Abstract
Background
Molecular tumor boards (MTBs) provide rational, genomics‐driven, patient‐tailored treatment recommendations. Worldwide, MTBs differ in terms of scope, composition, methods, and recommendations. This study aimed to assess differences in methods and agreement in treatment recommendations among MTBs from tertiary cancer referral centers in The Netherlands.
Materials and Methods
MTBs from all tertiary cancer referral centers in The Netherlands were invited to participate. A survey assessing scope, value, logistics, composition, decision‐making method, reporting, and registration of the MTBs was completed through on‐site interviews with members from each MTB. Targeted therapy recommendations were compared using 10 anonymized cases. Participating MTBs were asked to provide a treatment recommendation in accordance with their own methods. Agreement was based on which molecular alteration(s) was considered actionable with the next line of targeted therapy.
Results
Interviews with 24 members of eight MTBs revealed that all participating MTBs focused on rare or complex mutational cancer profiles, operated independently of cancer type–specific multidisciplinary teams, and consisted of at least (thoracic and/or medical) oncologists, pathologists, and clinical scientists in molecular pathology. Differences were the types of cancer discussed and the methods used to achieve a recommendation. Nevertheless, agreement among MTB recommendations, based on identified actionable molecular alteration(s), was high for the 10 evaluated cases (86%).
Conclusion
MTBs associated with tertiary cancer referral centers in The Netherlands are similar in setup and reach a high agreement in recommendations for rare or complex mutational cancer profiles. We propose a “Dutch MTB model” for an optimal, collaborative, and nationally aligned MTB workflow.
Implications for Practice
Interpretation of genomic analyses for optimal choice of target therapy for patients with cancer is becoming increasingly complex. A molecular tumor board (MTB) supports oncologists in rationalizing therapy options. However, there is no consensus on the most optimal setup for an MTB, which can affect the quality of recommendations. This study reveals that the eight MTBs associated with tertiary cancer referral centers in The Netherlands are similar in setup and reach a high agreement in recommendations for rare or complex mutational profiles. The Dutch MTB model is based on a collaborative and nationally aligned workflow with interinstitutional collaboration and data sharing.
Keywords: Molecular tumor board, Rare mutations, Molecular diagnostics, Decision making, Multidisciplinary
Short abstract
Worldwide, molecular tumor boards (MTBs) differ in terms of scope, composition, methods, and recommendations. This article assesses differences in methods and agreement in treatment recommendations among MTBs from tertiary cancer referral centers in The Netherlands.
Introduction
The emergence of DNA‐ and RNA‐based molecular cancer profiling techniques for predictive testing in the routine diagnostic setting has rapidly expanded the diagnostic guidance for targeted therapies [1]. Molecular tumor boards (MTB) support treating physicians in understanding the increasing complexity of molecular testing results, providing rational, genomics‐driven, patient‐tailored treatment recommendations with respect to the currently available targeted drugs [2, 3]. MTBs are hosted in cancer centers that offer extensive molecular profiling techniques [4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18]. Although the MTBs that have thus far published their methods all focus on translating molecular testing results into a therapeutic recommendation, there are major differences in terms of scope, composition, and methods [2, 3]. Notably, treatment recommendations provided by MTBs seem to vary widely. A recent comparison of five independent MTBs from four countries revealed that only two of five MTBs provided similar recommendations for four fictional cases with complex mutational profiles [19]. Regional and international differences in composition and logistics [2], method and prioritization [19], access to targeted drugs [3, 16], and molecular diagnostic workup [2] are potential sources of heterogeneity.
The incorporation of MTBs into standard‐of‐care cancer diagnostics necessitates mitigation of heterogeneity in MTB recommendations. Although perfect agreement in treatment recommendations may not be achievable because they are tailored to the patient at hand and dependent on drug and trial availability, discrepancies in target identification might be averted. For this purpose, the Predictive Analysis for Therapy project was initiated, which aims to optimize patient access to personalized cancer therapy in The Netherlands [20]. This includes optimizing MTBs by providing directives on the minimal requirements for hosting an MTB and achieving a recommendation, promoting the exchange of knowledge among MTBs through a shared database, and ensuring accessibility to MTBs for community hospitals and laboratories.
To identify the prerequisites for reaching a well‐informed MTB recommendation, this study aims to assess current similarities and differences in MTB methods and, secondly, to determine agreement in treatment recommendations among MTBs from tertiary cancer referral centers.
Materials and Methods
MTBs Included in Analysis
MTBs associated with tertiary cancer referral centers were identified through a digital survey among all Dutch molecular pathology laboratories. MTBs from all academic medical centers and one nonacademic tertiary cancer referral center were invited to participate.
Assessment of Similarities and Differences Between MTB Methods
All MTBs were invited to engage in one or more on‐site interviews. A survey to assess similarities and differences among MTBs was designed, covering scope and perceived value, logistics and composition, decision‐making method, reporting, and registration of MTB cases, and the interviewees’ view on harmonization and collaboration among MTBs (supplemental online Methods). The MTBs were asked to select interviewees representing multiple disciplines active in their MTB. Interviews and attendances of MTBs were performed by a medical researcher (B.K.) between June and September 2019. All interviewees consented to participation.
Comparison of Targeted Therapy Recommendations
MTB recommendations were compared using 10 cases representative of the MTBs’ setting in terms of cancer type, mutational profile, and inquiry. The participating MTBs were asked to submit an anonymized case (supplemental online Methods). The submitted cases were adjusted for further anonymization and formatting. Finalized cases were prepared as two‐page documents (example provided in the supplemental online Methods).
From September 26 until November 21, 2019, 10 cases were sent to participating MTBs. The MTBs were asked to handle these as routine MTB requests and provide recommendations according to their usual method. After the initial round, two cases with low rates of agreement with respect to the choice of inhibitor or its timing of use were selected. These cases were sent for a second time to the participants along with the recommendations received in the initial round for those cases, in a blinded fashion. The participating MTBs were asked if they would revise their initial answer when presented with the other MTBs’ recommendations and provide arguments for their decision.
The research ethics board of the department of Pathology at the University Medical Center Groningen (UMCG) approved the use of anonymous case descriptions for this study. The study protocol was consistent with the UMCG Research Code and national ethical and professional guidelines [21, 22].
Statistical Analysis
Descriptive statistics are provided. The consensus for a case was defined as the most frequently provided recommendation. Agreement was measured by the percentage of MTBs that provided a recommendation in accordance with this consensus. Agreement among MTB recommendations was based on which molecular alteration(s) was considered actionable with the next line of targeted therapy (next actionable target). The agreement rate was calculated from the initial recommendations.
Results
MTBs Included in the Analysis
MTBs from all eight Dutch tertiary cancer referral centers were included, representing Amsterdam University Medical Centers, Erasmus Medical Center, Leiden University Medical Center, Maastricht University Medical Center, Netherlands Cancer Institute, Radboud University Medical Center, University Medical Center Groningen, and University Medical Center Utrecht. On‐site interviews were held with 24 MTB members: nine clinical scientists in molecular pathology (CSMPs), six pathologists, five thoracic oncologists, two medical oncologists, and two (bio)medical researchers. Meetings of MTBs were attended when this could be combined with the on‐site interview.
