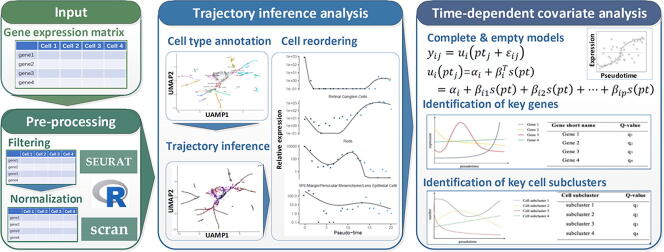
Graphical abstract
Keywords: scRNA-seq, Time series analysis, Trajectory inference analysis, Differential cell subclusters, Differentially expressed genes
Abstract
Time-series single-cell RNA sequencing (scRNA-seq) provides a breakthrough in modern biology by enabling researchers to profile and study the dynamics of genes and cells based on samples obtained from multiple time points at an individual cell resolution. However, cell asynchrony and an additional dimension of multiple time points raises challenges in the effective use of time-series scRNA-seq data for identifying genes and cell subclusters that vary over time. However, no effective tools are available. Here, we propose scTITANS (https://github.com/ZJUFanLab/scTITANS), a method that takes full advantage of individual cells from all time points at the same time by correcting cell asynchrony using pseudotime from trajectory inference analysis. By introducing a time-dependent covariate based on time-series analysis method, scTITANS performed well in identifying differentially expressed genes and cell subclusters from time-series scRNA-seq data based on several example datasets. Compared to current attempts, scTITANS is more accurate, quantitative, and capable of dealing with heterogeneity among cells and making full use of the timing information hidden in biological processes. When extended to broader research areas, scTITANS will bring new breakthroughs in studies with time-series single cell RNA sequencing data.
1. Introduction
With the development of technologies for cell separation and sequencing, single-cell RNA sequencing (scRNA-seq) makes it possible to characterize RNA molecules at a resolution of individual cells or nuclei on a genomic scale [1], and has opened new avenues for studies on human physiology and disease pathology [2], [3]. Based on a snapshot of the transcriptome of thousands of single cells in a cell population, scRNA-seq has proven powerful in detecting heterogeneity among individual cells and delineating cell maps [4], [5], [6]. With snapshots of single cells obtained at multiple time points, time-series scRNA-seq data is capable of providing large amount of invaluable information for understanding dynamic processes [7], [8]. This wealth of high-dimensional transcriptional information, however, presents many challenges in analyzing data. Until recently, no efficient tools were available for identifying genes and cell subclusters from time series scRNA-seq data that vary over time, which may be key to evolution or disease progression.
Recently, a few attempts have been made to identify differentially expressed genes (DEGs) and cell subclusters from time series scRNA-seq data. A novel microglia type and markers associated with neurodegenerative diseases (DAM) have been identified by Hadas et al. [9], and seven cell subclusters that differ during pancreatic organ development have been identified by Lauren et al. [10]. In these attempts, differential expression analysis of individual genes is often performed on discrete groups of cells in the developmental pathway, e.g., by comparing clusters of differentiated cell types. Such discrete differential expression approaches do not exploit the continuous expression resolution provided by multiple time points, and are incapable of taking full advantage of the timing information hidden in biological processes such as development, differentiation, aging, drug response, and disease development [11]. In contrast, it is often unclear with respect to cluster-based methods, as to which clusters should be compared and how to properly combine the results of several pairwise cluster comparisons. Moreover, a large number of studies indicate that cells captured at the same time point can vary considerably [12]. The resulting DEGs and differential cell subclusters identified by current attempts may be biased by cell asynchrony, since variations among cells obtained at different time points have not been considered during analysis. Therefore, development of a new method for time-series scRNA-seq data that is easy to use, quantitative, and capable of dealing with the heterogeneity among cells is urgently needed.
As to the heterogeneity among cells, a large number of studies indicate that cell asynchrony is not completely disordered [13], and that cell differentiation can be viewed as a continuous process [14]. When a sufficient number of individual cells are captured, it is possible to represent all cell states throughout a continuous process of development [15]. In this respect, trajectory inference (TI) methods are superior to discrete cluster-based approaches in that they can reduce the effect of cell asynchrony by pseudotemporal reordering of cells. Accordingly, multiple trajectory-based differential expression analysis methods such as Monocle [13], and tradeSeq [16] have been developed and successfully applied to exploit the continuous expression resolution along the trajectory for snapshot scRNA-seq data. On the other hand, time-series analysis [17] that can take into account the information generated at several time points at the same time, determine expression patterns (such as cyclical pattern), and identify regulatory factors in a dynamic biological process in a quantitative and knowledge-free way, will be a perfect choice for time series scRNA-seq data obtained from multiple snapshots. At present, time-series analysis is being successfully applied in studies focusing on human disease and drug development [18], [19]. Although not applied to scRNA-seq data, the advantage of time-series analysis in identifying DEGs in dynamic biological processes based on high-throughput RNA-seq data has also been revealed [20], [21]. Therefore, a strategy for time-series scRNA-seq data should be developed that can combine the advantages of TI and time-series analysis methods.
Here we propose scTITANS, a trajectory inference-based approach to identify DEGs and cell subclusters from time-series scRNA-seq data. After correcting the asynchrony of single cells based on TI analysis, a time-dependent covariate is introduced to identify the DEGs and cell subclusters in dynamic processes. Compared with current attempts, the method is quantitative and capable of dealing with the heterogeneity among cells and making full use of the timing information hidden in biological processes. Moreover, scTITANS achieves higher accuracy and has a wide range of applications.
2. Materials and methods
2.1. Workflow of scTITANS
Fig. 1 illustrates a schematic diagram of the scTITANS method. After data preprocessing with filtering and normalization, a TI method is first utilized to construct the trajectory and resolve the underlying pseudotime. Based on the pseudotime, all single cells are reordered. For DEGs, a curve is fitted for the dynamic expression of each gene along the pseudotime, and a time-dependent covariate is introduced to calculate the q-value that indicates the significance of the fluctuation of each gene over pseudotime. For differential cell subclusters, the pseudotime is first separated into several bins. After assigning single cells in each bin into corresponding clusters, a curve representing the number of cells falling into each interval is fitted for each subcluster. A time-dependent covariate is again utilized to evaluate the variations of cell numbers along pseudotime for each cluster. The lower the q-value is, the more significant the genes or cell subclusters will be.
Fig. 1.
A schema for the scTITANS method.
scTITANS can be performed in two modes. Users who have finished data preprocessing and TI analysis should provide the dataset, the detailed metadata for cells and genes resulting from trajectory analysis, and the root cell type. scTITANS also accepts raw digital gene expression matrices resulting from tools such as Cell Ranger (10X Genomics) or those filtered and normalized as input. A series of example datasets are then utilized to evaluate the performance of the scTITANS method.
