and colleagues assessed ctDNA using a tumor‐informed, multiplex, PCR‐based NGS approach before and after surgery and adjuvant chemotherapy in a prospective multicenter cohort study of patients with stage I–III CRC (n = 130) [32]. ctDNA positivity at postoperative day 30 was also associated with a sevenfold increased risk for relapse in this study, as compared with ctDNA‐negative status (recurrence rate 70% vs. 11.9%; HR = 7.2; p < .001). Following adjuvant chemotherapy, ctDNA‐positive patients had a 17‐fold increased risk for relapse (relapse rate 100% vs. 13.7%; HR = 17.5; p < .001), and monitoring of ctDNA during adjuvant treatment showed ctDNA clearance in 3 of 10 patients (30%) after adjuvant chemotherapy [32]. In addition, with serial ctDNA monitoring following treatment, ctDNA‐positive status was associated with markedly reduced recurrence‐free survival (RFS; recurrence rate 93.3% vs. 3.3%; HR = 43.5; p < .001); ctDNA was also the only factor significantly associated with RFS in multivariate analysis (HR = 39.9; p < .001). Of note, when comparing longitudinal analysis of the same surveillance population (n = 75), ctDNA showed higher sensitivity and specificity (88% and 98%, respectively) relative to carcinoembryonic antigen (CEA) monitoring (69% and 64%, respectively) [32]. Overall, the group was in agreement that, although these studies are limited by relatively small sample size, they strongly demonstrate the potential of ctDNA to risk stratify patients with CRC and to potentially inform adjuvant chemotherapy decisions, particularly in patients with stage II and III disease.
Table 3.
Impact of ctDNA‐positive status on recurrence risk [30]
| 3‐year RFI Kaplan‐Meier (%) | Hazard ratio | p value | |
|---|---|---|---|
| Post‐surgery | |||
| ctDNA+ | 47 | 3.8 | <.001 |
| ctDNA− | 76 | ||
| Post‐chemotherapy | |||
| ctDNA+ | 30 | 6.8 | <.001 |
| ctDNA− | 77 |
Abbreviations: ctDNA, circulating tumor DNA; RFI, relapse free interval.
Discussion
Given the significant and sometimes irreversible toxicity of chemotherapy, even when using the 3‐month approach with CAPOX as suggested by the IDEA collaboration update, the group expressed agreement regarding the need to improve risk stratification of patients receiving adjuvant chemotherapy [33]. The group acknowledged that, although compelling, there is insufficient data at present to support ctDNA‐based MRD assessments as a means to guide the use of adjuvant chemotherapy. For example, data presented by Henriksen et al. at ASCO GI 2021 showed that 4 of 20 patients initially ctDNA positive postoperatively, and all of whom received adjuvant chemotherapy, did not have recurrence, implying that residual disease may have been cleared with the use of adjuvant treatment in these patients [34]. In view of the small sample sizes and low numbers of events in these retrospective studies, the group is in agreement with recent discussion by Overman and colleagues emphasizing the need for randomized prospective studies to answer this question.
Some in the group stated that they were currently offering ctDNA testing at their center to all patients as a means of complementing clinical factors and further informing recurrence risk estimates [30]. The group expressed that it would be ideal to have a validated test in which one could have the option to withhold treatment until serial monitoring demonstrates ctDNA test positivity (as indicative of MRD). The group also emphasized that testing should not be performed in the immediate postsurgical period, to avoid false positives, which may create unnecessary anxiety for patients. In this regard, the group notes that timing of blood collection following resection is an especially important consideration. A recent study that included 453 patients with stage I–III CRC undergoing elective surgery found an approximately threefold increase in the level of cell‐free DNA following surgery (p ≤ .0001), and this elevation was noted to persist for approximately 4 weeks [35]. Therefore, it is suggested that later sampling would reduce the potential for contamination wild‐type (nontumor) cfDNA. The group suggests 4 weeks post surgery as a reasonable time frame for plasma sampling.
