Graphical abstract

Keywords: Caspase-6, Conformational transition, Perturbation-response scanning, Targeted molecular dynamics, H-bond interactions
Abstract
Caspase-6 participates in a series of neurodegenerative pathways, and has aroused widespread attentions as a promising molecular target for the treatment of neurodegeneration. Caspase-6 is a homodimer with 6 central-stranded β-sheets and 5 α-helices in each monomer. Previous crystallographic studies suggested that the 60′s, 90′s and 130′s helices of caspase-6 undergo a distinctive conformational transition upon substrate binding. Although the caspase-6 structures in apo and active states have been determined, the conformational transition process between the two states remains poorly understood. In this work, perturbation-response scanning (PRS) combined with targeted molecular dynamics (TMD) simulations was employed to unravel the atomistic mechanism of the dynamic conformational transitions underlying the substrate-induced activation process of caspase-6. The results showed that the conformational transition of caspase-6 from apo to active states is mainly characterized by structural rearrangements of the substrate-binding site as well as the conformational changes of 60′s and 130′s extended helices. The H-bond interactions between L1, 130′s helix and 90′s helix are proved to be key determinant factors for substrate-induced conformational transition. These findings provide valuable insights into the activation mechanism of caspase-6 as well as the molecular design of caspase-6 inhibitors.
1. Introduction
Caspase, also known as apoptotic protease Mch-2, is a cysteinyl aspartate-specific protease that plays essential roles in cell regulatory of apoptosis and inflammation [1], [2]. Up to now, a total of 11 functional caspase subtypes have been found in human, which are mainly divided into three classes: (i) apoptotic initiators (caspases-2, −8, −9, and −10); (ii) apoptotic executioners (caspases-3, −6, and −7); and (iii) inflammatory caspases (caspases-1, −4, −5 and −14) [3], [4], [5]. As an important member of caspase family, caspase-6 is involved in the process of axonal pruning and neuronal degeneration by cleaving neuronal substrates, e.g., microtubule-associated protein Tau [6], amyloid precursor protein (APP) [7], huntingtin protein (HTT) [8] and Parkinson disease protein 7 (PARK7) [9]. Recently, a series of substrate-mimic and allosteric molecules have been identified as caspase-6 inhibitors with nanomolar to micromolar potencies [10], [11]. Thus, caspase-6 has become one of the most important therapeutic targets for the treatments of neurodegenerative diseases [9], [12], [13], [14], [15], [16], [17], [18], [19].
Caspase-6 is firstly expressed as an inactive zymogen. The zymogen can be activated in two ways: auto-activation or caspase-3-mediated activation [20]. In the process of auto-activation, caspases-6 zymogen is initially cleaved at Asp193 in the inter-subunit linker. Then, it sequentially goes through proteolytic cleavages at Asp23 in the pro-domain and Asp179 in the inter-subunit linker, and finally inverts into full maturation (Fig. 1) [21], [22]. Caspase-6 is a homodimer and each monomer consists of 6 central-stranded β-sheets and 5 α-helices (Fig. 2). The core structure of caspase-6 is comprised of a large subunit, a small subunit and an inter-subunit linker. The large subunit contains active-site catalytic dyad residues (i.e., Cys163-His121), which can cleave natively folded proteins at a specified aspartic acid residue for apoptosis [23]. The small subunit comprises most of the dimer interface and an allosteric site [22], [23], [24]. Four loops (L1, L2, L3, and L4) protruding from the central β-sheet form the substrate-binding groove. All of the caspase subtypes can adopt a canonical conformation, where the substrates bound to the β-strand platform of the 130′s helix. Intriguingly, the lack of helix-breaking residues in the 130′s helix of caspase-6 leads to a non-canonical conformation solely presented in caspase-6 [25]. Notably, the canonical conformation was observed in the zymogen, ligand free (apo) and active states of caspase-6, while the non-canonical conformations exist in the apo, zinc-mediated allosteric and irreversible inhibitor Z-VAD-FMK-bound state, etc.. [22], [24], [26], [27], [28]. Different from the canonical conformation, the non-canonical conformation adopts two extended helices in atop of 60′s (residues 57–70) and 130′s (residues 128–142) regions. These conformational changes are also accompanied by an outward rotation of the 90′s helix (Fig. 2) [23], [24].
Fig. 1.
The activation process of caspase-6 zymogen. Five caspase-6 structures: full-length zymogen (PDB ID: 4YIR), △Asp193 zymogen, △pro-domain (PDB ID: 3NR2), non-canonical apo (PDB ID: 2WDP) and canonical active (PDB ID:3OD5) conformations. The pro-domain, L2 loop, large and small subunit are represented as orange, red, grey and cyan, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
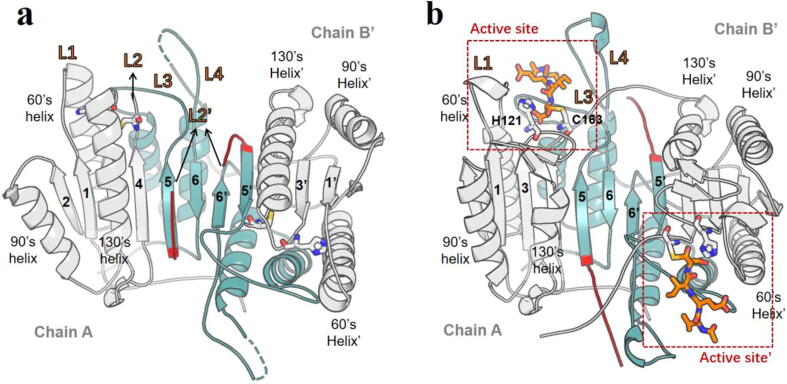
Fig. 2.
