Abstract
Background & Aims
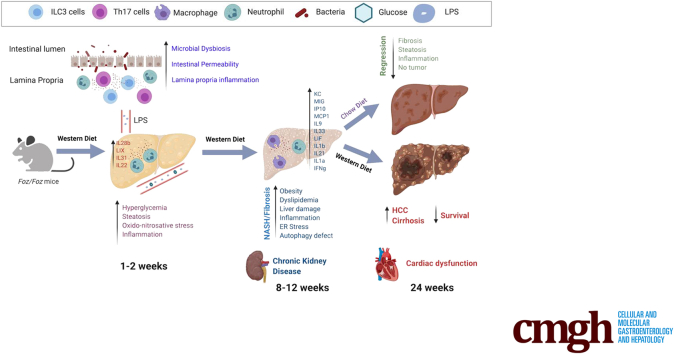
How benign liver steatosis progresses to nonalcoholic steatohepatitis (NASH), fibrosis, and hepatocellular carcinoma (HCC) remains elusive. NASH progression entails diverse pathogenic mechanisms and relies on complex cross-talk between multiple tissues such as the gut, adipose tissues, liver, and the brain. Using a hyperphagic mouse fed with a Western diet (WD), we aimed to elucidate the cross-talk and kinetics of hepatic and extrahepatic alterations during NASH–HCC progression, as well as regression.
Methods
Hyperphagic mice lacking a functional Alms1 gene (Foz/Foz) and wild-type littermates were fed WD or standard chow for 12 weeks for NASH/fibrosis and for 24 weeks for HCC development. NASH regression was modeled by switching back to normal chow after NASH development.
Results
Foz+WD mice were steatotic within 1 to 2 weeks, developed NASH by 4 weeks, and grade 3 fibrosis by 12 weeks, accompanied by chronic kidney injury. Foz+WD mice that continued on WD progressed to cirrhosis and HCC within 24 weeks and had reduced survival as a result of cardiac dysfunction. However, NASH mice that were switched to normal chow showed NASH regression, improved survival, and did not develop HCC. Transcriptomic and histologic analyses of Foz/Foz NASH liver showed strong concordance with human NASH. NASH was preceded by an early disruption of gut barrier, microbial dysbiosis, lipopolysaccharide leakage, and intestinal inflammation. This led to acute-phase liver inflammation in Foz+WD mice, characterized by neutrophil infiltration and increased levels of several chemokines/cytokines. The liver cytokine/chemokine profile evolved as NASH progressed, with subsequent predominance by monocyte recruitment.
Conclusions
The Foz+WD model closely mimics the pathobiology and gene signature of human NASH with fibrosis and subsequent HCC. Foz+WD mice provide a robust and relevant preclinical model of NASH, NASH-associated HCC, chronic kidney injury, and heart failure.
Keywords: Nonalcoholic Steatohepatitis, Hepatocellular Carcinoma, NASH Regression, Gut Inflammation, Liver Inflammation
Abbreviations used in this paper: ALT, alanine aminotransferase; APC, Allophycocyanin; CD-HFD, choline-deficient high-fat diet; CKD, chronic kidney disease; CVD, cardiovascular disease; CXCL2, C-X-C motif chemokine ligand 2; DSI, distal small intestine; eWAT, white adipose tissue (epididymal fat); FACS, fluorescence-activated cell sorter; FBS, fetal bovine serum; FITC, fluorescein isothiocyanate; HCC, hepatocellular carcinoma; HSC, hepatic stellate cell; IFN, interferon; IHC, immunohistochemistry; IL, interleukin; ILC, innate lymphoid cell; LBP, lipopolysaccharide binding protein; LPS, lipopolysaccharide; MIP2, Macrophage Inflammatory Protein 2; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; PBS, phosphate-buffered saline; PE, Phycoerythrin; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; RPCA, robust principal component analysis; rRNA, ribosomal RNA; SI, small intestine; TNF, tumor necrosis factor; WD, Western diet; WT, wild-type
Supplemental Graphical Summary
Summary.
