Graphical abstract
Keywords: COVID-19, Lung infection segmentation, Transfer learning, Computed tomography
Abstract
With the global outbreak of COVID-19 in early 2020, rapid diagnosis of COVID-19 has become the urgent need to control the spread of the epidemic. In clinical settings, lung infection segmentation from computed tomography (CT) images can provide vital information for the quantification and diagnosis of COVID-19. However, accurate infection segmentation is a challenging task due to (i) the low boundary contrast between infections and the surroundings, (ii) large variations of infection regions, and, most importantly, (iii) the shortage of large-scale annotated data. To address these issues, we propose a novel two-stage cross-domain transfer learning framework for the accurate segmentation of COVID-19 lung infections from CT images. Our framework consists of two major technical innovations, including an effective infection segmentation deep learning model, called nCoVSegNet, and a novel two-stage transfer learning strategy. Specifically, our nCoVSegNet conducts effective infection segmentation by taking advantage of attention-aware feature fusion and large receptive fields, aiming to resolve the issues related to low boundary contrast and large infection variations. To alleviate the shortage of the data, the nCoVSegNet is pre-trained using a two-stage cross-domain transfer learning strategy, which makes full use of the knowledge from natural images (i.e., ImageNet) and medical images (i.e., LIDC-IDRI) to boost the final training on CT images with COVID-19 infections. Extensive experiments demonstrate that our framework achieves superior segmentation accuracy and outperforms the cutting-edge models, both quantitatively and qualitatively.
1. Introduction
The outbreak of the 2019 coronavirus disease (COVID-19) has triggered a global public health emergency (Lancet, 2020). There are a total of 165,711,111 confirmed cases and 3,528,951 confirmed deaths worldwide as of May 20th, 2021. The COVID-19 pandemic has caused unprecedented hazards to public health, the global economy, and so on (Xiong et al., 2020). In this severe situation, rapid control of the spread of COVID-19 becomes particularly important.
Early diagnosis of COVID-19 plays a vital role in controlling the spread of the disease. Currently, reverse transcription-polymerase chain reaction (RT-PCR) is the most widely adopted approach for the diagnosis of COVID-19 (Guan et al., 2020). However, RT-PCR suffers from a number of limitations, including low efficiency, short of supply test kits, and low sensitivity (Fang, Zhang, Xie, Lin, Ying, Pang, Ji, 2020, Xie, Zhong, Zhao, Zheng, Wang, Liu, 2020). Compared with RT-PCR, chest computed tomography (CT) imaging allows effective COVID-19 screening with high sensitivity and is easy to access in a clinical setting (Xie et al., 2020). Besides, CT imaging gained increasing attention from the research community (Phelan et al., 2020), where efforts have been directed to investigate the COVID-19 induced pathological changes from the perspective of radiology.
Accurate segmentation of lung infections from CT images is crucial to the quantification and diagnosis of COVID-19 (Shan, Gao, Wang, Shi, Shi, Han, Xue, Shi, Shi, Wang, Shi, Wu, Wang, Tang, He, Shi, Shen, 2020, Tilborghs, Dirks, Fidon, Willems, Eelbode, Bertels, Ilsen, Brys, Dubbeldam, Buls, et al.). Traditional manual/semi-automatic segmentation techniques are time-consuming and require the intervention of clinical physicians. In addition, the segmentation results tend to be biased towards the expert’s experience. Therefore, automatic lung infection segmentation is greatly desired in a clinical setting. Significant efforts have been directed towards this direction (Vaishya et al., 2020). In particular, deep learning techniques have been widely employed and shown great potentials. For instance, Shan et al. (2020) proposed a deep learning model, called VB-Net, to segment lung lobes and lung infections from the CT scans of COVID-19 patients. In addition, a human-in-the-loop strategy is employed to refine the annotation of each CT scan. Elharrouss et al. (2020) developed a multi-task deep learning framework for lung infection segmentation from CT images. Qiu et al. (2020) proposed a lightweight deep learning model, called MiniSeg, for COVID-19 infection segmentation, aiming to resolve the issues of over-fitting and low computational efficiency.
However, accurate COVID-19 lung infection segmentation is still a challenging task due to three key factors, including (i) Low boundary contrast. The boundary between the COVID-19 infected regions and surrounding normal tissues suffers from the low contrast issue and is usually blurry (Fan et al., 2020). This induces significant difficulties for accurate lung infection segmentation. (ii) Large variation. The COVID-19 lung infection exhibits a large variety of morphological appearances, e.g., size, shape, etc., which aggregates the difficulty of accurate segmentation. Most importantly, (iii) Short of labeled data. Large-scale infection annotations provided by clinical doctors are extremely difficult to obtain, especially at an early stage of the disease outbreak. This is a major issue restricting the performance of deep learning segmentation models that rely on sufficient training data. To handle the low boundary contrast and large infection variation, a large receptive field is greatly desired since it can provide rich contextual information. In addition, the fusion of multi-level features is another key factor determining the success of infection segmentation. However, existing works usually overlook the importance of these two factors, which can result in unsatisfactory performance.
To tackle the shortage of labeled data, transfer learning has been adopted and gained increasing interest from the medical image analysis community (Shie, Chuang, Chou, Wu, Chang, 2015, Shin, Roth, Gao, Lu, Xu, Nogues, Yao, Mollura, Summers, 2016, Cheplygina, de Bruijne, Pluim, 2019). In general, there are two kinds of widely adopted transfer learning strategies: (i) Network Backbone. A network trained with large-scale datasets (e.g., ImageNet (Deng et al., 2009)) can be embedded into the medical image analysis models as a backbone for exacting informative features (Carneiro et al., 2015). (ii) Network Pre-training. Methods in this category pre-train the whole network using large-scale datasets, and then perform formal training with the target dataset (Shin, Roth, Gao, Lu, Xu, Nogues, Yao, Mollura, Summers, 2016, Chatfield, Simonyan, Vedaldi, Zisserman). Both strategies have shown promising performance in medical image analysis tasks. However, existing works usually focus on one aspect of these two kinds of strategies, which is unable to make full use of powerful transfer learning.
