Highlights
-
•
The adc1 disruption of Synechocystis sp. PCC 6803 did not inhibit cell growth, as evidently shown by its similar growth with wild type, as well as its intracellular pigments.
-
•
The Δadc1 mutant has the highest capacity to produce PHB up to 36.1 %w/DCW under nitrogen and phosphorus-deprived medium containing acetate (BG11-N-P+A).
-
•
Abundant PHB granules stained by Nile-Red are markedly visualized in Δadc1 mutant under BG11-N-P and BG11-N-P+A when compared to those of wild type.
-
•
Transcript levels of phaA and phaB are mainly responsible to PHB content whereas the changed proportion of phaC and phaE is noted when a certain phaC transcript level is decreased.
Keywords: Synechocystis sp. PCC6803, Polyhydroxybutyrate, Nutrient deprivation, Adc1 mutant
Abbreviations: ADC, arginine decarboxylase; DCW, dry cell weight; DMF, N,N-dimethylformamide; h, hour(s); HPLC, high pressure liquid chromatography; PCR, polymerase chain reaction; PHAs, polyhydroxyalkanoates; PHB, polyhydroxybutyrate; TAE, Tris-acetate-ethylene diamine tetraacetic acid; TCA, tricarboxylic acid
Abstract
Increased polyhydroxybutyrate production in cyanobacterium Synechocystis sp. PCC 6803 lacking adc1 gene (Δadc1) is first-timely reported in this study. We constructed the mutant by disrupting adc1 gene encoding arginine decarboxylase, thereby exhibiting a partial blockade of polyamine synthesis. This Δadc1 mutant had a proliferative growth and certain contents of intracellular pigments including chlorophyll a and carotenoids as similar as those of wild type (WT). Highest PHB production was certainly induced by BG11-N-P+A condition in both WT and Δadc1 mutant of about 24.9 %w/DCW at day 9 and 36.1 %w/DCW at day 7 of adaptation time, respectively. Abundant PHB granules were also visualized under both BG11-N-P and BG11-N-P+A conditions. All pha transcript amounts of Δadc1 mutant grown at 7 days-adaptation time were clearly upregulated corresponding to its PHB content under BG11-N-P+A condition. Our finding indicated that this adc1 perturbation is alternatively achieved for PHB production in Synechocystis sp. PCC 6803.
1. Introduction
Recently, bio-based and degradable bioplastics, in particular polyhydroxyalkanoates (PHAs) polymers, are better practically produced from prokaryotic organisms, such as well-known bacteria Alcaligenes eutrophus, Ralstonia eutropha [4, 21, 24] and cyanobacteria Spirulina sp., Nostoc muscorum, Synechocystis sp. PCC 6803 [1, 5, 32, 33, 46, 47]. The most common type of PHAs polymers in prokaryotic cells is polyhydroxybutyrate or PHB abundantly found as energy and carbon reserves, especially under starved condition [6]. Nowadays, three main strategies have been implemented to induce PHB production from prokaryotic organisms including nutrient modified medium, environmental stresses, and genetic engineering approaches. First strategy is generalized to accelerate the accumulation of energy reserves in forms of PHB granules inside living cells by generating deprived conditions of essential nutrients. Not only nutrient deficiencies of nitrogen (N) and/or phosphorus (P), but also carbon supplementation, such as acetate and glucose, could induce PHB accumulation in microorganisms via different patterns upon protein synthesis suppression and expression [10, 13, 32, 37]. The certain increase of PHB accumulation was consequently induced when Synechocystis sp. PCC 6803 cells pre-grown in BG11 medium containing 0.1% glucose were further adapted in a modified BG11 medium lacking phosphorus (P) and adding 0.4 % acetate [32]. Recent reports revealed that carbon pool via glycogen production and catabolism, as well as sugar catabolism, had influenced on PHB production under nitrogen deprivation periods in Synechocystis sp. PCC 6803 [22, 27]. Secondly, the environmental stresses could induce PHB production in microorganisms by supporting bacterial viability under adverse environments, such as heat stress, oxidative stress, high salinity [2, 3, 7, 28]. Third strategy relates to genetic and/or metabolic engineering approach. In addition, the integrated strategies are also promising potential for improving PHB quality and productivity. The enhanced PHB accumulation was previously achieved by overexpressing native PHB biosynthetic genes, including phaA (encoding beta-ketothiolase), phaB (encoding acetoacetyl-CoA reductase), phaEC (encoding PHA synthase), in cyanobacterium Synechocystis sp. PCC6803, in particular phaAB-overexpressing strain, by 2.6-fold increase of about 26% w/DCW PHB, and up to 35% PHB after adding 0.4% (w/v) acetate [20]. The metabolic engineering approach on competing metabolic pathways for enhancing acetyl-CoA supply to PHB production was employed in Synechocystis sp. PCC 6803 via the deletions of pta and ach genes, encoding phosphotransacetylase and acetyl-CoA hydrolase, respectively, which resulted to block the conversion of acetyl-CoA to acetate, and further combined with a heterologous expression of phosphoketolase encoded by xfpk from Bifidobacterium breve [8]. On the other hand, the metabolic influence from non-adjacent pathways also has impact on higher PHB production, such as a reduction of phosphate transport, a reduction of proline accumulation via nitrogen metabolism [19, 43]. Recently, the UV-randomly mutated Synechocystis sp. PCC 6714, a point mutation of phosphate-specific transport system integral membrane protein A (PstA) proved by genome sequencing, gave 2.5-fold higher PHB productivity than wild type by about 37 % w/DCW PHB under nitrogen and phosphorus starvation [19]. The transposon-mutated Synechocystis sp. PCC6803 strains with the disruptions of sll0461, encoding gamma-glutamyl phosphate reductase (ProA), and sll0565 encoding a hypothetical protein, contained higher PHB accumulation [43]. Interestingly, it was noted that after proline reduction via the sll0461 deletion would gain more glutamate production flowing to main TCA cycle, which not yet clear mechanism how to enhance PHB production. Since the certain knowledge has been known that proline and glutamate, as well as, polyamines tightly involve in stress response of living organisms via arginine utilization [15, 34], the knockout mutation of adc1 gene encoding arginine decarboxylase in polyamine biosynthesis (Δadc1 strain) was constructed in this study. Our result was first timely-evident finding that Δadc1 cells favored highest PHB accumulation after applying nitrogen and phosphorus starvation plus acetate addition for 7 days.