Structures of the caspase-6. (a): the non-canonical apo state (PDB ID:2WDP); (b) the canonical active state (PDB ID: 3OD5). The large and small subunits are colored in grey and cyan, respectively. The parts of the inter-domain linker (loops L2 and L2′ residues 198–205) are colored in red. The catalytic dyad residues (H121, C163) are represented as sticks. The dashed cyan line refers to the flexible part not determined in the crystal structure. The co-crystallized Ac-VEID-CHO, a tetrapeptide (Val-Glu-Ile-Asp) inhibitor, is represented as orange sticks. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A growing body of evidence suggested that caspase-6 undergoes distinctive conformational transitions upon substrate binding. By comparing the circular dichroism (CD) spectra between apo and substrate-bound states of caspase-6, Vaidya et al. [23] observed that the molar ellipticity (θ) ratio between 208 and 222 nm signals increased consistently, indicating a loss in α-helical content upon substrate binding. Furthermore, Dagbay et al. [29] employed hydrogen/deuterium exchange mass spectrometry (H/DX-MS) to explore the conformational flexibility of caspase-6. The results suggested that caspase-6 underwent a distinct helix-strand inter-conversion in the initial substrate recruitment, and then reached to a dynamic equilibrium between the helix-extended and canonical strand conformations. Although crystallographic studies have determined the helix-extended apo and active (bound by a substrate-mimic inhibitor) conformations [22], [28], the activation mechanism of caspase-6 is still unclear.
In this wok, taking full advantages of the crystal structures of caspase-6 in apo and active states, perturbation-response scanning (PRS) combined with targeted molecular dynamics (TMD) simulations was employed to elucidate the conformational transition in caspase-6 (Fig. 3). Firstly, 100 ns MD simulations were carried out to explore the intrinsic dynamics of the caspase-6 in apo and active states, respectively. Then, based on the MD trajectories, a total of 22 crucial residues for conformational transition were identified by PRS method. Finally, TMD simulations based on the Cα atoms of the 22 crucial residues identified were performed to explore atomic-level details in the process of the conformational transition. Collectively, our findings can provide valuable insights into the activation mechanism of caspase-6.
Fig. 3.
The flowchart for exploring the conformational transition during caspase-6 activation.
2. Methods
2.1. Structural preparation
In this work, the crystal structures of caspase-6 in apo (PDB ID: 2WDP) and active states (PDB ID: 3OD5, co-crystallized with a substrate-mimic inhibitor: Ac-VEID-CHO) were used to explore the conformational transition [22], [24]. The missing residues in L4 loop (residues 262–265) were modeled and refined by Modeller 9 software. The protonation states of Glu, Asp and His were assigned by using PDB2PQR web server at a pH value of 7.5 [30]. Glu135 was set to deprotonation state based on the experimental research [29]. After solvated by a TIP3P water box with the dimensions of 100 × 100 × 100 Å3 and neutralized by Cl− ions, 50 ps of steepest descent followed by 50 ps of conjugate gradient energy minimizations were performed before MD simulations.
2.2. Molecular dynamics simulations
MD simulations with periodic boundary conditions [31], [32] were performed by NAMD 2.12 with CHARMM36 force field [33]. First, each system was gradually heated from 0 to 310 K within 5000 ps in the NVT ensemble, where the backbone of caspase-6 was harmonically constrained by a force of 50 kcal/mol·Å2. Then, a 100 ns MD simulation was performed in the NPT ensemble (310 K, 1 atm) without any constraint. The integration time step was set to 2 fs and the cutoff value of van der Waals and electrostatic interactions was set to 10 Å. The particle mesh Ewald (PME) method was used to calculate the long-range electrostatic interactions [34] and the SHAKE algorithm [35] was used to constrain the covalent bonds with H atoms. For the structure in active state, duo to the Asp residue of Ac-VEID-CHO forms a covalent bond with the catalytic Cys163, the distance of C-S bond was thus harmonically constrained by a force of 250 kcal/mol·Å2 during MD simulations.
2.3. Perturbation-Response scanning
PRS is a linear response theory-based computational technique, which can be used to determine the residue-level effects of perturbed forces on the residues of a given protein [36], [37]. By monitoring the responses of the perturbed protein, PRS can depict dynamic couplings and link the directionality of the exerted forces to the protein response in functional movement [38], [39], [40]. In PRS, the motions of caspase-6 were fully defined by a 3 N × 3 N covariance matrix (C), derived from the equilibrated MD trajectories. The coordinates of Cα atoms of each residue were used to represent a node of the network. The idea of PRS technique is to exert an external force on each residue, and record the response of each node in the network. The vector of exerted force and the corresponding response of each residue i are represented as:
| (1) |
| (2) |
where, and denote residue i conformations in caspase-6 initial (apo state) and perturbed coordinates; and represent the Boltzmann constant and the temperature, respectively; vector contains the coordinates of the force vectors applied on the residues. C represents the cross-correlation matrix of the fluctuations of the nodes of the caspase-6 in apo state.