This study establishes a relevant mouse model of nonalcoholic steatohepatitis–fibrosis–hepatocellular carcinoma that closely recapitulates human disease. With this model, we elucidate multitissue dynamics of nonalcoholic steatohepatitis/hepatocellular carcinoma progression, as well as regression. We define the key signaling and cytokine pathways that are critical for disease development and resolution.
A sedentary lifestyle coupled with a Western diet (WD) induces obesity and nonalcoholic fatty liver disease (NAFLD).1 Although the majority of NAFLD patients only have simple steatosis, 10%–20% of people with NAFLD have nonalcoholic steatohepatitis (NASH), characterized by chronic liver inflammation, hepatocyte injury, and fibrosis. NASH progresses to cirrhosis, liver failure, and hepatocellular carcinoma (HCC).1 NASH is the underlying etiology of a third of HCC cases,2 and is the second leading cause of liver transplants.3 In addition, NASH is associated with cardiovascular disease (CVD) and chronic kidney disease (CKD).4
Several diet-induced models currently are being used to study NASH that have their unique challenges and drawbacks.5 Feeding wild-type (WT) mice a high-caloric diet or a WD fails to mimic the complete pathology of NASH–fibrosis–HCC. Adding a hepatotoxin such as CCl4 to a high-caloric diet6 introduces a mechanism of hepatocyte injury not relevant to human NASH pathophysiology. Similarly, mice with transgenes rendering hepatocytes more sensitive to injury develop NASH on a high-caloric diet, but the mechanism of the liver injury is different than human NASH.5 Leptin or leptin-receptor–deficient mice (ob/ob and db/db) on a high-caloric diet develop obesity, but because of the critical role of the leptin pathway, these mice do not develop NASH with fibrosis.5 Importantly, these models do not reflect the multitissue injuries characteristic of NASH and the metabolic syndrome.
Our study uses Alms1 mutated mice to optimize a robust model of the metabolic syndrome, including NASH with fibrosis, CKD, and CVD. Mutations in the human ALMS1 causes Alström syndrome, a rare genetic disorder with childhood-onset obesity, metabolic syndrome, NASH, and diabetes.7 Alms1 mutated mice (also known as fat Aussie or Foz/Foz) are hyperphagic, have reduced physical activity, and on a high-caloric diet develop NASH accompanied by obesity, diabetes, and hypercholesterolemia.8,9 Restriction of Foz/Foz mice to a diet equivalent to the average daily food intake of a normal-weight mouse resulted in weight stabilization at the level of WT littermates, indicating that obesity in these mice is driven by hyperphagia.7
Our study shows that this optimized NASH model of Foz/Foz mice on a WD develop liver fibrosis, which progresses to cirrhosis and HCC. Because the leptin pathway is intact in these mice, liver fibrosis is stimulated appropriately. Foz/Foz mice also develop NASH-associated cardiovascular and renal disease.10 Furthermore, we establish the temporal development of intestinal permeability, gut microbial dysbiosis, gut and liver inflammation, and fibrosis. Finally, switching the WD back to normal chow results in regression of NASH with fibrosis.