2. Materials and methods
2.1. Construction of Synechocystis sp. PCC 6803 mutant lacking adc gene (Δadc1)
Cyanobacterium Synechocystis sp. PCC 6803 mutant lacking adc1 gene (Δadc1) by knockout technique was constructed. To obtain this construct, the gene fragment of adc1 gene of about 2.4 kb was amplified by PCR from genomic DNA of Synechocystis sp. PCC 6803. Primer sequences were designed by adding enzyme restriction sequences shown by underlined letters. Forward primer was designed for about 200-bp upstream of adc1 gene (5′-GGAATTCCATATGCTCCTGCTGTCAACGGTTAA-3′) with added NdeI restriction enzyme sequences whereas reverse primer was about 200-bp downstream of this gene with added BamHI sequences (5′-CGGGATCCGCATCAAGTTACTATCTGAG-3′). Next, this amplified gene fragment was cloned into pGem®-T easy vector (Promega Corporation). Then, a kanamycin resistant cassette (about 1.8 kb) was inserted to interrupt adc1 gene which cut at one site by HindIII in that vector by blunt end ligation. After that, the obtained recombinant plasmid was transformed into Synechocystis sp. PCC 6803 wild type cells using natural transformation method [20]. Synechocystis host cells were freshy grown until their optical density (OD) at 730 nm of about 0.5. Fifty mL of cell culture was harvested and concentrated by centrifugation (2790 × g) and resuspended in 0.5 mL of new BG11 medium. About 10 μg of plasmid DNA was added into that concentrated cell suspension, and further incubated under normal growth condition for 6 hours before spreading cell suspension on 0.45 μm sterile mixed cellulose esters (MCE) membrane (MF-Millipore, Merck) placed over a BG11 agar plate. After 24 h incubation, that membrane filter was transferred to place on new BG11 agar containing 20 μg/mL kanamycin. Colonies were grown from that selective antibiotic agar plate under normal growth condition within 3 weeks. To confirm complete segregation of adc1 knockout strain (Δadc1), mutant cells were analyzed by PCR using forward and reverse primers of adc1.
2.2. Cell culture and modified nutrient conditions
Synechocystis sp. PCC 6803 cells, both wild type and mutant, were grown in BG11 medium [35] until late-log phase of growth for about 16 days. Growth condition was performed on the rotary shaker under a continuous light (40-50 μE/m2/s) at 28-32 °C. Cell growth was determined spectrophotometrically by measuring OD at 730 nm. Those late-log growing cells (10 days) were applied to various treatments of modified nutrient conditions under the same growth condition for 11 days. There were three modified nutrient media including the deprivation of both nitrogen (N) and phosphorus (P) (BG11-N-P), and the carbon supplementations of 0.4% (w/v) acetate (A) and 0.4% (w/v) glucose (G) into BG11-N-P medium, represented as BG11-N-P+A and BG11-N-P+G, respectively. For BG11-N-P medium, it was BG11 medium lacking NaNO3 in which ferric ammonium citrate was also replaced by equimolar concentrations of ferrous sulphate heptahydrate whereas phosphorus deprived condition was performed by replacing K2HPO4 with equimolar concentration of KCl.
2.3. Determinations of intracellular pigments
Intracellular chlorophyll a and carotenoid contents of Synechocystis sp. PCC 6803 cells were extracted by N,N-dimethylformamide (DMF) method [9, 16, 25]. Cell culture (1 mL) was harvested by centrifugation at 2790 × g at room temperature for 10 minutes. One mL of DMF was used to dissolve cell pellet fraction. After quick spinning, the DMF-extracted supernatant was spectrophotometically measured its absorbances at 461, 625, 664 nm, respectively, and subsequently calculated its intracellular contents according to equations [9, 25].
2.4. Determination of PHB granules by fluorescence microscopy
PHB granules in Synechocystis cells were stained by fluorescent dye Nile red, and monitored by fluorescence microscopy. One ml of Synechocystis cell culture was harvested by centrifugation at 2790 × g. Then, a small loop of cell pellets was resuspended into 3 μl of Nile red staining solution. Then, 0.9 % (w/v) normal saline (100 µL) was subsequently added, mixed and incubated overnight under darkness [20, 44]. To monitor the stained cells, fluorescent microscope equipped with a digital camera (Olympus DP72, Japan) was applied using a filter cup with 535 excitation wavelength, at magnification of 100X.
2.5. PHB extraction and HPLC analysis
In order to extract PHB from Synechocystis cells, cultured cells (about 50 mL) were harvested by centrifugation at 6000 × g for 10 minutes. Cell pellets were boiled at 100 °C for 60 minutes with 800 µL of concentrated sulfuric acid, and 200 µL of 20 mg/mL adipic acid (an internal standard). This hydrolysis of PHB polymer generated crotonic acid monomers which were detected by high pressure liquid chromatography (HPLC) instrument [44]. Boiled samples were filtered by 0.45 µm polypropylene membrane filter before detecting PHB content by HPLC instrument (Shimadzu HPLC LGE System, Japan) using C18 column, Inert Sustain 3-µm (GL Sciences, Japan). The flow rate was 1.0 ml per minute with UV detector set at 210 nm.. The running buffers were 30% (v/v) acetonitrile and 70% (v/v) of 10 mM KH2PO4 (pH 2.3). Authentic commercial PHB (Sigma) was used as standard which prepared as similar as cell sample. The yield of crotonic acid after hydrolysis detected by HPLC was used to generate PHB standard curve and estimated PHB content in cell sample. The unit of PHB content was % PHB weight per dry cell weight (% w/DCW). The dry cell weight was measured after drying harvested cell pellets in 80 °C oven until a constant weight was obtained [20].
2.6. Determination of relative transcript level
Synechocystis cell culture (50 – 100 mL) was harvested by centrifugation at 12,000 xg for 5 minutes and discarded supernatant. Cell pellets were extracted for total RNA by TRIzol Reagent® (Invitrogen). The cDNAs were synthesized from 5 μg of total RNAs later by using SuperScript® III First Strand Synthesis Kit (Invitrogen). After that, these synthesized cDNAs were used as the template for RT-PCR amplification using many specific primers listed in Table 1. The PCR condition for all pha genes consisted of 95 °C for 30 seconds, followed by 31 cycles of 95 °C for 30 seconds, 50 °C for 30 seconds, and 72 °C for 35 seconds, and the a final extension at 72 °C for 5 minutes. For 16S rRNA as a reference, the PCR condition consisted of 95 °C for 30 seconds, followed by 19 cycles of 95 °C for 30 seconds, 55 °C for 30 seconds, and 72 °C for 35 seconds, and the a final extension at 72 °C for 5 minutes. The PCR products were checked by 1.5% (w/v) agarose gel electrophoresis using in 0.5xTAE buffer.
Table 1.