Then, the Pearson correlation coefficients for each perturbed residue i was used to quantify the correlation between the theoretical and experimental conformational displacements averaged over all the affected residues j (Eq.3):
| (3) |
where, is the theoretical prediction of the displacement of residue j affected by perturbations; and the is the experimental displacements of residue j between the caspase-6 structures in the apo and active states. The overbar represents the average; and represent as the standard deviations of theoretical and experimental displacements, respectively. The value close to 1 means that a good agreement with the experimental conformational change, while the value close to 0 means that the lack of correlation between theoretical prediction and experimental displacements.
In this work, the thresholds of PRS are selected from the 100 perturbations based on following rules: (i) Firstly, rank and list the residues in descending order based on its value; (ii) If there is a sharp decrease in the correlation values, the top residues will be listed until that gap; (iii) If there is a smooth decrease in the correlation values, define the threshold equal to the Cimax value observed over all experiments less than 0.1 (Cimax-0.1) [41], [42]. Herein, the crystallized caspase-6 structures in apo and active states were used as inputs to perform PRS, and the equilibrated MD trajectory of caspase-6 in apo state was used to construct cross-correlation matrix of the fluctuations of the nodes. The PRS and correlation analysis for each residue of caspase-6 were carried out by MD-TASK [43].
2.4. Targeted molecular dynamic
As an enhanced sampling method, TMD can efficiently explore the conformational transition between two conformational states of a given biological system [44]. In this paper, caspase-6 structures in apo and active states were used as the initial and targeted conformations for the TMD simulation, respectively. Then, a steering force was applied to the Cα atoms of crucial residues identified by PRS, which accelerates the conformational transition from the initial apo state to the target active state. At each time step, the root-mean-square deviation (RMSD) between the current and target conformations was calculated, which was then used to determinate the instantaneous force on each atom at next time step (Eq.4).
| (4) |
Where ETMD represents the instantaneous potential applied to a given atom; is the steering force constant; M represents the number of steered atoms; RMSD(t) is the instantaneous RMSD between the current and the target coordinates; RMSD*(t) evolves linearly from the initial RMSD at the first TMD step to the final RMSD at the last TMD step.
3. Results and discussion
3.1. Structural flexibility of Caspase-6
In this paper, a 100-ns MD simulation was performed for caspase-6 structures in apo and active states, respectively. Both of the caspase-6 systems reached to equilibrium states in the last 80 ns, of which the mean RMSDs were 2.35 ± 0.18 Å for the apo state and 1.41 ± 0.09 Å for the active state (Fig. S1). The root-mean-square fluctuations (RMSFs) revealed that the structural flexibility of caspase-6 in active state is, in general, lower than that in apo state (Fig. 4a), which suggests a reduced structural flexibility upon the ligand binding. Also, it can be observed that the 90′s helix in active state exhibits smaller fluctuations than that in apo state. Moreover, it can be seen that, in both apo and active states, the fluctuations of the residues located in L1-L4 loops, 90′s and 130′s helices are more significant than other residues.
Fig. 4.
Conformational dynamics of caspase-6. (a) Cα-atom RMSF of the apo (grey) and active (cyan) states. The standard deviations of RMSF were calculated from 3 independent simulations. (b) Structural variations along the first PC mode. Directions of the first principal coordinates are represented as blue arrows. (c) Residue cross-correlation map. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Based on the last 80 ns equilibrium trajectories, principal component analysis (PCA) was carried out to investigate the structural variations and motion modes of caspase-6 in apo and active states. Fig. 4b shows the predominant movements of caspase-6 in the first PC mode. For the caspase-6 in apo state, it can be clearly observed that, the structural variance along the first PC mode is dominated by the motion of 90′s helix, the top half of 130′s helix, and loop residues. By contrast, the structural variance of the caspase-6 in active state is relatively small with the exception of residues in L1-L4 loops. Fig. 4c shows the residue cross-correlation map between the apo and active states. Notably, it can be seen that the 130′s helix has strong couplings with L1 loop in both apo and active states, which indicates the close concordance in the motions of L1 and 130′s extended helix. Collectively, significant motion couplings are observed among helix 90′s, helix130′s and L1-L4 regions.