2.2. Data pre-processing
For applications with raw digital gene expression matrices, data pre-processing is first carried out. For raw gene expression matrices (UMI counts per gene per cell), genes without any counts in any cell were filtered out. A gene present with two or more transcripts in at least 10 cells was defined as detected. For cell filtering, cells with the number of genes detected outside the 5th and 95th percentile were discarded. Moreover, cells with more than 10% of their UMIs assigned to mitochondrial genes were filtered out [22]. The matrix was further normalized with the scran R package using the default implementation of the pool and deconvolute normalization algorithm [23], [24]. All the parameters for data preprocessing can be adjusted as needed.
2.3. Trajectory inference analysis
Trajectory inference analysis is a key part of scTITANS. Over 70 TI methods are available, and the accuracy, scalability, stability, and usability of 45 TI methods have been thoroughly compared using 110 real and 229 synthetic datasets [25]. Although it has been proposed that the choice of TI method should depend mostly on dataset dimensions and trajectory topology [25], it is somewhat difficult to apply the principle in real applications where the actual dimensions and topologies are unclear. As a simplified application of the above principle in scTITANS, we first selected 10 out of the 45 previously evaluated TI methods [25], and evaluated their performance. The TI method with the best performance was then selected to be used in scTITANS for pseudotime reconstruction. Because topology is a key factor in the choice of TI method [25], we selected the 10 TI methods based on criteria such as topology accuracy higher than 0.8, topology stability higher than 0.5, and paper quality higher than 0.5 and so on. Paper quality was assessed using a transparent checklist of important scientific practices such as ‘publishing’, ‘peer review’, ‘evaluation on real data’ and ‘evaluation of robustness’ [25]. Considering the time and memory constrains in real-life applications, methods with less time consumption and with higher correlations between predicted and actual time on benchmarking datasets are to some extent preferred. The selected 10 methods include Monocle[13], Monocle2, Monocle3, TSCAN [26], SLICER [27], GPfates [28], Slingshot [29], Destiny [30], Mpath [31], and STEMNET [32]. Their performance was evaluated in terms of accuracy for identifying true branches and cell types based on three public datasets (HSMM [13], MEF [33], and CMP [34]) with definite timepoints and branches from the NCBI GEO database. After filtering and normalization in the database, the standardized datasets were downloaded and utilized. Concisely, HSMM (GSE52529) contained 271 cells with two branches from the Fluidigm C1 microfluidic system at four timepoints during the development of human primary myoblasts. MEF (GSE67310) contained 605 cells belonging to 10 cell types collected at five timepoints using the Fluidigm C1 microfluidic system. These cells were divided into three branches during the induced reprogramming from mature fibroblasts to neurons. CMP (GSE72857) was obtained from the myeloid progenitor cells of adult mice with FACS sorting and MARS-seq. CMP was a more complex dataset, with 2730 cells belonging to 19 cell types and eight branches.
After filtering and normalization, scTITANS utilizes Monocle3 [13], [35] to construct the trajectory and calculate the pseudotime for each cell. The Louvain method proposed in Monocle3 allows reasonable disconnection between trajectories when clustering cells. The dimension reduction method UMAP introduced in Monocle3 for the first time improves the accuracy of branch recognition and operating efficiency. The trajectory length was calculated as the total number of transcriptional changes experienced by the cell when it moved from the starting state to the end state. Pseudotime is the distance between the cell and the starting point along the shortest path, and was proposed to represent the real state of the cell in the dynamic process. Among the various cell type annotation methods [34], [36], scCATCH [34] was utilized to obtain cell type annotations.
2.4. Time-dependent covariate analysis
To identify DEGs and differential cell subclusters, single cells were reordered along the pseudotime resulting from TI analysis, and a time-dependent covariate [37] was introduced to evaluate the fluctuations of genes and cell numbers along pseudotime. First, a curve was fitted for the expression of each gene or the cell numbers in each interval for each subcluster along pseudotime (complete model). The null model was a corresponding flat line. A statistic named q-value was then calculated to compare the two models. It is a quantification of whether a gene or subcluster changes significantly with pseudotime. The smaller the q-value, the more significant the difference between the complete and null model is.
Taking DEGs as an example, the corresponding section of Fig. 1 illustrates a scatter plot of the expression of an example gene, where × and y axis represent pseudotime and the relative abundance, respectively. The solid curve is the complete model, which was fitted and optimized to minimize the difference between the fitted and actual expression values. The flat dashed line is the null model. Let yij be the expression level of gene i in cell j, where i = 1, 2, …, M genes and j = 1, 2, …, N cells. The expression level of gene i in cell j in the complete model is defined by equation (1):
| (1) |
where is the fitted average expression level of gene i at the pseudotime , and the random deviation. was assumed to be an independent random variable with mean zero and gene-dependent variance . is parameterized with an intercept plus a p-dimensional linear basis (equation (2)):
| (2) |
where is a prespecified p-dimensional basis [38], [39], [40], is the gene-specific intercept, and a p-dimensional vector of gene-specific parameters. The singular value decomposition method was used to automatically choose the dimension of the basis p and the p-dimensional vector of gene-specific parameter [38]. An F statistic is further calculated to compare the residuals () of the complete and null models. The F statistic of gene i is calculated as follows (equation (4)).
| (3) |
| (4) |
In equation (3), is the sum of the squared residuals obtained from the null model, and the sum of the squared residuals obtained from the complete model. quantifies in the increase in goodness of fit, and dividing this by provides the exchangeability of among genes. The null distribution of the statistic was calculated through a method named bootstrap [41]. The basic idea is that the data are resampled in a way that new versions of null data are randomly generated for each gene. Using these null data, statistics are formed exactly as before that simulate the case where there is no differential expression. Here, the null data are generated by re-sampling the residuals obtained under the alternative model fit and adding them back to the null model fit. Then, a p-value in equation (5) is formed for each gene by measuring the frequency by which the bootstrap null statistics exceed each observed statistic. Here, B and M represent the number of iterations and genes, respectively.
| (5) |
Finally, a statistic q-value is calculated to quantify the significance of the difference based on p-values [42]. The estimated q-value for the most significant gene is calculated in equation (6), where is the proportion of genes that are not differentially expressed.
| (6) |
The smaller the q-value, the more significant the difference is.
2.5. Evaluation of scTITANS in identifying DEGs and differential cell subclusters
The performance of scTITANS in identifying DEGs and differential cell subclusters was evaluated based on different examples of datasets in the following two aspects. Firstly, scTITANS was evaluated for the performance in identifying DEGs and differential cell subclusters reported as such in the literature. Then, it was revealed by example datasets that pseudotime correction in scTITANS helped to reconstruct the trends for genes and cell subclusters.