In this paper, we propose a novel two-stage cross-domain transfer learning framework for the accurate segmentation of COVID-19 lung infections from CT images. Our framework is based on a specially designed deep learning model, called nCoVSegNet, which segments lung infections with attention-aware feature fusion and large receptive fields. Specifically, we first feed the CT images to a backbone network to extract multi-level features. The features are then passed through our global context-aware (GCA) modules, which provide rich features from significantly enlarged receptive fields. Finally, we fuse the features using our dual-attention fusion (DAF) modules, which integrate the multi-level features with the guidance from spatial and channel attention mechanisms. Furthermore, we train our model with a two-stage transfer learning strategy for improved performance. The first stage takes advantage of a backbone network trained on ImageNet (Deng et al., 2009) and provides valuable cross-domain knowledge from the human perception of natural images. Since a large gap exists between natural images and COVID-19 CT images, we further perform a second stage transfer learning, where our model is pre-trained using LIDC-IDRI (Armato III et al., 2011), which is currently the largest CT dataset for pulmonary nodule detection and provides a large amount of chest CT images that share similar appearances with the COVID-19 CT images. The second stage transfer learning can fill the gap between two domains and provides vital knowledge from a neighboring domain to improve the segmentation accuracy. Finally, we train our model with COVID-19 CT images from the MosMedData (Morozov et al., 2020). Extensive experiments demonstrate the effectiveness of our nCoVSegNet and the two-stage transfer learning strategy, both quantitatively and qualitatively.
Our main contributions are summarized as follows:
-
1.
We develop a novel two-stage transfer learning framework for segmenting COVID-19 lung infections from CT images. Our framework learns valuable knowledge from both natural images and CT images with pulmonary nodules, allowing more effective network training for improved performance.
-
2.
We propose an effective infection segmentation network, nCoVSegNet, which consists of a backbone network along with our GCA and DAF modules. Our nCoVSegNet accurately segments lung infections from CT images by taking advantages of attention-aware feature fusion and large reception fields.
-
3.
Extensive experiments on two COVID-19 CT datasets demonstrate that our framework is able to segment lung infections accurately and outperforms state-of-the-art methods remarkably. Our framework, which provides vital information regarding lung infections, has great potentials to boost the clinical diagnosis and treatment of COVID-19.
Our paper is organized as follows: In Section 2, we introduce related works. Section 3 describes our COVID-19 lung infection segmentation framework in detail. In Section 4, we present the datasets and experimental results. Finally, we conclude our work in Section 5.
2. Related work
In this section, we review related works from two categories, including COVID-19 lung infection segmentation and transfer learning in medical imaging.
2.1. COVID-19 lung infection segmentation
Recently, deep learning has been actively employed for COVID-19 lung infection segmentation. Among various network architectures, U-Net (Ronneberger et al., 2015) is a popular network backbone in a large number of works. For instance, Müller et al. (2020) employed 3D U-Net for the segmentation of lungs and COVID-19 infected regions. Saeedizadeh et al. (2021) proposed TV-U-Net to promote connectivity of the segmentation map by adding a connectivity regularization term in the loss function. Meanwhile, attention mechanisms have also been incorporated in the encoder and/or decoder of the networks for COVID-19 infection segmentation (Zhou, Canu, Ruan, 2021, Chen, Yao, Zhang, Zhao, Li, Li, Chen, Zhao, Xingzhi, Liu, Zhao, 2020, Wu, Gao, Mei, Xu, Fan, Zhao, Cheng, 2021, Yan, Wang, Gong, Luo, Zhao, Shen, Shi, Jin, Zhang, You). A novel noise-robust framework (Wang et al., 2020a) was also proposed for COVID-19 pneumonia lesion segmentation, where a noise-robust Dice loss and a mean absolute error loss were used. The success of deep learning models relies on a large amount of labeled training data, which is, however, hard to guarantee for the COVID-19 infection segmentation, especially at the early break of the disease. To this end, effort has been dedicated to semi-supervised (Fan et al., 2020) and weakly-supervised (Xu et al., 2020) COVID-19 infection segmentation. In addition, Zhou et al. (2020a) resolved the data scarcity issue by fitting the dynamic change of real patients’ data measured at different time points. To alleviate the burden of data annotation, a label-free model was proposed in (Yao et al., 2021), where synthesized infections were embedded into normal lung CT scans for training the infection segmentation network. Bressem et al. (2021) proposed a transfer learning segmentation framework, where a 3D U-Net with an encoder pre-trained on Kinetics-400 dataset was employed for COVID-19 infection segmentation.
Different from existing works (Bressem, Niehues, Hamm, Makowski, Vahldiek, Adams, Zhou, Canu, Ruan, 2021, Fan, Zhou, Ji, Zhou, Chen, Fu, Shen, Shao, 2020), we propose a novel network with consideration of attention-aware feature fusion and enlarged receptive fields. In addition, we address the shortage of training data with an effective two-stage cross-domain transfer learning strategy.
2.2. Transfer learning in medical imaging
Transfer learning is particularly effective in resolving the shortage of training data for the deep learning models that are designed for medical image analysis tasks. In general, we classify the existing works into two categories. The first category is similar to our first-stage transfer learning, where a convolutional neural network (CNN) (e.g., VGGNet (Simonyan and Zisserman, 2015) and ResNet (He et al., 2016)) that has been pre-trained on a large-scale dataset (e.g., ImageNet (Deng et al., 2009)) is utilized as the backbone of a network for feature extraction. This strategy has been widely employed for a variety of medical image analysis tasks, such as lesion detection and classification (Byra, Galperin, Ojeda-Fournier, Olson, O’Boyle, Comstock, Andre, 2019, Khan, Islam, Jan, Din, Rodrigues, 2019), normal/abnormal tissue segmentation (Vu, Siddiqui, Zamdborg, Thompson, Quinn, Castillo, Guerrero, 2020, Bressem, Niehues, Hamm, Makowski, Vahldiek, Adams), disease identification (Bar, Diamant, Wolf, Greenspan, 2015, Shie, Chuang, Chou, Wu, Chang, 2015), etc. The other strategy is to pre-train the network with a large-scale dataset before the formal training using the target dataset with limited data. This strategy has been employed in brain tumor retrieval (Swati et al., 2019) and various disease diagnosis tasks (Tajbakhsh, Shin, Gurudu, Hurst, Kendall, Gotway, Liang, 2016, Liang, Zheng, 2020). In particular, Li et al. (2017) employed three different transfer learning strategies to conduct diabetic retinopathy fundus image classification, demonstrating that transfer learning is a promising technique for alleviating the shortage of training data.
In our work, we address the shortage of training data using a two-stage transfer learning strategy, which takes advantage of both model-level transfer learning (i.e., network backbone) and data-level transfer learning (i.e., network pre-training).
3. Method
In this section, we provide the details of our framework by first presenting the proposed segmentation model, nCoVSegNet, and then explaining the two-stage transfer learning strategy.
3.1. nCoVSegNet
Our segmentation model mainly consists of a backbone network and two key modules, i.e., the global context-aware (GCA) module and dual-attention fusion (DAF) module. The backbone network extracts multi-level features from the input CT images. Then, the GCA modules enhance the features before feeding them to the DAF modules for predicting the segmentation maps.