3.2. PRS reveals 22 crucial residues in conformational transition
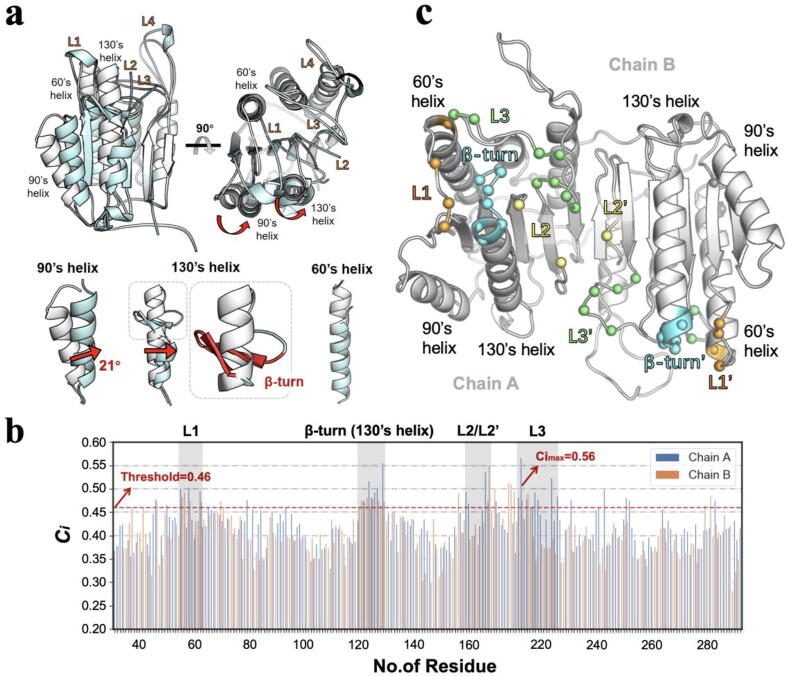
In comparison with apo state, the caspase-6 in active state undergoes significant structural changes: conformational transitions in L1-L4; β-turn formation in the top of 130′s helix (residues 125–142); rearrangements in the top of the extended 60′s helix (residues 57–70), and a 21° inward rotation of the 90′s helix (Fig. 5a) [24], [25]. Based on the last 80 ns MD trajectories of caspase-6 in apo state, the PRS with 100 times of perturbations was performed to detect the crucial residues affecting conformational transition. Fig. 5b shows the correlation coefficients (Ci) profiles between theoretical prediction and experimental displacements of each perturbed residue. According to the maximum Ci value obtained (Cimax = 0.56), the threshold of Ci was set to 0.46 (threshold = Cimax-0.1) for the following analysis. Herein, a total of 22 crucial residues with significant Ci were identified, which are mainly located in substrate-binding loops (L1-L3) and the top of 130′s helix (Table 1 and Fig. 5c).
Fig. 5.
Results of PRS. (a) Cα-atom-based structural superimposition of the caspase-6 in apo (gray) and active (cyan) states. The red arrows indicate the relative rotation movements. (b) Correlation coefficient (Ci) between theoretical prediction and experimental displacements for each perturbed residue. The threshold of 0.46 is shown by the red line. At least five replicates per PRS experiment. (c) The crucial residues identified by PRS. The Cα atoms of crucial residues located in L1/60′s helix, L2, L3, 130′s helix and others were colored in orange, yellow, green, cyan and gray, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Summary of hot residues of caspase-6.
| Location | Crucial residues | Refs |
|---|---|---|
| L1/60′s helix | Glu53,Phe55,Phe56, Trp57,His58,Pro62, Glu63*, Arg64*, Arg65* | [22], [29], [27] |
| L2/L2′ | Gln161*, Cys163*†, Arg164*,Gly165,His168, Asp179a, Asp193a,Leu200 | [22], [23] |
| L3 | Ser211,Val212,Glu214e,Gly215,Tyr217*e,Ser218*, His219*e, Arg220*, Glu221*,Thr222*,Val223, Ser226*, Trp227*, | [22], [24], [26], [46] |
| 130′s helix | His121*†,Gly122*e,Glu123e,Gly124,Asn125, His126e,Ile127,Tyr128, Asp131e, Lys133e, Glu135b | [23], [26], [29], [45] |
| Others | Asp23a, Arg42 f, Arg43 f, Arg44f, Lys36c, Arg65f, Gly66f,Ser242, Glu244c,Thr250, Leu251, Val261*,Leu282, His287c, Ser257d | [22], [23], [47], [48], [49] |
Bold: 22 crucial residues calculated by PRS (i.e., Phe55, Phe56, His58, Pro62, Gly165, Leu200, Ser211, Val212, Glu214, Gly215, Try217, Ser218, Thr222, Val223, His121, Gly122, Glu123, Gly124, Asn125, His126, Ile127 and Tyr128);
Italic: functional/allosterically relevant residues reported by previous researches;
† Catalytic dyad residues Cys163 and His121;
* Residues within 5 Å of active site;
Residues involved in caspase-6 events of a proteolysis, b rearrangement of 130′s helix, c zinc-mediated allosteric inhibition, d phosphorylation-mediated allosteric inhibition, e pep419 and Z-VAD-FMK-mediated allosteric inhibition, and f mutation-induced allosteric inhibition.
Previous studies revealed that there are strong inter-molecular interactions between the substrate-mimic inhibitor Ac-VEID-CHO and His121, Gly122, Tyr217, Ser218, His219, Arg220, Glu221 and Thr222 of the substrate-binding groove, especially the H-bond interactions with Arg64, His121, Ser218, Arg220, Glu221 [22], [24]. Recent researches suggested that residues located far from the substrate binding groove, e.g. Gly122, Glu123, Gly124, Asn125, His126, Ile127 and Tyr128 in the top of 130′s helix, also played important roles in the substrate-induced conformational transition process [23], [25]. Table 1 summarizes the recently reported hot residues affecting the conformational transition of caspase-6. It has been observed that most of the crucial residues determined by PRS are consistent with the identified functionally/allosterically relevant residues. For instance, His121-Tyr128 in 130′s helix and Ser211-Arg220 in L3 are pivotal to the binding of substrate-mimic inhibitors. In addition, it is worthy to note that Gly122, Glu123, His126, Glu214 and Tyr217 are located in allosteric sites for binding allosteric peptide pep419/Z-VAD-FMK [26], [45]. Collectively, the 22 crucial residues identified by PRS method agree well with the known hot residues, implying that PRS technique can provide useful information for exploring the key residues affecting conformational transition process.