With regard to identifying DEGs, a total of five example datasets deposited in NCBI GEO database, HSMM [13], HHD [43], MPD [10], CEED [44], and AD [9], were utilized. A short summary for each dataset is described below; details about the datasets such as species and number of cells and genes are summarized in Table 1. HSMM (GSE52529) was generated from a study on human primary myoblasts to identify new regulatory factors during cell differentiation. This dataset contained 271 cells obtained with the Fluidigm C1 microfluidic system at four timepoints and sequenced with a depth of approximately 4 million reads. HHD (GSE106118) was obtained from single-cell transcript sequencing on approximately 4,000 heart cells isolated from 18 human embryos at gestation periods of 5 to 25 weeks. Cells were captured from four areas: left atrium (LA), right atrium (RA), left ventricle (LV), and right ventricle (RV). Then, a modified single-cell labeling reverse transcription (STRT) protocol was used for scRNA-seq to obtain the gene expression profile of each cell. MPD (GSE101099) was obtained from single-cell transcript sequencing on embryonic murine pancreatic cells at days 12, 14, and 17 to explore pancreas development; live cells were collected by fluorescence-activated cell sorting (FACS) and sequenced with a depth of approximately 30,000 reads. CEED (GSE126954) was obtained from single-cell transcriptomes of Caenorhabditis elegans (C. elegans) embryos at approximately 300, 400, and 500 min after the first division. AD (GSE98969) was obtained from characterized immune cells involved in Alzheimer's disease; all immune cells (CD45+) in the mouse brain were classified by massive parallel single-cell RNA-seq (MARS-seq) at a sequencing depth of 50 K-100 K reads per cell. The standardized data were published on the NCBI GEO.
Table 1.
Details about the example datasets.
| Dataset | Category | Cell | Total gene | Average gene | Time points | Species |
|---|---|---|---|---|---|---|
| HSMM | tissue development | 384 | 47,193 | 7953 | 4 | human |
| HHD | organ development | 4898 | 24,153 | 4109 | 12 | human |
| MPD | organ development | 18,294 | 20,157 | 6076 | 3 | mouse |
| CEED | embryo development | 84,625 | 20,222 | 5402 | 3 | C. elegans |
| AD | disease development | 8016 | 34,016 | 5789 | 4 | mouse |
| MND | tissue development | 22,967 | 26,183 | 4435 | 5 | mouse |
| MEP | tissue development | 65,449 | 26,183 | 4231 | 5 | mouse |
| MRD | organ development | 120,804 | 24,016 | 3921 | 10 | mouse |
In addition to CEED [44], other three datasets MRD [45], MND [46], and MEP [46], were selected in this study to evaluate the performance of scTITANS in identifying differential cell subclusters. A short summary for each dataset is described as follows, with details about the datasets such as species and number of cells and genes being summarized in Table 1. MRD (GSE118614) was obtained from sequencing the Chx10-GFP (+) retinal progenitor cells across 10 timepoints of mouse retinal development to examine retinal progenitor cell heterogeneity during development. MND and MEP (GSE119945) were selected from a study that profiled over 2 million cells derived from 61 mouse embryos staged between 9.5 and 13.5 days of gestation by improved single-cell combinatorial indexing-based protocol (‘sci-RNA-seq3′) to investigate the transcriptional dynamics of mouse development during organogenesis at single-cell resolution. MND and MEP are subsets of datasets obtained from glial and epithelial tissues (MEP dataset).
3. Results and discussion
3.1. Monocle3 outperforms other trajectory inference methods
As a key aspect of scTITANS, we first evaluated the performance of multiple TI methods based on three benchmarking datasets, HSMM [13], MEF [33], and CMP [34]. Owing to the limitations of each method, different subsets of TI methods have been successfully applied to the three datasets. For HSMM, a dataset containing 271 cells obtained at four timepoints during the development of human primary myoblasts, results from six methods including Monocle [13], [35], Monocle2 [13], [35], Monocle3 [13], [35], Slingshot [29], GPfates [28] and Mpath [31] were obtained. Figure S1a shows that only Monocle, Monocle3, GPfates, and Mpath successfully reconstructed the two branches implicated in the 271 cells, although the trajectory from GPfates was apparently much less reasonable. It should also be noted that Mpath was able to provide the trajectory for only the centroid of each cell cluster instead of each single cell. Therefore, only Monocle and Monocle3 scored in dataset HSMM. For MEF, a dataset containing 605 cells collected at five timepoints during the reprogramming from mature fibroblasts to neurons, a total of seven methods, Monocle, Monocle2, Monocle3, Destiny [30], Slingshot, Slicer [27] and TSCAN [26] were applied. Figure S1b shows that only Monocle3 reconstructed the three branches implicated in this dataset. These results indicate that only Monocle3 scored in this case. For CMP, a dataset containing 2730 cells obtained from the myeloid progenitor cells of adult mice, eight methods, Monocle, Monocle2, Monocle3, SLICER, TSCAN, Destiny, Mpath and STEMNET [32], were successfully applied. Only Mpath accurately reconstructed the eight branches implicated in the database and also the trajectory path (Figure S1c). However, Mpath did not provide a trajectory for each single cell, which reduces its value in correcting the asynchrony of single cells for following time-series analysis. Although STEMNET identified the eight most mature cell types in the end stage of development with high accuracy, its value is reduced by its inability to provide information of cell-to-cell transformation during development. After excluding TSCAN, which failed to provide a reasonable trajectory, and Destiny, which resulted in two branches, the remaining four methods, Monocle, Monocle2, Monocle3, and SLICER, were used to reconstruct three branches from the dataset. However, it is apparent that the trajectory provided by SLICER was not consistent with prior knowledge. Moreover, it should be noted that Monocle3 surpassed Monocle and Monocle2 in the identification of the eight cell types with higher accuracy. Therefore, Monocle3 scored in this case, although its performance was somewhat less than satisfactory. Using the three example datasets, Monocle3 outperformed the other methods in reconstructing the true branches and cell types implicated in single cells, although its performance was less than perfect. Therefore, Monocle3 was selected as the TI method in scTITANS.
3.2. Performance of scTITANS in identifying differential genes
Datasets HHD [43], HSMM [13], MPD [10], CEED [44], and AD [9] were selected to demonstrate the performance of scTITANS in identifying differential genes. For simplicity, only the results of CEED are shown. Concisely, cells in the CEED dataset were clustered using the Louvain algorithm and annotated into 27 cell types based on the marker genes of each cell type described in the literature. Next, we reduced the dimensionality of the data, learned the trajectory graph, ordered the cells by pseudotime using Monocle3, and identified differential genes using scTITANS. The performance of scTITANS in identifying differential genes on datasets HSMM, MPD, CEED, and AD are shown in supplemental data and Table S1.