3.1.1. Network architecture
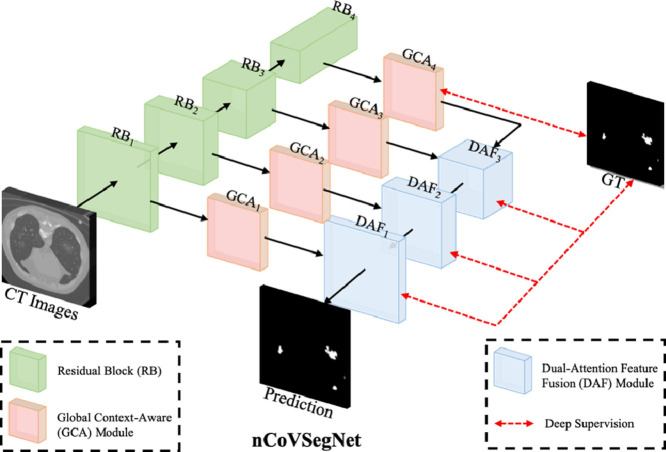
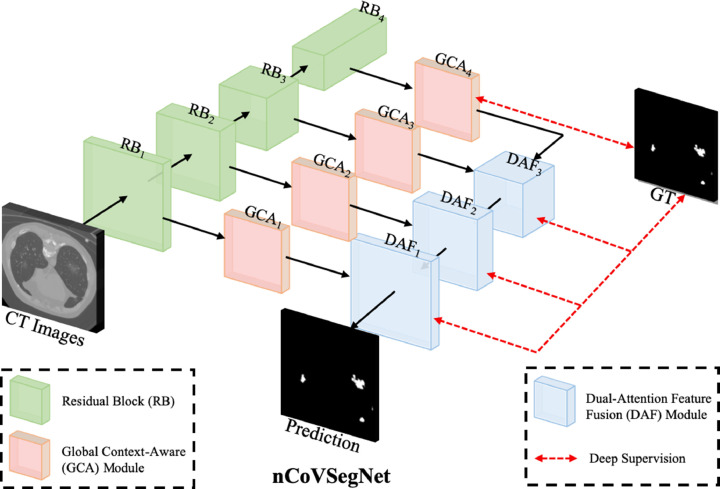
As shown in Fig. 1 , the multi-level features are first extracted from the hierarchical layers of the backbone network. Both of the low-level and high-level features are then fed to GCA modules for enhancement by enlarging the receptive fields. Note that the low-/high-level features denote the features closer to the beginning/end (i.e., input/output) of a backbone network (Lin, Dollár, Girshick, He, Hariharan, Belongie, 2017, He, Zhang, Ren, Sun, 2016). We then employ three DAF modules to perform feature fusion for predicting the segmentation maps. Furthermore, we employ a deep supervision strategy to supervise the outputs of three DAF modules and the output of the last GCA module. We use the first four layers of the pre-trained ResNet50 as the encoder for nCoVSegNet. The enhanced channel attention (ECA) component (Wang et al., 2020b) is embedded in each ResNet block (RB) to preserve as much useful texture information as possible in the original CT image and to filter out interference information, e.g., noise. Note that the size of the feature map is halved and the number of channels is doubled between two neighboring RBs.
Fig. 1.
An overview of our nCoVSegNet.
3.1.2. Global context-aware module
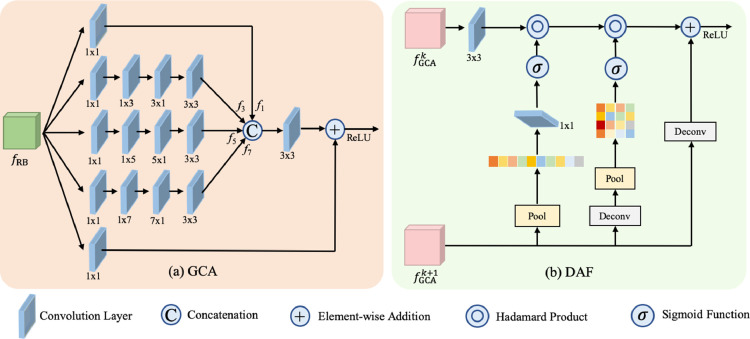
Inspired by Liu et al. (2018), we develop the GCA module, which exploits more informative features using enlarged receptive fields. As shown in Fig. 2 (a), a GCA module consists of five parallel branches, each of which is constituted of different convolutional layers. In particular, the three middle branches utilize asymmetric convolution layers with different kernel sizes, which provides rich multi-scale features from different receptive fields. After fusing the multi-scale features, we have more informative features, providing rich characteristics of image information. It is worth noting that, due to the use of asymmetric convolution layers, we reduce the computational amount and trainable parameters, while enlarging the receptive fields. Mathematically, the GCA module is defined as
| (1) |
where denotes the features from th branch with representing the size of the asymmetric convolutional kernel size; denotes the concatenation operation; and represent the convolutional unites with kernel sizes of and , respectively; denotes the features extracted from the backbone. For the last GCA module (i.e., in Fig. 1), we remove the branch for convolutional layers since the receptive field of high-level features is already large. This adjustment can save the memory and only has a marginal influence on the results.
Fig. 2.
Illustrations of GCA and DAF modules.
3.1.3. Dual-attention fusion module
To fuse the rich features from GCA modules, we propose a novel DAF module, which enhances the lower-level features by using the attention maps generated from the upper-level features, and then integrates the enhanced lower-level features with the upper-level features. Specifically, as shown in Fig. 2(b), we consider both channel attention (CA) (Chen et al., 2017) and spatial attention (SA) (Chen et al., 2017) mechanisms. Following (Woo et al., 2018), we employ global average pooling in the CA component and max pooling in the SA component. The fused feature is a summation of the upsampled upper-level features and the enhanced lower-level features provided by CA and SA components. Mathematically, we define the DAF module as
| (2) |
where and represent the features provided by th (lower-level) and th (upper-level) GCA modules with . denotes the Hadamard product, i.e., element-wise multiplication. represents the deconvolution operation with a kernel size of , which enlarges the size of the feature map. and are the CA and SA attention weight matrices, and are defined as
| (3) |
| (4) |
where denotes pooling operation and represents Sigmoid activation function.
3.1.4. Loss function
We employ a deep supervision strategy (Lee et al., 2015) to design the loss function. Specifically, as shown in Fig. 1, the supervision is added to each DAF module and the last GCA module, allowing better gradient flow and more effective network training. For each supervision, we consider two losses, i.e., the binary cross-entropy (BCE) loss and Dice loss (Milletari et al., 2016). The overall loss is therefore designed as
| (5) |
For the BCE loss, a modified version (Wei et al., 2020) is adopted to alleviate the imbalance problem between positive (lesion) and negative (normal tissue). For clarity, we omit the superscript . The definition of is as follows
| (6) |
where indicates two kinds of labels. and are the prediction and ground truth values at location in an CT image with a shape of width and height . represents all the parameters of the model, and denotes the predicted probability. is the weight of each pixel. Following (Wei et al., 2020), we define as
| (7) |
where represents the area surrounding the pixel , and denotes the computation of absolute value.