3.3. The atom-level details in conformational transition by TMD analysis
3.3.1. optimization
TMD simulation was performed to investigate the dynamic activation process of caspase-6 according to the 22 crucial residues determined by PRS (Table 1). In TMD simulations, the steering forces were only imposed on the Cα atoms of 22 crucial residues. Firstly, in order to determine an optimal value, 500 ps TMD simulations were performed twice by using different values as shown in Table 2. According to the TMD results (Table 2), it can be deduced that 70 kcal/mol·Å2 is an optimal value with a small RMSD obtained between the last TMD and targeted conformations by using the smallest possible steering force. From Fig. 6a, it can be observed that the TMD simulation with 70 kcal/mol·Å2 can reproduce preferably the target caspase-6 conformation in active state. Thus, 70 kcal/mol·Å2 was selected as the optimal value.
Table 2.
The average RMSDs between the last TMD snapshot and initial/targeted conformations.
| No. | (kcal/mol·Å2) | Time (ps) | RMSD between the last TMD and initial conformations (Å) | RMSD between the last TMD and targeted conformations (Å) |
|---|---|---|---|---|
| 1 | 35 | 500 | 5.38 | 2.98 |
| 2 | 50 | 500 | 5.86 | 2.71 |
| 3† | 70 | 500 | 6.28 | 2.59 |
| 4 | 100 | 500 | 6.56 | 2.46 |
| 5 | 120 | 500 | 6.91 | 2.96 |
The optimal .
Fig. 6.
The conformational transitions of 60′s and 130′s helices observed from TMD simulations. (a) The caspase-6 structures in different conformational states. Cyan: apo state (PDB ID: 2WDP); Gray: active state (PDB ID: 3OD5); Green: the last TMD snapshot. The conformational transitions of (b) 60′s helix, (c) 130′s helix, and (d) Glu135 during the TMD simulations. The green arrows indicate the movement direction of short loop regions; The co-crystallized inhibitor Ac-VEID-CHO VEID is represented as orange mesh; The residues within 3 Å and 5 Å distances to the inhibitor Ac-VEID-CHO are represented as pink and red stick models; H-bonds are represented as black dashed lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3.2. Structural rearrangements in active site facilitate the reorganization of 60′s and 130′s extended helices
Accumulated evidence suggested that 60′s and 130′s extended helices of caspase-6 must undergo structural reorganizations before substrate binding. Fig. 6b shows the snapshots of 60′s helix during the TMD simulation. It can be observed that Arg64 is important for structural rearrangement of the 60′s extended helix, which facilitates the conformational transition by forming a H-bond interaction with Thr67. Previous researches suggested that Arg64 is located in the primary specificity pocket and responsible for the strict substrate specificity of caspase-6 (Fig. S2) [50].
By contrast, the reorganization of the 130′s helix is more complicated. Ser120 and His121 located in the 130′s helix can form strong polar interactions with the inhibitor Ac-VEID-CHO by the formation of β turn motif, implying that Ser120 and His121 play important roles in the substrate-induced conformational rearrangement of 130′s helix (Fig. 6c and S3) [22], [23]. Besides, it can be also observed that the rearrangement of the extended 130′s helix involves a series of ruptures and formations of inter-helix H-bonds (Fig. 6c), e.g., the H-bonds interactions of Glu123, His126 and Lys133, etc.. It should be noted that Glu123, His126 and Lys133 are located in the allosteric site and maintain the helix-extended conformations of caspase-6 by forming polar interactions with inhibitor Z-VAD-FMK [26]. Thus, it can be deduced that the polar residues in 130′s helix contribute to the conformational transition during caspase-6 activation process and the molecules targeting these residues may stabilize the non-canonical conformation.
Previous research suggested that the protonation state of Glu135 is important for the conformational transition of the extended 130′s helices. At basic pH condition, deprotonated Glu135 can destabilize the 130′s extended helix by leading to electrostatic interactions with neighboring His52, Arg54 and Glu131 [29], [51]. In the substrate-induced conformational transition process, it can be observed that the deprotonated Glu135 can facilitate the formation of the β-turn through H-bond interactions with Lys133 and Ile134 (Fig. 6d).
3.3.3. The 90′s helix undergoes an inward rotation during TMD simulation
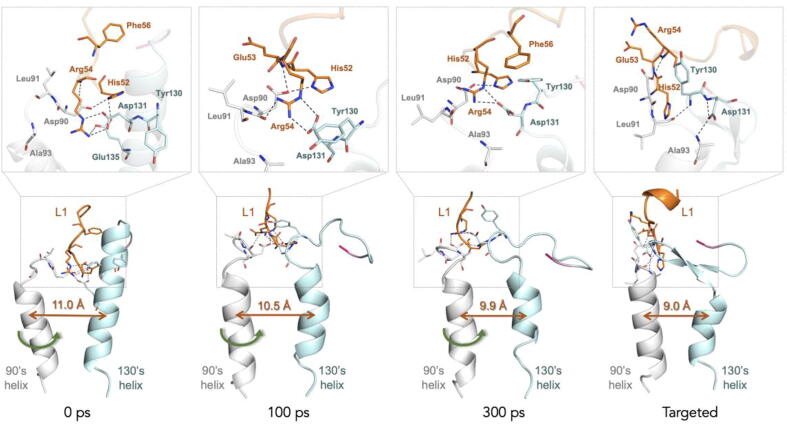
Crystallographic studies revealed that the 90′s helix of caspase-6 undergoes an inward rotation of 21° and thus moves close to the 130′s helix in the activation process (Fig. 5a) [23], [24], [25]. However, the conformational transition process of 90′s helix and determinant factors are still unclear.