3.2.1. scTITANS performs well in identifying differential genes
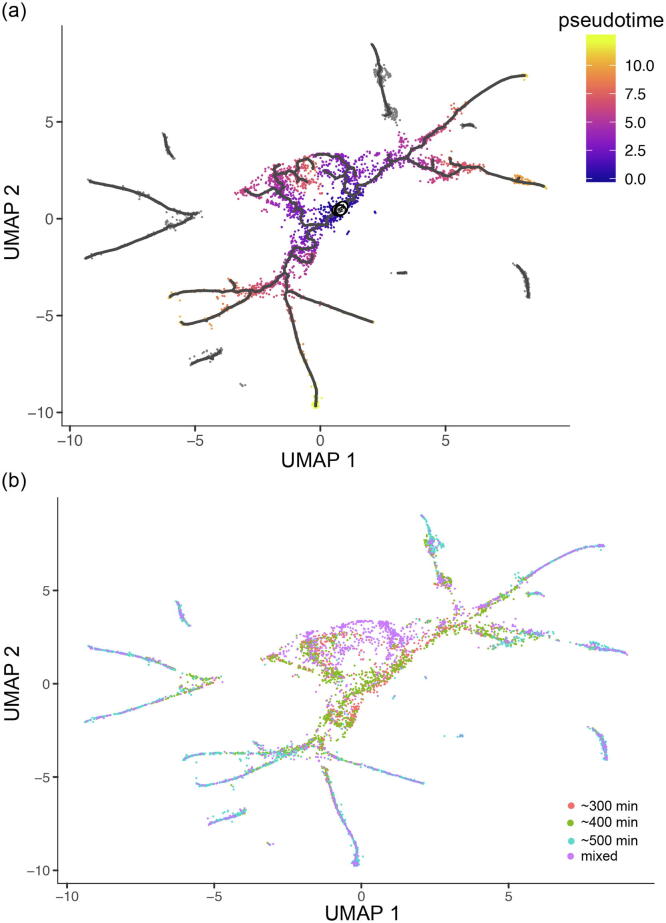
Most cells in the early developmental stage of the C. elegans nervous system are early embryonic developmental cells, which differentiate and develop into multiple types of mature cells along the developmental trajectory [44]. Fig. 2(a-b) illustrates the constructed trajectory with cells colored by pseudotime and time points. As shown in the figure, the gradual distribution of different cell types during pseudotime is consistent with those obtained during development time. Based on the pseudotime resulting from the above TI analysis, a curve representing the relative abundance of each gene along pseudotime was fitted, and the significance of the difference between the fitted and flat lines was evaluated and quantified with q-value. The smaller the q-value, the more significant the gene is. Using the scTITANS method, a series of genes were recognized as differential genes in the CEED dataset. Table 2 shows the top 20 differential genes.
Fig. 2.
The constructed trajectory with cells colored by (a) pseudotime and (b) time points for dataset CEED.
Table 2.
Top 20 differential genes identified in dataset CEED with scTITANS.
| Gene short name | q-value | Description | Verified by experiments or literatures |
|---|---|---|---|
| F18E9.1 | 1.78E-307 | hypothetical protein | |
| K07C5.9 | 1.76E-306 | hypothetical protein | |
| fbxb-70 | 1.16E-305 | FBA_2 domain-containing protein | |
| F22E5.20 | 6.03E-304 | hypothetical protein | |
| ZK792.7 | 7.73E-304 | hypothetical protein | |
| K04D7.6 | 1.24E-303 | hypothetical protein | |
| pab-1 | 2.07E-303 | Polyadenylate-binding protein | Y |
| F40G9.6 | 7.38E-302 | hypothetical protein | |
| odr-2 | 1.74E-300 | hypothetical protein | Y |
| sox-4 | 2.69E-299 | SOX family | Y |
| his-37 | 7.42E-298 | Histone H4 | Y |
| cla-1 | 1.67E-297 | Protein clarinet | Y |
| aqp-12 | 6.12E-295 | AQuaPorin or aquaglyceroporin related | Y |
| glb-18 | 2.27E-294 | GLOBIN domain-containing protein | Y |
| F53G12.4 | 8.72E-294 | hypothetical protein | |
| atf-5 | 2.05E-291 | ATF family | Y |
| oig-8 | 1.07E-286 | Ig-like domain-containing protein | Y |
| gcy-8 | 2.01E-196 | Receptor-type guanylate cyclase gcy-8 | Y |
| dac-1 | 2.75E-184 | Ski_Sno domain-containing protein | |
| hlh-4 | 1.75E-35 | Helix-loop-helix protein 4 | Y |
To evaluate the performance of the method in identifying true differential genes, the importance of the identified differential genes was further confirmed with literature surveys. Table 2 also illustrates whether the genes were verified through experiments or based on the literature. It should be noted that part of the differential genes is composed of hypothetical proteins, which are mostly inferred from computational analysis of genomic DNA sequencing by gene prediction software. Because the functions of hypothetical proteins cannot be easily determined, they were not considered in subsequent verifications. Overall, eleven of the top 12 differential genes have been successfully confirmed in this study. Pab-1 regulates the process of mRNA transportation and translation. It has been shown that pab-1 plays an important role in the development of nematodes, including establishment of mitotic spindles [47], regulation of the mitotic cycle [48] and germ cell proliferation [49]. Oig-8 is a previously uncharacterized transmembrane protein with a single immunoglobulin domain, and modulates the distinct, neuron-type-specific elaboration of ciliated endings of different olfactory neurons in nematode C. elegans [50]. Sox-4 is a member of the SOX family, which plays an important role in the regulation of embryonic development and cell differentiation [51]. The main function of cla-1 is the formation of synaptic vesicles, involving biological processes such as synaptic assembly and regulation of calcium-dependent exocytosis [52]. Atf-5 (activating transcription factor 5) is widely present in mammals, in adipocyte differentiation [53], and in regulation of transcription [54]. Gcy-8 encodes guanylate cyclase, and hlh-4 plays a key role in the development of the nervous system. It does make sense to recognize the above genes as differential genes in the development of the C. elegans nervous system. dac-1 is a Ski_Sno domain-containing protein that belongs to the Ski/Sno family, and a specific connection has been described between the Ski/Sno family and the TGF-β signaling pathway [55], the genes of which play a vital role in development and reproduction. Moreover, it has been reported that daf-5, another member of the Ski/Sno family, is a transcriptional regulator of genes in the TGF-β superfamily signaling pathway that play an important role in the development of the nervous system [56]. Therefore, it is also reasonable to recognize dac-1 and daf-5 as differential genes during growth and development.
The performance of scTITANS in identifying DEGs for time-series scRNA-seq data was further evaluated by the GO biological processes enriched from the top 20 DEGs for each dataset using ToppGene [57] (Table S2). CEED, a single-cell transcriptomic dataset of C. elegans embryos at approximately 300, 400 and 500 min after the first division, was used as an example dataset. Although only one of the top 20 DEGs was successfully annotated for CEED, the GO biological processes such as ascending aorta morphogenesis, ascending aorta development and septum primum development are typical processes during development. Corresponding results for the other four datasets further confirmed the performance of scTITANS in identifying differential genes from time-series scRNA-seq data. Details on the percentage of the top 20 genes verified by literature surveys and the top 10 enriched GO biological processes using ToppGene for CEED and other datasets are shown in Table S2. We also performed DEG analysis between time points for each of the five example datasets using a function suitable for bulk RNA-seq data provided in R package ‘edge’[58]. In this kind of analysis, cells from the same time point were considered as biological or technical replications, and the differences in genes among multiple time points were evaluated. Table S3 illustrated the top 20 DEGs identified by ‘edge’ for each dataset. Table S4 illustrated the percentages of genes among the top 20 DEGs verified with literatures using scTITANS and ‘edge’, the p-values and statistical power for the fisher exact tests comparing the performance of the above two methods for each example dataset. As shown in Table S4, the percentages of verified genes using scTITANS were significantly higher than that obtained using ‘edge’ for three datasets HSMM, HHD, and MPD with high confidence (p-value < 0.05, power greater than 0.9). The percentage of verified genes for scTITANS (92%) can still be considered to be higher than that of ‘edge’ (25%) for dataset CEED with a p-value slightly higher than 0.05 (p-value 0.06, power 0.98). Although the better performance of scTITANS over ‘edge’ (80% versus 40%) is not so significant for dataset AD (p-value 0.30), the corresponding low power (0.60) somehow implies that the p-value in this case may be not so reliable due to the limited number of genes considered. In other words, such results indicate that scTITANS outperforms ‘edge’ in that it picks up a higher percentage of verified genes.
