Abstract
COVID-19 is a disease caused by SARS-CoV-2, which has led to more than 4 million deaths worldwide. As a result, there is a worldwide effort to develop specific drugs for targeting COVID-19. Papain-like protease (PLpro) is an attractive drug target because it has multiple essential functions involved in processing viral proteins, including viral genome replication and removal of post-translational ubiquitination modifications. Here, we established two assays for screening PLpro inhibitors according to protease and anti-ISGylation activities, respectively. Application of the two screening techniques to the library of clinically approved drugs led to the discovery of tanshinone IIA sulfonate sodium and chloroxine with their IC50 values of lower than 10 μM. These two compounds were found to directly interact with PLpro and their molecular mechanisms of binding were illustrated by docking and molecular dynamics simulations. The results highlight the usefulness of the two developed screening techniques for locating PLpro inhibitors.
Keywords: Papain-like protease, Tanshinone IIA sulfonate sodium, Chloroxine
1. Introduction
Coronavirus Disease 2019 (COVID-19) is a disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In March 2020, the World Health Organization announced that the COVID-19 epidemic has constituted a global pandemic. COVID-19 has many similarities with severe acute respiratory syndrome (SARS) that broke out in China in 2003. Although the fatality rate of COVID-19 is far lower than SARS and Middle East respiratory syndrome (MERS), its infection rate and death toll far exceed the previous two diseases. Until now, few drugs are available for COVID-19 treatment.
SARS-CoV-2 is a linear single-stranded RNA virus with a full-length genome of about 30 kb. SARS-CoV-2 encodes an essential papain-like protease domain as part of its non-structural protein (nsp)-3, namely PLpro. PLpro is responsible for cutting the three N-terminal cleavage sites on ORF1a to yield product nsp1, nsp2 and nsp3 [1]. The cleavage specificity of PLpro corresponds to the pattern (R/K)L(R/K)GG↓X [2]. After protease digestion, these non-structural proteins participate in the assembly of the viral replicase complex, which initiates the replication and transcription of the viral genome [1], [3], [4].
Interferon Stimulated Gene 15 (ISG15) is a ubiquitin-like protein, which performs ISGylation by covalently binding to the target protein. As one of the strongest interferon-stimulating proteins, ISG15 can bind to lysine residues to induce the host antiviral response [5]. However, some viruses have evolved a variety of mechanisms to escape the host antiviral response by inhibiting or removing ISGylation. ISG15 has the unique sequence LRLRGG at the C-terminus, consistent with the coronavirus PLpro recognition motif [2]. Thus, PLpro should be able to effectively remove ISGylation and ubiquitylation modification [6], and subsequently inhibit inflammation and antiviral signal transduction [1], [7], [8], [9]. Multiple functions possessed by PLpro make it an attractive anti-SARS-CoV-2 target.
A fast way for locating a drug specific for COVID-19 treatment is to repurpose clinically approved drugs. Currently, remdesivir was approved to treat COVID-19 by FDA [10]. Here, we also focused on the question whether or not existing drugs can be used for inhibiting SARS-CoV-2 PLpro. To achieve this goal, we developed two screening techniques based on the protease and anti-ISGylation activities. Their application to a clinically approved drug library identified tanshinone IIA sulfonate sodium and chloroxine as PLpro inhibitors.
2. Materials and methods
2.1. Protein expression and purification
The SARS-CoV-2 PLpro coding sequence was codon optimized and synthesized by Genscript Co.,Ltd. and cloned into the pET28a vector. A 6 × His tag was added to the PLpro C-terminus. The recombinant plasmid, pET28a-PLpro-6His, was transformed into BL21 (DE3) cells. After culturing the cells at 37 °C for 4 h, isopropyl-beta-D-thiogalactopyranoside (IPTG) was added to induce PLpro expression at 16 °C for 16 h. The cells, harvested by centrifugation, were resuspended with a lysis buffer at pH 8.5 containing 20 mM Tris-HCl, 300 mM NaCl and 5 mM 2-Hydroxy-1-ethanethiol (β-ME). The resuspended cells were disrupted by sonication and the supernatant was centrifuged and loaded onto Ni-NTA resin (Genscript).
The elution from Ni-NTA resin was then purified on Q ion-exchange chromatography, and finally load onto gel filtration (Superdex200 Increase, GE Healthcare) with the buffer of 20 mM Tris-HCl, 300 mM NaCl and 5%(w/w) glycerol with pH 8.5. The protein fractions were concentrated by 10 kDa MWCO concentrator (Amicon Millipore) and stored at −80 °C.
The cloning, expression, and purification of the PLpro C111S mutant (PLpro-C111S) and human ISG15 were the same as the procedure for PLpro because they have a similar size and a similar predicted isoelectric point (PI = 6.9 for PLpro-WT, 6.9 for PLpro-C111S and 7.5 for ISG15). Moreover, they were all tagged by six of histidine residues for purification. Thus, we can purify those three proteins with the same protocol (the Ni-NTA resin, Q ion-exchange chromatography and gel filtration column on Superdex 200).
2.2. High-throughput screening (HTS)
2.2.1. Protease activity-based method for drug screening
A fluorogenic peptide, ALKGG-AMC, was designed based on the substrate sequence of the SARS-CoV-2 nsp3-nsp4 cleavage site. After cleavage, a free AMC will be released, and its fluorescence signal can be monitored to measure the protease activity of SARS-CoV-2 PLpro. This fluorogenic peptide was synthesized and purified to a > 95% purity by GenScript. The fluorogenic peptide was dissolved in Buffer A (50 mM HEPES, pH 7.5, 5 mM DTT and 0.1 mg/ml BSA) to a final concentration of 10 mM and stored at −20 °C. 250 μM ALKGG-AMC was loaded onto PLpro at concentrations ranging from 420 to 13.125 nM with 4-fold dilution in a black flat-bottom 96 well plate (Greiner). After 10 min of pre-incubation at room temperature, the fluorescence was monitored for 30 min on BioTek Synergy HTX (Ex: 340 nm, Em: 460 nm, Temperature: 25 °C). One negative control was made with a non-fluorescent peptide substrate. Another negative control was made by replacing PLpro with the blank Buffer A.