Fig. 7 shows the key snapshots of 90′s helix during the TMD simulations. It can be seen that Asp90 (90′s helix) forms stable H-bond interactions with His52 and Arg54 (L1 loop) during the TMD simulations. At the same time, Leu91 and Ala93 tend to gradually approach to 130′s helix by forming H-bond interactions with Tyr130 and Asp131 (130′s helix) at 300 ps. Moreover, intricate H-bond networks are also observed between L1 and 130′s helix during the TMD simulation.
Fig. 7.
The conformational transition of 90′s helix during TMD simulations. L1, 90′s and 130′s helix are colored in orange, gray and cyan, respectively. H-bonds are shown in black dashed lines. The green arrows indicate the movements of 90′s helix. The green line with arrow indicates the centroid distance between the 90′s and 130′s helices. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3 summarizes the pivotal residues involved in caspase-6 conformational transition. It can be seen that the overall conformational transitions of caspase-6 are mainly divided into three stages: (i) substrate-induced residue rearrangements in the active site (L1 and L3); (ii) the rearrangements of the active site facilitate the β-turn formation in 130′s extended helix; (iii) complicated H-bond network formed among L1, 130′s and 90′s helices result in the inward rotation of 90′s helices.
Table 3.
Pivotal residues involved in conformational transition of caspase-6.
| Regions | Residues | Roles |
|---|---|---|
| 60′s helix/L1 | His52, Glu53, Arg54 | β-turn formation in 130′s extended helix |
| Pro62, Glu63, Arg64, Arg65 | Residue rearrangement in active site | |
| Thr67 | Structural reorganization of 60′s helix | |
| 90′s helix | Asp90, Leu91, Ala93 | Inward rotation in 90′s helix |
| 130′s helix/L3 | Tyr130, Asp131, Glu135 | Inward rotation in 90′s helix |
| Ser120, His121 | Residue rearrangement in active site | |
| Glu123, Asn125, His126, Tyr128, Lys133 | β-turn formation in 130′s extended helix |
Bold: Residues within 5 Å distance to the co-crystallized inhibitor Ac-VEID-CHO.
4. Conclusions
Although intense efforts have been devoted to the treatment of neurodegenerative diseases over the past decades, the neurodegenerative diseases are clinically unmanageable due to the lack of success. Caspase-6 is involved in processes of axonal pruning and neuronal degenerations by cleaving a series of neuronal substrates, and thus has been considered as one of the most promising therapeutic targets. Accumulated evidences suggested that caspase-6 undergoes a distinctive conformational transition upon substrate binding. However, the conformational transition and activation mechanism of caspase-6 remains poorly understood, which limits the active-site oriented drug development, as well as the discovery of potential allosteric sites.
Herein, taking advantages of crystallized caspase-6 structures in apo and active states, PRS combined with TMD simulations was used to elucidate the conformational transition of caspase-6 at the atomic level. On the basis of MD simulations, a total of 22 residues were identified as crucial residues in caspase-6 activation process by PRS, which agree well with the functional/allosteric residues reported previously. Then, TMD simulation was performed to explore the conformational transition process of caspase-6. The results showed that the conformational transition of caspase-6 is mainly driven by the structural rearrangement of the active site as well as the 60′s and 130′s extended helices, which results in the inward rotation of 90′s helix. Generally, this work provides a dynamic perspective for the conformational transition of caspase-6 from the apo to the active states. However, the conformational transition of caspase-6 is an extremely complicated process and the biased TMD simulations in this paper only characterize some representative features in the conformational transition process. The precise mechanism and details require further validated by exploring free-energy landscapes.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors acknowledge the support of Key Projects of Technological Innovation and Application Development of Chongqing (Project No. csts2019jscx-gksbX0099), Collaborative Fund of Science and Technology Agency of Luzhou Government and Southwest Medical University (Project No. 2019LZXNYDZ05), Graduate Scientific Research and Innovation Foundation of Chongqing (Project No. CYB19042) and China Scholarship Council.
Author statement
All of the authors have seen and approved the final version of the manuscript being submitted. We warrant that the article is the authors' original work, hasn't received prior publication and isn't under consideration for publication elsewhere.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.07.017.
Contributor Information
Hu Mei, Email: meihu@cqu.edu.cn.
Laichun Lu, Email: lulaicq@cqu.edu.cn.