3.2.2. Pseudotime correction in scTITANS helps to reconstruct gene expression trends
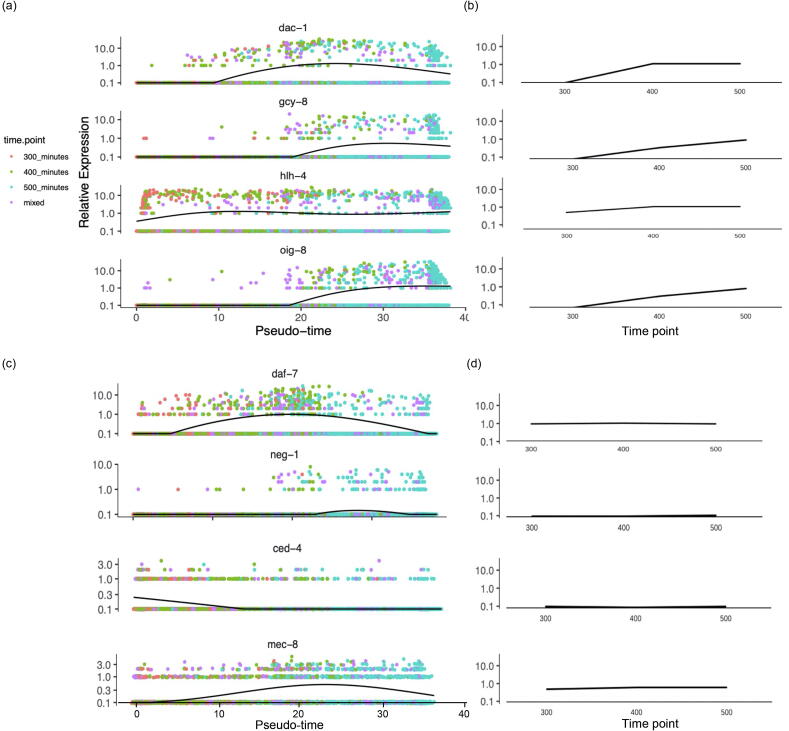
Gene expression trends implied in time-series scRNA-seq data are key to the performance of scTITANS. Thus, we further evaluated the contribution of pseudotime correction to scTITANS in reconstructing the true gene expression trends in time-series data. As shown in Fig. 3, pseudotime correction in scTITANS (Fig. 3a) reconstructed the trends of genes that were obviously differentially expressed among multiple time points (Fig. 3b). Gcy-8 encodes guanylate cyclase, which is critical to the thermal sensitivity of C. elegans. It has been reported that the expression of gcy-8 gradually increases with the development of the chemosensory neuron system [59], and then decreases with age and the reduction in the nematode's thermal sensitivity to the outside world. scTITANS can successfully characterize the abovementioned process, although the decrease of gcy-8 was somewhat marginal (Fig. 3a and 3b). Moreover, scTITANS reconstructed gene expression trends that are hidden by cell asynchrony, especially genes whose expression levels changed non-linearly during development. Genes daf-7 (q = 2.21E-32), neg-1 (q = 1.11E-42), ced-4 (q = 1.51E-59), and mec-8 (q = 1.90E-32) were recognized as differential genes by scTITANS, but showed no obvious perturbations along development. As shown in Fig. 3c, the expression of daf-7 increased first and then decreased, and was apparently different from the flat line fitted with real time points (Fig. 3d). Similar results apply for genes neg-1, ced-4 and mec-8. It has been reported that daf-7 is closely related to the development of the nervous system in that it is a development-regulated growth factor that regulates the transcription and apoptotic processes [60], plays an important role in larval development, and has a greater impact on the development of molting and excretory ducts [61]. Several lines of evidence support that neg-1 [62], ced-4 [63], and mec-8 [64] play important roles in the developmental process. Therefore, all the above results confirm that single-cell reordering based on TI analysis in scTITANS helps to reconstruct the true gene expression trends from real time-series data. Corresponding results for the other four datasets that support the same conclusion are provided in supplemental data.
Fig. 3.
The trends of genes along pseudotime time and real time points. (a-b) An illustration of the performance of pseudotime correction in scTITANS in reconstructing the trends of genes that are obviously differentially expressed along development. (c-d) An illustration of the performance of pseudotime correction in reconstructing the trends for genes which are masked by cell asynchrony.
3.3. Performance of scTITANS in identifying differential cell subclusters
Datasets MRD [45], MND [46], MEP [46], and CEED [44] were selected to demonstrate the performance of scTITANS in identifying differential cell subclusters. For simplicity, only the results of MRD are provided in the main manuscript. Concisely, cells were first clustered using the Louvain algorithm and annotated with the marker genes of each cell type reported in the literature. After filtering retinal pigment epithelium (RPE)/margin/periocular mesenchyme/lens epithelial cells owing to low cell counts, 14 cell types were considered for further analysis. Next, we reduced the dimensionality of the data, learned the trajectory graph, and ordered the cells by pseudotime using Monocle3. The performance of scTITANS in identifying differential cell subclusters on datasets MND, MEP and CEED are shown in supplemental data, Table S5, and Figures S1 and S2.