In addition, the weighted Dice loss is defined as
| (8) |
3.2. Two-stage cross-domain transfer learning
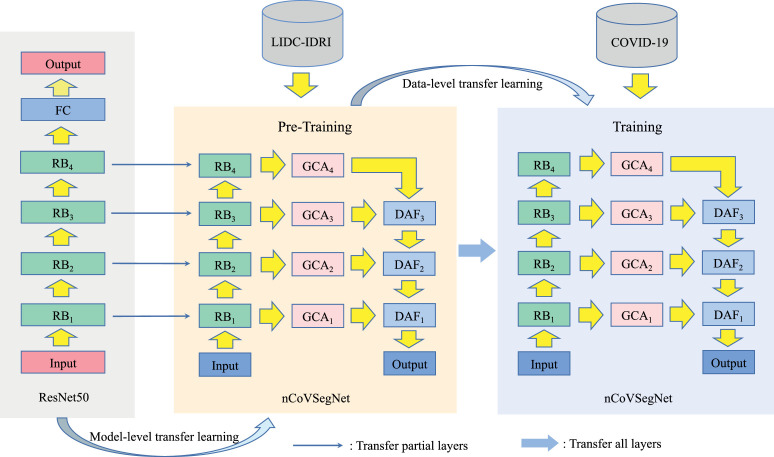
We train nCoVSegNet using an effective two-stage cross-domain transfer learning strategy. As shown in Fig. 3 , at the first stage, the knowledge learned by the backbone network, which is pre-trained using natural images, is transferred to our task at the model level. It is worth noting that this stage provides cross-domain learning, which transfers the knowledge from natural images to medical images. As aforementioned, we use a modified ResNet block pre-trained on ImageNet as the backbone of nCoVSegNet for transfer learning at this stage.
Fig. 3.
Diagram of two-stage cross-domain transfer learning.
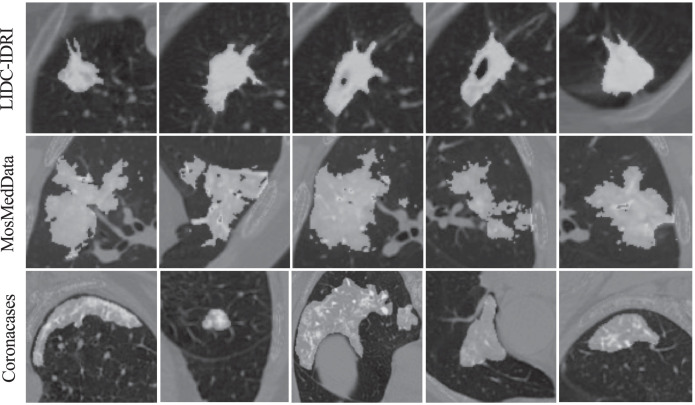
At the second stage, the CT images for lung nodule detection are utilized for transfer learning at the data level. Our motivation lies in two aspects. First, there exists a large gap between natural images and COVID-19 CT images, which raises significant demands to introduce a procedure for filling the cross-domain gap. Second, as shown in Fig. 4 , COVID-19 lung infections share similar appearances with the pulmonary nodules, such as ground-glass opacity at the early stage, and pulmonary consolidation at the late stage (Li, Wu, Wu, Guo, Chen, Fang, Li, 2020, Zhou, Wang, Zhu, Xia, 2020). Therefore, lung nodule segmentation is able to provide useful guidelines for COVID-19 lung infection segmentation. Motivated by these, we employ the current largest lung nodule CT dataset, LIDC-IDRI, to perform the second stage transfer learning, which provides vital knowledge from a neighboring domain to fill the cross-domain gap. Therefore, the LIDC-IDRI dataset is regarded as the source domain data to train the whole network for the model level transfer learning. After that, the COVID-19 dataset is employed for the final training of our model.
Fig. 4.
Visual comparison of the infection regions from pulmonary nodules in LIDC-IDRI and COVID-19 infections from MosMedData and Coronacases. The LIDC-IDRI is currently the largest lung nodule CT dataset. The MosMedData and Coronacases are clinical COVID-19 CT datasets and detailed in Section 4.1.
4. Experiments
In this section, we first provide detailed information on the datasets, followed by experimental settings and evaluation methods. Finally, we present the experimental results for our nCoVSegNet and the cutting-edge models.
4.1. Datasets
To conduct a data-level transfer learning, we use the largest dataset for lung nodule detection, i.e., LIDC-IDRI, for pre-training. In addition, the COVID-19 CT images from MosMedData dataset (Morozov et al., 2020) are used to train our model. In our experiments, 40 cases from MosMedData are used for training, while the remaining 10 subjects are used for testing. The COVID-19 CT images from https://coronacases.org/ with the annotations provided by (Ma et al., 2021) are used to test our trained model for evaluating its generalization performance. This dataset is denoted as “Coronacases”. Table 1 shows the statistics of our training and testing datasets.
Table 1.
Statistics of our training and testing datasets.
| Dataset | Total |
Train |
Test |
|||
|---|---|---|---|---|---|---|
| Cases | Slices | Cases | Slices | Cases | Slices | |
| LIDC-IDRI (Armato III et al., 2011) | 1010 | 244,527 | 875 | 13,916 | 0 | 0 |
| MosMedData (Morozov et al., 2020) | 50 | 2049 | 40 | 1640 | 10 | 409 |
| Coronacases (Ma et al., 2021) | 10 | 2581 | 0 | 0 | 10 | 2581 |
The data processing for three datasets is detailed as follows. LIDC-IDRI: According to the CSV files provided by the dataset, we select subjects with lung nodules. Next, we generate the nodule ground truth mask by referring to each patient’s XML file. Since each lung nodule is marked by four doctors, we take a 50% consensus as the choice to determine ground truth masks. MosMedData & Coronacases: Similar to (Fan et al., 2020), we extract the lung regions from CT images before training and testing. For this purpose, we first segment the lung using the unsupervised method in (Liao et al., 2019). Based on the 3D lung mask, we determine a bounding box for the region of the lung. This bounding box is then utilized to crop the 3D region of the lung from a CT scan.
