Abstract
Mammalian mitochondrial tRNA (mt-tRNA) plays a central role in the synthesis of the 13 subunits of the oxidative phosphorylation complex system (OXPHOS). However, many aspects of the context-dependent expression of mt-tRNAs in mammals remain unknown. To investigate the tissue-specific effects of mt-tRNAs, we performed a comprehensive analysis of mitochondrial tRNA expression across five mice tissues (brain, heart, liver, skeletal muscle, and kidney) using Northern blot analysis. Striking differences in the tissue-specific expression of 22 mt-tRNAs were observed, in some cases differing by as much as tenfold from lowest to highest expression levels among these five tissues. Overall, the heart exhibited the highest levels of mt-tRNAs, while the liver displayed markedly lower levels. Variations in the levels of mt-tRNAs showed significant correlations with total mitochondrial DNA (mtDNA) contents in these tissues. However, there were no significant differences observed in the 2-thiouridylation levels of tRNALys, tRNAGlu, and tRNAGln among these tissues. A wide range of aminoacylation levels for 15 mt-tRNAs occurred among these five tissues, with skeletal muscle and kidneys most notably displaying the highest and lowest tRNA aminoacylation levels, respectively. Among these tissues, there was a negative correlation between variations in mt-tRNA aminoacylation levels and corresponding variations in mitochondrial tRNA synthetases (mt-aaRS) expression levels. Furthermore, the variable levels of OXPHOS subunits, as encoded by mtDNA or nuclear genes, may reflect differences in relative functional emphasis for mitochondria in each tissue. Our findings provide new insight into the mechanism of mt-tRNA tissue-specific effects on oxidative phosphorylation.
Keywords: mitochondrial tRNA, tissue specific expression, translation, oxidative phosphorylation, murine
Abbreviations: DIG, digoxigenin; mRNA, messenger RNA; mt-aaRS, mitochondrial tRNA synthetases; mt-tRNA, mitochondrial tRNA; mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation complex system
Transfer RNAs (tRNA) function as adapter molecules that decode messenger RNAs (mRNAs) during protein translation by delivering amino acids to the ribosome. In mammal mitochondria, 22 tRNAs (mt-tRNA), together with two rRNAs and 13 mRNA coding 13 structural components of oxidative phosphorylation system (OXPHOS), are encoded by own mitochondrial genome (mtDNA) (1, 2, 3, 4). The mtDNA produces the polycistronic heavy (H)- and light (L)-strand transcripts that are catalyzed by mitochondrial transcription machinery (5, 6, 7, 8). The H-strand transcripts contain 12S rRNA, 16S rRNA, 12 mRNAs, and 14 tRNAs including tRNAHis, tRNALys, and tRNALeu(UUR), while L-strand transcripts harbor ND6 mRNA and eight tRNAs including tRNAGln and tRNASer(UCN) (5, 6, 7, 8, 9, 10). These polycistronic transcripts are then processed resulting in the release of 13 mRNAs, 2 rRNAs, and 22 tRNAs, which are catalyzed by RNase P and RNase Z, respectively (11, 12, 13). The formation of functional mt-tRNAs for the synthesis of 13 mtDNA encoding polypeptides requires extensive base modifications, CCA addition, and aminoacylation (14, 15, 16, 17, 18). Transcription control of mt-tRNA genes in the mammalian tissues plays an important role in specific oxidative phosphorylation capacities required to satisfy their metabolic and energetic demands (19, 20, 21, 22). However, little is known about the variations in the expression levels or physiological conditions of mt-tRNAs in differing types of both human and mouse tissues.
The abundance, nucleotide modification, and aminoacylation of tRNAs may reflect the tissue-specific differences in mitochondrial number, morphology, activity, and biogenesis (23, 24). In the present investigation, we performed systematic analysis of mt-tRNA expressions from mouse-derived brain, heart, liver, skeletal muscle, and kidney. The steady-state levels of 22 mt-tRNAs among these five tissue types were examined using tRNA Northern blot analysis (25, 26). The nucleotide modifications of tRNAs were assessed for the levels of 2-thiouridine modification at position U34 in tRNALys, tRNAGlu, and tRNAGln by isolating total mitochondrial RNAs from these tissues and then qualifying 2-thiouridine modifications by retardation of electrophoresis mobility in polyacrylamide gels containing N-acryloylamino phenyl mercuric chloride (27, 28, 29). Furthermore, we examined the aminoacylation levels of tRNAs using electrophoresis in an acidic urea polyacrylamide system to separate uncharged tRNA species from the correspondingly charged tRNAs among these tissues (18, 30). Moreover, we investigated the tissue-specific expression of 14 mitochondrial tRNA synthetases (mt-aaRS) and OXPHOS subunits that were encoded by mtDNA and nuclear genes.
Results
Variations in the steady-state levels of mt-tRNAs among mouse-derived tissues
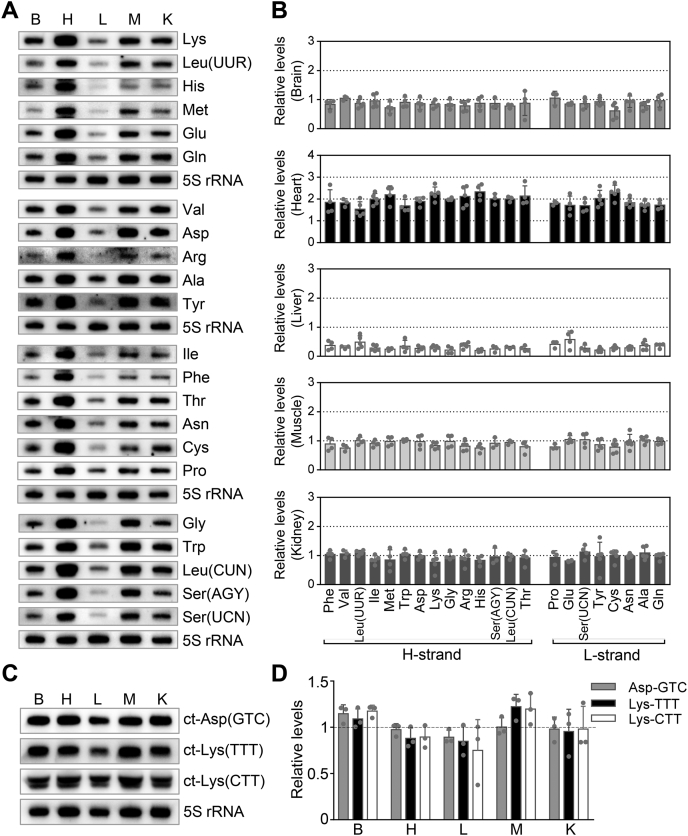