3.3.1. scTITANS performs well in identifying differential cell subclusters
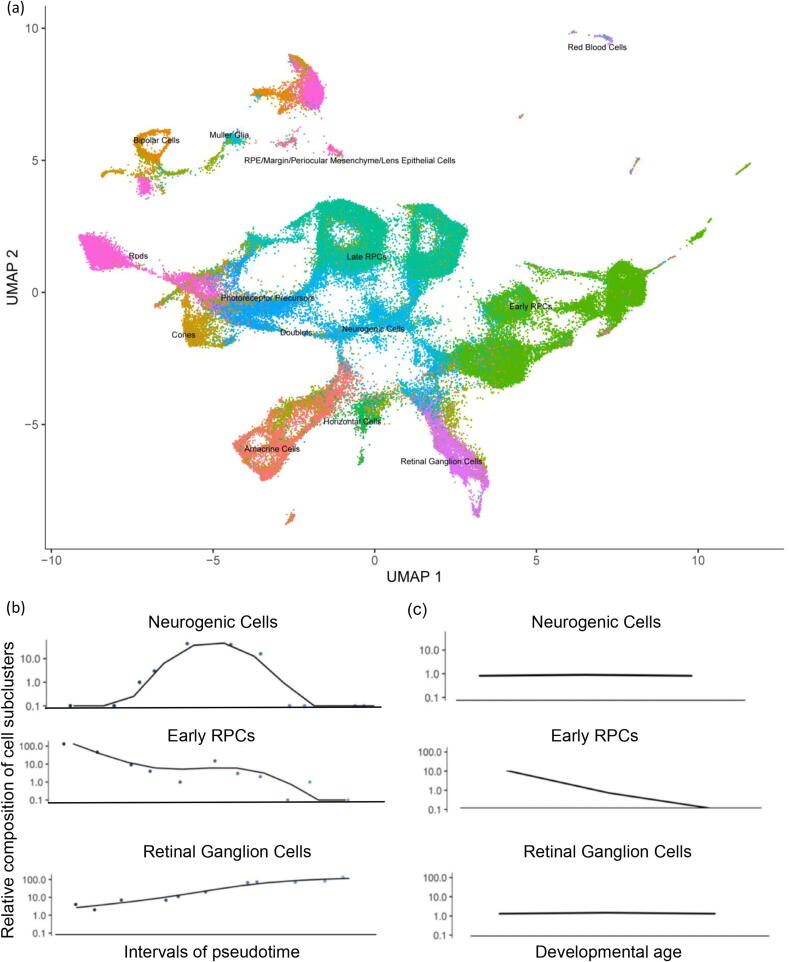
Fig. 4a illustrates the cell subclusters obtained in this study with Monocle3 for dataset MRD, a dataset sequenced from retinal progenitor cells across 10 timepoints of mouse retinal development. Consistent with experimental findings, retinal progenitor cells (RPCs), neurogenic Cells, and mature cell types such as amacrine cells, bipolar cells, and cones, were successfully clustered and annotated. After TI analysis and splitting the pseudotime into many time intervals, the curve of cell numbers in each interval was fitted along the pseudotime for each subcluster, and the q-value was calculated to identify differential subclusters. The q-values for the differential cell subclusters are provided in Table 3 in an ascending order.
Fig. 4.
(a) An illustration of the cell subclusters obtained in this study. (b-c) An illustration of the trends for cell subclusters along pseudotime and real time points.
Table 3.
Differential cell subclusters for dataset MRD.
| Cell type | q-value | Verified with literatures |
|---|---|---|
| Neurogenic Cells | 6.57E-87 | Y |
| Early RPCs | 1.31E-13 | Y |
| Retinal Ganglion Cells (RGCs) | 2.37E-06 | Y |
| Bipolar Cells | 7.14E-05 | |
| Cones | 7.14E-05 | |
| Late RPCs | 7.14E-05 |
We then performed literature verification on the identified differential subclusters (Table 3). RPCs are the earliest cells the arise during the development of the retina. Many studies indicate that RPCs have the ability to differentiate into different mature cell types during development [65]. It has been reported that early RPCs are capable of generating only radial glial cells (RGCs) and amacrine, horizontal and photoreceptor cells, whereas late RPCs can generate only rod, bipolar and muller cells [66]. Although the difference in competence between early and late RPCs is not clear, the progenitors are thought to progressively change their competence states. Neurogenic cells are reported to differentiate into all major retinal neuronal subtypes, with the exception of horizontal cells [66]. RGCs are transformed from epithelial cells, which directly or indirectly generate all neurons and produce glial cells in later stages of development [67]. Finally, the retina gradually differentiates into various mature cell types, such as amacrine cells, bipolar cells, cones, which play various roles in the formation and transmission of vision. Therefore, it is reasonable to classify early RPCs, late RPCs, neurogenic cells, and RGCs as differential subclusters. In this case, red blood cells were characterized as less critical during retinal development, although numerous scientific studies have shown that blood vessels are necessary to maintain the normal physiological activities of retina. Generation of neural tube walls accompanied by angiogenesis occurs early during retina development. Therefore, the number of blood cells does not change significantly during the differentiation process [68], which is consistent with the results obtained by our method.
3.3.2. Pseudotime correction in scTITANS helps to reconstruct the trends for cell subclusters
Pseudotime correction helps in the reconstruction of the trends for cell subclusters. Fig. 4b and 4c shows the fitted trends along pseudotime and real time points for three cell subclusters, respectively. scTITANS identified the cell clusters that were clearly differentially expressed among multiple time points, such as ‘early RPCs’. Moreover, scTITANS also identified differential cell clusters masked by cell heterogeneity, such as ‘neurogenic cells’ and ‘retinal ganglion cells’, that demonstrated no clear perturbations along real time points. It has been reported that retinal ganglion cells, responsible for integrating visual information and transmitting it to the central nervous system, should gradually increase during development [69]. The successful characterization of this trend confirms that cell reordering by pseudotime in scTITANS helps to reconstruct the trends for cell subclusters.
Based on several example datasets, scTITANS was confirmed quantitatively and performed well in identifying differential genes and cell subclusters by dealing with the heterogeneity among cells and making full use of the timing information hidden in biological processes. One major feature of scTITANS is that trajectory reference analysis is used to reconstruct pseudotime, based on which, single cells are reordered to preclude the potential biases resulting from cell asynchrony. Time-series analysis was then integrated with TI analysis to uncover the regulatory factors implied in dynamic biological processes in a quantitative way. Time-series analysis has been successfully applied in studies focusing on human disease and drug development [18], [19]. Therefore, time-series analysis clearly takes full advantage of the time-series information involved in dynamic processes. Moreover, its advantage in identifying differential genes in dynamic biological processes based on high-throughput RNA-seq data has also been revealed [20], [21]. Considering the fact that gene expression changes continuously along pseudotime, time-series analysis is a perfect choice to mine the information hidden behind the time-series single-cell sequencing data. Thus, ‘time-dependent covariate’ was then selected as the second crucial part of scTITANS.
The performance of scTITANS was evaluated and confirmed based on two aspects. On the one hand, literature verification was utilized to evaluate the capability of scTITANS in identifying genes and cell subclusters that have been shown to play key roles during development. On the other hand, pseudotime correction was revealed to be helpful in reconstructing the trends for genes and cell subclusters that were clearly differentially expressed or masked by cell asynchrony along real time points. Based on strict design and evaluation strategies, the outstanding performance of scTITANS in identifying differential genes and cell subclusters from time-series scRNA-seq data was confirmed with high reliability. It should be noted that scTITANS successfully identified odr-2 as a differential gene for dataset CEED, which was originally identified as a hypothetical protein during genome analysis by bioinformatic tools, and then confirmed to regulate AWC signaling within the neuronal network required for chemotaxis [70]. In this case, five of the top-ranked six differential genes are hypothetical proteins with p-values smaller than that of odr-2. Although no reports are available on their roles in the development of the C. elegans nervous system, the successful example of odr-2 indicates that special attention should be paid to them in future studies. Moreover, the performance of scTITANS was affected by the performance of TI analysis. Owing to the shortcomings involved in trajectory analysis [25], the performance of scTITANS may be less satisfactory in situations where only raw expression matrices are provided.