PCR primers for RT-PCR.
| Target gene | Name | Oligo sequences | Amplified fragment length (bp) |
|---|---|---|---|
| phaA | phaAF | 5’-CATGATGGTTTGACGGACAG- 3’ | 310 |
| phaAR | 5’-GACTACAGTTGCCCGCTGTT- 3’ | ||
| phaB | phaBF | 5’-ATGCCGGTATCACCAAAGAC- 3’ | 390 |
| phaBR | 5’-CAATTTCCTCCGGTTTACCA- 3’ | ||
| phaC | phaCF | 5’-GGGCACATTTAGCCTGTGTT- 3’ | 346 |
| phaCR | 5’-GTAAGTTTCCCCCGCTTGAT- 3’ | ||
| phaE | phaEF | 5’-GAGCAATATACCGCCACCAC- 3’ | 371 |
| phaER | 5’-TCTTCCATCAAAGCAGCAAA- 3’ | ||
| 16S rRNA | 16F | 5’-AGTTCTGACGGTACCTGATGA- 3’ | 521 |
| 16R | 5’-GTCAAGCCTTGGTAAGGTTCT- 3’ |
2.7. Results and discussion
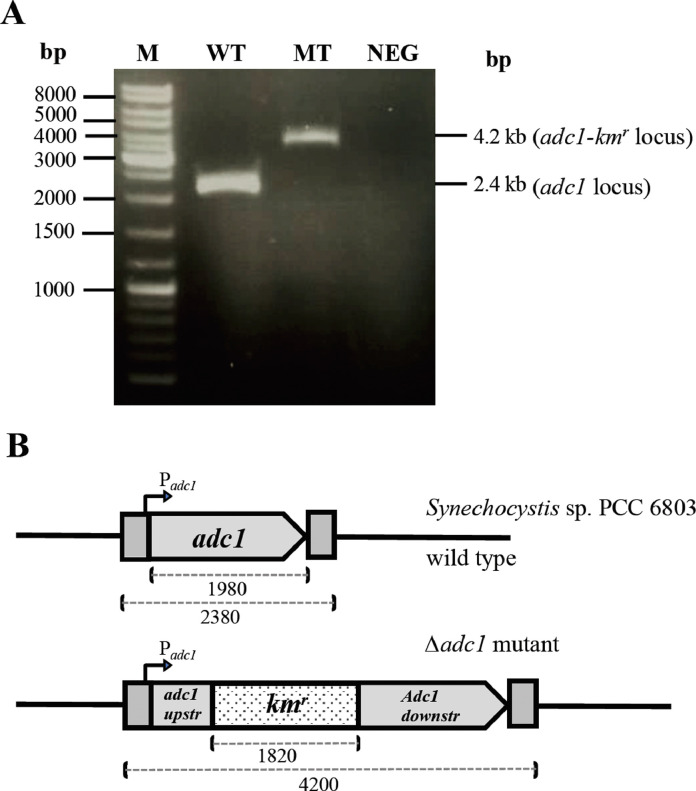
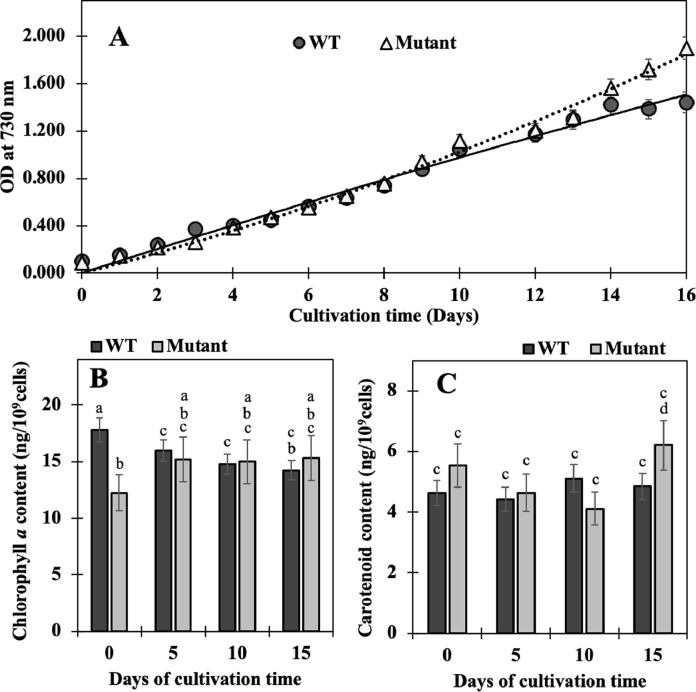
The adc1 gene was inactivated from the Synechocystis sp. PCC 6803 wild type, namely Δadc1 mutant which contained adc1-kmr locus (Fig. 1). Clearly, growth of Δadc1 mutant under normal growth condition was similar to that of wild type, except slightly higher during 14 - 16 days of cultivation (Fig. 2A). In general, cell growth at stationary phase was influenced by many substantial factors which directly related to nutrient-depleted medium and changed environment. The changed pH of medium was one of those important factors which impacted on cell growth in longer time period [18]. On the other hand, the similar accumulation of chlorophyll a was noted in both WT and mutant strains (Fig. 2B). For carotenoid content, the mutant strain contained significantly higher content than that in WT at 15 days of cultivation (Fig. 2C). These results indicate that the high polyamine accumulation was unnecessary under normal growth condition of Synechocystis sp. PCC 6803 as evidently supported by a slight amount of polyamines remained in Δadc1 mutant (data not shown). The induced polyamine content was responsive to environmental stresses, especially ionic and osmotic stresses, in Synechocystis cells [16]. The regulatory response of adc2 gene in Arabidopsis plant was mainly induced rather than adc1 by salt stress leading to increase putrescine accumulation [45] whereas the adc genes in Synechocystis sp. PCC 6803 including adc1 and adc2 had differential responses to stresses [17]. Accordingly, we suggested that the adc1 disruption of Synechocystis cells in this study did not inhibit growth and their intracellular pigment accumulations under normal growth condition since their adc2 gene had certainly functioned.
Fig. 1.
Inactivation of adc1 gene. (A)PCR analysis of the adc1 gene in the expected mutant in which a fragment of adc1 gene was interrupted and replaced with a kanamycin resistant cassette (kmr). Lane M: DNA marker, WT: wild type, MT: Δadc1 mutant, NEG: negative control without genomic DNA of Synechocystis 6803. (B) depiction of the recombinant plasmid used to generate Δadc1 mutant strain. The plasmid carries about 4.2 kb fragment of adc1 gene plus inserted kmr cassette.
Fig. 2.
Growth (A) and pigment contents including chlorophyll a (B) and carotenoids (C) of Synechocystis sp. PCC 6803 wild-type (WT) and Δadc1 mutant (Mutant) cells under normal condition for 16 days. Cells were grown in BG11 medium under normal growth condition. The error bars represent standard deviations of means (mean ± S.D., n = 6). In B and C, means with the same letter have nonsignificant differences at a significance level of P < 0.05.