To investigate the tissue-specific expression of mt-tRNAs in mice, we subjected total RNAs from brain, heart, liver, skeletal muscle, and kidney to Northern blots and hybridized them with digoxigenin (DIG)-labeled oligodeoxynucleotide probes for 14 mt-tRNAs from H-strand transcripts and 8 mt-tRNAs from L-strand transcripts, cytosolic (ct) tRNA (ct-tRNAAsp(GTC), ct-tRNALys(TTT), and ct-tRNALys(CTT)) as well as with reference 5S rRNA, a nuclear-encoded mitochondrial small RNA (31, 32). For comparison, the average levels of each tRNA among different tissues were then normalized to the average levels in the 5S rRNA of the same tissue. As shown in Figure 1A and Table S1, there were marked differences in the levels of each mt-tRNA among the five tissues. For example, the average levels of tRNAHis among the brain, heart, liver, skeletal muscle, and kidney were 0.87, 2.34, 0.20, 0.75, and 0.84-fold relative to average levels of this tRNA among five tissues, respectively. As shown in Figure 1A, various differing levels of these 22 mt-tRNAs were observed in the same tissue type. As shown in the Figure 1B and Table S1, the average levels of mt-tRNAs ranged from 1.54-fold (tRNALeu(UUR)) to 2.34-fold (tRNAHis) (relative to average of corresponding tRNA among five tissues) in the heart, 0.61-fold (tRNACys) to 1.05-fold (tRNAVal, tRNAPro) in the brain, 0.75-fold (tRNAHis) to 1.04-fold (tRNAGlu, tRNASer(UCN)) in the skeletal muscle, 0.77-fold (tRNALys) to 1.12-fold (tRNASer(UCN)) in the kidney, and 0.20-fold (tRNAHis) to 0.58-fold (tRNAGlu) in the liver. Strikingly, the heart exhibited the highest levels of average 22 mt-tRNAs (1.92-fold relative to average levels of 22 mt-tRNAs in five tissues). Brain, skeletal muscle, and kidney also revealed relatively higher levels of average 22 mt-tRNAs (0.86, 0.92, and 0.98 fold, respectively). The liver exhibited markedly lower levels of average 22 mt-tRNAs (0.31-fold relative to average levels of 22 mt-tRNAs in five tissues). However, there was no significant difference in the levels of ct-tRNAAsp(GTC), ct-tRNALys(TTT), and ct-tRNALys(CTT) among the brain, heart, liver, skeletal muscle, and kidney tissues (Fig. 1, C and D). These variations in the levels of mt-tRNAs may reflect the mtDNA contents among these tissues. We measured relative mtDNA copy numbers from five mouse-derived tissues by comparing the ratios of mtDNA to nDNA by real-time quantitative PCR (33). As shown in Fig. S1A, there were marked variations in the mtDNA copy numbers among five tissues. Notably, the mtDNA contents of the brain, heart, skeletal muscle, kidney, and liver were 1.01, 1.66, 1.05, 0.66, and 0.63-fold, relative to the overall average levels of those in five tissues combined, respectively. As shown in Fig. S1B, the variations in the levels of mt-tRNAs in five tissues showed significant correlations with the corresponding variations in mtDNA copy numbers (r2 = 0.78, p = 0.048). These results suggested that the differences of mitochondrial copy numbers contributed significantly to the variations of tRNA levels.
Figure 1.
Northern blot analysis of 22 mt-tRNAsteady-statelevels among five mouse tissues.A and C, four micrograms of total RNA from mouse-derived brain, heart, liver, skeletal muscle, and kidney tissues was electrophoresed through a denaturing polyacrylamide gel, electroblotted, and hybridized with DIG-labeled oligonucleotide probes specific for the 22 mt-tRNAs, ct-tRNAAsp(GTC), tRNALys(TTT) and tRNALys(CTT) and 5S rRNA, respectively. B and D, quantification of relative mt-tRNA levels of the five tissues. The content of each mt-tRNA was normalized to that of 5S rRNA in the five tissues. Relative tRNA levels for the brain, heart, liver, skeletal muscle, and kidney were normalized to the mean values of each tRNA among the five tissues. Calculations were based on 3 to 5 independent experiments. Error bars indicate two standard deviations (SD) of the means. B, Brain; H, Heart; K, Kidney; L, liver; M, skeletal Muscle.
Thiolation levels of mitochondrial tRNALys, tRNAGln, and tRNAGlu
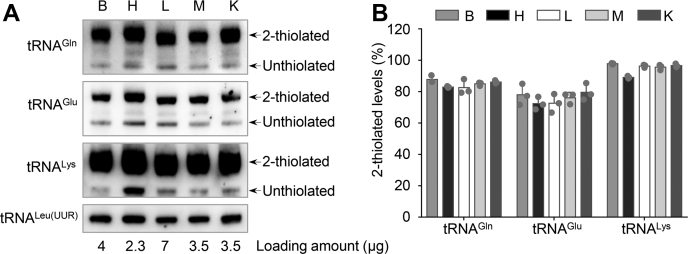
In the human mitochondrion, 18 types of nucleotide modification are present in the 137 positions of 22 mt-tRNA species (14). In particular, 5-taurinomethyl-2-thiouridine (τm5s2U) modification was only present in the tRNALys, tRNAGlu, and tRNAGln but not in other tRNAs (14). The presence of thiouridine modification in these tRNAs was examined using retardation of electrophoretic mobility in a polyacrylamide gel containing 0.05 mg/ml (N-acryloylaminophenyl) mercuric chloride (APM) (29). In this study, the levels in the 2-thiouridine modification at position U34 in tRNALys, tRNAGlu, and tRNAGln were determined by isolating total RNAs from mouse brain, heart, liver, skeletal muscle, and kidney tissues, then purifying tRNAs, qualifying the 2-thiouridine modification by retardation of electrophoresis mobility in APM polyacrylamide gel (27, 28, 29), and hybridizing DIG-labeled probes for mitochondrial tRNAGln, tRNAGlu and tRNALys and tRNALeu(UUR). In this system, the mercuric compound can specifically interact with the tRNAs containing a thiocarbonyl group such as tRNAGln, tRNAGlu, and tRNALys, thereby retarding tRNA migration. As shown in Figure 2, τm5s2U levels of tRNALys in the brain, heart, liver, skeletal muscle, and kidney tissues were 97.9%, 89.1%, 96.3%, 95.6%, and 96.6%, respectively; τm5s2U levels of tRNAGlu in the brain, heart, liver, skeletal muscle, and kidney were 78%, 72.4%, 72.6%, 76.1%, and 79.6%, respectively, and τm5s2U levels of tRNAGln in the brain, heart, liver, skeletal muscle, and kidney were 87.8%, 83%, 82.6%, 85%, and 86.1%, respectively. However, τm5s2U was absent in the mitochondrial tRNALeu(UUR). No significant differences in the τm5s2U levels of tRNALys, tRNAGlu, and tRNAGln were observed among these five tissues.