Xianchao Pan, Email: panxc@swmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Chang H.Y., Yang X.L. Proteases for cell suicide: Functions and regulation of caspases. Microbiol Mol Biol Rev. 2000;64:821. doi: 10.1128/mmbr.64.4.821-846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slee E.A., Adrain C., Martin S.J. Executioner caspase-3,-6, and-7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276(10):7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 4.McIlwain D.R., Berger T., Mak T.W. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denecker G., Ovaere P., Vandenabeele P., Declercq W. Caspase-14 reveals its secrets. J Cell Biol. 2008;180:451–458. doi: 10.1083/jcb.200709098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horowitz P.M., Patterson K.R., Guillozet-Bongaarts A.L., Reynolds M.R., Carroll C.A., Weintraub S.T., et al. Early N-terminal changes and caspase-6 cleavage of tau in Alzheimer's disease. J Neurosci. 2004;24:7895–7902. doi: 10.1523/JNEUROSCI.1988-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albrecht S., Bogdanovic N., Ghetti B., Winblad B., LeBlanc A.C. Caspase-6 Activation in Familial Alzheimer Disease Brains Carrying Amyloid Precursor Protein or Presenilin I or Presenilin II Mutations. J Neuropathol Exp Neurol. 2009;68(12):1282–1293. doi: 10.1097/NEN.0b013e3181c1da10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham R.K., Deng Y.u., Slow E.J., Haigh B., Bissada N., Lu G.e., et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125(6):1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 9.Giaime E., Sunyach C., Druon C., Scarzello S., Robert G., Grosso S., et al. Loss of function of DJ-1 triggered by Parkinson's disease-associated mutation is due to proteolytic resistance to caspase-6. Cell Death Differ. 2010;17(1):158–169. doi: 10.1038/cdd.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee H., Shin E.A., Lee J.H., Ahn D., Kim C.G., Kim J.-H., et al. Caspase inhibitors: a review of recently patented compounds (2013–2015) Expert Opin Ther Pat. 2018;28(1):47–59. doi: 10.1080/13543776.2017.1378426. [DOI] [PubMed] [Google Scholar]
- 11.Tubeleviciute-Aydin A., Beautrait A., Lynham J., Sharma G., Gorelik A., Deny L.J., et al. Identification of Allosteric Inhibitors against Active Caspase-6. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-41930-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X.-J., Cao Q., Zhang Y., Su X.-D. Activation and Regulation of Caspase-6 and Its Role in Neurodegenerative Diseases. Annu Rev Pharmacol Toxicol. 2015;55(1):553–572. doi: 10.1146/annurev-pharmtox-010814-124414. [DOI] [PubMed] [Google Scholar]
- 13.LeBlanc A., Liu H., Goodyer C., Bergeron C., Hammond J. Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer's disease. J Biol Chem. 1999;274(33):23426–23436. doi: 10.1074/jbc.274.33.23426. [DOI] [PubMed] [Google Scholar]
- 14.Klaiman G., Petzke T.L., Hammond J., LeBlanc A.C. Targets of Caspase-6 activity in human neurons and Alzheimer disease. Mol Cell Proteomics. 2008;7(8):1541–1555. doi: 10.1074/mcp.M800007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aharony I., Ehrnhoefer D.E., Shruster A., Qiu X., Franciosi S., Hayden M.R., et al. A Huntingtin-based peptide inhibitor of caspase-6 provides protection from mutant Huntingtin-induced motor and behavioral deficits. Hum Mol Genet. 2015;24:2604–2614. doi: 10.1093/hmg/ddv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham R.K., Deng Y., Carroll J., Vaid K., Cowan C., Pouladi M.A., et al. Cleavage at the 586 Amino Acid Caspase-6 Site in Mutant huntingtin Influences Caspase-6 Activation In Vivo. J Neurosci. 2010;30(45):15019–15029. doi: 10.1523/JNEUROSCI.2071-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riechers S.-P., Butland S., Deng Y.u., Skotte N., Ehrnhoefer D.E., Russ J., et al. Interactome network analysis identifies multiple caspase-6 interactors involved in the pathogenesis of HD. Hum Mol Genet. 2016;25(8):1600–1618. doi: 10.1093/hmg/ddw036. [DOI] [PubMed] [Google Scholar]
- 18.Warby S.C., Doty C.N., Graham R.K., Carroll J.B., Yang Y.Z., Singaraja R.R., et al. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet. 2008;17:2390–2404. doi: 10.1093/hmg/ddn139. [DOI] [PubMed] [Google Scholar]
- 19.Graham R.K., Ehrnhoefer D.E., Hayden M.R. Caspase-6 and neurodegeneration. Trends Neurosci. 2011;34(12):646–656. doi: 10.1016/j.tins.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9(3):459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 21.Klaiman G., Champagne N., LeBlanc A.C. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Bba-Mol Cell Res. 2009;1793(3):592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Wang X.-J., Cao Q., Liu X., Wang K.-T., Mi W., Zhang Y., et al. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010;11(11):841–847. doi: 10.1038/embor.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaidya S., Velázquez-Delgado E.M., Abbruzzese G., Hardy J.A. Substrate-Induced Conformational Changes Occur in All Cleaved Forms of Caspase-6. J Mol Biol. 2011;406(1):75–91. doi: 10.1016/j.jmb.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumgartner R., Meder G., Briand C., Decock A., D'Arcy A., Hassiepen U., et al. The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem J. 2009;423:429–439. doi: 10.1042/BJ20090540. [DOI] [PubMed] [Google Scholar]