There was also agreement that serial ctDNA testing further improves the sensitivity. Reinert et al., for example, found an improvement in sensitivity with two tests versus one, and sensitivity was further improved with three tests [32]. Therefore, if a patient is found to be ctDNA positive in the early postsurgery period, the group thought it would be reasonable to consider a second test before treatment is given. Although the time between blood draw and initial ctDNA test result takes longer, a second blood draw (which takes less time than the initial) could be performed immediately following receipt of the first. Most in the group felt that MRD testing is more commonly ordered in stage II cases and less often in stage III CRC, as it is not expected to change management in the latter until more data in this setting are available. It was also agreed that, outside the setting of a clinical trial, ctDNA testing would generally not be ordered if it would not impact or otherwise change their treatment decision based on clinicopathologic factors. The group also notes a recent publication of the National Cancer Institute (NCI) Task Force on the application of ctDNA testing; this report noted multiple potential applications of ctDNA testing in CRC, including the detection of MRD, management of rectal cancer, and as a means to monitor clonal evolution of the tumor and response to targeted therapies and other systemic treatments. Notably, the NCI task force called for harmonization around timepoints for the collection of ctDNA, standardization of sample collection and clinical validation of testing, a need for patient and provider education on the applications of the technology, and the incorporation of ctDNA testing into treatment guidelines [36].
ctDNA Testing: Potential Clinical Scenarios
Patient Case #1: What Should We Do When Clinical Risk Is Low but ctDNA Results Are Positive?
In a patient with stage II colon cancer with low‐risk clinical characteristics but with a positive ctDNA result, the group agreed that the best course of action would be to inform the patient of the high risk of recurrence, while noting that no data are yet available to ensure a benefit of treatment; in such a case, some patients might decide to forgo treatment until the disease becomes radiologically detectable. Some experts, however, considered discussing adjuvant chemotherapy, managing the patient as a high‐risk case, with monitoring of MRD to detect clearance or persistence of ctDNA. Some in the group also thought a confirmatory repeat of the ctDNA testing might be indicated.
Patient Case #2: What Should We Do When Clinical Risk Is High but ctDNA Results Are Negative?
In a patient with stage II colon cancer with high‐risk features who would normally be recommended adjuvant treatment but who has a negative ctDNA result, the group agreed that based on current data and guidelines, treatment should not be withheld for such a patient. It was recommended to monitor and consider discussion of CAPOX adjuvant therapy for 3 months versus 6 months. Some in the group also thought a repeat of the ctDNA test might be indicated here as well.
Patient Case #3: Relationship Between Molecular Markers and ctDNA Result?
The group then considered the case of a patient with stage II colon cancer with clinical low risk (i.e., well to moderately differentiated, Tumor size classification ≤T3, adequate lymph node harvest, no lymphovascular or perineural invasion, and no evidence of obstruction or perforation) but who on molecular testing was found to be BRAF wild type and MSI‐high. Although adjuvant treatment would not be typically recommended in this scenario, the group considered what they would do if ctDNA (Signatera) testing reported a positive ctDNA result, indicative of MRD. The group agreed in such a scenario that this would require shared decision‐making with the patient, and that the patient should be informed of the high risk of recurrence while at the same time noting the lack of data to clearly support a benefit from adjuvant treatment. Some in the group, however, considered discussing adjuvant chemotherapy, managing the patient as a high‐risk case, and monitoring MRD for clearance or persistence of ctDNA. The clinician might also consider any pre‐existent conditions (e.g., neuropathy), comorbidities, or other patient factors (e.g., age, performance status) that might also preclude the use of chemotherapy.
Role of ctDNA in Surveillance
Many in the group felt that ctDNA testing, especially in the current time of COVID‐19, may ultimately reduce the need for computed tomography (CT) testing for surveillance and its associated costs. For example, a strategy could be envisioned whereby one would only be CT scanned in the event of a positive ctDNA test; it was agreed such a strategy may completely change the established surveillance strategy. In this regard, the utility of routine CEA testing was also raised, given that CEA levels may be impacted by multiple factors unrelated to CRC. As such, CEA elevations may sometimes be harder to interpret as a definitive surrogate marker of disease recurrence in the absence of radiological relapse. The group also highlighted potential limitations of ctDNA testing, including lack of data supporting the impact of earlier treatment (e.g., for recurrent metastatic disease) on modifying the overall disease trajectory in CRC, a lack of guidance regarding persistent ctDNA positivity after adjuvant treatment, uncertainty regarding the frequency of ctDNA testing, and a lack of consensus regarding the kinetics of ctDNA relative to clinically actionable (radiological) recurrence. Similar issues were raised in a recently convened National Cancer Institute task force on colon cancer and remain important questions for the field [36].