Figure 2.
APM gel electrophoresis combined with Northern blot analysis of mt-tRNAs.A, four, 2.3, 7, 3.5, and 3.4 μg of total RNAs from the brain, heart, liver, skeletal muscle, and kidney tissues were separated by polyacrylamide gel electrophoresis containing 0.05 mg/ml APM, electroblotted onto a positively charged membrane, and hybridized with a DIG-labeled oligonucleotide probe specific for mt-tRNAGln, tRNAGlu, tRNALys, and tRNALeu(UUR), respectively. B, quantification of 2-thiolated tRNA levels. Calculations were based on more than three independent experiments. Error bars indicate SD of the means.
Aminoacylation levels of mitochondrial tRNAs
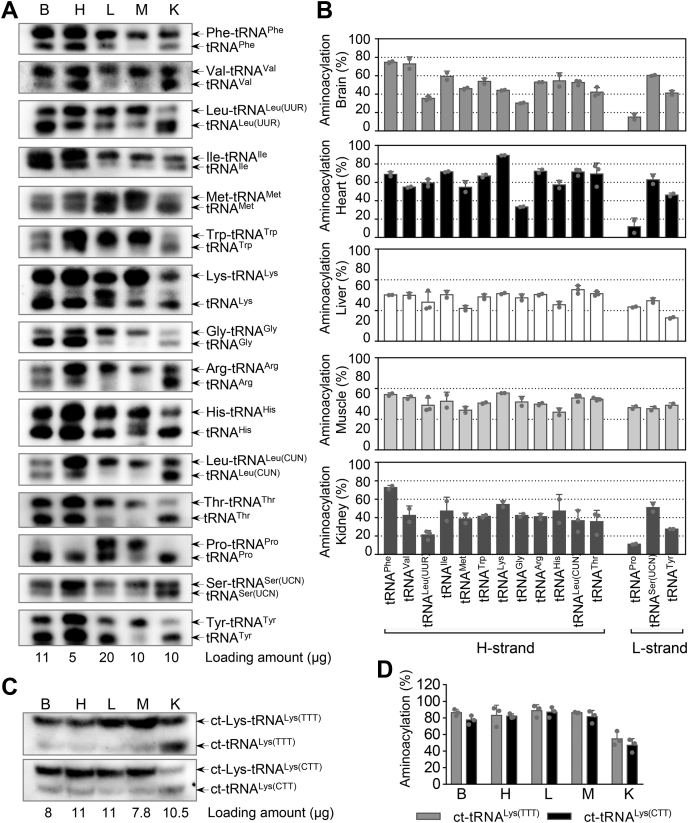
Aminoacylation is an essential step in the synthesis of proteins (17). We examined the aminoacylation levels of 15 mt-tRNAs including tRNAMet, tRNALeu(UUR), tRNALeu(CUN), tRNAIle, tRNAHis, ct-tRNALys(TTT), and ct-tRNALys(CTT) in the five tissues, by the use of electrophoresis in an acidic urea PAGE system to separate uncharged tRNA species from the corresponding charged tRNA, electroblotting and hybridizing with the tRNA probes described above (18). As shown in Figure 3 and Table S2, various aminoacylation levels of each tRNA were observed among the five tissues. There were marked differences of aminoacylation levels in these mt-tRNAs such as tRNALeu(UUR), tRNALeu(CUN), tRNAGly, tRNAPro, and tRNATyr among five tissues. In particular, the average levels of tRNALeu(UUR) aminoacylation among the brain, heart, liver, skeletal muscle, and kidney were 35.6%, 59.4%, 63.7%, 72.7%, and 21.3%, respectively. As shown in Figure 3A and Table S2, the various aminoacylation levels of the 15 mt-tRNAs were observed in the same tissue. Specially, the average aminoacylation levels of 15 mt-tRNAs varied from 15.2% in tRNAPro to 74.6% in tRNAPhe with an average 51% of 15 tRNAs in the brain, 11.7% in tRNAPro to 89.3% in tRNALys with average 59.3% of 15 tRNAs in the heart, 38.1% in tRNATyr to 84.1% in tRNALeu(CUN) with average 69.1% of 15 tRNA in the liver, 61.5% in tRNAHis to 92.5% in tRNALys with average 77.9% of 15 tRNAs in the skeletal muscle, and 11.2% in tRNAPro to 72.6% in tRNAPhe with average 40.3% of 15 mt-tRNAs in the kidney. As shown in Figure 3, C and D, there were no significant differences in aminoacylation levels of ct-tRNALys(TTT) and ct-tRNALys(CTT) among the brain, heart, liver, and skeletal muscle. However, there were relatively higher aminoacylation levels of cytosolic ct-tRNALys(TTT) and ct-tRNALys(CTT) in the kidney tissues.
Figure 3.
Aminoacylation analysis of tRNAs.A, Eleven, 5, 20, 10, or 10 μg and C, 8, 11, 11, 7.8, or 10.5 μg of total RNAs from mouse-derived brain, heart, liver, skeletal muscle, and kidney tissues, respectively, were electrophoresed at 4 °C through an acid (pH 5.0) 8.5% polyacrylamide with 8 M urea gel, electroblotted, and hybridized with a DIG-labeled oligonucleotide probes-specific for 15 mt-tRNAs and two ct-tRNAs. The charged (upper band) and uncharged (lower band) forms of different mt-tRNAs were separated by the gel system. B and D, quantification of aminoacylated proportions of tRNAs. Calculations were based on 2 to 3 independent determinations. Error bars indicate SD of the means.