Results
Foz/Foz Mice on WD Develop Obesity, Dyslipidemia, and Insulin Resistance
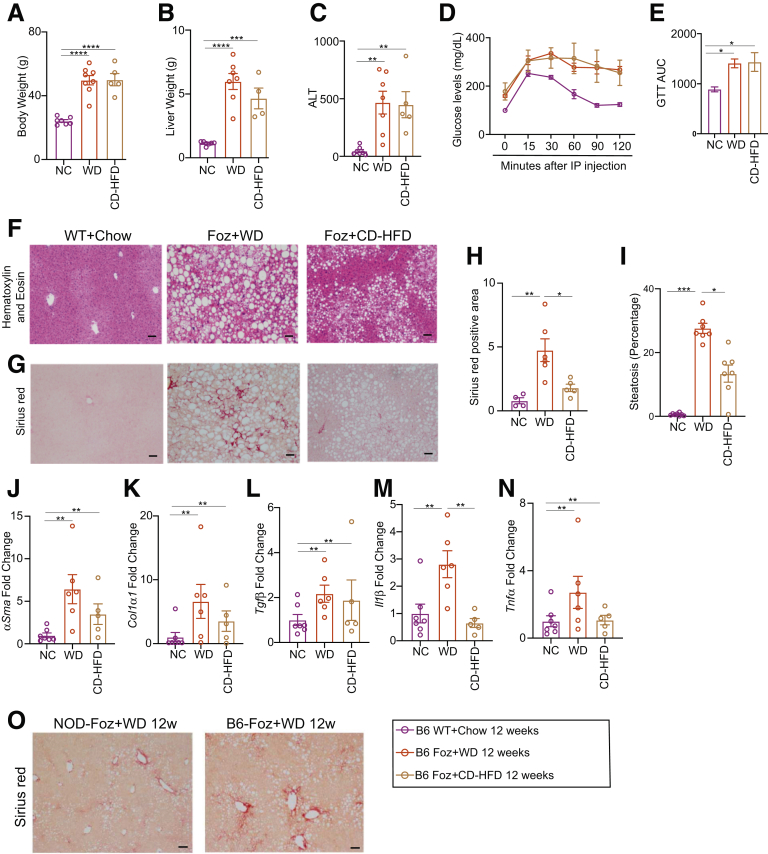
To identify the diet that generates better NASH liver pathology in Foz/Foz mice, we performed a head-to-head comparison of 2 widely used diets in the field: choline-deficient high-fat diet (CD-HFD) and WD. Foz/Foz mice on both diets developed equivalent obesity, liver weight, liver damage (Figure 1A–C), and glucose intolerance (Figure 1D and E). However, compared with CD-HFD, WD induced more severe steatosis, fibrosis, and inflammation (Figure 1F–N). We also found that WD-induced liver fibrosis is greater when the Alms1 mutation is in the B6 compared with the Non-obese Diabetic (NOD) B10 strain (Figure 1O). Therefore, our studies used Foz/Foz mice on a C57BL6 background on a WD.
Figure 1.
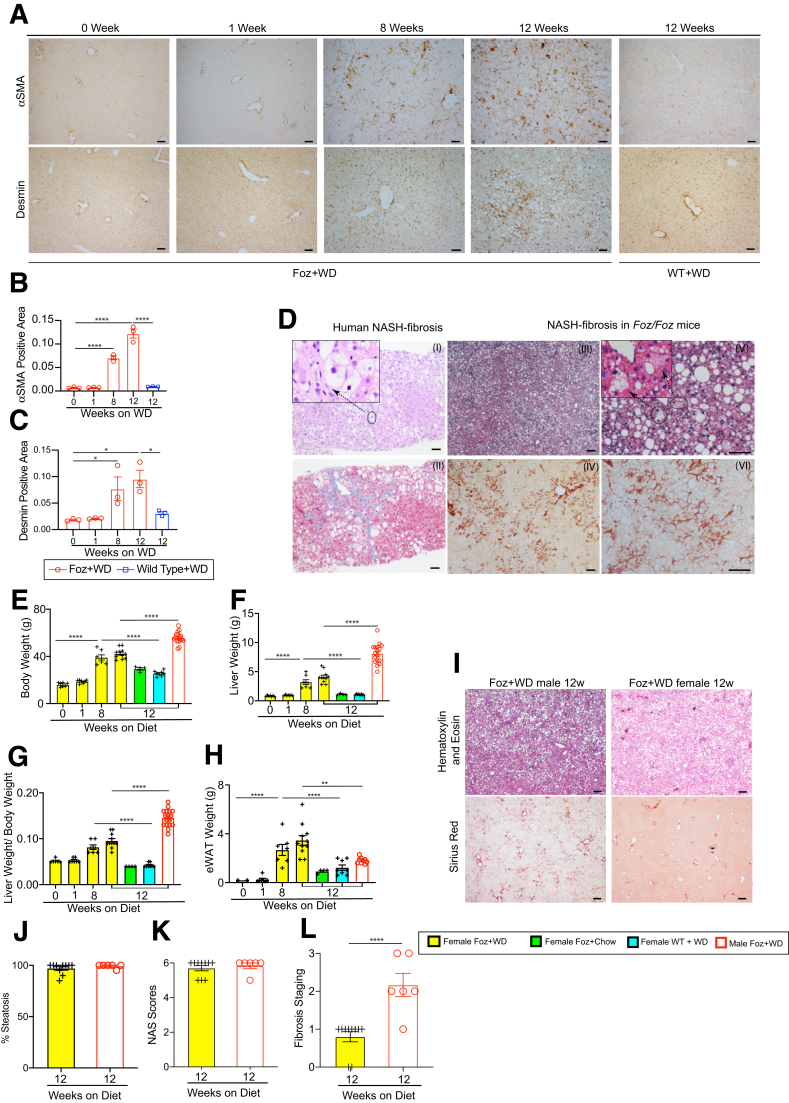
Foz+WD mice on a B6 background developed severe fibrotic NASH. (A–N) Foz/Foz mice on a B6 background were fed a normal chow diet (NC), WD, or CD-HFD for up to 12 weeks. Changes in (A) body weight and (B) liver weight were plotted. (C) Plasma ALT levels were measured for the indicated groups. (D) A glucose tolerance test (GTT) was performed. (E) GTT results are represented as the area under the curve (AUC) from panel D. (F–I) Formalin-Fixed Paraffin-Embedded (FFPE) liver sections were either (F) H&E stained or (G) stained with Sirius