The compound library, purchased from APExBio, consists of 1971 compounds of clinically approved drugs. Each well was added 50 μl of PLpro in Buffer A at a concentration of 100 nM, followed by 1 μl of each compound (~10 mM in DMSO). After 10 min of pre-incubation, an addition of 50 μl of 125 μM fluorogenic peptide was made to the plates. Then, after 10 min of incubation at room temperature, the reaction was stopped after adding 50 μl of 0.2 M sodium acetate and the end-point fluorescence signals were read by the plate reader. One negative control (compound-negative control) was made with no compounds added. Another negative control (PLpro-negative control) was made with no PLpro added. All the controls were performed in triplicates. We employed the percentage of effect and a Z score to select the compounds for further studies. The percentage of effect is the inhibition rate of the sample against the PLpro activity at a certain concentration. The following formula was used to calculate the percentage of effect (Eq. (1)):
| (1) |
where μ c1- is the mean value of compound-negative controls and μ c2- is the mean value of PLpro-negative controls.
We calculated Z' factor to evaluate the quality of the assay. The Z' factor can be calculated as follows (Eq. (2)).
| (2) |
where μ c1- and μ c2- are the means, and σc1- and σ c2- are the standard deviations of the two negative controls, respectively.
Z score [11] is a value to evaluate the deviation from the normal distribution of the fluorescence value of a sample on a plate. Ninety-nine percent of fluorescence values of the sample are within 3 standard deviations from the mean. Thus, Z score > 3, indicating a statistically significant finding. Z score is calculated by using the equation below (Eq. (3)).
| (3) |
where x is the fluorescence of a sample, x̅ is the mean of all samples on each plate and standard deviation is denoted as S i.
2.2.2. Fluorescence polarization for drug screening (FPA)
For ISG15 labeling, ISG15 (0.5 mg/ml) was treated with extra FITC (molar ratio of ISG15: FITC = 1:3) in a buffer containing 100 mM NaCO3, 150 mM NaCl at pH 9.0 and incubated in the dark at 4 °C for 8 h. Desalting column (PD MiniTrap G-25, GE) was utilized to remove the unreacted FITC following manufacturer's instructions. To avoid over-labeling, we measured the absorbance ratio of A495/A280, and ensured that the ratio was between 0.3 and 1.0.
For the FP-based assay, 50 μl of PLpro at different concentrations (0.024–25 μM) was added to 50 μl of 200 nM ISG15-FITC with a pH -7.6 buffer containing 25 mM Na3PO4, 150 mM NaCl, 0.1 mg/ml BSA and 7.5 μM ZnCl2 in the 96-well plate. After 30 min incubation, the fluorescence polarization signals were measured by Tecan Spark (Ex: 485 nm, Em: 535 nm).
In order to establish the competitive assay, we firstly employed unlabeled ISG15 to mimic a competitive inhibitor. 50 μl of 10 μM PLpro was pre-incubated with 10 μl of unlabeled ISG15 at different concentrations (0.0976–10,000 μM) for 30 min. Then, 40 μl of 250 nM ISG15-FITC were added and the reaction system was incubated at room temperature for 1 h. The fluorescence polarization signals were measured by Tecan Spark using the same parameters as above. The IC50 value was calculated based on the dose-dependent fluorescence polarization curve using GraphPad Prism 6.0.
The procedures of the HTS based on FPA was similar to the competitive assay by using the testing compound rather than unlabeled ISG15. After adding 50 μl of 10 μM PLpro to the 96-well plate, 1 μl compound was added to each well and pre-incubated with PLpro for 30 min. In this screening, we have one positive control based on unlabeled ISG15 and two negative controls (compound-negative and PLpro -negative with no PLpro-C111S added).
2.3. Biolayer interferometry (BLI)
The BLI assay was performed on Fortebio Octet K2 and the data was analyzed with the Octet systems software. The streptavidin (SA) sensor was first equilibrated with the BLI assay buffer (Buffer BLI) of pH 7.5 containing 20 mM Tris-HCl, 150 mM NaCl, and 0.02% Tween 20. Then, the SA sensor was loaded with the biotin-labeled PLpro or PLpro-C111S for 5 min at 50 μg/ml concentration, which was labeled with NHS-PEG12-Biotin (Thermo Fisher Cat. No. 21312) by following manufacturer's instruction. Next, the sensors were dipped into the compound solution in Buffer BLI at different concentrations for 3 min to allow the association of the testing compound with PLpro, then dipped into the blank Buffer BLI for 3 min allowing the bound compound to dissociate. The control sensor was loaded with the Buffer BLI only.
2.4. Thermal-shift assay (TSA)
10 μl of 50 μM PLpro in different pH buffers was mixed with 10 μl of 50 μM compounds in the corresponding pH buffer or 1 μl of DMSO. After 10 min pre-incubation, 1 μl SYPRO Orange solution (Sigma Cat. No. S5692) was added into each sample. All samples were performed in triplicates. The samples were placed in eight-strip PCR tubes, and the fluorescence signals were monitored on the Bio-Rad CFX1000 instrument with heating from 25 °C to 95 °C at a rate of 1 °C/min, and ROX as the fluorescence channel. In this assay, the reference melting temperature was measured under the same condition but without the testing compound.
2.5. Docking and molecular dynamics
2.5.1. Protein and ligands
The 3D structures of tanshinone IIA sulfonate (CID: 23669322) and chloroxine (CID:2722) were downloaded from PubChem. The 3D structures of rac5c and ergotamine were obtained from PubChem (CID: 76853649) and ZINC 15 database (ZINC ID ZINC000052955754), respectively. It is noted that the rac5c from PubChem is the S-enantiomer. The PLpro chains from two X-ray crystal structures of SARS-CoV-2 PLpro in complex with mouse ISG15 C-terminal domain propargylamide [12] (PDB: 6XA9) and with ubiquitin propargylamide [12] (PDB: 6XAA) were used as the templates of SARS-CoV-2 PLpro.
2.5.2. Docking
All protein and ligand structures were prepared with MGLTools [13] v1.5.6 and submitted to AutoDock Vina [14] v1.1.2 for in silico docking. The original solvent and ligands in the crystal structures were removed to determine the docking grids of proteins. The docking grids were set to 52 Å × 82 Å × 66 Å for the PDB structure 6XA9 and 60 Å × 82 Å × 70 Å for the PDB structure 6XAA with grid spacing of 1 Å to cover all potential binding sites on the protein surface. Polar hydrogens were added to protein and ligand structures, and rotatable bonds of ligands were determined by the AutoDockTools [13] module included in MGLTools. In each docking attempt, AutoDock Vina reported 10 most possible binding conformations, and 5 repeats of attempts were performed for each PLpro-ligand pair. A total of 200 binding conformations were obtained and inspected as the candidates of further molecular dynamics analysis.