Assessment of Similarities and Differences Between MTB Methods
Scope and Value of the MTBs
The eight MTBs were founded between 2014 and 2017 (Table 1). Reciprocal improvement of expertise for attendees was considered a core value of an MTB and an important reason for founding MTBs. The MTBs reviewed cases with molecular‐oriented questions, such as the clinical consequences of molecular findings, the availability of therapeutic options, or the most appropriate test(s) to perform for a case. Seven MTBs reviewed only selected rare or complex cases (two to eight cases average per meeting); one MTB reviewed all molecular testing results (estimated 5–15 cases per meeting). The most common molecular findings eligible for review included somatic mutations, copy number variants, and fusion genes detected by targeted panel next‐generation sequencing, fluorescence in situ hybridization, immunohistochemistry, or RNA‐based fusion transcript analysis. Results from whole‐genome sequencing (WGS) or whole‐exome sequencing (WES) were uncommon, as these methods were not used routinely at the time of this study, although all MTBs were open to interpret these results.
Table 1.
MTB demographics as attained by on‐site interviews
| Demographic | MTBs, n (%) |
|---|---|
| MTBs participating in the on‐site interviews | 8 (100%) |
| Type of hospital that the MTB is associated with | |
| University medical center (academic) | 7 (87.5) |
| Nonacademic tertiary cancer referral hospital | 1 (12.5) |
| Year in which the MTB was established | |
| 2014 (active for 6 years) | 1 (12.5) |
| 2015 (active for 5 years) | 3 (37.5) |
| 2016 (active for 4 years) | 2 (25) |
| 2017 (active for 3 years) | 2 (25) |
| Cancer types eligible for review by the MTB | |
| Any type of cancer | 5 (62.5) |
| Thoracic oncology | 3 (37.5) |
| Frequency of MTB meetings | |
| MTB meets once every week | 4 (50) |
| MTB meets once in every 2 weeks | 4 (50) |
| Internal reporting of recommendation | |
| Report in the patient's electronic health record | 7 (87.5) |
| Recommendation is included in the pathology report | 1 (12.5) |
| Communication of recommendation to external applicants | |
| Directly to applicant (videoconferencing, e‐mail, telephone call) | 5 (62.5) |
| By means of a medical letter | 2 (25) |
| Directly to applicant by videoconferencing and through pathology report | 1 (12.5) |
| Registration of cases reviewed by the MTB | |
| Cases, recommendations, and follow‐up are registered in a local database | 2 (25) |
| Only basic case information is registered in a local database | 2 (25) |
| No registration in local database | 4 (50) |
| Regional collaboration by the MTB | |
| Cases from peripheral hospitals are reviewed | 7 (87.5) |
| Cases from other tertiary cancer referral centers are reviewed | 2 (25) |
| External specialists attend MTB meetings through videoconferencing | 4 (50) |
| Experts participating in MTB meetings | |
| Clinical scientists in molecular pathology | 8 (100) |
| Pathologists | 8 (100) |
| Thoracic oncologists | 8 (100) |
| Medical oncologists | 5 (62.5) |
| Postdocs, PhD students, researchers | 5 (62.5) |
| Clinical geneticists | 3 (37.5) |
| Clinical chemists | 2 (25) |
| Laboratory technicians | 2 (25) |
| Hemato‐oncologists | 1 (12.5) |
| Nurse practitioners | 1 (12.5) |
| Pharmacists | 1 (12.5) |
| Radiation oncologists | 1 (12.5) |
| Structural biologists | 1 (12.5) |
Abbreviation: MTB, molecular tumor board.
Five MTBs reviewed any type of cancer (“cancer agnostic”), whereas three MTBs only reviewed thoracic oncology. Thoracic oncology cases were most common in six MTBs because of the diversity of relevant actionable biomarkers and because the Dutch national guideline for non‐small cell lung cancer (NSCLC) recommends referral to an MTB in case of rare mutational profiles [23]. The most common reason for reviewing other cancer types was to assess eligibility for trials such as the Drug Rediscovery Protocol (DRUP) [24].
All MTBs took place as individual meetings, outside of conventional, cancer type–specific multidisciplinary team (MDT) meetings. Differentiation between primary treatment modalities was considered the responsibility of the conventional MDT, whereas MTBs were considered more proficient in guiding decision making on the most appropriate treatment when targetable alterations are present. Two thoracic oncology MTBs were hosted sequentially with a conventional MDT, selecting cases for either meeting depending on the cancer stage. Six MTBs were not tuned to conventional MDTs, although attending oncologists and pathologists were also involved in conventional MDTs.
Logistics and Composition of the MTBs
In all MTBs, cases could be submitted by the treating physician, CSMP, or pathologist. Participants in all MTBs included oncologists (thoracic, medical, or hemato‐oncologists, depending on the cancer type reviewed), CSMPs, and pathologists. Other common attendees were (bio)medical researchers (five MTBs) and clinical geneticists (three MTBs, of which two consulted geneticists on request) (Table 1). Seven MTBs received inquiries from peripheral hospitals or pathology laboratories, of which four facilitated attendance of external specialists through videoconferencing.
Decision‐Making Method of the MTBs
In preparing a case, a variety of (online) resources were consulted (Fig. 1). Nine resources were used by all MTBs: the 1000 Genomes Browser [25], cBioPortal [26], ClinVar [27], COSMIC [28], dbSNP [29], Ensembl [30], JAX‐CKB [31], OncoKB [32], and PubMed [33]. Trial overviews were not systematically consulted prior to the MTB meetings because of time restrictions. In some MTBs, dedicated trial‐coordinating oncologists were consulted after evaluation of actionability by the MTB. Interviewees regarded awareness of the trial availability variable and dependent on which oncologist(s) attended meetings.
Figure 1.