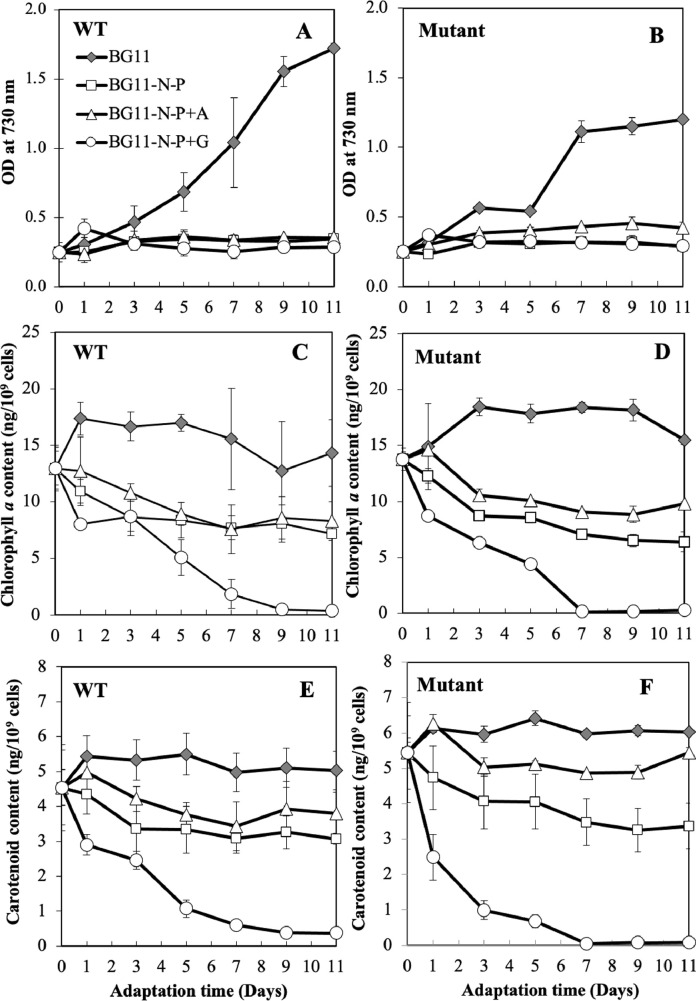
Recent reports revealed that the maximum PHB production in Synechocystis cells was started at the late-log and stationary phases of growth [20, 32]. In this study, we then adapted the harvested Synechocystis cells at late-log phase of growth (10 days of cultivation) into various modified media including BG11-N-P, BG11-N-P+A, and BG11-N-P+G for 11 days (Fig. 3). Unexpectedly, after transferring the mutant cells which were grown till late-log phase of cell growth into new normal BG11 medium, their growth recovery was notably less active, as evidently shown in stable level after 7 days of adaptation time when compared with WT (Fig. 3A and B). Our results indicated that growth of both WT and mutant strains was dramatically decreased under BG11 medium lacking N and P nutrients (Fig. 3A and B). The supplementation of acetate could slightly increase cell growth of Δadc1 mutant when compared to those cells adapted in BG11-N-P and BG11-N-P+G (Fig. 3B). Glucose addition into BG11-N-P medium did not abolish cell growth but severely inhibited chlorophyll a and carotenoid accumulations within 7 days of adaptation time (Fig. 3C-F). These results suggested that Synechocystis cells would assimilate glucose as carbon source for maintaining cell growth with less photosynthesis, as evident by lower contents of intracellular pigments. On the other hand, we also found that the acetate addition could significantly induce the accumulations of chlorophyll a and carotenoids, in particular in Δadc1 mutant, when compared with those under BG11-N-P and BG11-N-P+G (Fig. 3D and F). The supplemented acetate under heterotrophic growth was thoughtfully assimilated by Synechocystis cells via acetyl-CoA-derived metabolic intermediate which located in bottleneck flowing to many crucial pathways including Krebs’ cycle, PHB, glycogen and fatty acid syntheses [8, 20, 41, 42]. In Synechocystis, two mechanism models were considerably proposed for driving the conversion of acetate into acetyl-CoA via either a catalysis of ATP-driven acetyl-CoA synthetase (ACS encoded by sll0542 gene), or catalyzed reactions of acetate kinase (ACK encoded by sll1299 gene) and phosphotransacetylase (PTA encoded by slr2132) [39, 41].
Fig. 3.
Growth (A) and pigment contents including chlorophyll a (B) and carotenoids (C) of Synechocystis sp. PCC 6803 wild-type (WT) and Δadc1 mutant (Mutant) cells under adaptation period in various nutrient conditions for 11 days. Cells were grown in various nutrient conditions including normal BG11 medium as control, BG11 without nitrogen and phosphorus nutrients (or BG11-N-P), BG11-N-P medium supplemented by 0.4% (w/v) acetate (or BG11-N-P+A), BG11-N-P medium supplemented by 0.4% (w/v) glucose (or BG11-N-P+G). The error bars represent standard deviations of means (mean ± S.D., n = 3).
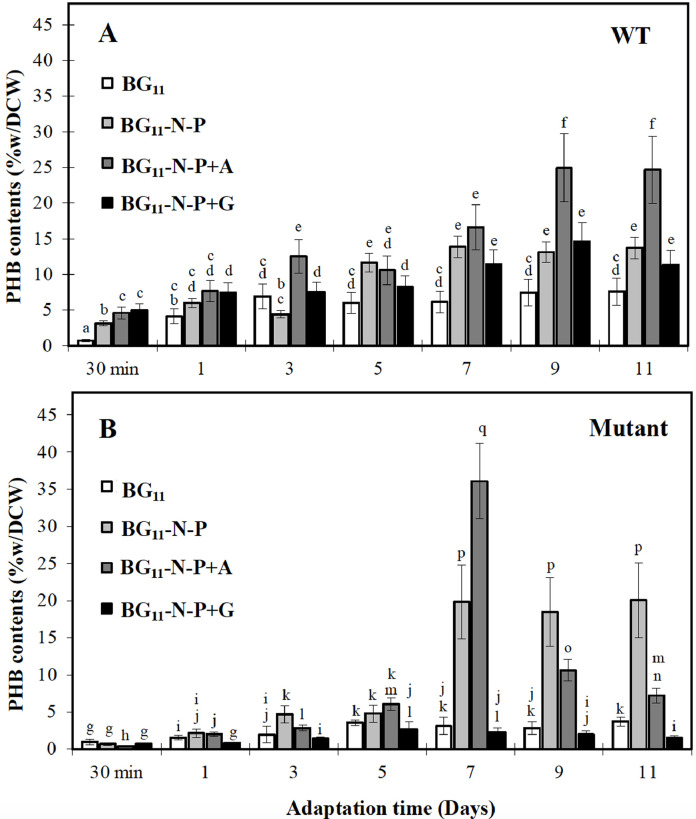
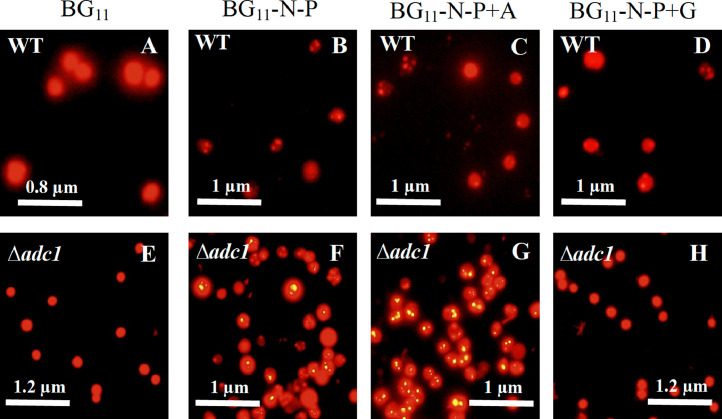
We demonstrated the PHB production in Synechocystis cells adapted in different modified media (Fig. 4). In Synechocystis WT cells, it was found that higher PHB accumulation was significantly induced by all modified media at day 7 of adaptation time (Fig. 4A) whereas the highest increase pointed to PHB contents of about 24.9 %w/DCW at day 9 of adaptation time under BG11-N-P+A condition, and stayed constant till day 11 of adaptation time. On the other hand, a sharp increase was shown by about 36.1 %w/DCW in Δadc1 mutant at day 7 of adaptation time under BG11-N-P+A condition (Fig. 4B) following by that in BG11-N-P condition of about 19.8 %w PHB/DCW. These produced PHB inductions occurred faster in Δadc1 mutant when compared with WT. It is worth to note that the certain PHB decrease of Δadc1 mutant after day 7 of adaptation time occurred in cells which had slight increases of its growth and intracellular pigments at the same time under BG11-N-P+A condition. It ruminatively suggested that this cellular condition would result a lack of fatal situation to enhance PHB when compared to WT. Moreover, the substantial decrease of PHB could represent a stored carbon mobilization for survival under scarce conditions, such as carbon limitation, thermal or oxidative stresses [26]. We also showed the Nile-Red stained PHB granules in Synechocystis Δadc1 mutant (Fig. 5). Abundant PHB granules were