Negative correlation between mt-aaRS expression and tRNA aminoacylation
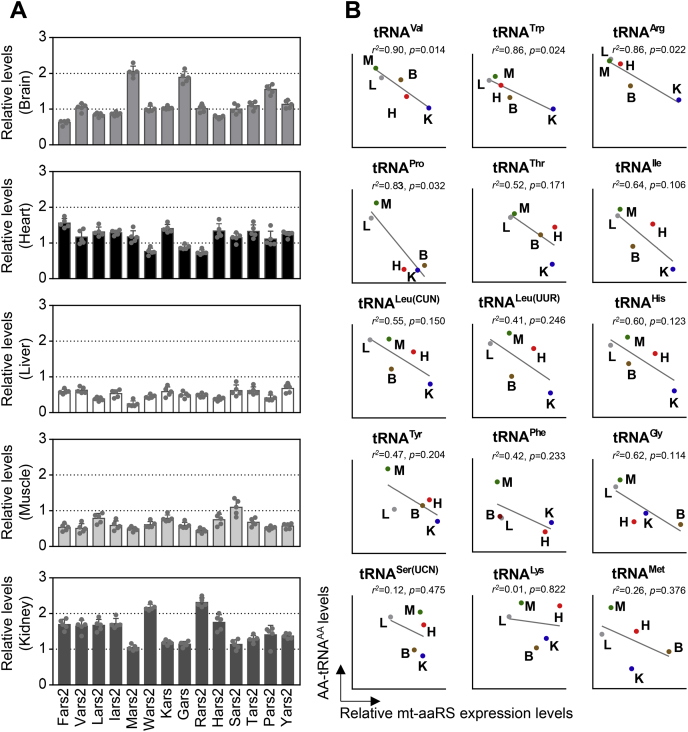
A specific cognate amino acid is charged or aminoacylated to each tRNA catalyzed by mt-aaRSs (17). We measured the mRNA expression levels of 14 genes encoding mt-aaRSs among the five tissues using quantitative PCR for these genes and 18S rRNA as normalization. The average mRNA level of each gene in various tissues was then normalized to the average levels in the same tissue for the reference 18S rRNA. As shown in Figure 4A and Table S3, there were marked differences in the mRNA levels of each genes among the five tissues. For example, the average mRNA levels of Fars2 among the brain, heart, liver, skeletal muscle, and kidney were 0.62, 1.56, 0.59, 0.53, and 1.7-fold relative to average mRNA levels of 15 genes in five tissues, respectively. As shown in Figure 4A and Table S3, the varied mRNA levels of these 15 genes were observed in the same tissue, with the average mRNA levels of the 15 genes ranging from 0.62-fold (Fars2) to 2.05-fold (Mars2) (relative to average of 15 genes among five tissues) in the brain, 0.74-fold (Rars2) to 1.56-fold (Fars2) in the heart, 0.24-fold (Mars2) to 0.68-fold (Yars2) in the liver, 0.45-fold (Rars2) to 1.1-fold (Sars2) in the skeletal muscle, and 1.05-fold (Mars2) to 2.31-fold (Rars2) in the kidney.
Figure 4.
Mt-aaRS gene expression analysis.A, the relative expression levels of mt-aaRS genes from mouse-derived brain, heart, liver, skeletal muscle, and kidney tissues using RT-qPCR. Relative levels were normalized to the mean values of each mt-aaRS gene among five tissues. n =5 per group. B, Pearson correlation was used to measure the extent of correlation between the mean level of mt-aaRS expression and corresponding mt-tRNA aminoacylation in each of the five tissues. The r2 and p values are indicated in each graph.
We then investigated if there was any potential correlation between the mt-aaRS mRNA levels and mt-tRNA aminoacylation levels in various tissues. As shown in Figure 4B, the variations in the aminoacylation levels in the mt-tRNAs among the five tissues exhibited negative correlations with the corresponding variations in the aaRS mRNA levels. In particular, the aminoacylation levels in the tRNAVal, tRNATrp, tRNAArg, and tRNAPro revealed significantly negative correlation with the corresponding variations in aaRS expression levels in these five tissues (r2 = 0.90, p = 0.014; r2 = 0.86, p = 0.024; r2 = 0.86, p = 0.022; r2 = 0.83, p = 0.032, respectively). However, the correlations between aminoacylation levels of other ten tRNAs and mRNA levels of their corresponding aaRS were not statistically significant.
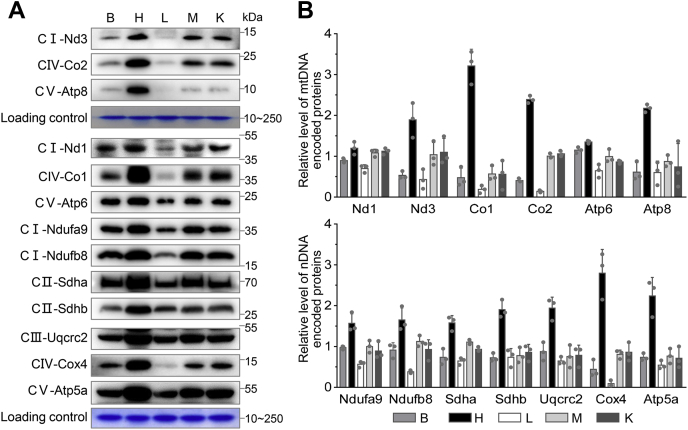
Tissue-specific expression of OXPHOS subunits
To assess the tissue-specific expression of OXPHOS subunits, we examined the levels of the subunits of OXPHOS in the five tissues using western blot analysis. These subunits included six mtDNA encoding polypeptides (Nd1, Nd3, Co1, Co2, Atp6, and Atp8), seven nucleus encoding proteins: Ndufa9 and Ndufb8 [subunits of NADH dehydrogenase (complex I)], Sdha and Sdhb [subunits of succinate dehydrogenase (complex II)], Uqcrc2 [subunit of ubiquinol-cytochrome c reductase (complex III)], Cox4 (subunit of complex IV), and Atp5a [subunit of H+-ATPase (complex V)] (3). As shown in Figure 5 and Table S4, there were varying levels of each subunit among the five tissues. Co1, Co2, Cox4, subunits of complex IV exhibited marked differences in the levels of these proteins among the five tissues, especially extremely high expression in the heart. The Nd1, Nd3, Ndufa9, and Ndufb8 subunits of complex I revealed relatively low variations in the levels of these proteins among these five tissues. As shown in Figure 5 and Table S4, the average levels of 13 subunits ranged from 0.41-fold (Co2) to 1.15-folds (Atp6) with average of 0.73-fold (relative to average of 13 subunits among five tissues) in the brain, 1.2-fold (Nd1) to 3.21-fold (Co1) with average of 1.99 fold in the heart, 0.09-fold (Cox4) to 0.73-fold (Sdhb) with the average of 0.49-fold in the liver, 0.57-fold (Co1) to 1.13-fold (Ndufb8) with average of 0.92-fold in the skeletal muscle, and 0.56-fold (Co1) to 1.12-fold (Nd1) with average of 0.88-fold in the kidney. Notably, the variations in the average levels of mtDNA encoding proteins among five tissues showed significant correlations with the corresponding variations in mtDNA copy numbers (r2 = 0.81, p = 0.039) or with mt-tRNA levels (r2 = 0.95, p = 0.048) (Fig. S1, C and D). These data suggested that the mtDNA contents and mt-tRNA levels may reflect the tissue-specific expressions of OXPHOS subunits.
Figure 5.
Western blotting analysis of mitochondrial proteins.A, eight micrograms of total cellular proteins from five mouse-derived tissue types was electrophoresed through a denaturing polyacrylamide gel, electroblotted, and corresponding Coomassie Brilliant Blue stained gel were used as loading control. The blots were hybridized with antibodies for 11 subunits of OXPHOS (six encoded by mtDNA and seven encoded by nDNA). B, quantification of OXPHOS subunits levels in the five tissues. The content of each proteins was normalized to that of total cellular protein in the five tissues. Relative protein levels for the brain, heart, liver, skeletal muscle, and kidney tissues were normalized to the mean values of each protein among the five tissues. Calculations were based on three independent determinations. Error bars indicate SD of the means.