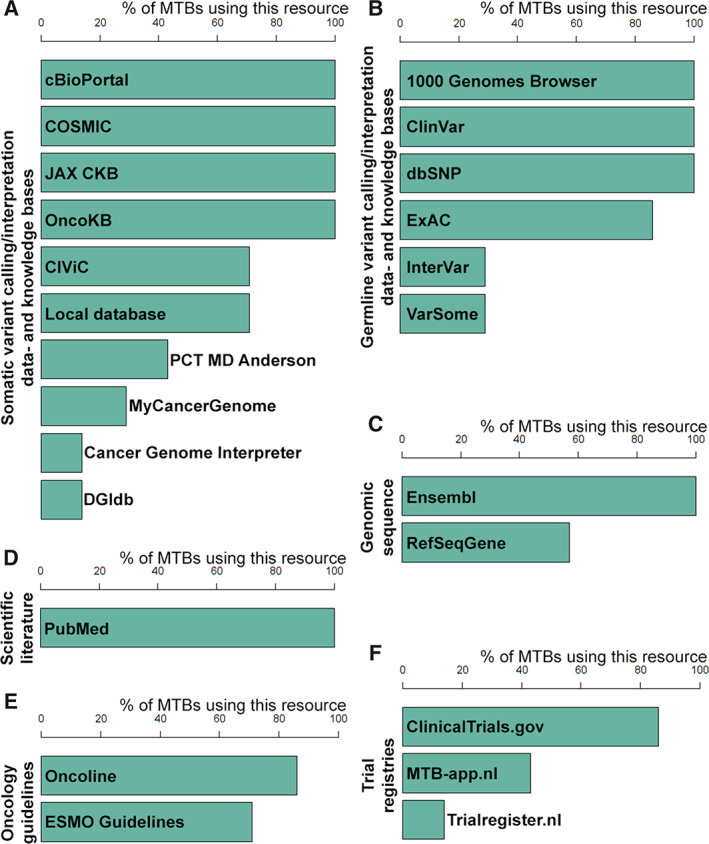
Online information resource usage for MTB decision making. Bar graphs depicting the usage of online resources for decision making within MTBs. A survey was filled in by representatives of seven MTBs. (A): Data‐ and knowledge bases for somatic variant calling and/or interpretation. (B): Data‐ and knowledge bases for germline variant calling and/or interpretation. (C): Online resources for genomic sequences. (D): Online resources for scientific literature. (E): Online resources for oncology guidelines. (F): Trial registries.Abbreviations: CIViC, Clinical Interpretation of Variants in Cancer; COSMIC, Catalogue of Somatic Mutations in Cancer; dbSNP, Short Genetic Variations database; DGIdb, Drug Gene Interaction Database; ESMO, European Society for Medical Oncology; ExAC, Exome Aggregation Consortium; JAX CKB, the Jackson Laboratory Clinical Knowledge Base; OncoKB, Precision Oncology Knowledge Base; PCT MD Anderson, Personalized Cancer Therapy Knowledge Base; MTB, molecular tumor board.
The MTBs served as a unifying platform to achieve a profile‐based, patient‐tailored consensus recommendation based on the identification/prioritization of genomic alterations and potential drug actionability (prepared by CSMPs/pathologists) and the assessment of availability of clinical trials or compassionate use drugs for eligible patients (prepared by oncologists). A recommendation could be diagnostic (such as which additional tests to perform), therapeutic, or both (Fig. 2).
Figure 2.
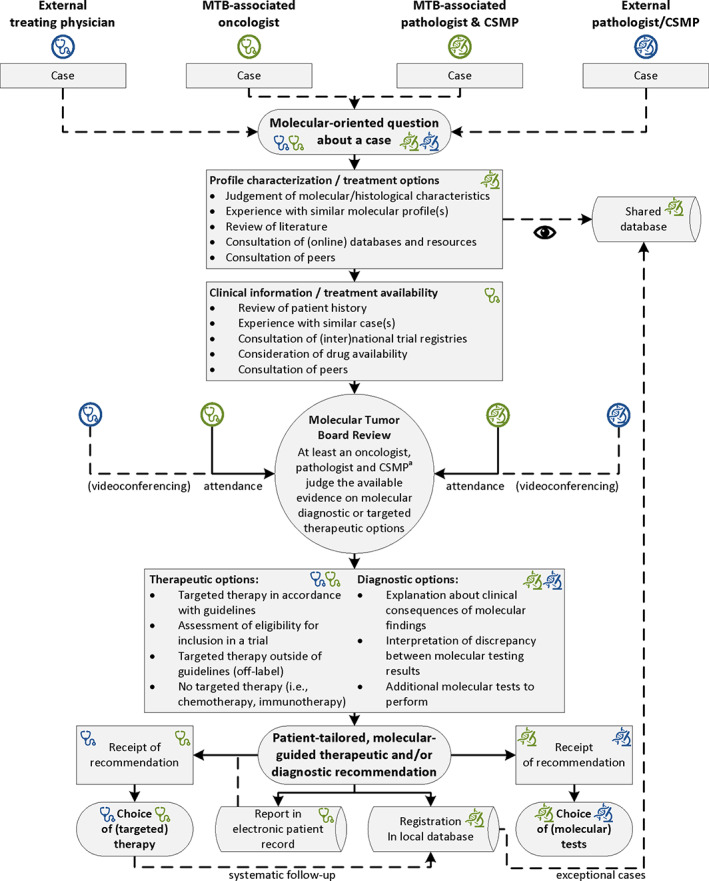
The Dutch MTB model. Flow diagram depicting an optimal MTB workflow as recommended by the eight molecular tumor boards participating in this study. Responsibilities of each party are annotated for external (gray) or MTB‐associated (black) physicians/oncologists (stethoscopes) and CSMPs/pathologists (microscopes). All four parties can submit molecular‐oriented questions about their cases to the MTBs. The MTB‐associated oncologist is responsible for the clinical case preparation, and the CSMP and pathologist are jointly responsible for characterization of the molecular profile. During review in an MTB meeting, a diagnostic and/or therapeutic recommendation is formulated, which is communicated to the requestor, recorded in the patient's electronic health record, and registered in a local database. The requestor can then use this recommendation in their choice of (molecular) tests or (targeted) therapy. aIn case of biomarkers with both germline and somatic implications, attendance of a clinical geneticist is recommended. In case of discussing whole‐exome or whole‐genome sequencing results, attendance of a bioinformatician is recommended.Abbreviations: CSMP, clinical scientist in molecular pathology; MTB, molecular tumor board.
Reporting and Registration of MTB Cases
For reviewed cases, MTBs created a report in the individual patient's electronic health record (seven MTBs) or in the pathology report (one MTB). Recommendations to external applicants were communicated through videoconferencing, e‐mail, an official written letter, or the pathology report. Four MTBs maintained a local database for registration of new cases and searching prior cases.
View on Harmonization and Collaboration Between MTBs
Two MTBs occasionally received inquiries from other tertiary referral centers harboring an MTB. As personal experience with a similar case, including treatment results, was considered an important factor in rationalizing a recommendation, facilitation of access to data on comparable cases from other MTBs was acknowledged as a valuable addition to their toolset.
Comparison of Targeted Therapy Recommendations
All eight MTBs participated in a study to compare MTB recommendations based on a selection of anonymized cases. Each MTB submitted one case description. The composition of cases was based on information gathered in the interviews: six NSCLC cases, two melanoma cases, one colorectal cancer (CRC) case, and one gastrointestinal stromal tumor (GIST) case (supplemental online Table 1).
As three MTBs were restricted to thoracic oncology, 68 answers were expected (six NSCLC cases with eight responses each and four non‐NSCLC cases with five answers each). One MTB was unable to review the last three non‐NSCLC cases because of lack of availability of a medical oncologist, leaving 65 responses eligible for analysis.
Agreement Between MTB Recommendations
The agreement between MTBs in identifying the foremost actionable target ranged between 60% and 100% (Table 2). Seven of eight MTBs (87.5%) identified BRAF/MEK as actionable targets in case 1A (osimertinib‐resistant NSCLC, resistance by BRAF p.(V600E)). The majority of MTBs recommended osimertinib/dabrafenib/trametinib combination therapy. In case 1B (CRC, KRAS p.(L19F)), all five MTBs agreed that the mutation could potentially inhibit response to anti‐EGFR antibodies, but only three MTBs (60%) considered the evidence sufficient to recommend against anti‐EGFR antibodies.
Table 2.