apparently found under BG11-N-P and BG11-N-P+A conditions for 7 days in Δadc1 mutant when compared to those in WT (Fig. 5B, C, F and G). According to modified medium, the nutrient deficiency of either nitrogen or phosphorus, herein or both, enabled to efficiently induce higher accumulation of PHB granules in Synechocystis sp. PCC 6803, and those granules were further increased by supplying more carbon source of acetate (0.4 %w/v), as recently reported in wild type [32] and engineered strains (Table 2). Recently, large PHB granules were noted by about 81 %w/DCW PHB under nutrient limiting conditions in a metabolically engineered ΔpirC-REphaAB Synechocystis sp. PCC6803 strain, with heterologous overexpression of Cupriavidus necator phaAB and a disruption of pirC gene encoding phosphoglycerate mutase [23]. The pirC deletion resulted in certain increase of glycogen degradation and lower glycolysis under nitrogen starvation in which PHB accumulation was augmented [23, 29]. On the other hand, we considered that the genetic engineering on any transcription factors may give less impact to induce PHB production whereas the metabolic engineering of competing/neighboring pathways would gain more promising for increasing PHB (Table 2), such as agp knockout [48], and Xfpk expresstion with double pta_ach deletion [8]. Importantly, our finding demonstrated that the polyamine synthesis disturbance dramatically induced higher PHB production in Synechocystis sp. PCC 6803 up to 36.1% w/DCW under modified nutrient condition. In general, the polyamine pathway in cyanobacteria relates to arginine catabolism, one of amino acids utilized as a source of nitrogen for cell growth [34]. Some arginine amino acids could be converted to polyamines by arginine decarboxylase (encoded by adc genes) whereas the main direction of arginine mostly flows to TCA cycle via proline-glutamate pathway [11, 15]. Altogether, we proposed that the adc1 mutation with trace polyamines (data not shown) might enhance a fast flow of more arginine to TCA cycle, thereby increasing energy metabolism for growth and generating carbon pool, as evident by the higher growth of Δadc1 mutant (Fig. 2). Excess acetyl-CoA or carbon pool might then contribute to PHB accumulation, instead of TCA cycle direction, which efficiently induced by our modified nutrient conditions used in this study. On the other hand, as previously noted, Tyo et al. [43] discussed that Synechocystis 6803 Δsll0461 (or proA) strain which represented lower proline was attacked by other stresses which subsequently induced higher PHB accumulation. Rationally, we would then speculate that not only other stresses attacking when cells have less polyamines but also the increase flow of amino acids, herein arginine, to energy metabolism, which favor PHB production in Synechocystis sp. PCC 6803, particularly under stressed condition. For prospective direction, additional experiments on specific gene manipulations related to arginine catabolism and neighboring pathways, as well as up-to-date technology of omics analyses, might verify and gain more understanding whether polyamine pathway disturbance literally influences on PHB production via the increase of the acetyl-CoA pool.
Fig. 4.
Polyhydroxybutyrate (PHB) contents of Synechocystis sp. PCC 6803 (A) wild-type (WT) and (B) Δadc1 mutant (Mutant) cells under adaptation period in various nutrient conditions for 11 days. PHB content (%w/dry cell weight (DCW)) was measured from cells grown in various nutrient conditions including normal BG11 medium as control, BG11 without nitrogen and phosphorus nutrients (or BG11-N-P), BG11-N-P medium supplemented by 0.4% (w/v) acetate (or BG11-N-P+A), BG11-N-P medium supplemented by 0.4% (w/v) glucose (or BG11-N-P+G). The error bars represent standard deviations of means (mean ± S.D., n = 3). Means with the same letter have nonsignificant differences at a significance level of P < 0.05.
Fig. 5.
Images of Synechocystis 6803 wild type (A-D) and Δadc1 mutant (E-H) adapted for 7 days under various nutrient conditions. Images are from fluorescence microscopy showing PHB granules as bright gold particles with 100x magnification.
Table 2.
PHB production in engineered Synechocystis sp. PCC6803 (or S6803) strains which directly and indirectly related to its synthetic pathway.