Conclusion
The group identified at least two key trials underway that will help to further clarify the role of ctDNA in MRD testing and evaluate the potential benefit of MRD status to inform use of adjuvant chemotherapy (Table 4). The group strongly encourages clinical trial enrollment for patients to further advance the genomic technologies described herein and support the validation of ctDNA MRD testing and other genomic assays in CRC. The group was also in agreement that routine incorporation of ctDNA testing might create a new paradigm for risk stratification as outlined by Yang and colleagues [37]. ctDNA genomic testing may also hold promise as a single blood draw screen for CRC, although it was agreed that a positive test would still be followed by a diagnostic colonoscopy. For patients with CRC facing adjuvant treatment decisions, a sensitive and specific MRD test reflecting biologic tumor behavior may ultimately guide the use or avoidance of potentially toxic adjuvant therapies. Lastly, further refinements in the use of ctDNA to detect MRD in patients with successfully resected CRC may dramatically reduce patient treatment intensity and cumulative side effects as well as the associated costs of imaging for disease recurrence. The group is optimistic on the future of MRD testing in CRC (Table 5) and anticipates that this type of testing will emerge as a standard component of clinical decision‐making as data continue to rapidly emerge.
Table 4.
Genomic testing for MRD: relevant clinical trials
| Clinical Trial | Relevant results |
|---|---|
| BESPOKE [38] NCT04264702 |
|
|
COBRA [39] NRG GI005 NCT04068103 |
|
Abbreviations: CRC, colorectal cancer; ctDNA, circulating tumor DNA; FFPE, formalin‐fixed, paraffin‐embedded; MRD, molecular/minimal residual disease; OS, overall survival; QoL, quality of life; RFS, recurrence‐free survival.
Table 5.
Use of molecular assays in CRC: summary of GICTEG practice points
| Molecular assay | Summary |
|---|---|
| In CRC screening |
|
| In early‐stage CRC |
|
| In surveillance for CRC recurrence |
|
Abbreviations: CRC, colorectal cancer; ctDNA, circulating tumor DNA; GICTEG, Gastrointestinal Cancer Therapy Expert Group; MRD, molecular/minimal residual disease.
Author Contributions
Conception/design: Arturo Loaiza‐Bonilla, Al B. Benson III, Axel Grothey, Misagh Karimi, Samuel J. Klempner, Daniel Lin, Reshma Mahtani, Heloisa P. Soares
Collection and/or assembly of data: Arturo Loaiza‐Bonilla, Al B. Benson III, Axel Grothey, Misagh Karimi, Samuel J. Klempner, Daniel Lin, Reshma Mahtani, Heloisa P. Soares
Data analysis and interpretation: Arturo Loaiza‐Bonilla, Al B. Benson III, Axel Grothey, Misagh Karimi, Samuel J. Klempner, Daniel Lin, Reshma Mahtani, Heloisa P. Soares
Manuscript writing: Arturo Loaiza‐Bonilla, Al B. Benson III, Axel Grothey, Misagh Karimi, Samuel J. Klempner, Daniel Lin, Reshma Mahtani, Heloisa P. Soares
Final approval of manuscript: Arturo Loaiza‐Bonilla, Al B. Benson III, Axel Grothey, Misagh Karimi, Samuel J. Klempner, Daniel Lin, Reshma Mahtani, Heloisa P. Soares
Disclosures
Samuel J. Klempner: Eli Lilly, Merck, Bristol‐Myers Squibb, Pieris, Foundation Medicine, Natera Inc. (C/A), Turning Point Therapeutics (OI); Heloisa P. Soares: Ipsen, Lexicon, Natera, QED Therapeutics, Advanced Accelerator Applications and Exelixis (C/A); Al B. Benson III: Bristol‐Myers Squibb, Therabionic, Guardant, Merck, Lexicon, AMGEN, Artemida Pharma, Array (Pfizer), AbbVie, Apexigen, Tyme, SynCore, Natera, Samsung, HalioDx, Janssen (C/A), Bristol‐Myers Squibb (C/A), Astellas (C/A), AMGEN (C/A), SynCore (C/A), Tyme (C/A), Celegene, Infinity Pharmaceuticals, Merck Sharp and Dohme, Taiho Pharmaceutical, Rafael Pharmaceuticals, Medimmune/AstraZeneca, Xencor, ST Pharm, Elevar Therapeutics, Lexicon Pharmaceuticals/TerSera (RF). Loaiza‐Bonilla: PSI CRO, Bayer, Blueprint, Astra‐Zeneca, Medidata, Taiho, QED, Cardinal Health, BrightInsight, The Lynx Group, Boston Biomedical (C/A), Roche/Genentech, Amgen, Bayer, Guardant, Natera, Eisai, Ipsen, Merck (H), Ipsen (RF), Massive Bio (OI). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Acknowledgments
An unrestricted educational grant for this activity was provided by Natera Oncology to GICTEG. Medical writing support was provided by SciavoTECH Research and Consultancy services, Inc., and was funded by Total Health Conferencing. All research funds were provided directly to Northwestern University.