Agreement between MTB recommendations a
| Case ID | Cancer type | Setting | Mutational profile | Answers received | Recommendations | Next actionable target b | Agreement c |
|---|---|---|---|---|---|---|---|
| 1A | NSCLC | Resistance to osimertinib |
EGFR p.(L747_A750delinsP) BRAF p.(V600E) |
n = 8 |
BRAF/MEK inhibition (dabrafenib + trametinib) and osimertinib, in combination or sequential treatment (n = 6) Switch to chemotherapy; if that is not an option, BRAF/MEK inhibition (dabrafenib + trametinib) (n = 1) Switch to chemotherapy (n = 1) |
BRAF/MEK (n = 7) No target (n = 1) |
87.5% |
| 1B | CRC | Primary stage IV diagnosis | KRAS p.(L19F) | n = 5 |
Do NOT treat with anti‐EGFR antibodies (n = 3) No contraindication for anti‐EGFR antibodies in current guidelines (n = 2) |
No target (n = 3) EGFR (n = 2) |
60% |
| 2A | Melanoma | Exhaustion of standard treatment options |
BRAF p.(G464E) NRAS p.(Q61K) CDKN2A p.(R80*) |
n = 4 |
First exhaust treatment options with immunotherapy, then treat with palbociclib within the DRUP trial (if biallelic CDKN2A inactivation is proven) (n = 2) Treat with palbociclib within the DRUP trial (if biallelic CDKN2A inactivation is proven) (n = 1) MEK inhibition in compassionate use (n = 1) |
CDK4/6 (n = 3) MEK (n = 1) |
75% |
| 2B | NSCLC | Primary stage IV diagnosis |
EGFR p.(G719A) ERBB2 p.(S310F) |
n = 8 |
Treat with agent that may inhibit both mutations, such as afatinib (n = 5) Switch to chemotherapy; treat with afatinib at progression (n = 1) Treat with erlotinib; relevance of ERBB2 mutation is unknown (n = 1) ERBB2 mutation may induce resistance; consider lapatinib (ERBB2 inhibitor) (n = 1) |
EGFR/ERBB2 (n = 6) EGFR (n = 1) ERBB2 (n = 1) |
75% |
| 3A | NSCLC | Resistance to osimertinib |
EGFR p.(E746_A750del) TP53 p.(P278R) TRIM24‐BRAF fusion gene |
n = 8 |
EGFR inhibitor (osimertinib) combined with a MEK inhibitor (trametinib), preferentially within a clinical trial (if available) (n = 6) Consider treatment with MEK inhibitor (n = 1) No other options than chemotherapy. Pan‐RAF inhibitor such as sorafenib may be beneficial, but no options for trials (n = 1) |
MEK (n = 7) RAF (n = 1) |
87.5% |
| 3B | Colorectal cancer | Exhaustion of standard treatment options |
MET p.(R988C) TP53 p.(R158H) TP53 p.(R196*) ERBB2 amplification |
n = 4 | Anti‐ERBB2 antibodies (trastuzumab + pertuzumab) within the DRUP trial (n = 4) | ERBB2 (n = 4) | 100% |
| 4A | GIST | Primary stage IV diagnosis | BRAF p.(V600E) | n = 4 | BRAF/MEK inhibition (dabrafenib + trametinib or vemurafenib + cobimetinib) within the DRUP trial (n = 4) | BRAF/MEK (n = 4) | 100% |
| 4B | NSCLC | Primary stage IV diagnosis |
KRAS p.(A146T) BRAF p.(G469A) CDKN2A p.(H83Y) TERT promoter: C228T |
n = 8 |
Depending on PD‐L1, immunotherapy or chemotherapy combined with immunotherapy (n = 7) d No targets available. Additional diagnostics for revision of diagnosis because of unexpected combination of mutations (n = 1) |
No targets (n = 7) RAF or MEK (n = 1) d |
87.5% |
| 5A | NSCLC | Resistance to osimertinib |
EGFR p.(E746_A750del) TP53 p.(F328Ifs*18) MET amplification EGFR amplification |
n = 8 |
Continue osimertinib; combine with MET inhibitor (n = 4) MET inhibitor or chemotherapy combined with immunotherapy (n = 1) Continue osimertinib; combine with anti‐EGFR antibodies (necitumumab) (n = 1) Chemotherapy combined with immunotherapy; treat with combination of MET inhibitor and EGFR inhibitor at progression (n = 1) Chemotherapy; treat with MET inhibitor at progression (n = 1) |
MET (n = 7) EGFR (n = 1) |
87.5% |
| 5B | NSCLC | Resistance to alectinib |
ALK fusion gene ALK p.(G1202R) |
n = 8 |
Lorlatinib (or brigatinib within an available trial) (n = 7) Brigatinib within an available trial or chemotherapy; lorlatinib at progression (n = 1) |
ALK (n = 8) | 100% |
| Total: | n = 65 | Average: | 86% | ||||
Full description of cases available in supplemental online Table 1.
For the next actionable target, the identified consensus recommendation used to determine agreement is indicated in bold.
Agreement after first recommendation; the provided percentage does not include recommendations after revision.
One MTB suggested targeting RAF or MEK at a later line of therapy; the other MTBs did not identify a target.
Abbreviations: CRC, colorectal cancer; DRUP, Drug Rediscovery Protocol; GIST, gastrointestinal stromal tumor; MTB, molecular tumor board; NSCLC, non‐small cell lung cancer.
For case 2A (melanoma, mutations in BRAF, NRAS, and CDKN2A), three of four MTBs (75%) recognized CDK4/6 as actionable targets of palbociclib within the DRUP trial (if CDKN2A biallelic inactivation was proven). Two MTBs recommended immunotherapy first, in line with the national NSCLC guideline. In case 2B (NSCLC, mutations in EGFR and ERBB2), six of eight MTBs (75%) acknowledged the potential inhibitory effect of afatinib on cancers harboring the ERBB2 mutation—thus identifying both EGFR and ERBB2 as actionable by afatinib—although one MTB recommended chemotherapy prior to treatment with afatinib.
In case 3A (osimertinib‐resistant NSCLC, resistance caused by a TRIM24‐BRAF fusion), seven of eight MTBs (87.5%) recognized MEK as a target. The majority of MTBs recommended osimertinib/trametinib combination treatment. Case 3B (CRC, ERBB2 amplification) achieved 100% agreement: inclusion in the DRUP trial for treatment with anti‐ERBB2 antibodies.
One hundred percent agreement was also achieved for case 4A (GIST, BRAF p.(V600E)): inclusion in the DRUP trial for treatment with BRAF/MEK inhibition. In case 4B (NSCLC, mutations in KRAS, BRAF, CDKN2A, and the TERT promoter region), seven of eight MTBs (87.5%) did not identify a target. Only one MTB commented on the potential actionability of the BRAF non‐V600 mutation with a MEK inhibitor in a late line of treatment. Seven MTBs recommended immunotherapy or chemotherapy combined with immunotherapy, all noting the omitted PD‐L1 status.
For case 5A (osimertinib‐resistant NSCLC, mutations in EGFR and TP53 and amplification of MET and EGFR), seven MTBs (87.5%) acknowledged MET as the actionable target in the next or subsequent line of targeted treatment. However, the prioritization of treatment options varied, with some MTBs preferring exhaustion of treatment options with chemotherapy and disagreement on continuation of EGFR inhibition. In case 5B (ALK inhibitor–resistant NSCLC, ALK fusion, ALK p.(G1202R)), all MTBs (100%) acknowledged that ALK was still actionable with a third‐generation ALK inhibitor. Lorlatinib was recommended by seven of eight MTBs.