| Genetic engineering approach | PHB-producing condition(s) | PHB content | References |
|---|---|---|---|
| Engineered S6803 strain directly relating to PHB synthetic pathway | |||
| Heterologous expression of a gene encoding PHA synthase from Alcaligenes eutrophus | BG11 containing deprived-N and supplemented acetate, phototrophic growth for 14 days | 11 %w/DCW | Sudesh et al. [38] |
| Heterologous expression of PHA biosynthetic operon from Microcystis aeruginosa NIES-843 | BG11 containing deprived-N, phototrophic growth for 8 days | 7 %w/DCW | Hondo et al. [14] |
| The native overexpression of phaAB | BG11 containing deprived-N and supplemented acetate, phototrophic growth for 9 days | 35 %w/DCW | Khetkorn et al. [20] |
| The overexpression of Cupriavidus necator phaAB combined with PirC disruption | BG11 containing deprived-N-P and supplemented acetate, phototrophic growth for 20 days | 81 %w/DCW | Koch et al. [23] |
| Engineered S6803 strain indirectly relating to PHB synthetic pathway | |||
| The overexpression of native SigE gene encoding RNA polymerase sigma factor | BG11 containing deprived-N, phototrophic growth for 9 days | 1.4 %w/DCW | Osanai et al. [30] |
| The overexpression of native rre37 gene encoding OmpR-type response regulator | BG11 containing deprived-N, phototrophic growth | 1.2 %w/DCW | Osanai et al. [31] |
| The knockout of sll0783 gene encoding transcription factor NtcA | BG11 containing deprived-N, phototrophic growth for 120h | > 1 %w/DCW | Schlebusch and Forchhammer [36] |
| The knockout of agp gene encoding ADP-glucose pyrophosphorylase (related to glycogen synthesis) | BG11 containing deprived-N, phototrophic growth for 120 hours | 18.6 %w/DCW | Wu et al. [48] |
| Inverse metabolic engineering; transposon random mutation library; mutagenized strains at sll0461, and sll0565 genes | BG11 containing 10% phosphate, phototrophic growth for 14 days | 9 %w/DCW (Δsll0461), 10 %w/DCW (Δsll0565) | Tyo et al. [43] |
| the expression of a heterologous phosphoketolase (XfpK) from Bifidobacterium breve in a double pta and ach deletion background | Photoautotrophic growth with CO2 bubbling in medium deprived-N-P, for about 32 days | ∼12 %w/DCW | Carpine et al. [8] |
| The knockout of adc1 gene encoding arginine decarboxylase related to polyamine synthesis | BG11 containing deprived-N-P and supplemented acetate, phototrophic growth for 7 days | 36.1 %w/DCW | This study |
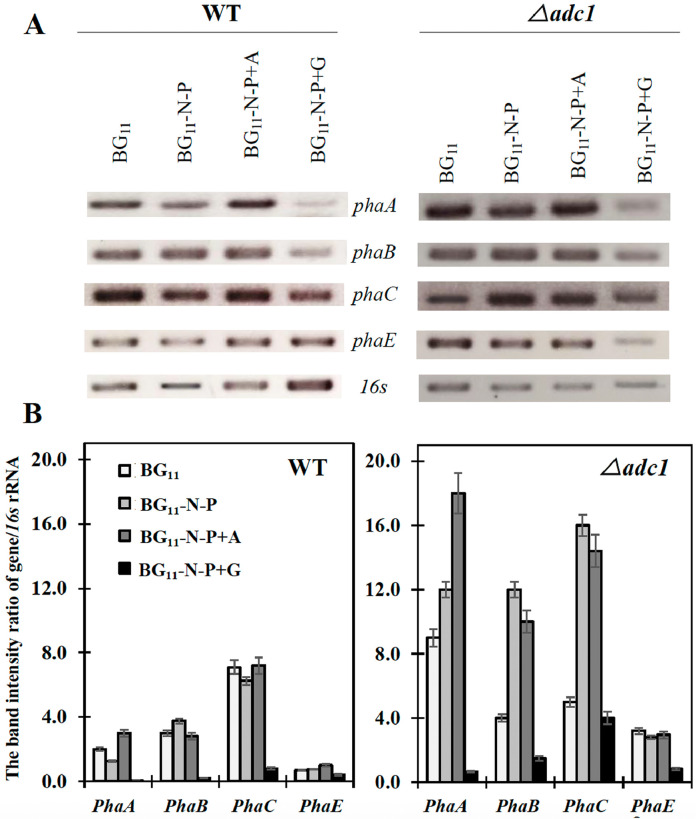
We also demonstrated the changed transcript levels of each gene in PHB biosynthetic pathway under all conditions at day 7 of adaptation time (Fig. 6). Up to date, it is known that there are a cluster of two open reading frames for phaA and phaB [40], and phaE and phaC which both responsible to a polyhydroxyalkanoic acid (PHA) synthase [12]. We showed that Synechocystis WT had high transcript amounts of phaA, phaB and phaC whereas phaE transcript level was lowest under normal growth condition (BG11) (Fig. 6A and B). In previous work, the double overexpression of phaAB in Synechocystis 6803 had influenced on a certain increase of PHB production [20]. Intriguingly, the Δadc1 mutant contained higher transcript level of all pha genes, except phaC, when compared to those of WT (Fig. 6). Modified medium of BG11-N-P+A could dramatically induce all pha genes of Δadc1 mutant among other conditions corresponding to PHB production (Figs. 4 and 5). However, we found that WT cells under BG11-N-P+G condition contained lower transcript levels of phaA and phaB that did not show a tight connection with PHB content whereas the transcript amounts of both phaC and phaE did (Fig. 6). Our results indicated that gene expressions of phaA and phaB mainly occurred in similar tendency whereas the changed transcript proportion of phaC and phaE was alternately regulated once phaC transcript amount was apparently decreased.
Fig. 6.
Relative transcript levels (A) of phaA, phaB, phaC and phaE performed by RT-PCR and the band intensity ratio of each gene/16s rRNA (B) in WT and Δadc1 mutant strains grown under various nutrient conditions for 7 days. The 16S rRNA was used as reference control.
3. Conclusion
Synechocystis sp. PCC 6803 lacking adc1 gene (Δadc1) had the highest capacity to synthesize PHB under nutrient modified media. This Δadc1 mutant could grow as similar as Synechocystis wild type. Its highly accumulated PHB occurred up to 36.1 %w/DCW after adaptation in a nitrogen and phosphorus-deprived BG11 medium containing 0.4 %(w/v) acetate for 7 days. To understand the actual connection between lower polyamine synthesis and high PHB production in cyanobacteria, further specific gene manipulations on arginine catabolism and neighboring pathways, as well as omics analysis, would be promising in bioenergy and biomaterial fields of Biotechnology. Last but not least, the metabolic engineering approach and product recovery system for preventing intracellular PHB degradation in cyanobacterial cells would draw a spotlight guidance for sustained PHB production from algal resource in practical application.
Authors' contribution
Suthira Utharn: study conception, main experimenter, data collection, analysis. Panutda Yodsang: experimenter (mutant construction). Aran Incharoensakdi: study conception, research discussion. Saowarath Jantaro: Funding acquisition, project administration, conceptualization, methodology, supervision, validation, writing – review and editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was funded by the Ratchadapisek Sompoch Endowment Fund (2016), Chulalongkorn University (CU-59-018-FW), and Chulalongkorn University grant (2019) (CU_GR_62_25_23_08) to S.J.