2.5.3. Molecular dynamics simulations
Protein topologies were prepared under Amber99SB-ildn [15], [16], [17] forcefield with Ambertools20 [18] and converted to GROMACS formats with ParmED [19]. ZAFF parameters [20] were applied to the zinc finger of PLpro. Topologies for all ligands were prepared with ACPYPE [21]. Protein structures complexed with a ligand were selected from the representative docking results. The system was then solvated with TIP3P water in a dodecahedron box extending 2 nm from the solute. Na+ and Cl− ions were added to neutralize the system's net charge and to reach a salt concentration of 150 mM. All simulations were set up with the running parameters described in previous study [22] and with GROMACS 2019.3 [23] on the high performance cluster of Shenzhen Bay Laboratory. Each simulation was performed for at least 200 ns.
2.5.4. Luciferase-based IFN-β reporter assay
In the luciferase-based IFN-β reporter assay, the effect of a compound on suppression of IFN-β promoter activity by SARS-CoV2 PLpro was investigated. HEK293T cells grown to 80% confluency in a 12-wells plate were co-transfected with 10 ng of plasmid pRL-SV40P (Addgene #27163), 50 ng of IFN-β-pGL3 firefly reporter plasmid (Addgene #102597), 50 ng of MAVS WT (Addgene #52135) and 50 ng of SARS-CoV2 PLpro with Lipofectamine 2000 (Invitrogen, 11,668,019). After 6 h of transfection, the medium was changed to a medium containing a testing compound. At 24 h post-transfection, cells were lysed in 1× passive lysis buffer (Promega, E1941). Firefly and Renilla luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega, E1910) on a Synergy HTX plate reader (BioTek). Experiments were performed in triplicates. Firefly luciferase activity was normalized by Renilla luciferase activity, and the statistical significance was determined using an unpaired two-tailed Student's t-test. A value of <0.05 was considered statistically significant.
2.5.5. Immunoblotting for detection of ISGylation
HEK293T cells were co-transfected with plasmids encoding Myc-tagged ISG15, SARS-CoV2 PLpro-EGFP, pcDNA3,1-UBE1L (E1), UBCH8 (E2), or 3XFlag-HERC5 (E3) [24]. These plasmids were kindly gifted by Dr. Hao Huang. After 6 h of transfection, the medium was changed to a medium containing a testing compound. At 24 h post-transfection, cells were lysed in 1× passive lysis buffer (Promega, E1941). Protein concentrations were determined by BCA assay (Beyotime, P0012). Equal protein amounts (30 μg) were subjected to SDS-PAGE, followed by transferring (200 mA 60 min) to PVDF membranes (Millipore, ISEQ00010). Membranes were blocked with TBST containing 5% skim-milk for 1 h at room temperature and then overnight with anti-ISG15 (1:1000, CST, 2758), anti-GFP (1:1000, CST, 2956), anti-GAPDH (1:1000, Biodragon, B1034) antibodies at 4 °C. After three washing steps, membranes were incubated with HRP-conjugated secondary antibodies (1:1000, Biodragon, BF03008) for 1 h at room temperature and then visualized with ECL substrate (Beyotime, P0018AFT) using a ChemiDoc MP Imaging System (Bio-Rad).
3. Results
3.1. Expression and purification of the proteins
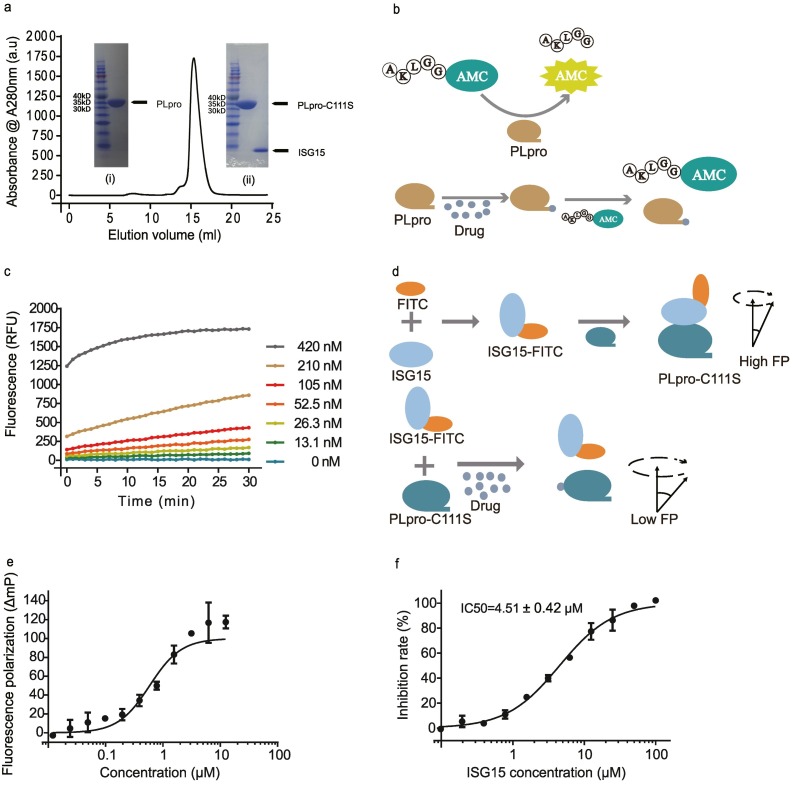
PLpro and PLpro-C111S were expressed by E.coli BL21(DE3) and purified by nickel column affinity, anion exchange, and gel filtration chromatography as shown by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Fig. 1a). PLpro-C111S is a PLpro mutant, in which the catalytic cysteine is replaced by serine to inactivate the protease activity but retain the binding capacity. ISG15 was also expressed and purified by nickel column (Fig. 1a).
Fig. 1.
Two assays established for screening inhibitors targeting SARS-CoV-2 PLpro. a) Size exclusion chromatography curve of PLpro purification. In the inset (i) the PLpro protein band is present at ~36.8kD on 12% SDS-PAGE, (ii) the PLpro-C111S protein band is present at ~36.8 kD on 12% SDS-PAGE and the ISG15 protein band at ~9.5 kD. b) The schematic diagram for the PLpro protease activity-based assay for inhibitor screening. Cleavage of a fluorogenic peptide by PLpro will release a free AMC fluorophore with its fluorescence signal correlated to the protease cleavage kinetics. The cleavage kinetics will be changed upon inhibition of the protease activity by an inhibitor (i.e., a drug candidate). c) Dependence of reaction kinetics (shown as fluorescence changes) on PLpro concentrations at a constant concentration of ALKGG-AMC (125 μM). d) The schematic diagram for the competitive fluorescence polarization-based assay for inhibitor screening. The binding between inactive C111S mutant of SARS-CoV-2 PLpro with FITC labeled ISG15 will restrict the rotation of ISG15-FITC. This restriction will be indicated with higher fluorescence polarization signals. The competition between an inhibitor and PLpro for binding with ISG15-FITC will be reflected from the concentration dependence of the fluorescence polarization signal. e) Florescence polarization changes in the reaction between PLpro and ISG15-FITC. f) Florescence polarization changes in competition with an unlabeled ISG15.