Discussion
Mammalian tissues differ in many aspects in their metabolic profiles and energy demands during development, physiological adaptation, and pathology. In mammalian cell, mitochondria produce energy through the process of oxidative phosphorylation, and the extent of their occurrence in tissues varies depending on age, organ, and/or physiological condition (34). The copy number of mtDNA reflects the abundance of mitochondria within a cell and is regulated by transcriptional and translational factors (33, 34, 35). Essential for the synthesis of essential subunits of OXHPOS, mt-tRNAs play central roles in cellular functions in health and disease. However,the tissue-specific expression and physiological conditions of mammalian mt-tRNAs and the mechanisms underlying tissue-specific effects of the mt-tRNA mutations linked to clinical presentations are largely unknown. In this study, we demonstrated the striking differences in the tissue-specific expression of 22 mt-tRNAs in mouse-derived brain, heart, liver, skeletal muscle, and kidney tissues. In particular, the heart exhibited the highest levels of average 22 mt-tRNAs, the brain, skeletal muscle, and kidney revealed relatively higher levels of average 22 mt-tRNAs, and the liver exhibited markedly lower levels of average 22 mt-tRNAs. These variations in the levels of mt-tRNAs may reflect the mtDNA contents among these tissues, as the mtDNA contents of the heart were much higher than those in the brain, liver, skeletal muscle, and kidney (33). In particular, the variations in the levels of mt-tRNAs in the five tissues showed a significant correlation with the corresponding variations in mtDNA copy numbers. These results suggested that the differences of mtDNA copy number contributed to the variations of tRNA levels among these five tissues. Alternatively, these wide variations of tRNA abundances suggested that tRNAs may be dynamically related to specific activities, not just to general accumulation levels among these tissues. The tRNA, with traditional functions in translation, plays a role in the tissue-specific posttranscriptional regulations and translational heterogeneity via availability of certain tRNAs (20, 36). By contrast, the relatively mild differences in those cytosolic tRNAs (ct-tRNALys(TTT), ct-tRNALys(CTT), and ct-tRNAAsp(GTC)) among these tissues were comparable with those of previous studies (19).
There were 18 types of nucleotide modifications present in the 137 positions of human 22 mt-tRNAs (14). These nucleotide modifications included pseudouridine (Ψ) at position 55 at the TΨC loop, nucleotides at positions 34 and 37 at the anticodon loop of tRNAs (4, 14, 37). Posttranscriptional modifications of tRNAs affect all aspects of tRNA structure and function (38). Using APM gel, we focused on the analysis of the levels of 2-thiouridine modification at position 34 in mitochondrial tRNALys, tRNAGln, and tRNAGlu among five mouse-derived tissues. There were no significant differences in the τm5s2U levels of tRNALys, tRNAGlu, and tRNAGln among these five tissues. These suggested that there may be no tissue-specific τm5s2U modifications of tRNAs among these tissues, in contrast with the marked variations in the steady-state levels of tRNAs among the mouse organs represented, as described above.
Aminoacylation, the attachment of an amino acid to an mt-tRNA, is catalyzed by mitochondrial aminoacyl-tRNA synthetases (4, 30). During protein synthesis, charged mt-tRNAs deliver amino acids to translating ribosomes and are then recharged by tRNA synthetases. Notably, the Wars2 and Dars2 mutant mice displayed tissue-specific effects of mitochondrial deficiencies (22, 39). In this study, we examined the aminoacylation levels of 15 mt-tRNAs including tRNAMet, tRNALeu(UUR), tRNALeu(CUN), tRNAIle, and tRNAHis among the five mouse-derived tissues. We demonstrated wide ranges in aminoacylation levels between each mt-tRNA type among these five tissues and of these 15 tRNAs in the same tissue. In fact, 15 tRNAs including tRNALeu(UUR), tRNALeu(CUN), tRNAGly, tRNAPro, and tRNATyr displayed striking differences in aminoacylation levels among these tissues. In particular, the average aminoacylation levels of tRNAPro among the brain, heart, liver, skeletal muscle, and kidney were 15.2%, 11.7%, 55.8%, 68.9%, and 11.2%, respectively. In contrast, tRNAHis, tRNAThr, and tRNAPhe revealed relatively low discrepancies of aminoacylation levels among these tissues. Especially, the average aminoacylation levels of tRNAThr among the brain, heart, liver, skeletal muscle, and kidney were 60.3%, 62.8%, 65.9%, 67.1%, and 50.8%, respectively. Furthermore, the overall average aminoacylation levels of the 15 mt-tRNAs in the brain, heart, liver, skeletal muscle, and kidney were 51%, 59.3%, 69.1%, 77.9%, and 40.3%, respectively. However, such variations in aminoacylation levels of the 15 mt-tRNAs were not significantly correlated with the steady-state levels of these tRNAs among five tissues and may instead reflect the translation efficiencies for the heterogeneity of protein synthesis among tissues. Furthermore, we measured the mRNA expression levels of the 14 mt-aaRSs encoding genes among the five tissues. Marked differences in the levels of each genes were observed among the five tissues, the average expression levels of these genes among the brain, heart, liver, skeletal muscle, and kidney being 1.14, 1.18, 0.51, 0.64, and 1.54-fold, respectively, relative to average levels of these five tissues combined. Remarkably, the variations in the aminoacylation levels of mt-tRNAs among the five tissues exhibited negative correlations with the corresponding variations in their mt-aaRS expression levels. These may be due to a negative feedback mechanism that matches translational demand for aminoacylated mt-tRNA to the aaRS recharging capacity (40). In fact, a reduced abundance of mt-aaRS could interfere with the normal ability of the synthetase to sequester the uncharged tRNA pool. Alternatively, T-box riboswitches in mitochondria may regulate the expression of aminoacyl-tRNA synthetases and other proteins in response to fluctuating transfer RNA aminoacylation levels under various nutritional or physiological states (41).