References
- 1.Abed R.M., Dobretsov S., Sudesh K. Applications of cyanobacteria in biotechnology. J. Appl. Microbiol. 2009;106:1–12. doi: 10.1111/j.1365-2672.2008.03918.x. [DOI] [PubMed] [Google Scholar]
- 2.Al Rowaihi I.S., Paillier A., Rasul S., Karan R., Grötzinger S.W., Takanabe K., Eppinger J. Poly(3-hydroxybutyrate) production in an integrated electromicrobial setup: investigation under stress-inducing conditions. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0196079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alves L.P.S., Santana-Filho A.P., Sassaki G.L., de Oliveira Pedrosa F.O., de Souza E.M., Chubatsu L.S., Müller-Santos M. 3-Hydroxybutyrate derived from poly-3 hydroxybutyrate (PHB) mobilization alleviates protein aggregation in heat-stressed Herbaspirillum seropedicae SmR1. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.01265-20. e01265-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson A.J., Dawes E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. PMID: 2087222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ansari S., Fatma T. Cyanobacterial polyhydroxybutyrate (PHB): screening, optimization and characterization. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaji S., Gopi K., Muthuvelan B. A review on production of poly β hydroxybutyrates from cyanobacteria for the production of bio plastics. Algal Res. 2013;2:278–285. doi: 10.1016/j.algal.2013.03.002. [DOI] [Google Scholar]
- 7.Batista M.B., Teixeira C.S., Sfeir M.Z.T., Alves L.P.S., Valdameri G., de Oliveira Pedrosa F., Sassaki G.L., Steffens M.B.R., de Souza E.M., Dixon R., Müller-Santos M. PHB biosynthesis counteracts redox stress in Herbaspirillum seropedicae. Front. Microbiol. 2018;9:472. doi: 10.3389/fmicb.2018.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpine R., Du W., Olivieri G., Pollio A., Hellingwerf K.J., Marzocchella A., Branco dos Santos F. Genetic engineering of Synechocystis sp. PCC6803 for poly-β-hydroxybutyrate overproduction. Agal Res. 2017;25:117–127. doi: 10.1016/j.algal.2017.05.013. [DOI] [Google Scholar]
- 9.Chamovitz D., Sandmann G., Hirschberg J. Molecular and biochemical-characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J. Biol. Chem. 1993;268:17348–17353. PMID: 8349618. [PubMed] [Google Scholar]
- 10.Dutt V., Srivastava S. Novel quantitative insights into carbon sources for synthesis of poly hydroxybutyrate in Synechocystis PCC 6803. Photosynth Res. 2018;136:303–314. doi: 10.1007/s11120-017-0464-x. [DOI] [PubMed] [Google Scholar]
- 11.Flores E, Herrero A. Assimilatory nitrogen metabolism and its regulation. In: Bryant D.A., editor. The Molecular Biology of Cyanobacteria. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1994. pp. 487–517. [Google Scholar]
- 12.Hein S., Tran H., Steinbüchel A. Synechocystis sp. PCC6803 possesses a two-component polyhydroxyalkanoic acid synthase similar to that of anoxygenic purple sulfur bacteria. Arch. Microbiol. 1998;170:162–170. doi: 10.1007/s002030050629. [DOI] [PubMed] [Google Scholar]
- 13.Hirai K., Nojo M., Sato Y., Tsuzuki M., Sato N. Contribution of protein synthesis depression to poly-β-hydroxybutyrate accumulation in Synechocystis Sp. PCC 6803 under nutrient-starved conditions. Sci. Rep. 2019;9:19944. doi: 10.1038/s41598-019-56520-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hondo S., Takahashi M., Osanai T., Matsuda M., Hasunuma T., Tazuke A., Nakahira Y., Chohnan S., Hasegawa M., Asayama M. Genetic engineering and metabolite profiling for overproduction of polyhydroxybutyrate in cyanobacteria. J. Biosci. Bioeng. 2015;120:510–517. doi: 10.1016/j.jbiosc.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Jantaro S., Kanwal S. Low molecular weight nitrogenous compounds (GABA and polyamines) in blue green algae. In: Rastogi R.P., Madamwar D., Pandey A., editors. Algal green chemistry: recent progress in biotechnology. Elsevier B.V.; 2017. pp. 149–169. [DOI] [Google Scholar]
- 16.Jantaro S., Maenpaa P., Mulo P., Incharoensakdi A. Content and biosynthesis of polyamines in salt and osmotically stressed cells of Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 2003;228:129–135. doi: 10.1016/S0378-1097(03)00747-X. [DOI] [PubMed] [Google Scholar]
- 17.Jantaro S., Pothipongsa A., Khanthasuwan S., Incharoensakdi A. Short-term UV-B and UV-C radiations preferentially decrease spermidine contents and arginine decarboxylase transcript levels of Synechocystis sp. PCC 6803. Curr. Microbiol. 2011;62:420–426. doi: 10.1007/s00284-010-9724-0. [DOI] [PubMed] [Google Scholar]
- 18.Jiang H.-B., Cheng H.-M., Gao K., Qiu B.-S. Inactivation of Ca2+/H+ exchanger in Synechocystis sp. strain PCC 6803 promotes cyanobacterial calcification by upregulating CO2-concentrating mechanisms. Appl. Environ. Microbiol. 2013;79:4048–4055. doi: 10.1128/AEM.00681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamravamanesh D., Kovacs T., Stefan Pflügl S., Druzhininab I., Kroll P., Maximilian Lackner M., Herwig C. Increased poly-β-hydroxybutyrate production from carbon dioxide in randomly mutated cells of cyanobacterial strain Synechocystis sp. PCC 6714: Mutant generation and characterization. Bioresour. Technol. 2018;266:34–44. doi: 10.1016/j.biortech.2018.06.057. [DOI] [PubMed] [Google Scholar]
- 20.Khetkorn W., Incharoensakdi A., Lindblad P., Jantaro S. Enhancement of poly-3-hydroxybutyrate production in Synechocystis sp. PCC 6803 by overexpression of its native biosynthetic genes. Bioresour. Technol. 2016;214:761–768. doi: 10.1016/j.biortech.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Kichise T., Fukui T., Yoshida Y., Doi Y. Biosynthesis of polyhydroxyalkanoates (PHA) by recombinant Ralstonia eutropha and effects of PHA synthase activity on in vivo PHA biosynthesis. Int. J. Biol. Macromol. 1999;25:69–77. doi: 10.1016/S0141-8130(99)00017-3. [DOI] [PubMed] [Google Scholar]
- 22.Koch M., Doello S., Gutekunst K., Forchhammer K. PHB is produced from glycogen turn-over during nitrogen starvation in Synechocystis sp. PCC 6803. Int. J. Mol. Sci. 2019;20:1942. doi: 10.3390/ijms20081942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch M., Bruckmoser J., Scholl J., Hauf W., Reiger B., Forchhammer K. Maximizing PHB content in Synechocystis sp. PCC 6803: a new metabolic engineering strategy based on the regulator PirC. Microb. Cell Fact. 2020;19:231. doi: 10.1186/s12934-020-01491-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liebergesell M., Sonomoto K., Madkour M., Mayer F., Steinbuchel A. Purification and characterization of the poly(hydroxyalkanoic acid) synthase from Chromatium vinosum and localization of the enzyme at the surface of poly(hydroxyalkanoic acid) granules. Eur. J. Biochem. 1994;226:71–80. doi: 10.1111/j.1432-1033.1994.tb20027.x. [DOI] [PubMed] [Google Scholar]
- 25.Moran R. Formulae for determination of chlorophyllous pigments extracted with N,N-dimethylformamide. Plant Physiol. 1982;69:1376–1381. doi: 10.1104/pp.69.6.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller-Santos M., Koskimäki J.J., Alves L.P.S., de Souza E.M., Jendrossek D., Pirttilä A.M. The protective role of PHB and its degradation products against stress situations in bacteria. FEMS Microbiol. Rev. 2021;45:1–13. doi: 10.1093/femsre/fuaa058. [DOI] [PubMed] [Google Scholar]
- 27.Nakaya Y., Iijima H., Takanobu J., Watanabe A., Hirai M.Y., Osanai T. One day of nitrogen starvation reveals the effect of sigE and rre37 overexpression on the expression of genes related to carbon and nitrogen metabolism in Synechocystis sp. PCC 6803. J. Biosci. Bioeng. 2015;120:128–134. doi: 10.1016/j.jbiosc.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 28.Obruca S., Sedlacek P., Koller M., Kucera D., Pernicova I. Involvement of polyhydroxyalkanoates in stress resistance of microbial cells: Biotechnological consequences and applications. Biotechnol. Adv. 2018;36:856–870. doi: 10.1016/j.biotechadv.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Orthwein, T., Scholl, J., Spät, P., Lucius, S., Koch, M., Macek, B., Hagemann, M., Forchhammer, K., 2020. The novel PII-interacting regulator PirC (Sll0944) identifies 3-phosphoglycerate mutase (PGAM) as central control point of carbon storage metabolism in cyanobacteria. bioRxiv. https://doi.org/10.1101/2020.09.11.292599.