3.2. Establishing two assays for high-throughput drug screening targeting SARS-CoV-2 PLpro
We established two assays for inhibitor screening. The first one is based on the protease activity of SARS-CoV-2 PLpro. We designed a fluorogenic peptide ALKGG-AMC as the substrate. PLpro will recognize the peptide and specifically cleave the peptide bond after ALKGG. After cleavage, the AMC fluorophore will be released to its free state and emit fluorescence (Fig. 1b). If a compound binds to the corresponding active site of PLpro, the fluorogenic peptide ALKGG-AMC will not be cleaved by PLpro, thus, a lower fluorescence signal will be observed (Fig. 1b).
The second screening assay is based on fluorescence polarization. The binding activity between PLpro and ISG15 can be determined by monitoring the change of fluorescence polarization signal of fluorescein 5-isothiocyanate (FITC) labeled ISG15 (ISG15-FITC) [25]. Because the speed of molecular rotation of ISG15-FITC is faster in its free state than in its bound state with PLpro-C111S, the fluorescence polarization signal will decrease if an inhibitor can compete with ISG15-FITC in PLpro-C111S binding and render ISG15-FITC in its free state. We tested the competitive assay using unlabeled ISG15, which should compete with itself in PLpro binding. We employed 5 μM PLpro-C111S to react with various concentrations of unlabeled ISG15 for a period of time, and then added ISG15-FITC to a final concentration of 100 nM. We found that the half inhibitory concentration (IC50) of unlabeled ISG15 on PLpro-C111S and ISG15-FITC is 4.51 ± 0.42 μM (Fig. 1e).
3.3. Identification of two potential drugs targeting SARS-CoV-2 PLpro from the clinically approved drugs
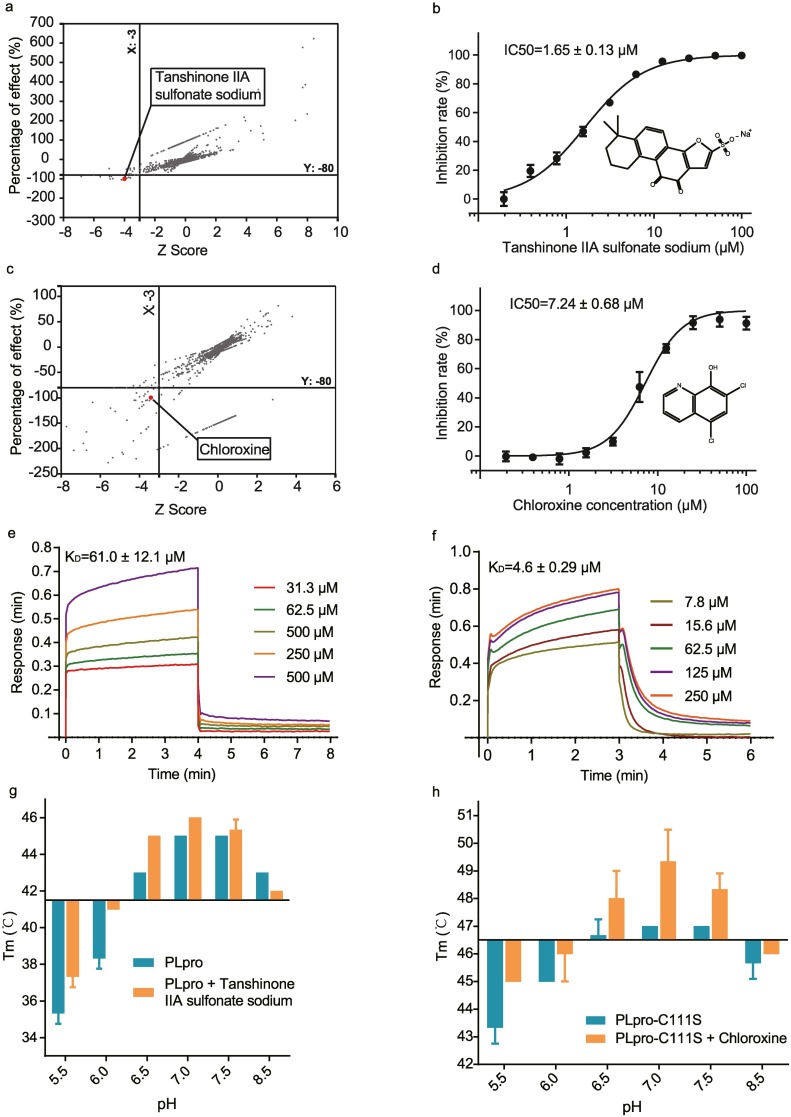
In the PLpro protease activity-based assay, all plates Z' factor values ranged from 0.60–0.98, confirming the reliability of the assay as 0.5 is considered as the threshold for a reliable assay [26]. There were 10 compounds whose inhibitory-effect values are greater than 80% against PLpro and Z scores are lower than −3 (Fig. 2a). We performed dose-dependent inhibition assays on PLpro to determine the IC50s of these 10 compounds. Only tanshinone IIA sulfonate sodium showed a lower than 10 μM IC50, with an IC50 of 1.65 ± 0.13 μM.
Fig. 2.
Tanshinone IIA sulfonate sodium and chloroxine were identified as the inhibitors of SARS-CoV-2 PLpro. a) Screening the clinically approved drugs by the PLpro protease-activity-based assay. Z scores indicate the significance of potential inhibitors discovered. The percentage of effect indicates the inhibition rate of a compound against the PLpro protease activity. b) The dose-dependent inhibition curve of tanshinone IIA sulfonate sodium in the PLpro protease-activity-based assay. c) Screening clinically approved drugs based on the fluorescence polarization assay. d) The dose-dependent inhibition curve of chloroxine in the competitive PLpro binding-based assay. e,f) Sensorgrams of tanshinone IIA sulfonate sodium (e) and chloroxine (f) in the BLI assay at different compound concentrations. Both compounds can bind to PLpro in the BLI assay. g,h) Melting temperatures (Tm) of PLpro in the absence or presence of tanshinone IIA sulfonate sodium (g) and chloroxine (h) in the thermal shift assay at different pHs. Both compounds can alter Tm of PLpro at various pHs, which is consistent with their specific binding with PLpro.