Overall, among 65 MTB recommendations on the foremost actionable target for these 10 representative cases, the overall agreement was 86% (56 of 65 responses) after the first round of recommendations.
Revision of Cases with a Low Rate of Agreement
Of cases with responses from all MTBs, 2B and 5A had the lowest rates of agreement with respect to the choice of inhibitor or its timing of use. These cases were sent back to the MTBs along with the other (blinded) MTBs’ recommendations. For case 2B (NSCLC, mutations in EGFR and ERBB2), two MTBs changed their answer based on the other MTBs’ motivations, acknowledging the provided evidence for targeting ERBB2 with afatinib. For case 5A (NSCLC with mutations in EGFR and TP53 and amplifications of MET and EGFR), one MTB changed its answer to include continued targeting of EGFR with osimertinib with its previous recommendation for targeting MET. In addition, one MTB changed its answer because a trial combining osimertinib with MET inhibitor tepotinib had opened in its center.
In total, four MTBs changed their initial answer for case 2B or 5A, either based on evidence provided by other MTBs or because new center‐specific treatment possibilities had become available.
Discussion
This study describes similarities and differences between eight MTBs from tertiary cancer referral centers in The Netherlands. Despite differences in terms of scope and methods, MTBs had similar compositions of experts and reached a high level of agreement in identifying actionable targets. Based on these results and the perspectives of participating MTBs, a consensus was formed on an optimized, collaborative workflow for an MTB hosted by a tertiary cancer referral center (Fig. 2). This workflow, which we designated the “Dutch MTB model” (Table 3), integrates the similarities identified in this study and was approved by representatives of the eight participating MTBs.
Table 3.
Definition of a molecular tumor board according to the Dutch MTB model
| A | An MTB is the primary source of information for interpreting (rare or complex) molecular diagnostic results in oncology and serves as a reciprocal educative platform for clinicians, oncologists, and molecular pathology specialists. |
| B | An MTB focuses on differentiating between targeted therapeutic options (standard of care, within a clinical trial, or off label) or other therapeutic options based on molecular diagnostic results. |
| C | An MTB can be cancer agnostic and operates independently of the conventional cancer type–specific MDTs but features oncologists and pathologists who also participate in conventional MDTs. |
| D | An MTB features at least (a) oncologists (thoracic, medical or hematological, depending on the type of cancer discussed), (b) (molecular‐oriented) pathologists, and (c) clinical scientists in molecular pathology. A clinical geneticist is recommended in case of discussing biomarkers with both germline and somatic implications. |
| E | An MTB is hosted in a tertiary cancer referral center and is open for participation by experts from peripheral hospitals and pathology laboratories, preferably with access through videoconferencing. It is a responsibility of the MTB's network to ensure that all patients in its region have access to an MTB recommendation. |
| F | An MTB ensures its recommendation is accessible in the patient's electronic health record. |
| G | An MTB maintains a local registry of reviewed cases, preferentially with systematic follow‐up of cases within the MTB to evaluate effectiveness of the recommendations. |
Abbreviations: MDT, multidisciplinary team; MTB, molecular tumor board.
One of the MTBs from this study had previously published on the treatment outcome resulting from their workflow [4]. Several individual MTBs in other countries have also reported their scope, methods, recommendations, and treatment outcomes [5, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18]. Comparing these MTBs, the scope and observed value of an MTB, as well as its place within the health care process, vary. Some MTBs assess eligibility for clinical trials [5]. Others also include surgeons, radiation oncologists, and radiologists and are effectively expanded conventional MDTs [8, 16]. In this setup, complex molecular cases are discussed in addition to the nonmolecular cases typically discussed in conventional cancer type–specific MDTs. MTBs adhering to the Dutch MTB model, however, review (complex) molecular cases only, differentiate between treatment options based on molecular alteration(s), and indicate whether the provided (targeted) therapy recommendation is standard of care, within a clinical trial, or off label. This is similar to other previously published MTBs [4, 6, 7, 9, 10, 13, 15, 18]. The main reason for not incorporating complex molecular results into an “enhanced” conventional MDT is the highly specialized and complex nature of the MTB. A low case load, enabled by selectivity in the cases that are discussed, allows time to elaborately discuss biological and clinical consequences of molecular results for a patient. This leads to a reciprocal improvement of expertise of all parties involved on the clinical consequences of molecular test results and the biological rationalization of targeted therapy options. In addition, the possibility to organize an MTB in a cancer‐agnostic fashion allows for the interdisciplinary exchange of molecular diagnostic and therapeutic knowledge, as well as the proper assessment of eligibility for trials that are not limited to a single type of cancer.
The Dutch MTB model encompasses a comprehensive approach toward a case, featuring the identification/prioritization of genomic alterations, potential drug actionability, assessment of drug availability, and the patient's eligibility for a targeted treatment (Fig. 2). MTBs feature experts that are directly involved in differentiation between targeted therapy options: oncologists (thoracic, medical, or hematological, depending on the cancer type discussed), molecular‐oriented pathologists, and CSMPs. These are considered the minimal attendees for reaching a consensus expert recommendation. Treating physicians, pathologists, and CSMPs can all submit cases. The CSMP has an essential role with the primary responsibility to interpret genomic variants in routine molecular diagnostics [34]. The attendance of CSMPs is a major distinguishing factor from conventional MDTs, which was also the case for multiple previously published MTBs [9, 10, 11, 12, 14, 16, 17, 18].
In addition to these three core members, a majority of previously published MTBs also incorporate geneticists or bioinformaticians [5, 7, 8, 10, 11, 12, 13, 14, 15, 16, 17]. In a previous effort to assess differences between MTBs in The Netherlands, attendance of geneticists and bioinformaticians was recommended when discussing large‐scale sequencing results, such as WES and WGS [2]. However, geneticists were members of only three out of eight MTBs in our study, and bioinformaticians were absent. This is because current MTBs rarely discuss WES/WGS, as these techniques are at present not performed in routine molecular diagnostics. Thus, although geneticists and bioinformaticians are valuable, we do not currently consider them mandatory. For now, consultation on demand is deemed sufficient, unless the MTB frequently reviews biomarkers with germline implications, such as BRCA1 and BRCA2 (geneticists), or biomarkers detected with WES/WGS such as complex genomic rearrangements (bioinformaticians).