Thirteen polypeptides synthesized by mitochondrial translations were the essential subunits of OXPHOS machinery, while other 77 OXPHOS subunits were encoded by nuclear genes and synthesized in cytosol and then imported into mitochondria (3). The specific oxidative phosphorylation capacities of cells or tissues depend on their metabolic and energetic demands (42, 43). To assess the tissue-specific impacts of mitochondrial tRNA metabolism on translation and OXPHOS biogenesis across tissues, we examined the levels of six mtDNA encoding OXPHOS subunits (Nd1, Nd3, Co1, Co2, Atp6, and Atp8) and seven nucleus encoding OXPHOS subunits (Ndufa9, Ndufb8, Sdha, Sdhb, Uqcrc2, Cox4, and Atp5a), noting the various levels of these 13 proteins according to tissue type. All of these OXPHOS subunits were expressed at much higher levels in the heart than in any other tissues. The average levels of 13 OXPHOS subunits ranged from 0.73-fold in the brain, 2.04-fold in the heart, 0.47-fold in the liver, 0.9-fold in the skeletal muscle, and 0.86-fold in the kidney. Furthermore, we revealed the various levels of each subunit among the five tissues. The Co1, Co2, and Cox4 subunits of complexes IV exhibited marked differences in the levels of these proteins among the five tissues, with especially high expressions in the heart. Conversely, the Nd1, Nd3, Ndufa9, and Ndufb8 subunits of complex I revealed relatively low variations in the levels of these proteins among these five tissues. Notably, the variations in levels of these proteins were significantly correlated with the steady levels of mitochondrial tRNAs (r2 = 0.9498, p = 0.0048) but not with the aminoacylation levels of these tRNAs among these tissues. These various expressions of these OXPHOS subunits may reflect the relative functional emphasis of mitochondria among different tissues (44). Mitochondria in the brain are specialized to the neurotransmitter metabolism, while in the heart mitochondria are specialized for constant and effective production of ATP by the means of β-oxidation (45). These are also inherently different from mitochondria in the liver and kidney, which are more specialized for the various reactions of anabolic and catabolic metabolism (45). Therefore, the tissue-specific metabolic aspects of mitochondrial tRNA and related expression levels of these OXPHOS subunits are accounted for by these tissue-specific requirements and facilitated by their specialized mitochondrial features in different tissues.
In summary, we demonstrated the tissue-specific metabolisms of 22 mt-tRNAs. We showed that the steady-state levels of 22 tRNAs varied as much as tenfold among these tissues. The heart, in particular, exhibited the highest levels of tRNAs. There were no significant differences in the 2-thiouridylation levels of tRNALys, tRNAGlu, and tRNAGln among these tissues. We demonstrated a wide range of aminoacylation levels of each tRNA among these tissues, with skeletal muscle revealing the highest aminoacylation levels of tRNAs. Aminoacylation levels of mt-tRNAs exhibited negative correlations with the variations in mitochondrial tRNA synthetase expressions among these tissues, while the variations of tRNA abundances were significantly correlated with the levels of OXPHOS subunits. Our findings may provide new insights into the mechanism of mt-tRNA tissue-specific effects on physiological conditions.
Experimental procedures
Mice
All animal care protocols were approved by the Animal Care and Use Committee of Zhejiang University. C57BL/6 mice were purchased from Shanghai Slac Animal Inc. Mice were housed in a 12-h light/dark cycle and provided food and water ad libitum in the Experimental Animal Center of Zhejiang University. The 8∼12-week-old male or virgin female mice were sacrificed by cervical dislocation. For liver samples, only the largest lobe of the mouse liver was used. Whole organs were then dissected in the following order: brain, heart, kidney, and skeletal muscle (tibialis anterior).
Mitochondrial tRNA analysis
Total RNAs from various mice tissues were obtained by using TOTALLY RNA kit (Ambion), as detailed elsewhere (46). For tRNA Northern blot analysis, 4 μg of total RNAs was electrophoresed through a 10% polyacrylamide/8 M urea gel in 1×TBE buffer after heating the samples at 95 °C for 5 min and then electroblotted onto a positively charged nylon membrane (Millipore) for hybridization analysis with the respective DIG-labeled oligodeoxynucleotide probes. The set of DIG-labeled probes of 22 mt-tRNAs, 5S rRNA, and ct-tRNALys(TTT), tRNALys(CTT), and tRNAAsp(GTC) used for this study were listed in the Table S5 (47). The specificities of these tRNA probes were analyzed using nucleotide BLAST (www.ncbi.nlm.nih.gov/BLAST/). The hybridization and quantification of density for each band were performed as detailed previously (25, 48).
The aminoacylation assays were carried out as detailed previously (18, 25). To further distinguish nonaminoacylated tRNA from aminoacylated tRNA, total RNAs were heated for 10 min at 60 °C at pH 9.0 and then run in parallel (18, 25). DIG-labeled oligodeoxynucleotide probes for mt-tRNAs and ct-tRNAs were as described above. The quantification for density in each band was performed as detailed previously (18, 25).
Thiouridine modification in the tRNAs was analyzed using the retardation of electrophoretic mobility in a polyacrylamide gel containing 0.05 mg/ml APM (27, 28, 29). Total RNAs were separated using polyacrylamide gel electrophoresis and blotted onto positively charged membrane (Roche Applied Science). Each tRNA was detected with the specific DIG-oligodeoxynucleoside probe at the 3′ termini as detailed elsewhere (29, 49). Oligonucleotide probes for mitochondrial tRNALys, tRNAGlu, tRNAGln, and tRNALeu(UUR) were described as above. DIG-labeled oligodeoxynucleosides were generated by using the DIG Oligonucleoside Tailing Kit (Roche). APM gel electrophoresis and quantification of 2-thiouridine modification in tRNAs were conducted as detailed (27, 49).
Western blotting analysis
Western blotting analysis was performed as detailed previously (9, 25, 50). Twenty micrograms of total cell proteins obtained from each of the various tissues was denatured and loaded on sodium dodecyl sulfate (SDS) polyacrylamide gels. The gels were electroblotted onto a polyvinylidene difluoride (PVDF) membrane for hybridization. The antibodies obtained from different companies were as follows: Abcam [Nd1 (ab74257), Co1 (ab14705), Co2 (ab198286), Ndufb8 (ab110242), Ndufa9 (ab14713), Sdhb (ab14714), Uqcrc2 (ab14745), and Atp5a (ab14748)], Diagbio [Cox4 (db15)] and Proteintech [Atp6 (55313-1-AP), Atp8 (26723-1-AP), and Sdha (14865-1-AP)]. Peroxidase Affini Pure goat anti-mouse IgG and goat anti-rabbit IgG (Jackson) were used as secondary antibodies and protein signals were detected using the ECL system (CWBIO). Quantification of the density of each band was performed as detailed previously (9, 25, 49, 50).
Measurement of mtDNA/nDNA
Total DNA was isolated from each of the various mouse tissues using a Tissue gDNA kit (Biomiga) according to the manufacturer’s protocol. Primers for mouse mtDNA (Mito) and mouse nDNA (B2M) were used to amplify the respective products from mouse genomic DNA (Table S6). MtDNA content was determined by comparing the ratio of mtDNA to nDNA by real-time quantitative PCR (33). The relative ratio was analyzed on a 7900HT system (Applied Biosystems) using FastStart Universal SYBR Green Master Mix (Roche Diagnostics GmbH).