In fluorescence polarization-based assay, the Z'-factor values ranged from 0.7–0.95. There were 20 compounds whose inhibitory effect values are greater than 80% against PLpro-C111S, Z scores are lower than −3 and fluorescence polarization lower than the positive control. (Fig. 2c). For further studies, we selected these compounds to determine their IC50s. Only chloroxine has an IC50 lower than 10 μM (7.24 ± 0.68 μM).
3.4. Validating binding of SARS-CoV-2 PLpro with tanshinone IIA sulfonate sodium and chloroxine
We confirmed the interactions between PLpro and the above two compounds by biolayer interferometry (BLI) assay. In this assay, a protein (PLpro or PLpro-C111S) labeled with biotin was loaded on the SA sensor, and different concentrations of the testing compound was placed in a 96-well plate for association and dissociation. We found that KD is 145 ± 8.5 μM between PLpro and tanshinone IIA sulfonate sodium (Fig. 2e) and 4.6 ± 0.29 μM between PLpro-C111S and chloroxine (Fig. 2f).
We performed the thermal shifting assay to further examine the interaction between PLpro and these two compounds. In this assay, SYPRO Orange, a fluorogenic dye binding to hydrophobic residues, was utilized to monitor the denaturation of PLpro as the temperature increases and calculate the melting temperature (Tm) [27]. When a compound binds to the protein, the stability of the protein may change (i.e. a shift of Tm). For tanshinone IIA sulfonate sodium, the thermal shifting assay was conducted with a mixture of 25 μM PLpro and tanshinone IIA sulfonate sodium at a molar ratio of 1:1 under six different pH and using the buffer without tanshinone IIA sulfonate sodium as a control. PLpro and PLpro/tanshinone IIA sulfonate sodium exhibited the highest stability at pH = 7.0. Tm was increased by 1 °C for PLpro mixed with tanshinone IIA sulfonate sodium (Fig. 2g). PLpro-C111S and PLpro-C111S/chloroxine also exhibited the highest stability at pH = 7.0. Mixing PLpro with chloroxine increases Tm by 2.5 °C (Fig. 2h). Interestingly, the Tm value (47 °C) of PLpro-C111S is 2 °C higher than that of PLpro (45 °C).
3.5. Interaction mechanism of SARS-CoV-2 PLpro with tanshinone IIA sulfonate sodium and chloroxine
Docking and molecular dynamics (MD) were employed to investigate the molecular mechanisms of the interaction of SARS-CoV-2 PLpro with tanshinone IIA sulfonate sodium and chloroxine.
First, tanshinone IIA sulfonate sodium and chloroxine were individually docked to two template PLpro structures in substrate-bound state. For each PLpro-ligand pair, 5 repeats of docking attempts were made with AutoDock Vina [14], resulting a total of 200 predicted binding conformations.
We found that the docked conformations of tanshinone IIA sulfonate sodium are dominantly located at the catalytically active pocket of PLpro. Thus, three representative conformations with predicted affinities of −8.6, −8.2 and − 8.0 kcal/mol (marked as tanshinone IIA sulfonate sodium-a, tanshinone IIA sulfonate sodium-b, tanshinone IIA sulfonate sodium-c, respectively) were selected for further molecular dynamics verifications. By comparison, docking conformations of chloroxine were located near the PLpro-ISG15 binding interface, in addition to the catalytically active pocket. We selected four representative conformations: two in the active pocket with predicted affinities −5.9 and − 5.7 kcal/mol (marked as chloroxine-a and chloroxine-b) and two at the interface site (near Arg65 with predicted affinity −5.6 kcal/mol, marked as chloroxine-c and near Phe31 with predicted affinity −5.1 kcal/mol, marked as chloroxine-d).
The above-selected binding conformations were subjected to MD simulations for 200 ns. Among all the simulations, the ones started from tanshinone IIA sulfonate sodium-a and chloroxine-c reached stable binding.
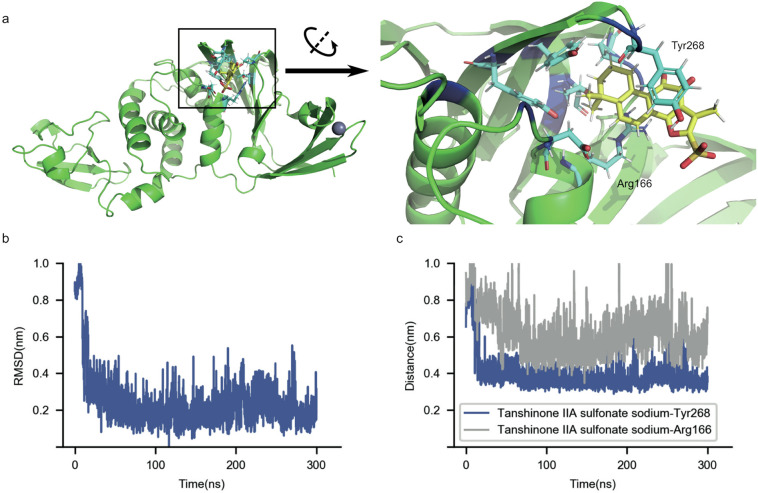
The stable binding of tanshinone IIA sulfonate sodium (Fig. 3b, c) attributes to a stable conformation in the active pocket (Fig. 3a) with the hydrophilic sulfonate group exposed to the solvent and the hydrophobic end inserted into the pocket. The stability is further strengthened by the interaction between the aromatic rings of tanshinone IIA sulfonate sodium and nearby residues (Fig. 3c): π-π stacking to Tyr268 side chain and cation-π interaction on the other side to Arg166 side chain. These aromatic-related interactions clamp the ligand in a relatively rigid conformation (Fig. 3b). To further confirm the stability of this binding mode, we extended the simulation to 300 ns and the binding conformation persisted throughout the simulation. Thus, tanshinone IIA sulfonate sodium inhibits the PLpro activity by occupying the active pocket of PLpro.
Fig. 3.