Assessment and Harmonization of MTB Recommendations
Although our results reveal some differences in the methods used to reach a recommendation, all MTBs operate in accordance with the Dutch MTB model presented here. To evaluate the agreement in treatment recommendations from MTBs adhering to the Dutch MTB model, we compared recommendations provided for 10 typical complex MTB cases. A prior study by Rieke and colleagues used a comparable setup, comparing five MTBs from four countries based on four fictional cases [19]. The authors found a poor agreement, with comparable recommendations from two out of five MTBs (40%) for three cases and three out of five MTBs (60%) for one case. The authors attributed this heterogeneity to differences in interpretation of tumor and germline aberrations and standards of prioritization. The higher overall rate of agreement in our study may in part be explained by a lower number of alterations than was observed in the study of Rieke et al., who included more unknown alterations and had an average of 8 alterations compared with 2.6 in our study [19]. In addition, our comparison was performed with MTBs within the same health care system and thus subject to the same rules and regulations with respect to the availability of drugs. Molecular pathology laboratories associated with these MTBs were already collaborating in a national consortium to achieve uniformity in the interpretation of genomic aberrations in cancer [20] and are unified in The Netherlands Society for Pathology [35]. Dutch thoracic oncologists and medical oncologists collaborate within respective professional networks [36, 37]. These networks have long histories of collaborating in developing national guidelines, organizing joint educational and consensus meetings, and shared national training programs. A Dutch MTB is thus effectively a meeting of local experts representing these existing cooperative efforts. We consider this combination of national connection and interpretative kinship through close collaboration among tertiary cancer referral centers a key factor in achieving optimal diagnostic and/or therapeutic recommendations with a high agreement.
In calculating agreement, we considered recommendations comparable based on the next identified actionable target. This is because differentiating between treatment options based on molecular alterations is the core task of an MTB distinguishing it from conventional MDTs. Our comparison revealed that the MTBs were generally able to independently gather the available evidence on a molecular aberration (Table 2). Yet, access to the same evidence does not necessarily lead to identical recommendations: for example, three of five MTBs cited two preclinical studies on the relatively rare KRAS p.(L19F) mutation (case 1B) [38, 39], but one out of these three did not regard the evidence sufficient to recommend against anti‐EGFR antibodies. This translates to an inevitable limitation in agreement rates between MTBs, as the role of the MTB is to interpret the available evidence and weigh this against patient factors, guidelines, rules and regulations, and the availability of therapeutic choices. Continuous access to and revision of the new scientific evidence is imperative to an adequate performance of MTBs. We consider data sharing between MTBs and regularly comparing treatment recommendations for specific complex cases key in achieving this.
The final treatment recommendations (Table 2) were more heterogeneous than represented by the calculated agreement for the identification of the next actionable target for three major reasons. First, the prioritization of targeted therapy versus other treatment options differed: in cases 1A, 2A, 2B, and 5A, several MTBs recommended nontargeted treatment modalities prior to targeted therapy. Second, there were differences in the exact inhibitor recommended. Drugs were available both within or outside of clinical trials (for example, ALK inhibitors in case 5B), and MTBs tended to recommend the trial available in their own centers. Finally, there were slight differences in whether or not to combine treatments when multiple alterations could be targeted (cases 3A and 5A). Considering our proposed definition of an MTB as primarily responsible for differentiation between targeted therapeutic options (Table 3), the last difference is most significant to address. A revision for two cases (2B and 5A) revealed that MTBs are adaptable when presented with new evidence: four MTBs revised their recommendations based on evidence provided by other MTBs. In case 5A, specifically, the MTB not suggesting combination treatment changed its recommendation when presented with the recommendations of the other MTBs. Thus, the rate of agreement between MTBs may be improved by ensuring MTBs maintain local databases of reviewed cases, preferentially with systematic follow‐up of cases, and sharing exceptional cases with other MTBs in a secure database to allow exchange of knowledge [20, 40].
We used cases submitted by the MTBs to ensure a representative case mix routinely discussed by Dutch MTBs. As the MTBs had all been incorporated into the routine diagnostic setting of their respective institutions, the selected cases represented cancer types with established benefit of targeted molecular testing. In other words, these MTBs primarily facilitate expansion of “established” precision oncology programs, with a greater chance of alterations that may be targeted with off‐label therapy, in compassionate use or in clinical trials. In contrast, MTBs focusing on discovering novel treatment options with pancancer WES or WGS primarily facilitate “explorative” precision oncology programs. In these MTBs, the discussed cases harbor more alterations with unknown significance, decreasing the chance of benefit for patients. Ideally, an MTB should encompass both types of programs.
Further improvement in the Dutch MTB model may be especially gained by achieving homogeneity in gathering the increasing variant‐level evidence to explore treatment options based on the molecular profile. This includes grading the actionability of variants, for which various classification systems have been proposed [41, 42, 43], and access to knowledge bases such as cBioPortal [26], JAX‐CKB [31], and OncoKB [32]. Grading of actionability was not standard procedure for participating MTBs and thus not analyzed. This is a limitation of the current Dutch MTB model, and efforts are ongoing to harmonize the classification of pathogenicity and actionability in The Netherlands. Steps requiring harmonization include how to distinguish germline variants from somatic variants, how to interpret variants of unknown significance, which variant‐level data‐ and knowledge bases need to be consulted to identify potential treatment options, which classification system(s) to apply, and exact criteria for allocating variants to the different classes.
Conclusion
This study shows that the eight Dutch MTBs reach targeted therapy recommendations in high agreement even with differences in scope, logistics, and methods. A high agreement (86%) was achieved especially in identifying the principal actionable targets for cases with rare or complex molecular testing results. Regional connection and data sharing among MTBs and interpretative kinship through collaboration among pathology departments were identified as key factors to achieve a high rate of agreement between MTB recommendations. An MTB, although hosted in a tertiary cancer referral center, should be accessible for all patients with cancer, which requires active participation of health care professionals from peripheral hospitals and pathology laboratories in a regional MTB network. We recommend using our proposed Dutch MTB model for an optimal, collaborative, and nationally aligned MTB workflow to transform precision medicine from retrospective anecdotal evidence to successful prospective evidence.