Gene expression analysis of 14 genes encoding mt-aaRSs
Total RNA was extracted from each of the various mouse using TRIzol reagent (Ambion) and reverse transcripted into cDNA using PrimeScript II first Strand cDNA Synthesis Kit (Takara). Quantitative PCR was performed on the Applied Biosystems 7900HT Fast Real-Time PCR System. Data were analyzed using the 7900 System SDS RQ Manager Software, and the gene expression levels of the 14 genes encoding mt-aaRSs were determined using the 2−ΔΔCt method using nucleus encoded 18S rRNA as normalization. Primer sequences for this analysis were listed in Table S6.
Statistical analysis
All statistical analysis was performed using unpaired, two-tailed Student’s t test contained in the Graphpad prism 8 program (Graphpad software) and Microsoft-Excel program (version 2016). A p value <0.05 was considered statistically significant.
Data availability
Representative experiments are shown in the figures and supplemental materials. For any additional information, please contact the corresponding author.
Supporting information
This article contains supporting information.
Conflict of interest
All authors declare that they have no conflict of interest with contents of this article.
Acknowledgments
Author contributions
M.-X. G. and Y. C. conceptualization; Q. H., X. H., Y. X., Q. Z., Z. Y., L. C., and M.-X. G. data curation; Q. H. formal analysis; M.-X. G. funding acquisition; Q. H., X. H., and M.-X. G. investigation; Q. H., X. H., and Y. X. methodology; M.-X. G. project administration; M.-X. G. resources; Y. C. and M.-X. G. supervision; Q. H. and Q. Z. validation; Q. H. and Y. C. writing-original draft; M.-X. G. writing-review and editing.
Funding and additional information
This research was supported by grants from Grant 2018C03026 from Ministry of Science and Technology of Zhejiang Province, the National Key Technologies R&D Program Grant 2018YFC1004802 from the Ministry of Science and Technology of China (to M.-X. G.), 82030028 (M.-X. G.), and 31671305 (Y. C.) from the National Natural Science Foundation of China.
Edited by Karin Musier-Forsyth
Contributor Information
Ye Chen, Email: yechency@zju.edu.cn.
Min-Xin Guan, Email: gminxin88@zju.edu.cn.
Supporting information
References
- 1.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., Schreier P.H., Smith A.J., Staden R., Younge I.G. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Bibb M.J., Van Etten R.A., Wright C.T., Walberg M.W., Clayton D.A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 3.Wallace D.C. A mitochondrial bioenergetic etiology of disease. J. Clin. Invest. 2013;123:1405–1412. doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki T., Nagao A., Suzuki T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 5.Montoya J., Gaines G.L., Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 6.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 7.Mercer T.R., Neph S., Dinger M.E., Crawford J., Smith M.A., Shearwood A.M., Haugen E., Bracken C.P., Rackham O., Stamatoyannopoulos J.A., Filipovska A., Mattick J.S. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 9.Xiao Y., Wang M., He Q., Xu L., Zhang Q., Meng F., Jia Z., Zhang F., Wang H., Guan M.X. Asymmetrical effects of deafness-associated mitochondrial DNA 7516delA mutation on the processing of RNAs in the H-strand and L-strand polycistronic transcripts. Nucleic Acids Res. 2020;48:11113–11129. doi: 10.1093/nar/gkaa860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X., Cui L., Xiao Y., Mao Q., Aishanjiang M., Kong W., Liu Y., Chen H., Hong F., Jia Z., Wang M., Jiang P., Guan M.X. Hypertension-associated mitochondrial DNA 4401A>G mutation caused the aberrant processing of tRNAMet, all 8 tRNAs and ND6 mRNA in the light-strand transcript. Nucleic Acids Res. 2019;47:10340–10356. doi: 10.1093/nar/gkz742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez M.I., Mercer T.R., Davies S.M., Shearwood A.M., Nygard K.K., Richman T.R., Mattick J.S., Rackham O., Filipovska A. RNA processing in human mitochondria. Cell Cycle. 2011;10:2904–2916. doi: 10.4161/cc.10.17.17060. [DOI] [PubMed] [Google Scholar]
- 12.Brzezniak L.K., Bijata M., Szczesny R.J., Stepien P.P. Involvement of human ELAC2 gene product in 3' end processing of mitochondrial tRNAs. RNA Biol. 2011;8:616–626. doi: 10.4161/rna.8.4.15393. [DOI] [PubMed] [Google Scholar]
- 13.Holzmann J., Frank P., Loffler E., Bennett K.L., Gerner C., Rossmanith W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T., Yashiro Y., Kikuchi I., Ishigami Y., Saito H., Matsuzawa I., Okada S., Mito M., Iwasaki S., Ma D., Zhao X., Asano K., Lin H., Kirino Y., Sakaguchi Y. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020;11:4269. doi: 10.1038/s41467-020-18068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng F., Zhou M., Xiao Y., Mao X., Zheng J., Lin J., Lin T., Ye Z., Cang X., Fu Y., Wang M., Guan M.X. A deafness-associated tRNA mutation caused pleiotropic effects on the m1G37 modification, processing, stability and aminoacylation of tRNAIle and mitochondrial translation. Nucleic Acids Res. 2021;49:1075–1093. doi: 10.1093/nar/gkaa1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Y.M. CCA addition to tRNA: Implications for tRNA quality control. IUBMB Life. 2010;62:251–260. doi: 10.1002/iub.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sissler M., González-Serrano L.E., Westhof E. Recent advances in mitochondrial aminoacyl-tRNA synthetases and disease. Trends Mol. Med. 2017;23:693–708. doi: 10.1016/j.molmed.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Enriquez J.A., Attardi G. Analysis of aminoacylation of human mitochondrial tRNAs. Methods Enzymol. 1996;264:183–196. doi: 10.1016/s0076-6879(96)64019-1. [DOI] [PubMed] [Google Scholar]
- 19.Torres A.G., Reina O., Stephan-Otto Attolini C., Ribas de Pouplana L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. U. S. A. 2019;116:8451–8456. doi: 10.1073/pnas.1821120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dittmar K.A., Goodenbour J.M., Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lant J.T., Berg M.D., Heinemann I.U., Brandl C.J., O'Donoghue P. Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 2019;294:5294–5308. doi: 10.1074/jbc.REV118.002982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agnew T., Goldsworthy M., Aguilar C., Morgan A., Simon M., Hilton H., Esapa C., Wu Y., Cater H., Bentley L., Scudamore C., Poulton J., Morten K.J., Thompson K., He L. A Wars2 mutant mouse model displays OXPHOS deficiencies and activation of tissue-specific stress response pathways. Cell Rep. 2018;25:3315–3328. doi: 10.1016/j.celrep.2018.