Binding of tanshinone IIA sulfonate sodium in the PLpro active pocket. a) The representative conformation of the stable binding mode found by docking and MD simulations. The pocket region is rotated and zoomed on the right panel for a clearer view. PLpro is shown in green, pocket residues in blue, tanshinone IIA sulfonate sodium in yellow and the PLpro residues within 3 Å of tanshinone IIA sulfonate sodium in cyan. b) The heavy-atom RMSD of tanshinone IIA sulfonate sodium with the representative conformation as the reference. c) The distance between the center of mass (COM) of tanshinone IIA sulfonate sodium and the COM of Tyr268 side chain, and between the COM of tanshinone IIA sulfonate sodium and the CZ (the terminal carbon) atom of Arg166 side chain. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
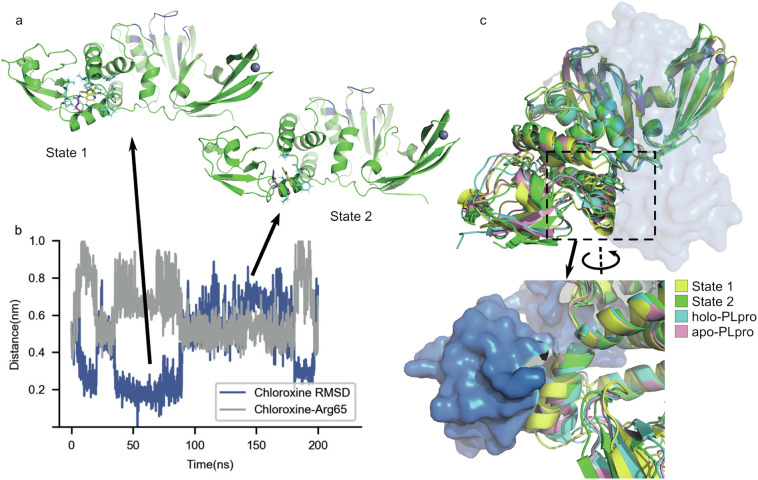
By comparison, chloroxine did not show stable binding to the active pocket but has a unique binding site at the PLpro-ISG15 binding interface, near Arg65. The trajectory initiated from chloroxine at the active pocket (chloroxine-a) showed an interesting event of relocation of the ligand from the active pocket to the Arg65 site (data not shown). MD simulations revealed a two-state manner at this binding site (Fig. 4a, b). Chloroxine can either pack in the hydrophobic pocket (State 1) or be anchored by Arg65 through cation-π interaction (State 2). In both states, binding of chloroxine can cause significant change in the orientation of the helix where Arg65 resides (Fig. 4c). This helix is critical for the binding of the N-terminal domain of ISG15 (Fig. 4c). Additionally, residues Arg65, Pro77, Thr75 and Phe69 are all important interface residues for ISG15 binding. These residues can be re-orientated to interact with chloroxine in State 2. Binding of chloroxine could have a direct impact on interrupting the PLpro-ISG15 binding interface.
Fig. 4.
Interruption of the PLpro-ISG15 binding interface by binding of chloroxine near Arg65 in a two-state manner. a) The representative conformations of State 1 and State 2 with the same coloring scheme as in Fig. 3a. Arg65 is individually colored magenta. b) The heavy-atom RMSD of chloroxine with State 1 as the reference and the distance between the center of mass of chloroxine and the CZ (the terminal carbon) atom of Arg65 side chain. c) Apo-PLpro, the structures of State 1 and State 2 aligned to the holo-PLpro (PLpro with ISG15 bound) structure. The region of the helix where Arg65 resides is zoomed and rotated 180°for a clearer view. ISG15 in the holo-PLpro structure is colored marine and shown as surface. The structures of apo-PLpro and holo-PLpro were taken from PDB 6XAA[12] and 6YVA [35], respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
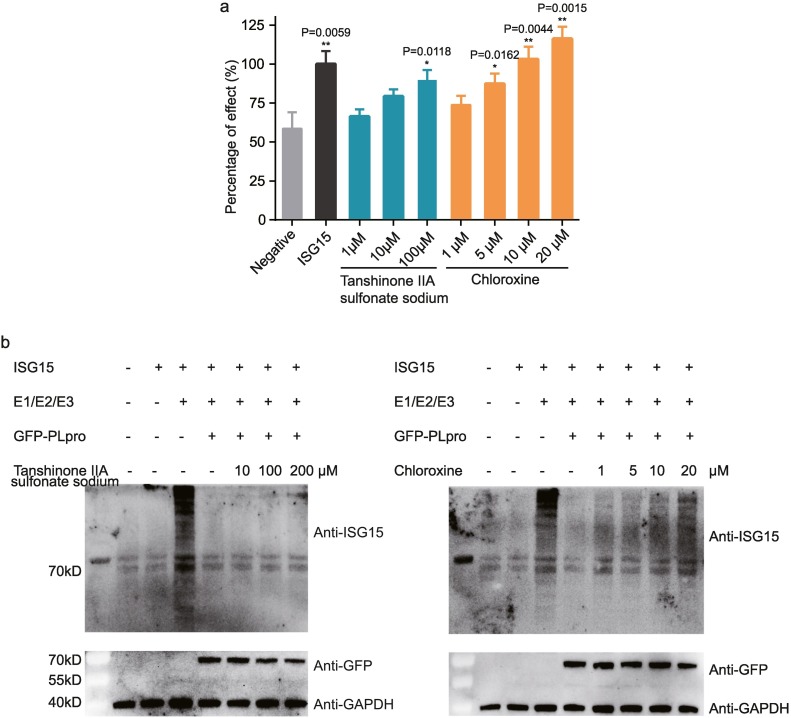
To investigate the inhibitory effects of tanshinone IIA sulfonate sodium and chloroxine on the deISGylation activity of PLpro inside cells, two cell-based methods, luciferase-based IFN-β reporter assay and anti-ISG15 immunoblotting, were employed to observe whether the two compounds can recover the cellular ISGylation level inhibited by PLpro. Consistent with their activities in the two drug-screening assays, tanshinone IIA sulfonate sodium and chloroxine can recover the cellular ISGylation level with a dose-dependent manner in both cell-based methods (Fig. 5 , also See Supplementary Fig. S2, S3 and Table S1, S2 for the original data), indicating that both compounds can enter cells to inhibit SARS-CoV-2 PLpro. Meanwhile, chloroxine showed a higher potency to recover the cellular ISGylation level than tanshionone IIA sulfonate sodium, suggesting that chloroxine directly targets the PLpro-ISG15 binding interface, as shown from the protease activity assay, the binding assay, the docking results and the MD simulations.
Fig. 5.
Tanshinone IIa sulfonate sodium and chloroxine can recover the anti-viral activity of ISG15 according to cell-based assays. a) Reduced suppression of IFN-β promoter activity by SARS-CoV-2 PLpro in the presence of tanshinone IIa sulfonate sodium and chloroxine according to luciferase-based IFN-β reporter assay. The negative control is treated with blank plasmid. The sample that treated with ISG15 expressed plasmid is selected as positive control, and its IFN-β promoter activity is set as 100% percentage of effect. Other samples are presented by scaling to positive control. Significance relative to the negative control was calculated by an unpaired two-tailed Student's t-test and labeled above the histogram bars. b) Immunoblotting for detection of ISGylation with or without tanshinone IIa sulfonate sodium or chloroxine treatment at various concentrations, with the plasmids encoding ISG15, E1/E2/E3 enzymes, and GFP-PLpro in combination.