Author Contributions
Conception/design: Bart Koopman, Harry J.M. Groen, Marjolijn J.L. Ligtenberg, Katrien Grünberg, Ed Schuuring, Léon C. van Kempen
Provision of study material or patients: Bart Koopman, Harry J.M. Groen, Marjolijn J.L. Ligtenberg, Katrien Grünberg, Kim Monkhorst, Adrianus J. de Langen, Mirjam C. Boelens, Marthe S. Paats, Jan H. von der Thüsen, Winand N.M. Dinjens, Nienke Solleveld, Tom van Wezel, Hans Gelderblom, Lizza E. Hendriks, Ernst Jan M. Speel, Tom E. Theunissen, Leonie I. Kroeze, Niven Mehra, Berber Piet, Anthonie J. van der Wekken, Arja ter Elst, Wim Timens, Stefan M. Willems, Ruud W.J. Meijers, Wendy W.J. de Leng, Anne S.R. van Lindert, Teodora Radonic, Sayed M.S. Hashemi, Daniëlle A.M. Heideman, Ed Schuuring, Léon C. van Kempen
Collection and/or assembly of data: Bart Koopman, Harry J.M. Groen, Marjolijn J.L. Ligtenberg, Katrien Grünberg, Kim Monkhorst, Adrianus J. de Langen, Mirjam C. Boelens, Marthe S. Paats, Jan H. von der Thüsen, Winand N.M. Dinjens, Nienke Solleveld, Tom van Wezel, Hans Gelderblom, Lizza E. Hendriks, Ernst Jan M. Speel, Tom E. Theunissen, Leonie I. Kroeze, Niven Mehra, Berber Piet, Anthonie J. van der Wekken, Arja ter Elst, Wim Timens, Stefan M. Willems, Ruud W.J. Meijers, Wendy W.J. de Leng, Anne S.R. van Lindert, Teodora Radonic, Sayed M.S. Hashemi, Daniëlle A.M. Heideman, Ed Schuuring, Léon C. van Kempen
Data analysis and interpretation: Bart Koopman, Harry J.M. Groen, Ed Schuuring, Léon C. van Kempen
Manuscript writing: Bart Koopman, Harry J.M. Groen, Marjolijn J.L. Ligtenberg, Katrien Grünberg, Kim Monkhorst, Adrianus J. de Langen, Mirjam C. Boelens, Marthe S. Paats, Jan H. von der Thüsen, Winand N.M. Dinjens, Nienke Solleveld, Tom van Wezel, Hans Gelderblom, Lizza E. Hendriks, Ernst Jan M. Speel, Tom E. Theunissen, Leonie I. Kroeze, Niven Mehra, Berber Piet, Anthonie J. van der Wekken, Arja ter Elst, Wim Timens, Stefan M. Willems, Ruud W.J. Meijers, Wendy W.J. de Leng, Anne S.R. van Lindert, Teodora Radonic, Sayed M.S. Hashemi, Daniëlle A.M. Heideman, Ed Schuuring, Léon C. van Kempen
Final approval of manuscript: Bart Koopman, Harry J.M. Groen, Marjolijn J.L. Ligtenberg, Katrien Grünberg, Kim Monkhorst, Adrianus J. de Langen, Mirjam C. Boelens, Marthe S. Paats, Jan H. von der Thüsen, Winand N.M. Dinjens, Nienke Solleveld, Tom van Wezel, Hans Gelderblom, Lizza E. Hendriks, Ernst Jan M. Speel, Tom E. Theunissen, Leonie I. Kroeze, Niven Mehra, Berber Piet, Anthonie J. van der Wekken, Arja ter Elst, Wim Timens, Stefan M. Willems, Ruud W.J. Meijers, Wendy W.J. de Leng, Anne S.R. van Lindert, Teodora Radonic, Sayed M.S. Hashemi, Daniëlle A.M. Heideman, Ed Schuuring, Léon C. van Kempen
Disclosures
Kim Monkhorst: Roche, AstraZeneca, PGDx (RF), Merck Sharp & Dohme, Roche, AstraZeneca (H), Pfizer, Bristol‐Myers Squibb, Roche, Merck Sharp & Dohme, Abbvie, AstraZeneca, Diaceutics (C/A), Roche, Takeda, Pfizer (other); Lizza E. Hendriks: Roche, Boehringer Ingelheim, Bristol‐Myers Squibb, Eli Lilly & Co., Pfizer, Takeda, Merck Sharp & Dohme (SAB), Roche, AstraZeneca (RF), Roche (other—travel, fees for interview sessions), AstraZeneca (other—mentorship program), Merck Sharp & Dohme (other—speaker educational session, travel), Quadia (other—fees for educational webinars); Ernst Jan M. Speel: AstraZeneca, Merck Sharp & Dohme, Bristol‐Myers Squibb, Novartis (RF), Bristol‐Myers Squibb, Merck Sharp & Dohme, AbbVie, Pfizer, Roche, Bayer (C/A), AbbVie (other); Niven Mehra: Roche, Merck Sharp & Dohme, Bristol‐Myers Squibb, Astellas, Janssen (C/A), Astellas, Janssen, Pfizer, Roche, Sanofi, Genzyme (RF), Astellas, Merck Sharp & Dohme (other—travel support); Wim Timens: Roche Diagnostics‐Ventana, AbbVie, Bristol‐Myers Squibb (C/A), Merck Sharp & Dohme (SAB); Stefan M. Willems: AstraZeneca, Roche, Merck Sharp & Dohme, Bristol‐Myers Squibb, Bayer, Amgen, Nextcure (RF); Wendy W.J. de Leng: Roche, Bristol‐Myers Squibb, Pfizer (RF); Sayed M.S. Hashemi: AbbVie, AstraZeneca, Bristol‐Myers Squibb, Eli Lilly & Co., GlaxoSmithKline, Loxo, Merck Sharp & Dohme, Novartis, Roche, Takeda, Xcovery (RF, SAB); Daniëlle A.M. Heideman: Pfizer, Bristol‐Myers Squibb (SAB), Self‐Screen B.V. (OI), Qiagen (other—speakers bureau); Ed Schuuring: AstraZeneca, Roche, Pfizer, Novartis, Merck Sharp & Dohme/Merck, Bayer, Bristol‐Myers Squibb, BioCartis, Illumina, Agena Bioscience, Janssen Cilag (Johnson&Johnson), Diaceutics (C/A), Abbott, Pfizer, Biocartis, Bristol‐Myers Squibb, Bio‐Rad, Roche, Agena Bioscience, CC Diagnostics, Boehringer Ingelheim, Qiagen, Promega, TATAA (RF), Bio‐Rad, Novartis, Roche, Biocartis, Agena Bioscience, Illumina, Pfizer, AstraZeneca (H). Léon van Kempen: NanoString (C/A), Roche, Bayer, NanoString (RF), Merck Sharpe & Dohme, AstraZeneca, Bristol‐Myers Squibb, Pfizer (H). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Appendix S1. Supporting Information.
Appendix S2. Supporting Information.
Acknowledgments
We are grateful to Anke van den Berg, Matthew Groves, Geke Hospers, Birgitta Hiddinga, Hilde Jalving, Lucie Hijmering‐Kappelle, Joost Kluiver, Michel van Kruchten, Elise van der Logt, Maarten Niemantsverdriet, Sjoukje Oosting, Juliana Vilacha, Michel van den Heuvel, Lieneke Steeghs, Ingrid Vogelaar, Linda Bosch, Liudmila Kodach, Egbert Smit, Annette Bijsmans, Maxime van Berge Henegouwen, Hans Morreau, Lisa Hillen, Xiao Fei Li, John Hinrichs, Anne Jansen, Joyce Radersma‐van Loon, and Aryan Vink and all other members of the molecular tumor boards in The Netherlands for their contributions to the conduct and successful completion of this study. This work was supported by ZonMW (The Netherlands Organization for Health Research) within the Personalized Medicine Program (grant number 846001001).
Disclosures of potential conflicts of interest may be found at the end of this article.