- 25.Vaidya S., Hardy J.A. Caspase-6 Latent State Stability Relies on Helical Propensity. Biochemistry. 2011;50(16):3282–3287. doi: 10.1021/bi2001664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller I., Lamers M.B.A.C., Ritchie A.J., Dominguez C., Munoz-Sanjuan I., Kiselyov A. Structure of human caspase-6 in complex with Z-VAD-FMK: New peptide binding mode observed for the non-canonical caspase conformation. Bioorg Med Chem Lett. 2011;21(18):5244–5247. doi: 10.1016/j.bmcl.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 27.Cao Q., Wang X.-J., Li L.-F., Su X.-D. The regulatory mechanism of the caspase 6 pro-domain revealed by crystal structure and biochemical assays. Acta Crystallogr D. 2014;70(1):58–67. doi: 10.1107/S1399004713024218. [DOI] [PubMed] [Google Scholar]
- 28.Liu X., Zhang H., Wang X.-J., Li L.-F., Su X.-D., Mayer C. Get Phases from Arsenic Anomalous Scattering: de novo SAD Phasing of Two Protein Structures Crystallized in Cacodylate Buffer. PLoS One. 2011;6(9):e24227. doi: 10.1371/journal.pone.0024227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dagbay K.B., Bolik-Coulon N., Savinov S.N., Hardy J.A. Caspase-6 Undergoes a Distinct Helix-Strand Interconversion upon Substrate Binding. J Biol Chem. 2017;292(12):4885–4897. doi: 10.1074/jbc.M116.773499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stennicke H.R., Salvesen G.S. Biochemical characteristics of caspases-3, -6, -7, and -8. J Biol Chem. 1997;272(41):25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 31.MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 32.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J., MacKerell A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J Comput Chem. 2013;34(25):2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darden T., York D., Pedersen L. Particle Mesh Ewald - an N. Log(N) Method for Ewald Sums in Large Systems. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 35.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- 36.Atilgan C., Atilgan A.R., Nussinov R. Perturbation-response scanning reveals ligand entry-exit mechanisms of ferric binding protein. PLoS Comput Biol. 2009;5(10):e1000544. doi: 10.1371/journal.pcbi.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jalalypour F., Sensoy O., Atilgan C. Perturb-Scan-Pull: A Novel Method Facilitating Conformational Transitions in Proteins. J Chem Theory Comput. 2020;16(6):3825–3841. doi: 10.1021/acs.jctc.9b01222. [DOI] [PubMed] [Google Scholar]
- 38.Verkhivker G.M. Molecular Simulations and Network Modeling Reveal an Allosteric Signaling in the SARS-CoV-2 Spike Proteins. J Proteome Res. 2020;19(11):4587–4608. doi: 10.1021/acs.jproteome.0c00654. [DOI] [PubMed] [Google Scholar]
- 39.Asciutto E.K., Gedeon P.C., General I.J., Madura J.D. Structure and Dynamics Study of LeuT Using the Markov State Model and Perturbation Response Scanning Reveals Distinct Ion Induced Conformational States. J Phys Chem B. 2016;120(33):8361–8368. doi: 10.1021/acs.jpcb.6b02053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdizadeh H., Guven G., Atilgan A.R., Atilgan C. Perturbation response scanning specifies key regions in subtilisin serine protease for both function and stability. J Enzyme Inhib Med Chem. 2015;30(6):867–873. doi: 10.3109/14756366.2014.979345. [DOI] [PubMed] [Google Scholar]
- 41.Abdizadeh H., Atilgan C. Predicting long term cooperativity and specific modulators of receptor interactions in human transferrin from dynamics within a single microstate. PCCP. 2016;18(11):7916–7926. doi: 10.1039/c5cp05107j. [DOI] [PubMed] [Google Scholar]
- 42.Penkler D., Sensoy Ö., Atilgan C., Tastan Bishop Ö. Perturbation-Response Scanning Reveals Key Residues for Allosteric Control in Hsp70. J Chem Inf Model. 2017;57(6):1359–1374. doi: 10.1021/acs.jcim.6b00775. [DOI] [PubMed] [Google Scholar]
- 43.Brown D.K., Penkler D.L., Amamuddy O.S., Ross C., Atilgan A.R., Atilgan C., et al. MD-TASK: a software suite for analyzing molecular dynamics trajectories. Bioinformatics. 2017;33:2768–2771. doi: 10.1093/bioinformatics/btx349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlitter J., Engels M., Krüger P. Targeted Molecular-Dynamics - a New Approach for Searching Pathways of Conformational Transitions. J Mol Graph. 1994;12(2):84–89. doi: 10.1016/0263-7855(94)80072-3. [DOI] [PubMed] [Google Scholar]
- 45.Stanger K., Steffek M., Zhou L., Pozniak C.D., Quan C., Franke Y., et al. Allosteric peptides bind a caspase zymogen and mediate caspase tetramerization. Nat Chem Biol. 2012;8(7):655–660. doi: 10.1038/nchembio.967. [DOI] [PubMed] [Google Scholar]
- 46.Fu G., Chumanevich A.A., Agniswamy J., Fang B., Harrison R.W., Weber I.T. Structural basis for executioner caspase recognition of P5 position in substrates. Apoptosis. 2008;13(11):1291–1302. doi: 10.1007/s10495-008-0259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velázquez-Delgado E.M., Hardy J.A. Zinc-mediated Allosteric Inhibition of Caspase-6. J Biol Chem. 2012;287(43):36000–36011. doi: 10.1074/jbc.M112.397752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tubeleviciute-Aydin A., Zhou L., Sharma G., Soni I.V., Savinov S.N., Hardy J.A., et al. Rare human Caspase-6-R65W and Caspase-6-G66R variants identify a novel regulatory region of Caspase-6 activity. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-22283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacPherson D.J., Mills C.L., Ondrechen M.J., Hardy J.A. Tri-arginine exosite patch of caspase-6 recruits substrates for hydrolysis. J Biol Chem. 2019;294(1):71–88. doi: 10.1074/jbc.RA118.005914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark A.C. Caspase Allostery and Conformational Selection. Chem Rev. 2016;116(11):6666–6706. doi: 10.1021/acs.chemrev.5b00540. [DOI] [PubMed] [Google Scholar]
- 51.Muñoz V., Serrano L. Elucidating the Folding Problem of Helical Peptides Using Empirical Parameters. Nat Struct Biol. 1994;1(6):399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.