4. Discussion
Drug development is a time-consuming and costly process because of unknown toxicity and efficacy of chemical compounds. As a result, repurposing existing drugs for new applications becomes increasingly attractive. This is particularly true as there is an urgent need for COVID-19 therapeutics. Here, we established two screening techniques for locating potential inhibitors of PLpro protease and ISG15-binding activities from clinically approved drugs. Two inhibitors (tanshinone IIA sulfonate sodium and chloroxine) were found with IC50 < 10 μM in our assays. Other studies also found several PLpro inhibitors with IC50 values at 1–6 μM [28]. Those inhibitors provided potential drugs or lead compounds for treating COVID-19.
Recently, Klemm et al. [4] also performed a fluorescence polarization-based assay to screen potential PLpro inhibitors from 5576 compounds for inhibiting the protease cleavage function. They did not find any hits as genuine PLpro inhibitors. In our study, we screened potential PLpro inhibitors by developing two different assays based on the protease activity (Assay I) and the deISGylation activity via interacting with ISG15 (Assay II), respectively. Our Assay II is a new assay. Assay I differs from Klemm et al. in the substrates used for the functional assay. Different substrates may have different sensitivities and specificities on the PLpro protease activity, which may lead to different screening results. We also noted that tashinone IIA sulfonate sodium, the hit from our Assay I, is not contained in the 5576 compounds screened by Klemm et al. That is, the ability to discover PLpro inhibitors in this work is due to a different compound library and new assays.
Two PLpro-ligand systems were also previously investigated by Huynh et al. [29]. Ligand rac5c was reported to stably bind to the active pocket of PLpro in MD simulations, while ergotamine, although having a good docking score, dissociated from the PLpro pocket during the MD simulation. To confirm our docking and MD protocols employed in our work, we utilized the same protocols on these two PLpro-ligand systems. Our docking results found the best score of −7.9 kcal/mol for rac5c and − 9.2 kcal/mol for ergotamine, respectively. Considering the fact that our docking protocol is rigid docking to the crystal structure of PLpro, the score for ergotamine was consistent with the reported rigid docking score (−9.2 kcal/mol) in the previous work. The docking conformation of ergotamine was not compared because such a conformation was not provided in the previous work. For rac5c, our best-score docking conformation was similar to the docking structure of PLpro-rac5c complex in the previous work, excepting the flipped naphthalene orientation due to different ligand chirality. The best scored structures were then subjected to MD simulations following the same setup as of tanshinone IIA sulfonate and chloroxine, with the exception of resetting the temperature to 300 K in order to be consistent with the previous work. During the simulation, ergotamine escaped from the pocket after 23.6 ns, whereas the binding conformation of rac5c was initially distorted during heating, but restored in 22 ns and the conformation remained stable throughout the rest of the simulation. The RMSD of rac5c is shown in Supplementary Fig. S1a. Thus, although different initial protein conformations (an experimentally determined structure versus a hypothetical structure) and different forcefields (Amber99SB-ildn versus CHARMM36 for PLpro and GAFF versus SwissParam generated forcefield for rac5c) were employed in this and previous work, the two resulting trajectories displayed binding features essentially the same: the naphthalene fragment (with the attached methyl group) is stably surrounded by P247, P248, T301, Y268 and the more distant M208 (Supplementary Fig. S1b); the piperidine fragment interacts with nearby Y264, Y273 and Y268 (Supplementary Fig. S1c) and the pyridine fragment interacts with Q269 and L162 in a flexible manner (Supplementary Fig. S1d). The hydrogen bond between the peptide-backbone of rac5c and the backbone oxygen of Y268 is also formed and persist throughout the simulation (Supplementary Fig. S1a). The sidechain of Y268 is found repeatedly flipping up and down in a two-state manner (Supplementary Fig. S1b,c). A similar behavior can also be observed in the trajectory of the previous work, although the flipping was not repeated in the previous work and the authors suggested that the down-state is energetically more favorable. The overall consistency between this work and Huynh et al. on the simulation results of the same two ligands confirms the robustness of the simulation protocols employed in this work.
Tanshinone IIA sulfonate sodium discovered here is a derivative of tanshinone IIA, which is very promising in the development of cardioprotective drugs. Chloroxine is an analogue of chloroquine. Since the SARS-CoV outbreak in 2003, chloroquine was reported to have an anti-SARS-CoV activity [30], [31]. SARS-CoV could enter a host cell through the pH-dependent mechanism and the low pH of lysosome could trigger the fusion between the virus and cell membrane [32]. It was suggested that chloroquine increases the endosomal pH [33] and thus prevents the virus from entering the cell. Chloroquine also interfered the terminal glycosylation of ACE2 on the host cell membrane, affecting Spike protein to recognize ACE2. Because of the high toxicity of chloroquine, the concentration range suitable for clinical treatment is narrow. Chloroquine phosphate, a derivative of chloroquine, was also used to treat COVID-19 in China. Here we showed that chloroxine, as the analogue of chloroquine, has IC50 with 7.24 ± 0.68 μM targeting PLpro, compared to EC50 of 5.47 μM for chloroquine in vitro [34]. Thus, the potential usefulness of chloroxine as a drug should be further investigated by performing antiviral activity experiments at the cellular and animal levels.
The current μM level activity of tanshinone IIA sulfonate sodium and chloroxine indicates the room for further improvement. Docking and molecular dynamics of PLpro with tanshinone IIA sulfonate sodium and chloroxine provided reasonable binding conformations that can be employed for improving potency of these inhibitors by chemical modifications. Stronger binders would also allow determination of complex structures to confirm binding poses obtained from docking and MD simulations.
CRediT authorship contribution statement
Yaoqi Zhou, Jian Zhan and Wei Xu: Conceptualization, Methodology, Supervision, Writing – Reviewing and Editing, Project administration and Funding acquisition. Yunxia Xu, Ke Chen, Juanli Pan: Investigation, Data curation, Writing - Original draft preparation. Yingshou Lei, Danting Zhang, Lipei Fang, Jinle Tang, Xin Chen, Yanhong Ma: Visualization, Investigation, Validation. Yi Zheng, Bao Zhang: Resources.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgements
We appreciate the grants of National Natural Science Foundation of China (32041005), Guangzhou Science and Technology Program key projects (201904010477, 202008040001). This work is also supported by Key Area Research and Development Program of Guangdong Province (2020A111128015), Shenzhen Science and Technology Program (Grant No. KQTD20170330155106581), and Major Program of Shenzhen Bay Laboratory (S201101001). The support of Shenzhen Bay supercomputing facility is also acknowledged. We would like to thank Dr. Hao Huang lab at Peking University Shenzhen Graduate School for the plasmids of ISGylation assay.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2021.07.184.