References
- 1. Garraway LA. Genomics‐driven oncology: Framework for an emerging paradigm. J Clin Oncol 2013;31:1806–1814. [DOI] [PubMed] [Google Scholar]
- 2. van der Velden DL, van Herpen CML, van Laarhoven HWM et al. Molecular tumor boards: Current practice and future needs. Ann Oncol 2017;28:3070–3075. [DOI] [PubMed] [Google Scholar]
- 3. Willemsen AECAB, Krausz S, Ligtenberg MJL et al. Molecular tumour boards and molecular diagnostics for patients with cancer in The Netherlands: Experiences, challenges, and aspirations. Br J Cancer 2019;121:34–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koopman B, van der Wekken AJ, ter Elst A et al. Relevance and effectiveness of molecular tumor board recommendations for patients with non–small‐cell lung cancer with rare or complex mutational profiles. JCO Precis Oncol 2020;4:393–410. [DOI] [PubMed] [Google Scholar]
- 5. Basse C, Morel C, Alt M et al. Relevance of a molecular tumour board (MTB) for patients’ enrolment in clinical trials: Experience of the Institut Curie. ESMO Open 2018;3:e000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryce AH, Egan JB, Borad MJ et al. Experience with precision genomics and tumor board, indicates frequent target identification, but barriers to delivery. Oncotarget 2017;8:27145–27154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dalton WB, Forde PM, Kang H et al. Personalized medicine in the oncology clinic: Implementation and outcomes of the Johns Hopkins molecular tumor board. JCO Precis Oncol 2017;1:PO.16.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harada S, Arend R, Dai Q et al. Implementation and utilization of the molecular tumor board to guide precision medicine. Oncotarget 2017;8:57845–57854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaderbhai CG, Boidot R, Beltjens F et al. Use of dedicated gene panel sequencing using next generation sequencing to improve the personalized care of lung cancer. Oncotarget 2016;7:24860–24870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knepper TC, Bell GC, Hicks JK et al. Key lessons learned from Moffitt's molecular tumor board: The Clinical Genomics Action Committee experience. The Oncologist 2017;22:144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee B, Tran B, Hsu AL et al. Exploring the feasibility and utility of exome‐scale tumour sequencing in a clinical setting. Intern Med J 2018;48:786–794. [DOI] [PubMed] [Google Scholar]
- 12. Marks LJ, Oberg JA, Pendrick D et al. Precision medicine in children and young adults with hematologic malignancies and blood disorders: The Columbia University experience. Front Pediatr 2017;5:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moore DA, Kushnir M, Mak G et al. Prospective analysis of 895 patients on a UK genomics review board. ESMO Open 2019;4:e000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ortiz M V, Kobos R, Walsh M et al. Integrating genomics into clinical pediatric oncology using the molecular tumor board at the Memorial Sloan Kettering Cancer Center. Pediatr Blood Cancer 2016;63:1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rolfo C, Manca P, Salgado R et al. Multidisciplinary molecular tumour board: A tool to improve clinical practice and selection accrual for clinical trials in patients with cancer. ESMO Open 2018;3:e000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwaederle M, Parker BA, Schwab RB et al. Molecular tumor board: The University of California‐San Diego Moores Cancer Center experience. The Oncologist 2014;19:631–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tafe LJ, Gorlov IP, de Abreu FB et al. Implementation of a molecular tumor board: The impact on treatment decisions for 35 patients evaluated at Dartmouth‐Hitchcock Medical Center. The Oncologist 2015;20:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trédan O, Wang Q, Pissaloux D et al. Molecular screening program to select molecular‐based recommended therapies for metastatic cancer patients: Analysis from the ProfiLER trial. Ann Oncol 2019;30:757–765. [DOI] [PubMed] [Google Scholar]
- 19. Rieke DT, Lamping M, Schuh M et al. Comparison of treatment recommendations by molecular tumor boards worldwide. JCO Precis Oncol 2018;2:PO.18.00098. [DOI] [PubMed] [Google Scholar]
- 20. Nederlandse organisatie voor gezondheidsonderzoek en zorginnovatie (ZonMW) . Predictive Analysis for Therapy: PATH to Optimising Access to Personalised Cancer Therapy in The Netherlands. 2016. Available at https://www.zonmw.nl/nl/onderzoek‐resultaten/geneesmiddelen/programmas/project‐detail/personalised‐medicine/predictive‐analysis‐for‐therapy‐path‐to‐optimising‐access‐to‐personalised‐cancer‐therapy‐in‐the‐net/. Accessed August 8, 2019.
- 21. Universitair Medisch Centrum Groningen . Research Code. 2007. Available at https://www.umcg.nl/SiteCollectionDocuments/English/Researchcode/umcg‐research‐code‐2018‐en.pdf. Accessed May 5, 2020.
- 22. Dutch Federation of Biomedical Scientific Societies . Code of Conduct for Medical Research. 2004. Available at https://www.federa.org/sites/default/files/bijlagen/coreon/code_of_conduct_for_medical_research_1.pdf. Accessed May 5, 2020.
- 23. Nederlandse Vereniging voor Artsen voor Longziekten en Tuberculose . Behandeling patiënten met een zeldzame mutatie bij NSCLC. 2020. Available at https://richtlijnendatabase.nl/richtlijn/niet_kleincellig_longcarcinoom/systemische_behandeling_stadium_iv_nsclc/behandeling_pati_nten_met_een_zeldzame_mutatie_bij_nsclc.html. Accessed April 23, 2020.
- 24. van der Velden DL, Hoes LR, van der Wijngaart H et al. The Drug Rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature 2019;574:127–131. [DOI] [PubMed] [Google Scholar]
- 25. 1000 Genomes Project Consortium ; Auton A, Brooks LD, Durbin RM et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao J, Aksoy BA, Dogrusoz U et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Landrum MJ, Lee JM, Riley GR et al. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42:D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forbes SA, Beare D, Boutselakis H et al. COSMIC: Somatic cancer genetics at high‐resolution. Nucleic Acids Res 2017;45:D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sherry ST, Ward MH, Kholodov M et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res 2001;29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cunningham F, Achuthan P, Akanni W et al. Ensembl 2019. Nucleic Acids Res 2019;47:D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patterson SE, Liu R, Statz CM et al. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum Genomics 2016;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chakravarty D, Gao J, Phillips SM et al. OncoKB: A precision oncology knowledge base. JCO Precis Oncol 2017;2017:513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. PubMed. U.S. National Library of Medicine Web site . Available at https://pubmed.ncbi.nlm.nih.gov/.
- 34. Deans ZC, Costa JL, Cree I et al. Integration of next‐generation sequencing in clinical diagnostic molecular pathology laboratories for analysis of solid tumours; an expert opinion on behalf of IQN Path ASBL. Virchows Arch 2017;470:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nederlandse Vereniging voor Pathologie (NVVP) . Available at https://pathology.nl/. Accessed April 28, 2020.
- 36. Nederlandse Vereniging van Artsen voor Longziekten en Tuberculose (NVALT) . Available at https://www.nvalt.nl/. Accessed April 28, 2020.
- 37. Nederlandse Vereniging voor Medische Oncologie (NVMO) . Available at https://www.nvmo.org/. Accessed April 28, 2020.
- 38. Akagi K, Uchibori R, Yamaguchi K et al. Characterization of a novel oncogenic K‐ras mutation in colon cancer. Biochem Biophys Res Commun 2007;352:728–32. [DOI] [PubMed] [Google Scholar]
- 39. Smith G, Bounds R, Wolf H et al. Activating K‐Ras mutations outwith “hotspot” codons in sporadic colorectal tumours ‐ implications for personalised cancer medicine. Br J Cancer 2010;102:693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Health RI: Enabling data driven health . Available at https://www.health-ri.nl/services/cbioportal. Accessed March 3, 2020.
- 41. Mateo J, Chakravarty D, Dienstmann R et al. A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol 2018;29:1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li MM, Datto M, Duncavage EJ et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19:4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leichsenring J, Horak P, Kreutzfeldt S et al. Variant classification in precision oncology. Int J Cancer 2019;145:2996–3010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Appendix S1. Supporting Information.
Appendix S2. Supporting Information.