4.2. Experimental settings
Our model is implemented using PyTorch and trained on an NVIDIA GeForce RTX 2080 Ti GPU. The code is publicly available at https://github.com/Jiannan-Liu/nCoVSegNet. We first pre-train the model using LIDC-IDRI with the settings as follows: 100 epochs, initial learning rate of 1e-4 with a decay of 0.1 every 50 epochs, batch size of 4, and image size of . Note that a quarter of LIDC-IDRI training data is used for validation to search for the best hyperparameters. The resulting hyperparameters are employed for the formal training with all training data. An Adam optimizer (Kingma and Ba, 2014) is employed for optimization. At this stage, 875 subjects with a total of 13,916 slices are fed into nCoVSegNet to pre-train the model for filling the gap between natural images and medical images. Next, MosMedData, which contains 40 subjects with a total of 1640 slices, is utilized to train the model for COVID-19 infection segmentation. The learning rate is reduced to 5e-5 and the number of epochs is set to 50, and an early stopping strategy is employed to prevent over-fitting. Similar to LIDC-IDRI, a validation procedure is employed to determine the best hyperparameters for training with MosMedData. For data augmentation, we use the random horizontal flip, random crop, and multi-scale resizing with different ratios {0.75,1,1.25}. The input images are normalized to (0,1) before the training. Finally, the remaining subjects in MosMedData and the whole Coronacases are used for the testing.
4.3. Evaluation metrics
Six widely adopted metrics are used for measuring the performance of segmentation models.
Dice similarity coefficient (DSC): The DSC is used to measure the similarity between the predicted lung infections and the ground truth (GT). DSC is defined as
| (9) |
where and represent the voxel sets of the infection segmentation and GT, respectively. denotes the operation of cardinality computation, which provides the number of elements in a set.
Sensitivity (SEN): The SEN reflects the percentage of lung infections that are correctly segmented. Its definition is as follows
| (10) |
Specificity (SPE): The SPE reflects the percentage of non-infection regions that are correctly segmented and is defined as
| (11) |
where denotes the voxel set of the whole CT volume.
Positive Predictive Value (PPV): The PPV reflects the accuracy of the segmented lung infections and is defined as
| (12) |
Volumetric Overlap Error (VOE): The VOE represents the error rate of the segmentation result. VOE equals zero for a perfect segmentation and one for the worst case that there is no overlap between the segmentation result and GT (Heimann et al., 2009). VOE is defined as:
| (13) |
Relative Volume Difference (RVD): The RVD is used to measure the volume difference between the predicted lung infections and GT, which is defined in Eq. (14) (Heimann et al., 2009). We take the absolute value of RVD and report the corresponding results in this work.
| (14) |
4.4. Baseline methods
Our proposed model is compared with various state-of-the-art deep learning models, including two popular ones, U-Net (Ronneberger et al., 2015) and U-Net++ (Zhou et al., 2018), as well as three recently developed CE-Net (Gu et al., 2019), UNet (Qin et al., 2020), and U-Net3+ (Huang et al., 2020). All baseline methods are trained with default settings based on the codes provided in the literature.
4.5. Results
4.5.1. Quantitative results
We show the quantitative results in Table 2 . As can be observed, our nCoVSegNet performs the best in terms of DSC, SEN, SPE, VOE, and RVD. Specifically, it outperforms the second-best model, CE-Net, by a large margin of 4.3% in terms of DSC, which is the key evaluation metric in segmentation. In addition, it significantly improves the SEN by a large margin of 12%, while maintaining a high SPE. Furthermore, it provides the best performance in terms of VOE and RVD with 9.1% and 15.5% improvement in comparison with CE-Net. To further demonstrate the effectiveness of our nCoVSegNet, we performed paired student t-test between our results and those provided by CE-Net. The t-test results show that our improvement is statistically significant with -values smaller than 0.05. Our nCoVSegNet performs the best in terms of all evaluation metrics, except PPV, where nCoVSegNet ranks the third-best method. The underlying reason is that there are many independent and scattered pulmonary nodules in the MosMedData, which posed challenges in segmentation. It is worth noting that our nCoVSegNet consistently outperforms the second best method, CE-Net, in terms of PPV.
Table 2.
Quantitative segmentation results on MosMedData (Morozov et al., 2020). The best, second best, and third best results are marked by italic, boldface, and underline, respectively.
| Method | DSC | SEN | SPE | PPV | VOE | RVD |
|---|---|---|---|---|---|---|
| U-Net (Ronneberger et al., 2015) | 0.5419 | 0.4585 | 0.9971 | 0.7425 | 0.6283 | 0.3667 |
| U-Net (Zhou et al., 2018) | 0.5454 | 0.4546 | 0.9971 | 0.7674 | 0.6250 | 0.3856 |
| UNet (Qin et al., 2020) | 0.6022 | 0.5813 | 0.9971 | 0.6867 | 0.5691 | 0.0803 |
| U-Net3+ (Huang et al., 2020) | 0.6265 | 0.5724 | 0.9975 | 0.7653 | 0.5438 | 0.1885 |
| CE-Net (Gu et al., 2019) | 0.6416 | 0.6295 | 0.9973 | 0.7276 | 0.5276 | 0.0297 |
| nCoVSegNet (Ours) | 0.6843 | 0.7480 | 0.9976 | 0.7525 | 0.4798 | 0.0251 |
To verify the generalization capability of our nCoVSegNet, we further evaluate the segmentation performance on the Coronacases dataset. Note that none of Coronacases’ data is used during the training process. The quantitative segmentation results, shown in Table 3 , indicate that our nCoVSegNet outperforms the cutting-edge methods in terms of all the evaluation metrics, demonstrating that nCoVSegNet achieves promising generalization capability.
Table 3.
Quantitative segmentation results on Coronacases (Ma et al., 2021). The best, second best, and third best results are marked by italic, boldface, and underline, respectively.
| Method | DSC | SEN | SPE | PPV | VOE | RVD |
|---|---|---|---|---|---|---|
| U-Net (Ronneberger et al., 2015) | 0.5184 | 0.3925 | 0.9848 | 0.8829 | 0.6501 | 0.5463 |
| U-Net+ (Zhou et al., 2018) | 0.5371 | 0.4231 | 0.9850 | 0.8470 | 0.6328 | 0.4788 |
| UNet (Qin et al., 2020) | 0.5311 | 0.4134 | 0.9851 | 0.8690 | 0.6384 | 0.5129 |
| U-Net3+ (Huang et al., 2020) | 0.6097 | 0.4869 | 0.9871 | 0.8883 | 0.5614 | 0.4441 |
| CE-Net (Gu et al., 2019) | 0.6712 | 0.5501 | 0.9885 | 0.8905 | 0.4948 | 0.3903 |
| nCoVSegNet (Ours) | 0.6894 | 0.5918 | 0.9886 | 0.8949 | 0.4739 | 0.3270 |
4.5.2. Qualitative results
The visual comparison of segmentation results is shown in Fig. 5 . It can be observed that U-Net and U-Net++ fail in providing complete infection segmentation. A large amount of missing segmentation confirms the low sensitivity in Table 2. The other three baseline methods, CE-Net, UNet, and U-Net3+, improve the results, but are still not good as our nCoVSegNet, especially for the regions marked by red arrows. In contrast, nCoVSegNet consistently provides the best performance in the visual comparison, demonstrating its effectiveness sufficiently.