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., Gygi S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchner S., Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 25.Zhou M., Xue L., Chen Y., Li H., He Q., Wang B., Meng F., Wang M., Guan M.X. A hypertension-associated mitochondrial DNA mutation introduces an m1G37 modification into tRNAMet, altering its structure and function. J. Biol. Chem. 2018;293:1425–1438. doi: 10.1074/jbc.RA117.000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M., Peng Y., Zheng J., Zheng B., Jin X., Liu H., Wang Y., Tang X., Huang T., Jiang P., Guan M.X. A deafness-associated tRNAAsp mutation alters the m1G37 modification, aminoacylation and stability of tRNAAsp and mitochondrial function. Nucleic Acids Res. 2016;44:10974–10985. doi: 10.1093/nar/gkw726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng F., Cang X., Peng Y., Li R., Zhang Z., Li F., Fan Q., Guan A.S., Fischel-Ghosian N., Zhao X., Guan M.X. Biochemical evidence for a nuclear modifier allele (A10S) in TRMU (methylaminomethyl-2-thiouridylate-methyltransferase) related to mitochondrial tRNA modification in the phenotypic manifestation of deafness-associated 12S rRNA mutation. J. Biol. Chem. 2017;292:2881–2892. doi: 10.1074/jbc.M116.749374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q., Zhang L., Chen D., He X., Yao S., Zhang Z., Chen Y., Guan M.X. Deletion of Mtu1 (Trmu) in zebrafish revealed the essential role of tRNA modification in mitochondrial biogenesis and hearing function. Nucleic Acids Res. 2018;46:10930–10945. doi: 10.1093/nar/gky758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K., Suzuki T. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. [DOI] [PubMed] [Google Scholar]
- 30.Enriquez J.A., Chomyn A., Attardi G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNALys and premature translation termination. Nat. Genet. 1995;10:47–55. doi: 10.1038/ng0595-47. [DOI] [PubMed] [Google Scholar]
- 31.Magalhaes P.J., Andreu A.L., Schon E.A. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol. Biol. Cell. 1998;9:2375–2382. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jühling F., Mörl M., Hartmann R.K., Sprinzl M., Stadler P.F., Pütz J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuyama M., Iida R., Takatsuka H., Yasuda T., Matsuki T. Quantitative change in mitochondrial DNA content in various mouse tissues during aging. Biochim. Biophys. Acta. 2005;1723:302–308. doi: 10.1016/j.bbagen.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Muir R., Diot A., Poulton J. Mitochondrial content is central to nuclear gene expression: Profound implications for human health. Mol. Cell. Dev. Biol. 2016;38:50–156. doi: 10.1002/bies.201500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barazzoni R., Short K.R., Nair K.S. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J. Biol. Chem. 2000;275:3343–3347. doi: 10.1074/jbc.275.5.3343. [DOI] [PubMed] [Google Scholar]
- 36.Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M., Liu H., Zheng J., Chen B., Zhou M., Fan W., Wang H., Liang X., Zhou X., Eriani G., Jiang P., Guan M.X. A deafness and diabetes associated tRNA mutation caused the deficient pseudouridinylation at position 55 in tRNAGlu and mitochondrial dysfunction. J. Biol. Chem. 2016;291:21029–21041. doi: 10.1074/jbc.M116.739482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Yacoubi B., Bailly M., de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 39.Dogan S.A., Pujol C., Maiti P., Kukat A., Wang S., Hermans S., Senft K., Wibom R., Rugarli E.I., Trifunovic A. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab. 2014;19:458–469. doi: 10.1016/j.cmet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 40.McFarland M.R., Keller C.D., Childers B.M., Adeniyi S.A., Corrigall H., Raguin A., Romano M.C., Stansfield I. The molecular aetiology of tRNA synthetase depletion: Induction of a GCN4 amino acid starvation response despite homeostatic maintenance of charged tRNA levels. Nucleic Acids Res. 2020;48:3071–3088. doi: 10.1093/nar/gkaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J. Unboxing the T-box riboswitches-A glimpse into multivalent and multimodal RNA-RNA interactions. Wiley Interdiscip. Rev. RNA. 2020;11 doi: 10.1002/wrna.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McManus M.J., Picard M., Chen H.W., De Haas H.J., Potluri P., Leipzig J., Towheed A., Angelin A., Sengupta P., Morrow R.M., Kauffman B.A., Vermulst M., Narula J., Wallace D.C. Mitochondrial DNA variation dictates expressivity and progression of nuclear DNA mutations causing cardiomyopathy. Cell Metab. 2019;29:78–90. doi: 10.1016/j.cmet.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbers E., Kekäläinen N.J., Hangas A., Pohjoismäki J.L., Goffart S. Tissue specific differences in mitochondrial DNA maintenance and expression. Mitochondrion. 2019;44:85–92. doi: 10.1016/j.mito.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Johnson D.T., Harris R.A., French S., Blair P.V., You J., Bemis K.G., Wang M., Balaban R.S. Tissue heterogeneity of the mammalian mitochondrial proteome. Am. J. Phys. Cell Phys. 2007;292:C689–C697. doi: 10.1152/ajpcell.00108.2006. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez-Vizarra E., Enriquez J.A., Perez-Martos A., Montoya J., Fernandez-Silva P. Tissue-specific differences in mitochondrial activity and biogenesis. Mitochondrion. 2011;11:207–213. doi: 10.1016/j.mito.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 46.King M.P., Attardi G. Post-transcriptional regulation of the steady-state levels of mitochondrial tRNAs in HeLa cells. J. Biol. Chem. 1993;268:10228–10237. [PubMed] [Google Scholar]
- 47.Bayona-Bafaluy M.P., Acín-Pérez R., Mullikin J.C., Park J.S., Moreno-Loshuertos R., Hu P., Pérez-Martos A., Fernández-Silva P., Bai Y., Enríquez J.A. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji Y., Nie Z., Meng F., Hu C., Chen H., Jin L., Chen M., Zhang M., Zhang J., Liang M., Wang M., Guan M.X. Mechanistic insights into mitochondrial tRNAAla 3'-end metabolism deficiency. J. Biol. Chem. 2021;297:100816. doi: 10.1016/j.jbc.2021.100816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan M.X., Yan Q., Li X., Bykhovskaya Y., Gallo-Teran J., Hajek P., Umeda N., Zhao H., Garrido G., Mengesha E., Suzuki T., del Castillo I., Peters J.L., Li R., Qian Y. Mutation in TRMU related to transfer RNA modification modulates the phenotypic expression of the deafness-associated mitochondrial 12S ribosomal RNA mutations. Am. J. Hum. Genet. 2006;79:291–302. doi: 10.1086/506389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu J., Liang X., Ji Y., Ai C., Liu J., Zhu L., Nie Z., Jin X., Wang C., Zhang J., Zhao F., Mei S., Zhao X., Zhou X., Zhang M. PRICKLE3 linked to ATPase biogenesis manifested Leber's hereditary optic neuropathy. J. Clin. Invest. 2020;130:4935–4946. doi: 10.1172/JCI134965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Representative experiments are shown in the figures and supplemental materials. For any additional information, please contact the corresponding author.