Disclosures of potential conflicts of interest may be found at the end of this article.
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact commercialreprints@wiley.com. For permission information contact permissions@wiley.com.
References
- 1. NCT04136002 . Available at Clinicaltrials.gov. Accessed February 5, 2021.
- 2. NCT04136002 . Available at Clinicaltrials.gov. Accessed February 5, 2021.
- 3. NCT03934866 . Available at Clinicaltrials.gov. Accessed February 5, 2021.
- 4. NCT04241796 . Available at Clinicaltrials.gov. Accessed February 5, 2021.
- 5. Putcha G, Liu T‐Y, Ariazi E et al. Blood‐based detection of early‐stage colorectal cancer using multiomics and machine learning. Abstract presented at: the American Society of Clinical Oncology (ASCO) GI Symposium 2020; January 23–25, 2020; San Francisco, CA.
- 6. NCT04369053 . Available at Clinicaltrials.gov. Accessed February 5, 2021.
- 7. Imperiale TF, Ransohoff DF, Itzkowitz SH et al. Multitarget stool DNA testing for colorectal‐cancer screening. N Engl J Med 2014;370:1287–1297. [DOI] [PubMed] [Google Scholar]
- 8. NCT01397747 . Available at Clinicaltrials.gov. Accessed February 5, 2021.
- 9. Lennon AM, Buchanan AH, Kinde I et al. Feasibility of blood testing combined with PET‐CT to screen for cancer and guide intervention. Science 2020;369:eabb9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gray RG, Quirke P, Handley K et al. Validation study of a quantitative multigene reverse transcriptase‐polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol 2011;29:4611–4619. [DOI] [PubMed] [Google Scholar]
- 11. Venook AP, Niedzwiecki D, Lopatin M et al. Biologic determinants of tumor recurrence in stage II colon cancer: Validation study of the 12‐gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol 2013;31:1775–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yothers G, O'Connell MJ, Lee M et al. Validation of the 12‐gene colon cancer recurrence score in NSABP C‐07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol 2013;31:4512–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamanaka T, Oki E, Yamazaki K et al. 12‐gene recurrence score assay stratifies the recurrence risk in stage II/III colon cancer with surgery alone: The SUNRISE study. J Clin Oncol 2016;34:2906–2913. [DOI] [PubMed] [Google Scholar]
- 14. Galon J, Costes A, Sanchez‐Cabo F et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960–1964. [DOI] [PubMed] [Google Scholar]
- 15. Mlecnik B, Tosolini M, Kirilovsky A et al. Histopathologic‐based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol 2011;29:610–618. [DOI] [PubMed] [Google Scholar]
- 16. Mlecnik B, Bindea G, Angell HK et al. Integrative analyses of colorectal cancer show Immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 2016;44:698–711. [DOI] [PubMed] [Google Scholar]
- 17. Kirilovsky A, Marliot F, El Sissy C et al. Rational bases for the use of the Immunoscore in routine clinical settings as a prognostic and predictive biomarker in cancer patients. Int Immunol 2016;28:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sinicrope FA, Shi Q, Hermitte F et al. Association of immune markers and Immunoscore with survival of stage III colon carcinoma (CC) patients (pts) treated with adjuvant FOLFOX: NCCTG N047 (Alliance). J Clin Oncol 2017;35(suppl15):3579. [Google Scholar]
- 19. Pagès F, Mlecnik B, Marliot F et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018;391:2128–2139. [DOI] [PubMed] [Google Scholar]
- 20. Pagès F, Kirilovsky A, Mlecnik B et al. In situ cytotoxic and memory T cells predict outcome in patients with early‐stage colorectal cancer. J Clin Oncol 2009;27:5944–5951. [DOI] [PubMed] [Google Scholar]
- 21. NCCN Guidelines Version 1.2020 Colon Cancer . Available at www.nccn.org.