Fig. 5.
Visual comparison of segmentation results for different methods.
4.5.3. Effectiveness of two-stage cross-domain transfer learning
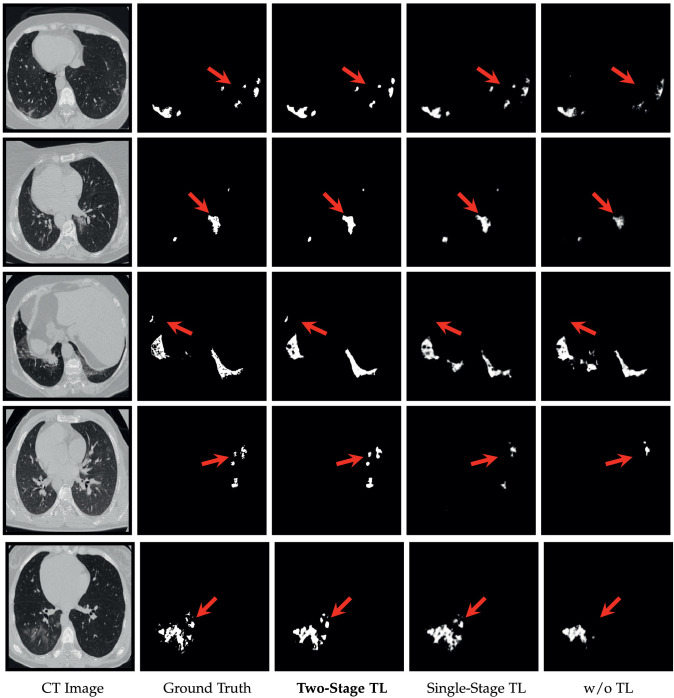
We further investigate the effectiveness of two-step cross-domain transfer learning strategy on MosMedData by comprising three versions of our method, including “w/o TL”: Without transfer learning, “Single-Stage TL”: One-stage transfer learning with ImageNet pre-trained model, and “Two-Stage TL”: The full version of two-stage transfer learning, i.e., considering both ImageNet and LIDC-IDRI.
The results, shown in Table 4 , indicate that “Single-Stage TL” outperforms “w/o TL” in terms of all evaluation metrics, demonstrating that the knowledge learned from the natural image can be used to improve the performance effectively. In addition, “Two-Stage TL” further improves the performance, which outperforms the other two versions with respect to all the metrics. This finding demonstrates that pre-training with LIDC-IDRI can fill the cross-domain gap and further improve the performance by using the knowledge from a neighboring domain.
Table 4.
Quantitative comparison of different transfer learning results. The best and second best results are marked by italic, boldface, respectively.
| Method | DSC | SEN | SPE | PPV | VOE | RVD |
|---|---|---|---|---|---|---|
| w/o TL | 0.6411 | 0.6802 | 0.9973 | 0.6562 | 0.5282 | 0.1626 |
| Single-Stage TL | 0.6680 | 0.7007 | 0.9975 | 0.6977 | 0.4984 | 0.1299 |
| Two-Stage TL | 0.6843 | 0.7480 | 0.9976 | 0.7525 | 0.4798 | 0.0251 |
Furthermore, we show some representative visual results in Fig. 6 . As can be observed, the “Two-Stage TL” provides the best segmentation results that are close to the ground truth. In contrast, “Single-Stage TL” and “w/o TL” show unsatisfactory performance and are unable to provide segmentation results with complete infection regions. Overall, the results, shown in Table 4 and Fig. 6, sufficiently demonstrate that the two-stage transfer learning strategy is particularly effective in the COVID-19 infection segmentation task.
Fig. 6.
Visual comparison of segmentation results provided by different transfer learning strategies.
4.5.4. Ablation study
Finally, we perform an extensive ablation study on MosMedData to investigate the effectiveness of each component in the network. In general, we consider the effectiveness of GCA module, DAF module, and the deep supervision (DS) strategy. We summarize the quantitative results in Table 5 . The comparison baseline is denoted by “(A) Backbone”, which is a U-Net-like architecture with the backbone network (ResNet50 with ECA components) as an encoder.
Table 5.
Quantitative results for ablation studies. The best results are in boldface.
| Model | Backbone | GCA | DAF | DS | DSC | SEN | SPE | PPV |
|---|---|---|---|---|---|---|---|---|
| (A) | 0.5908 | 0.5758 | 0.9972 | 0.6536 | ||||
| (B) | 0.6055 | 0.5870 | 0.9973 | 0.6691 | ||||
| (C) | 0.6283 | 0.6445 | 0.9973 | 0.6656 | ||||
| (D) | 0.6119 | 0.6577 | 0.9971 | 0.6129 | ||||
| (E) | 0.6270 | 0.6640 | 0.9973 | 0.6377 | ||||
| (F) | 0.6355 | 0.6651 | 0.9974 | 0.6846 | ||||
| (G) | 0.6453 | 0.6711 | 0.9974 | 0.6809 | ||||
| (H) | 0.6680 | 0.7007 | 0.9975 | 0.6977 |
Effectiveness of GCA: The ablated version (B) is the backbone with our GCA modules. As shown in Table 5, (B) outperforms (A) in terms of all evaluation metrics, demonstrating that GCA modules can improve the performance effectively.
Effectiveness of DAF: We then investigate the effectiveness of the DAF module. The ablated version (C) is the backbone with our DAF modules. Compared with (A), our proposed DAF module improves the performance in terms of all metrics, sufficiently demonstrating its effectiveness.
Effectiveness of DS: As shown in Table 5, the ablated version with DS, i.e., (D), shows improved performance over (A) in terms of DSC, SEN, and PPV. In addition, it provides a comparable SPE in comparison with (A). All of these results demonstrate that DS is an effective component capable of improving performance effectively.
Effectiveness of Module Combinations: Finally, we investigate the effectiveness of different module combinations, including (E), (F), and (G). As shown in Table 5, each module combination outperforms the corresponding ablated versions with a single module. The results demonstrate that the module combination can improve the results. In addition, the full version of our nCoVSegNet , i.e., (H), outperforms (E), (F), and (G), indicating that jointly incorporating GCA, DAF, and DS into the network provides the best performance.