- 30.Osanai T., Numata K., Oikawa A., Kuwahara A., Iijima H., Doi Y., Tanaka K., Saito K., Hirai M.Y. Increased bioplastic production with an RNA polymerase sigma factor SigE during nitrogen starvation in Synechocystis sp. PCC 6803. DNA Res. 2013;20:525–535. doi: 10.1093/dnares/dst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osanai T., Oikawa A., Numata K., Kuwahara A., Iijima H., Doi Y., Saito K., Hirai M.Y. Pathway-level acceleration of glycogen catabolism by a response regulator in the cyanobacterium Synechocystis species PCC 6803. Plant Physiol. 2014;164:1831–1841. doi: 10.1104/pp.113.232025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panda B., Jain P., Sharma L., Mallick N. Optimization of cultural and nutritional conditions for accumulation of poly-beta-hydroxybutyrate in Synechocystis sp. PCC 6803. Bioresour. Technol. 2006;97:1296–1301. doi: 10.1016/j.biortech.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Panda B., Mallick N. Enhanced poly-beta-hydroxybutyrate accumulation in a unicellular cyanobacterium Synechocystis sp. PCC 6803. Lett. Appl. Microbiol. 2007;44:194–198. doi: 10.1111/j.1472-765X.2006.02048.x. [DOI] [PubMed] [Google Scholar]
- 34.Quintero M.J., Muro-Pastor A.M., Herrero A., Flores E. Arginine catabolism in the cyanobacterium Synechocystis sp. strain PCC 6803 involves the urea cycle and arginase pathway. J. Bacteriol. 2000;182:1008–1015. doi: 10.1128/jb.182.4.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 36.Schlebusch M., Forchhammer K. Requirement of the nitrogen starvation induced protein Sll0783 for polyhydroxybutyrate accumulation in Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2010;76:6101–6107. doi: 10.1128/AEM.00484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma L., Mallick N. Accumulation of poly-beta-hydroxybutyrate in Nostoc Muscorum: regulation by pH, light-dark cycles, N and P status and carbon sources. Bioresour. Technol. 2005;96:1304–1310. doi: 10.1016/j.biortech.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Sudesh K., Taguchi K., Doi Y. Effect of increased PHA synthetase activity on polyhydroxyalkanoates biosynthesis in Synechocystis sp. PCC 6803. Int. J. Biol. Macromol. 2002;30:97–104. doi: 10.1016/s0141-8130(02)00010-7. [DOI] [PubMed] [Google Scholar]
- 39.Summers M.L., Denton M.C., McDermott T.R. Genes coding for phosphotransacetylase and acetate kinase in Sinorhizobium meliloti are in an operon that is inducible by phosphate stress and controlled by PhoB. J. Bacteriol. 1999;181:2217–2224. doi: 10.1128/JB.181.7.2217-2224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taroncher-Oldenburg G., Nishina K., Staphanopoulos G. Identification and analysis of the polyhydroxyalkanoate-specific β-ketothiolase and acetoacetyl coenzyme A reductase genes in the cyanobacterium Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 2000;66:4440–4448. doi: 10.1128/aem.66.10.4440-4448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiel K., Vuorio E., Aro E.-M., Kallio P.T. The effect of enhanced acetate influx on Synechocystis sp. PCC 6803 metabolism. Microb. Cell Fact. 2017;16:21. doi: 10.1186/s12934-017-0640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Towijit U., Songruk N., Lindblad P., Incharoensakdi A., Jantaro S. Co-overexpression of native phospholipid-biosynthetic genes plsX and plsC enhances lipid production in Synechocystis sp. PCC 6803. Sci. Rep. 2018;8:13510. doi: 10.1038/s41598-018-31789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyo K.E., Jin Y.S., Espinoza F.A., Stephanopoulos G. Identification of gene disruptions for increased poly-3-hydroxybutyrate accumulation in Synechocystis PCC 6803. Biotechnol. Prog. 2009;25:1236–1243. doi: 10.1002/btpr.228. [DOI] [PubMed] [Google Scholar]
- 44.Tyo K.E., Zhou H., Stephanopoulos G.N. High-throughput screen for poly-3-hydroxybutyrate in Escherichia coli and Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 2006;72:3412–3417. doi: 10.1128/AEM.72.5.3412-3417.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urano K., Yoshiba Y., Nanjo T., Ito T., Yamaguchi-Shinozaki K., Shinozaki K. Arabidopsis stress inducible gene for arginine decarboxylase AtADC2 is required for accumulation of putrescine in salt tolerance. Biochem. Biophys. Res. Commun. 2004;313:369–375. doi: 10.1016/j.bbrc.2003.11.119. [DOI] [PubMed] [Google Scholar]
- 46.Vincenzini M., Sili C., Philippis R.D., Ena A., Materassi R. Occurrence of poly-b-hydroxybutyrate in Spirulina species. J. Bacteriol. 1990;172:2791–2792. doi: 10.1128/jb.172.5.2791-2792.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang B., Pugh S., Nielsen D.R., Zhang W., Meldrum D.R. Engineering cyanobacteria for photosynthetic production of 3-hydroxybutyrate directly from CO2. Metab. Eng. 2013;16:68–77. doi: 10.1016/j.ymben.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Wu G.F., Shen Z.Y., Wu Q.Y. Modification of carbon partitioning to enhance PHB production in Synechocystis sp. PCC 6803. Enzyme Microb. Technol. 2002;30:710–715. doi: 10.1016/S0141-0229(02)00044-3. [DOI] [Google Scholar]