- 22. Pagès F, André T, Taieb J et al. Prognostic and predictive value of the Immunoscore in stage III colon cancer patients treated with oxaliplatin in the prospective IDEA France PRODIGE‐GERCOR cohort study. Ann Oncol 2020;31:921–929. [DOI] [PubMed] [Google Scholar]
- 23. Taieb J, Zaanan A, Le Malicot K et al. Prognostic effect of BRAF and KRAS mutations in patients with stage III colon cancer treated with leucovorin, fluorouracil, and oxaliplatin with or without cetuximab: A post hoc analysis of the PETACC‐8 Trial. JAMA Oncol 2016;2:643–653. [DOI] [PubMed] [Google Scholar]
- 24. Sinicrope FA, Mahoney MR, Smyrk TC et al. Prognostic impact of deficient DNA mismatch repair in patients with stage III colon cancer from a randomized trial of FOLFOX‐based adjuvant chemotherapy. J Clin Oncol 2013;31:3664–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Binefa G, Rodríguez‐Moranta F, Teule A et al. Colorectal cancer: From prevention to personalized medicine. World J Gastroenterol 2014;20:6786–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan GHJ, Chee CE. Making sense of adjuvant chemotherapy in colorectal cancer. J Gastrointest Oncol 2019;10:1183–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morris V, Kopetz S. Circulating Tumor DNA as a Surrogate for Minimal Residual Disease in Colorectal Cancer. Available at https://dailynews.ascopubs.org/do/10.1200/adn.18.190034/full/. [PubMed]
- 28. Corcoran RB, Chabner BA. Application of cell‐free DNA analysis to cancer treatment. N Engl J Med 2018;379:1754–1765. [DOI] [PubMed] [Google Scholar]
- 29. Parikh AR, Van Seventer EE, Boland GM et al. A plasma‐only integrated genomic and epigenomic circulating tumor DNA (ctDNA) assay to inform recurrence risk in colorectal cancer (CRC). J Clin Oncol 2019;37(suppl15):3602. [Google Scholar]
- 30. Tie J, Cohen JD, Wang Y et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol 2019;5:1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tie J, Wang Y, Tomasetti C et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reinert T, Henriksen TV, Christensen E et al. Analysis of plasma cell‐free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sobrero AF, Andre T, Meyerhardt JA et al. Overall survival (OS) and long‐term disease‐free survival (DFS) of three versus six months of adjuvant (adj) oxaliplatin and fluoropyrimidine‐based therapy for patients (pts) with stage III colon cancer (CC): Final results from the IDEA (International Duration Evaluation of Adj chemotherapy) collaboration. J Clin Oncol 2020;38(suppl 15):4004. [Google Scholar]
- 34. Henriksen TV, Tarazona N, Reinert T et al. Circulating tumor DNA analysis for assessment of recurrence risk, benefit of adjuvant therapy, and early relapse detection after treatment in colorectal cancer patients. J Clin Oncol 2021;39(suppl 3):11. [Google Scholar]
- 35. Henriksen TV, Reinert T, Christensen E et al. The effect of surgical trauma on circulating free DNA levels in cancer patients‐implications for studies of circulating tumor DNA. Mol Oncol 2020;14:1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dasari A, Morris VK, Allegra CJ et al. ctDNA applications and integration in colorectal cancer: An NCI Colon and Rectal‐Anal Task Forces whitepaper. Nat Rev Clin Oncol 2020;17:757–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang M, Forbes ME, Bitting RL et al. Incorporating blood‐based liquid biopsy information into cancer staging: Time for a TNMB system? Ann Oncol 2018;29:311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. NCT04264702 . Available at Clinicaltrials.gov. Accessed February 5, 2021.
- 39. NCT04068103 . Available at Clintrials.gov. Accessed February 5, 2021.