Appendix A. Supplementary data
Supplementary figures
Supplementary tables
References
- 1.Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79(24):15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Báez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E., Blackmore T.R., van der Heden G.J., van Noort P.P., Geurink H., Ovaa J., Newman A., Riboldi-Tunnicliffe P.E., Czabotar J.P., Mitchell R., Feltham B.C., Lechtenberg K.N., Lowes G., Dewson M., Pellegrini G., Lessene D.Komander. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39(18) doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W., Bailey-Elkin B.A., Knaap R.C.M., Khare B., Dalebout T.J., Johnson G.G., van Kasteren P.B., McLeish N.J., Gu J., He W., Kikkert M., Mark B.L., Sidhu S.S. Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants. PLoS Pathog. 2017;13(5) doi: 10.1371/journal.ppat.1006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K., Baez-Santos Y.M., Wang J., Takayama J., Ghosh A.K., Li K., Mesecar A.D., Baker S.C. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 2010;84(9):4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindner H.A., Lytvyn V., Qi H., Lachance P., Ziomek E., Ménard R. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 2007;466(1):8–14. doi: 10.1016/j.abb.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siemieniuk R., Rochwerg B., Agoritsas T., Lamontagne F., Leo Y.S., Macdonald H., Agarwal A., Zeng L., Lytvyn L., Appiah J.A., Amin W., Arabi Y., Blumberg L., Burhan E., Bausch F.J., Calfee C.S., Cao B., Cecconi M., Chanda D., Cooke G., Du B., Dunning J., Geduld H., Gee P., Hashimi M., Hui D.S., Kabra S., Kanda S., Kawano-Dourado L., Kim Y.J., Kissoon N., Kwizera A., Laake J.H., Machado F.R., Mahaka I., Manai H., Mino G., Nsutedu E., Pshenichnaya N., Qadir N., Sabzwari S., Sarin R., Sharland M., Shen Y., Ranganathan S.Sri, Souza J., Ugarte S., Venkatapuram S., Dat V.Quoc, Vuyiseka D., Stegemann M., Wijewickrama A., Maguire B., Zeraatkar D., Bartoszko J., Ge L., Brignardello-Petersen R., Owen A., Guyatt G., Diaz J., Jacobs M., Vandvik P.O. A living WHO guideline on drugs for covid-19. BMJ (Clinical research ed.) 2020;370 doi: 10.1136/bmj.m3379. [DOI] [PubMed] [Google Scholar]
- 11.Brideau C., Gunter B., Pikounis B., Liaw A. Improved statistical methods for hit selection in high-throughput screening. J. Biomol. Screen. 2003;8(6):634–647. doi: 10.1177/1087057103258285. [DOI] [PubMed] [Google Scholar]
- 12.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E., Blackmore T.R., van der Heden G.J., van Noort P.P., Geurink H., Ovaa J., Newman A., Riboldi-Tunnicliffe P.E., Czabotar J.P., Mitchell R., Feltham B.C., Lechtenberg K.N., Lowes G., Dewson M., Pellegrini G., Lessene D.Komander. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39(18) doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17(1):57–61. [PubMed] [Google Scholar]
- 14.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Case D.A., Cheatham T.E., 3rd, Darden T., Gohlke H., Luo R., Merz K.M., Jr., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 18.Moriarty N.W., Janowski P.A., Swails J.M., Nguyen H., Richardson J.S., Case D.A., Adams P.D. Improved chemistry restraints for crystallographic refinement by integrating the Amber force field into phenix. Acta Crystallogr. D Struct. Biol. 2020;76(Pt 1):51–62. doi: 10.1107/S2059798319015134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirts M.R., Klein C., Swails J.M., Yin J., Gilson M.K., Mobley D.L., Case D.A., Zhong E.D. Lessons learned from comparing molecular dynamics engines on the SAMPL5 dataset. J. Comput. Aided Mol. Des. 2017;31(1):147–161. doi: 10.1007/s10822-016-9977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters M.B., Yang Y., Wang B., Füsti-Molnár L., Weaver M.N., Merz K.M., Jr. Structural survey of zinc containing proteins and the development of the zinc AMBER force field (ZAFF) J. Chem. Theory Comput. 2010;6(9):2935–2947. doi: 10.1021/ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sousa da Silva A.W., Vranken W.F. ACPYPE - AnteChamber PYthon parser interfacE. BMC Res. Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoemark D.K., Colenso C.K., Toelzer C., Gupta K., Sessions R.B., Davidson A.D., Berger I., Schaffitzel C., Spencer J., Mulholland A.J. Molecular simulations suggest vitamins, retinoids and steroids as ligands of the free fatty acid pocket of the SARS-CoV-2 spike protein*. Angew. Chem. Int. Ed. Engl. 2021;60(13):7098–7110. doi: 10.1002/anie.202015639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 24.Fu Z., Huang B., Tang J., Liu S., Liu M., Ye Y., Liu Z., Xiong Y., Zhu W., Cao D., Li J., Niu X., Zhou H., Zhao Y.J., Zhang G., Huang H. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021;12(1):488. doi: 10.1038/s41467-020-20718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao T., Frey G., Fu Q., Lavine C.L., Scott D.A., Seaman M.S., Chou J.J., Chen B. HIV-1 fusion inhibitors targeting the membrane-proximal external region of env spikes. Nat. Chem. Biol. 2020;16(5):529–537. doi: 10.1038/s41589-020-0496-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 27.Huynh K., Partch C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015;79 doi: 10.1002/0471140864.ps2809s79. 28.9.1-28.9.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y., Du X., Duan Y., Pan X., Sun Y., You T., Han L., Jin Z., Shang W., Yu J., Guo H., Liu Q., Wu Y., Peng C., Wang J., Zhu C., Yang X., Yang K., Lei Y., Guddat L.W., Xu W., Xiao G., Sun L., Zhang L., Rao Z., Yang H. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein & Cell. 2021 doi: 10.1007/s13238-021-00836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huynh T., Cornell W., Luan B. In silico exploration of inhibitors for SARS-CoV-2's papain-like protease. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.624163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323(1):264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z.Y., Huang Y., Ganesh L., Leung K., Kong W.P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78(11):5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Bari M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5(1) doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., Geurink P.P., Wilhelm A., van der Heden G.J., van Noort H., Ovaa S., Müller K.P., Knobeloch K., Rajalingam B.A., Schulman J., Cinatl G., Hummer S., Ciesek I.Dikic. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587(7835):657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplementary tables