5. Conclusion
In this paper, we have proposed a two-stage cross-domain transfer learning framework for COVID-19 lung infection segmentation from CT images. Our framework includes an effective infection segmentation model, nCoVSegNet, which is based on attention-aware feature fusion and enlarged receptive fields. In addition, we train our model using an effective two-stage transfer learning strategy, which takes advantage of valuable knowledge from both ImageNet and LIDC-IDRI. Extensive experiments on COVID-19 CT datasets indicate that our model achieves promising performance in lung infection segmentation and outperforms cutting-edge segmentation models. The results also demonstrate the effectiveness of the two-stage transfer learning strategy, the generalization of our model, and the effectiveness of proposed modules.
CRediT authorship contribution statement
Jiannan Liu: Software, Validation, Visualization, Data curation, Methodology, Investigation, Writing – original draft. Bo Dong: Software, Methodology. Shuai Wang: Writing – review & editing. Hui Cui: Writing – review & editing. Deng-Ping Fan: Writing – review & editing. Jiquan Ma: Supervision, Writing – review & editing. Geng Chen: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Armato III S.G., McLennan G., Bidaut L., McNitt-Gray M.F., Meyer C.R., Reeves A.P., Zhao B., Aberle D.R., Henschke C.I., Hoffman E.A., et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 2011;38(2):915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar Y., Diamant I., Wolf L., Greenspan H. Medical Imaging 2015: Computer-Aided Diagnosis. vol. 9414. International Society for Optics and Photonics; 2015. Deep learning with non-medical training used for chest pathology identification; p. 94140V. [Google Scholar]
- Bressem, K. K., Niehues, S. M., Hamm, B., Makowski, M. R., Vahldiek, J. L., Adams, L. C., 2021. 3D U-Net for segmentation of COVID-19 associated pulmonary infiltrates using transfer learning: state-of-the-art results on affordable hardware. arXiv preprint arXiv:2101.09976.
- Byra M., Galperin M., Ojeda-Fournier H., Olson L., O’Boyle M., Comstock C., Andre M. Breast mass classification in sonography with transfer learning using a deep convolutional neural network and color conversion. Med. Phys. 2019;46(2):746–755. doi: 10.1002/mp.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro G., Nascimento J., Bradley A.P. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. Unregistered multiview mammogram analysis with pre-trained deep learning models; pp. 652–660. [Google Scholar]
- Chatfield, K., Simonyan, K., Vedaldi, A., Zisserman, A., 2014. Return of the devil in the details: delving deep into convolutional nets. arXiv preprint arXiv:1405.3531.
- Chen L., Zhang H., Xiao J., Nie L., Shao J., Liu W., Chua T.-S. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. SCA-CNN: spatial and channel-wise attention in convolutional networks for image captioning; pp. 5659–5667. [Google Scholar]
- Chen, X., Yao, L., Zhang, Y., 2020. Residual attention U-Net for automated multi-class segmentation of COVID-19 chest CT images. arXiv preprint arXiv:2004.05645.
- Cheplygina V., de Bruijne M., Pluim J.P. Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 2019;54:280–296. doi: 10.1016/j.media.2019.03.009. [DOI] [PubMed] [Google Scholar]
- Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2009. ImageNet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- Elharrouss, O., Subramanian, N., Al-Maadeed, S., 2020. An encoder-decoder-based method for COVID-19 lung infection segmentation. arXiv preprint arXiv:2007.00861. [DOI] [PMC free article] [PubMed]
- Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. Inf-Net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020:200432. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Cheng J., Fu H., Zhou K., Hao H., Zhao Y., Zhang T., Gao S., Liu J. CE-Net: context encoder network for 2D medical image segmentation. IEEE Trans. Med. Imaging. 2019;38(10):2281–2292. doi: 10.1109/TMI.2019.2903562. [DOI] [PubMed] [Google Scholar]
- Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S., et al. Clinical characteristics of 2019 novel coronavirus infection in China. MedRxiv. 2020 [Google Scholar]
- He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- Heimann T., Van Ginneken B., Styner M.A., Arzhaeva Y., Aurich V., Bauer C., Beck A., Becker C., Beichel R., Bekes G., et al. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans. Med. Imaging. 2009;28(8):1251–1265. doi: 10.1109/TMI.2009.2013851. [DOI] [PubMed] [Google Scholar]
- Huang H., Lin L., Tong R., Hu H., Zhang Q., Iwamoto Y., Han X., Chen Y.-W., Wu J. ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing. IEEE; 2020. UNet3+: a full-scale connected UNet for medical image Segmentation; pp. 1055–1059. [Google Scholar]
- Khan S., Islam N., Jan Z., Din I.U., Rodrigues J.J.C. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett. 2019;125:1–6. [Google Scholar]
- Kingma, D. P., Ba, J., 2014. Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980.
- Lancet T. Emerging understandings of 2019-nCoV. Lancet. 2020;395(10221):311. doi: 10.1016/S0140-6736(20)30186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-Y., Xie S., Gallagher P., Zhang Z., Tu Z. Artificial Intelligence and Statistics. 2015. Deeply-supervised nets; pp. 562–570. [Google Scholar]
- Li K., Wu J., Wu F., Guo D., Chen L., Fang Z., Li C. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest. Radiol. 2020 doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Pang T., Xiong B., Liu W., Liang P., Wang T. 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics. IEEE; 2017. Convolutional neural networks based transfer learning for diabetic retinopathy fundus image classification; pp. 1–11. [Google Scholar]
- Liang G., Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput. Methods Programs Biomed. 2020;187:104964. doi: 10.1016/j.cmpb.2019.06.023. [DOI] [PubMed] [Google Scholar]
- Liao F., Liang M., Li Z., Hu X., Song S. Evaluate the malignancy of pulmonary nodules using the 3D deep leaky noisy-or network. IEEE Trans. Neural Netw. Learn. Syst. 2019;30(11):3484–3495. doi: 10.1109/TNNLS.2019.2892409. [DOI] [PubMed] [Google Scholar]
- Lin T.-Y., Dollár P., Girshick R., He K., Hariharan B., Belongie S. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Feature pyramid networks for object detection; pp. 2117–2125. [Google Scholar]
- Liu S., Huang D., et al. Proceedings of the European Conference on Computer Vision. 2018. Receptive field block net for accurate and fast object detection; pp. 385–400. [Google Scholar]
- Ma J., Wang Y., An X., Ge C., Yu Z., Chen J., Zhu Q., Dong G., He J., He Z., Cao T., Zhu Y., Nie Z., Yang X. Towards data-efficient learning: a benchmark for COVID-19 CT lung and infection segmentation. Med. Phys. 2021;48(3):1197–1210. doi: 10.1002/mp.14676. [DOI] [PubMed] [Google Scholar]
- Milletari F., Navab N., Ahmadi S.-A. 2016 fourth International Conference on 3D Vision. IEEE; 2016. V-Net: fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- Morozov S., Andreychenko A., Pavlov N., Vladzymyrskyy A., Ledikhova N., Gombolevskiy V., Blokhin I., Gelezhe P., Gonchar A., Chernina V., et al. Mosmeddata: chest CT scans with COVID-19 related findings. medRxiv. 2020 [Google Scholar]
- Müller, D., Rey, I. S., Kramer, F., 2020. Automated chest CT image segmentation of COVID-19 lung infection based on 3D U-Net. arXiv preprint arXiv:2007.04774. [DOI] [PMC free article] [PubMed]
- Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in Wuhan, China: challenges for global health governance. JAMA. 2020;323(8):709–710. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- Qin X., Zhang Z., Huang C., Dehghan M., Zaiane O.R., Jagersand M. U-Net: going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020;106:107404. [Google Scholar]
- Qiu, Y., Liu, Y., Xu, J., 2020. MiniSeg: an extremely minimum network for efficient COVID-19 segmentation. arXiv preprint arXiv:2004.09750. [DOI] [PubMed]
- Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-Net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Saeedizadeh N., Minaee S., Kafieh R., Yazdani S., Sonka M. COVID TV-UNet: segmenting COVID-19 chest CT images using connectivity imposed U-Net. Comput. Methods Programs Biomed. Update. 2021;1:100007. doi: 10.1016/j.cmpbup.2021.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, F., Gao, Y., Wang, J., Shi, W., Shi, N., Han, M., Xue, Z., Shi, Y., 2020. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655.
- Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- Shie C.-K., Chuang C.-H., Chou C.-N., Wu M.-H., Chang E.Y. 2015 37th annual international conference of the IEEE Engineering in Medicine and Biology Society. IEEE; 2015. Transfer representation learning for medical image analysis; pp. 711–714. [DOI] [PubMed] [Google Scholar]
- Shin H.-C., Roth H.R., Gao M., Lu L., Xu Z., Nogues I., Yao J., Mollura D., Summers R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K., Zisserman A. 2015. Very deep convolutional networks for large-scale image recognition. [Google Scholar]
- Swati Z.N.K., Zhao Q., Kabir M., Ali F., Ali Z., Ahmed S., Lu J. Content-based brain tumor retrieval for MR images using transfer learning. IEEE Access. 2019;7:17809–17822. doi: 10.1016/j.compmedimag.2019.05.001. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh N., Shin J.Y., Gurudu S.R., Hurst R.T., Kendall C.B., Gotway M.B., Liang J. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans. Med. Imaging. 2016;35(5):1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- Tilborghs, S., Dirks, I., Fidon, L., Willems, S., Eelbode, T., Bertels, J., Ilsen, B., Brys, A., Dubbeldam, A., Buls, N., et al., 2020. Comparative study of deep learning methods for the automatic segmentation of lung, lesion and lesion type in CT scans of COVID-19 patients. arXiv preprint arXiv:2007.15546.
- Vaishya R., Javaid M., Khan I.H., Haleem A. Artificial intelligence (AI) applications for COVID-19 pandemic. Diabetes Metabolic Syndrome. 2020 doi: 10.1016/j.dsx.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu C.C., Siddiqui Z.A., Zamdborg L., Thompson A.B., Quinn T.J., Castillo E., Guerrero T.M. Deep convolutional neural networks for automatic segmentation of thoracic organs-at-risk in radiation oncology–use of non-domain transfer learning. J. Appl. Clin. Med. Phys. 2020;21(6):108–113. doi: 10.1002/acm2.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., Meng T., Li K., Huang N., Zhang S. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans. Med. Imaging. 2020;39(8):2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu B., Zhu P., Li P., Zuo W., Hu Q. Proceedings of the IEEE/Computer Vision Foundation Conference on Computer Vision and Pattern Recognition. 2020. ECA-Net: efficient channel attention for deep convolutional neural networks; pp. 11534–11542. [Google Scholar]
- Wei J., Wang S., Huang Q. Proceedings of the AAAI Conference on Artificial Intelligence. vol. 34. 2020. FNet: fusion, feedback and focus for salient object detection; pp. 12321–12328. [Google Scholar]
- Woo S., Park J., Lee J.-Y., So Kweon I. Proceedings of the European Conference on Computer Vision. 2018. CBAM: convolutional block attention module; pp. 3–19. [Google Scholar]
- Wu Y.-H., Gao S., Mei J., Xu J., Fan D.-P., Zhao C., Cheng M.-M. JCS: an explainable COVID-19 diagnosis system by joint classification and segmentation. IEEE Trans. Image Process. 2021;30:3113–3126. doi: 10.1109/TIP.2021.3058783. [DOI] [PubMed] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCov pneumonia: relationship to negative RT-PCR testing. Radiology. 2020:200343. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J., Lipsitz O., Nasri F., Lui L.M., Gill H., Phan L., Chen-Li D., Iacobucci M., Ho R., Majeed A., et al. Impact of COVID-19 pandemic on mental health in the general population: a systematic review. J. Affect. Disord. 2020 doi: 10.1016/j.jad.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z., Cao, Y., Jin, C., Shao, G., Liu, X., Zhou, J., Shi, H., Feng, J., 2020. GASNet: weakly-supervised framework for COVID-19 lesion segmentation. arXiv preprint arXiv:2010.09456.
- Yan, Q., Wang, B., Gong, D., Luo, C., Zhao, W., Shen, J., Shi, Q., Jin, S., Zhang, L., You, Z., 2020. COVID-19 chest CT image segmentation–a deep convolutional neural network solution. arXiv preprint arXiv:2004.10987. [DOI] [PMC free article] [PubMed]
- Yao Q., Xiao L., Liu P., Zhou S.K. Label-Free segmentation of COVID-19 lesions in lung CT. IEEE Trans. Med. Imaging. 2021 doi: 10.1109/TMI.2021.3066161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Li Y., Li Z., Chen Y., Zhao W., Xingzhi X., Liu J., Zhao D. SCOAT-Net: A novel network for segmenting COVID-19 lung opacification from CT images. medRxiv. 2020 doi: 10.1016/j.patcog.2021.108109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Li Z., Zhou J., Li H., Chen Y., Huang Y., Xie D., Zhao L., Fan M., Hashmi S., et al. A rapid, accurate and machine-agnostic segmentation and quantification method for CT-based COVID-19 diagnosis. IEEE Trans. Med. Imaging. 2020;39(8):2638–2652. doi: 10.1109/TMI.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Wang Y., Zhu T., Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. Am. J. Roentgenol. 2020;214(6):1287–1294. doi: 10.2214/AJR.20.22975. [DOI] [PubMed] [Google Scholar]
- Zhou T., Canu S., Ruan S. Automatic COVID-19 CT segmentation using U-Net integrated spatial and channel attention mechanism. Int. J. Imaging Syst. Technol. 2021;31(1):16–27. doi: 10.1002/ima.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. UNet: a nested U-Net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]