4. Conclusion
scTITANS combines the advantage of TI and time-series analyses for the first time to identify DEGs and differential cell subclusters for time-series scRNA-seq data. By reordering single cells along pseudotime resulting from TI analysis, the biases associated with asynchrony of single cells can be fully excluded from subsequent time-series analysis. Compared with current attempts, scTITANS is more accurate, quantitative, knowledge-free, and capable of dealing with heterogeneity among cells and making full use of the timing information hidden in biological processes. When extended to larger datasets and broader research areas, scTITANS will cause new breakthroughs in studies using single-cell sequencing.
5. Availability
All code is available at https://github.com/ZJUFanLab/scTITANS. Supplementary Data are available online.
Funding
The present study was funded by grants from the National Natural Science Foundation of China (No. 31870839), the Natural Science Foundation of Zhejiang Province (LZ20H290002), and the HangZhou Medical and Health Technology Project (Z20200052).
Author contributions
X.F. conceived and designed the study. R.X., L.S., X.L. J.L., and X.S. collected the single-cell RNA-seq data and developed the algorithm. R.X. and L.S. developed the package of scTITIANS. All authors wrote, read and approved the final manuscript.
CRediT authorship contribution statement
Xiaohui Fan: Conceptualization, Project administration, Writing- review & editing, Funding acquisition. Li Shao: Methodology, Formal analysis, Funding acquisition, Writing – review & editing. Rui Xue: Formal analysis, Writing – original draft. Xiaoyan Lu: Writing - original draft. Jie Liao: Writing - original draft, Software. Xin Shao: Writing - original draft, Software.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Alibaba Cloud.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.07.016.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Haque A., Engel J., Teichmann S.A., Lonnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9(1):75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao J., Lu X., Shao X., Zhu L., Fan X. Uncovering an organ's molecular architecture at single-cell resolution by spatially resolved transcriptomics. Trends Biotechnol. 2021;39(1):43–58. doi: 10.1016/j.tibtech.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Shao X., Lu X., Liao J., Chen H., Fan X. New avenues for systematically inferring cell-cell communication: through single-cell transcriptomics data. Protein & Cell. 2020;11(12):866–880. doi: 10.1007/s13238-020-00727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Losic B., Craig A.J., Villacorta-Martin C., Martins-Filho S.N., Akers N., Chen X. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat Commun. 2020;11(1) doi: 10.1038/s41467-019-14050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell. 2019;179(4):829–845. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Weinreb C., Rodriguez-Fraticelli A., Camargo F.D., Klein A.M. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science. 2020;367(6479):eaaw3381. doi: 10.1126/science.aaw3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davidson S., Efremova M., Riedel A., Mahata B., Pramanik J., Huuhtanen J. Single-Cell RNA Sequencing Reveals a Dynamic Stromal Niche That Supports Tumor Growth. Cell Reports. 2020;31(7):107628. doi: 10.1016/j.celrep.2020.107628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voigt A.P., Binkley E., Flamme-Wiese M.J., Zeng S., DeLuca A.P., Scheetz T.E. Single-Cell RNA Sequencing in Human Retinal Degeneration Reveals Distinct Glial Cell Populations. Cells. 2020;9(2):438. doi: 10.3390/cells9020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169(7):1276–1290. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 10.Byrnes L.E., Wong D.M., Subramaniam M., Meyer N.P., Gilchrist C.L., Knox S.M. Lineage dynamics of murine pancreatic development at single-cell resolution. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-06176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandel E.M., Grosschedl R. Transcription control of early B cell differentiation. Curr Opin Immunol. 2010;22(2):161–167. doi: 10.1016/j.coi.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Schlitzer A., Sivakamasundari V., Chen J., Sumatoh H.R.B., Schreuder J., Lum J. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. 2015;16(7):718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- 13.Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendall S., Davis K., Amir E.-A., Tadmor M., Simonds E., Chen T. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157(3):714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alemany A., Florescu M., Baron C.S., Peterson-Maduro J., van Oudenaarden A. Whole-organism clone tracing using single-cell sequencing. Nature. 2018;556(7699):108–112. doi: 10.1038/nature25969. [DOI] [PubMed] [Google Scholar]
- 16.Van den Berge K., Roux de Bézieux H., Street K., Saelens W., Cannoodt R., Saeys Y. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-14766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Härdle W., Lütkepohl H., Chen R. A review of nonparametric time series analysis. International statistical review. 1997;65(1):49–72. [Google Scholar]
- 18.Vasey B., Shankar A.H., Herrera B.B., Becerra A., Xhaja K., Echenagucia M. Multivariate time-series analysis of biomarkers from a dengue cohort offers new approaches for diagnosis and prognosis. PLoS NeglTrop Dis. 2020;14(6):e0008199. doi: 10.1371/journal.pntd.0008199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu F., Warren J., Exeter D.J. Interrupted time series analysis on first cardiovascular disease hospitalization for adherence to lipid-lowering therapy. Pharmacoepidemiol Drug Saf. 2020;29(2):150–160. doi: 10.1002/pds.4916. [DOI] [PubMed] [Google Scholar]
- 20.Spies D., Renz P.F., Beyer T.A., Ciaudo C. Comparative analysis of differential gene expression tools for RNA sequencing time course data. Briefings Bioinf. 2019;20(1):288–298. doi: 10.1093/bib/bbx115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spies D., Ciaudo C. Dynamics in Transcriptomics: Advancements in RNA-seq Time Course and Downstream Analysis. Comput Struct Biotechnol J. 2015;13:469–477. doi: 10.1016/j.csbj.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grubman A., Chew G., Ouyang J.F., Sun G., Choo X.Y., McLean C. A single-cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell-type-specific gene expression regulation. Nat Neurosci. 2019;22(12):2087–2097. doi: 10.1038/s41593-019-0539-4. [DOI] [PubMed] [Google Scholar]
- 23.Lun A.T., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 2016;5:2122. [DOI] [PMC free article] [PubMed]
- 24.Lun A.T., Bach K., Marioni J.C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol. 2019;37(5):547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- 26.Ji Z., Ji H. TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res. 2016;44(13):e117. doi: 10.1093/nar/gkw430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welch J.D., Hartemink A.J., Prins J.F. SLICER: inferring branched, nonlinear cellular trajectories from single cell RNA-seq data. Genome Biol. 2016;17(1):106. doi: 10.1186/s13059-016-0975-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lonnberg T., Svensson V., James K.R., Fernandez-Ruiz D., Sebina I., Montandon R. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Science immunology. 2017;2(9):eaal2192. doi: 10.1126/sciimmunol.aal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19(1) doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angerer P., Haghverdi L., Büttner M., Theis F.J., Marr C., Buettner F. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32(8):1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 31.Chen J., Schlitzer A., Chakarov S., Ginhoux F., Poidinger M. Mpath maps multi-branching single-cell trajectories revealing progenitor cell progression during development. Nat Commun. 2016;7:11988. doi: 10.1038/ncomms11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velten L., Haas S.F., Raffel S., Blaszkiewicz S., Islam S., Hennig B.P. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol. 2017;19(4):271–281. doi: 10.1038/ncb3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treutlein B., Lee Q.Y., Camp J.G., Mall M., Koh W., Shariati S.A.M. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature. 2016;534(7607):391–395. doi: 10.1038/nature18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao X., Liao J., Lu X.Y., Xue R., Ai N., Fan X.H. scCATCH: Automatic Annotation on Cell Types of Clusters from Single-Cell RNA Sequencing Data. iScience. 2020;23 doi: 10.1016/j.isci.2020.100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14(3):309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang A.W., O’Flanagan C., Chavez E.A., Lim J.L.P., Ceglia N., McPherson A. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nature Methods. 2019;16:1007–1015. doi: 10.1038/s41592-019-0529-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diggle P.J., Heagerty P., Liang K.Y., Zeger S.L. Analysis of longitudinal data. 2nd ed Oxford University Press.
- 38.Storey J.D., Xiao W., Leek J.T., Tompkins R.G., Davis R.W. Significance analysis of time course microarray experiments. PNAS. 2005;102(36):12837–12842. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice J.A., Wu C.O. Nonparametric mixed effects models for unequally sampled noisy curves. Biometrics. 2001;57(1):253–259. doi: 10.1111/j.0006-341x.2001.00253.x. [DOI] [PubMed] [Google Scholar]
- 40.Irizarry R.A., Tankersley C., Frank R., Flanders S. Assessing homeostasis through circadian patterns. Biometrics. 2001;57(4):1228–1237. doi: 10.1111/j.0006-341x.2001.01228.x. [DOI] [PubMed] [Google Scholar]
- 41.Roger W.J. An introduction to the bootstrap. Teaching Statistics. 2001;23(2):49–54. [Google Scholar]
- 42.Storey J.D. A direct approach to false discovery rates. J R Statist Sco B. 2002;64:479–498. [Google Scholar]
- 43.Cui Y., Zheng Y., Liu X., Yan L., Fan X., Yong J. Single-Cell Transcriptome Analysis Maps the Developmental Track of the Human Heart. Cell Reports. 2019;26(7):1934–1950. doi: 10.1016/j.celrep.2019.01.079. [DOI] [PubMed] [Google Scholar]
- 44.Packer J.S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 2019;365(6459):eaax1971. doi: 10.1126/science.aax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark B.S., Stein-O’Brien G.L., Shiau F., Cannon G.H., Davis-Marcisak E., Sherman T. Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron. 2019;102(6):1111–1126. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stubenvoll MD, Medley JC, Irwin M, Song MH. ATX-2, the C. elegans Ortholog of Human Ataxin-2, Regulates Centrosome Size and Microtubule Dynamics. PLoS genetics 2016;12(9):e1006370. [DOI] [PMC free article] [PubMed]
- 48.Ko S., Kawasaki I., Shim Y.-H., Antoniewski C. PAB-1, a Caenorhabditis elegans poly(A)-binding protein, regulates mRNA metabolism in germline by interacting with CGH-1 and CAR-1. PLoS ONE. 2013;8(12):e84798. doi: 10.1371/journal.pone.0084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciosk R., DePalma M., Priess J.R. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131(19):4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- 50.Howell K., Hobert O. Morphological Diversity of C. elegans Sensory Cilia Instructed by the Differential Expression of an Immunoglobulin Domain Protein. Curr Biol. 2017;27(12):1782–1790. doi: 10.1016/j.cub.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 51.She Z.-Y., Yang W.X. SOX family transcription factors involved in diverse cellular events during development. Eur J Cell Biol. 2015;94(12):547–563. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 52.Xuan Z., Manning L., Nelson J., Richmond J.E., Colon-Ramos D.A., Shen K. Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. Elife. 2017;6 doi: 10.7554/eLife.29276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y., Zhang Y.D., Zhang Y.Y., Qian S.W., Zhang Z.C., Li S.F. p300-dependent acetylation of activating transcription factor 5 enhances C/EBPbeta transactivation of C/EBPalpha during 3T3-L1 differentiation. Mol Cell Biol. 2014;34(3):315–324. doi: 10.1128/MCB.00956-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madarampalli B., Yuan Y., Liu D., Lengel K., Xu Y., Li G. ATF5 Connects the Pericentriolar Materials to the Proximal End of the Mother Centriole. Cell. 2015;162(3):580–592. doi: 10.1016/j.cell.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 55.Liu X., Sun Y., Weinberg R.A., Lodish H.F. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 2001;12(1):1–8. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 56.Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A.G. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38(8):896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 57.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leek J.T., Monsen E., Dabney A.R., Storey J.D. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22(4):507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- 59.Inada H., Ito H., Satterlee J., Sengupta P., Matsumoto K., Mori I. Identification of guanylyl cyclases that function in thermosensory neurons of Caenorhabditis elegans. Genetics. 2006;172(4):2239–2252. doi: 10.1534/genetics.105.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGehee A.M., Moss B.J., Juo P. The DAF-7/TGF-beta signaling pathway regulates abundance of the Caenorhabditis elegans glutamate receptor GLR-1. Molecular and cellular neurosciences. 2015;67:66–74. doi: 10.1016/j.mcn.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crook M., Grant W.N. Dominant negative mutations of Caenorhabditis elegans daf-7 confer a novel developmental phenotype. Developmental dynamics : an official publication of the American Association of Anatomists. 2013;242(6):654–664. doi: 10.1002/dvdy.23963. [DOI] [PubMed] [Google Scholar]
- 62.Elewa A., Shirayama M., Kaymak E., Harrison P.F., Powell D.R., Du Z. POS-1 Promotes Endo-mesoderm Development by Inhibiting the Cytoplasmic Polyadenylation of neg-1 mRNA. Dev Cell. 2015;34(1):108–118. doi: 10.1016/j.devcel.2015.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang W., Jiang T., Choi W., Qi S., Pang Y., Hu Q. Mechanistic insights into CED-4-mediated activation of CED-3. Genes Dev. 2013;27(18):2039–2048. doi: 10.1101/gad.224428.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan J.H., Fraser A.G. The combinatorial control of alternative splicing in C. elegans. PLoS genetics. 2017;13(11) doi: 10.1371/journal.pgen.1007033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morikawa M., Derynck R., Miyazono K. TGF-beta and the TGF-beta Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb Perspect Biol. 2016;8(5) doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu S., Liu X., Li S., Huang X., Qian H., Jin K. Foxn4 is a temporal identity factor conferring mid/late-early retinal competence and involved in retinal synaptogenesis. Proc Natl Acad Sci U S A. 2020;117(9):5016–5027. doi: 10.1073/pnas.1918628117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paridaen J.TML., Huttner W.B. Neurogenesis during development of the vertebrate central nervous system. EMBO Rep. 2014;15(4):351–364. doi: 10.1002/embr.201438447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atan D. Immunohistochemical Phenotyping of Mouse Amacrine Cell Subtypes. Methods Mol Biol. 2018;1753:237–248. doi: 10.1007/978-1-4939-7720-8_16. [DOI] [PubMed] [Google Scholar]
- 69.Sanes J.R., Masland R.H. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci. 2015;38(1):221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- 70.Chou J.H., Bargmann C.I., Sengupta P. The Caenorhabditis elegans odr-2 gene encodes a novel Ly-6-related protein required for olfaction. Genetics. 2001;157(1):211–224. doi: 10.1093/genetics/157.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.