red. Quantification of the (H) Sirius red–positive area and (I) amount of steatosis (ImageJ). (J–N) Total RNA from the liver was isolated and qRT-PCR for the indicated genes was performed (fold change with respect to WT+NC 12 weeks). (O) Sirius red–stained tissue sections from WD-treated Foz/Foz mice on a NOD and B6 background. Data are expressed as the means ± SEM; 1-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. Scale bar: 200 μm. IP, intraperitoneal.
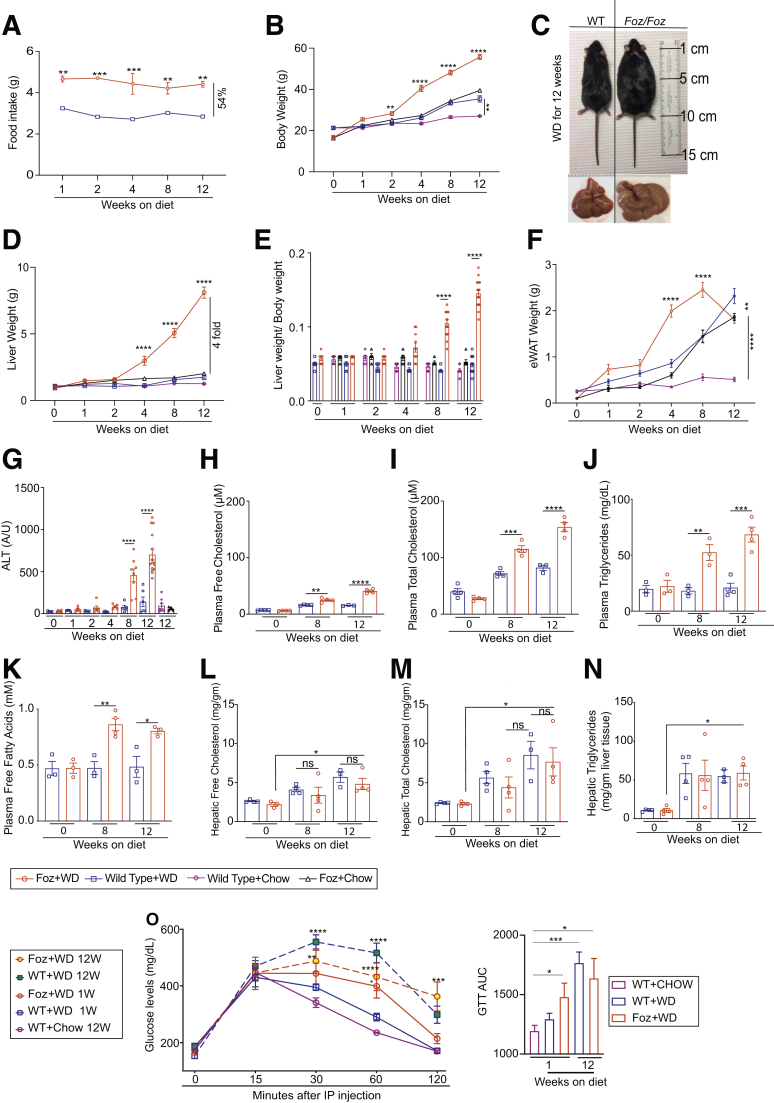
Foz/Foz mice are hyperphagic,9 consuming more than 50% more calories than their WT littermates (Figure 2A). When fed either chow (Foz+chow vs WT+chow) or a WD (Foz+WD vs WT+WD), Foz/Foz mice gained more weight than their WT counterparts (Figure 2B and C). The liver weight (Figure 2D) and liver:body weight ratio (Figure 2E) of Foz+WD mice increased within 8 weeks, and by 12 weeks their average liver weight was 4 times that of the other experimental groups (Figure 2D). White adipose tissue (epididymal fat [eWAT]) weight increased in Foz+WD, WT+WD, as well as Foz+chow mice compared with WT+chow mice (Figure 2F). eWAT weight in Foz+WD mice peaked within the first 8 weeks on WD, with a subsequent decrease from 8 to 12 weeks (Figure 2F). Foz mice had significantly more liver damage compared with all the other groups by 8 to 12 weeks of WD consumption, as evidenced by increased plasma alanine aminotransferase (ALT) levels (Figure 2G). Plasma levels of free and total cholesterol (Figure 2H and I), triglycerides (Figure 2J), and free fatty acids (Figure 2K) also were significantly higher in Foz+WD mice at 8 to 12 weeks compared with the WT+WD littermates. However, hepatic free and total cholesterol (Figure 2L and M) and triglyceride levels (Figure 2N) increased similarly in both of these groups. Foz+WD mice became glucose-intolerant within 1 week of WD feeding, and continued to be glucose intolerant at 12 weeks (Figure 2O).
Figure 2.
Foz+WD mice develop obesity, liver injury, and dyslipidemia. (A) Weekly WD intake. (B) Changes in body weight. (C) Representative gross images of WT+WD and Foz+WD 12-week mice and livers. Changes in (D) liver weight, (E) liver to body weight ratio, and (F) eWAT weight. Plasma was isolated at the indicated time points and subjected to the following analyses: (G) ALT, (H) free cholesterol, (I) total cholesterol, (J) triglycerides, and (K) free fatty acids. Liver extract from the indicated times was analyzed for (L) hepatic free cholesterol, (M) total cholesterol, and (N) triglycerides. (O) A glucose tolerance test (GTT) was performed in Foz+WD mice and their WT-B6 counterparts. Results also are represented as the area under the curve for the indicated groups. Data are expressed as the means ± SEM; ordinary 1-way analysis of variance (ANOVA). ∗P < .05, ∗∗P < .01,∗∗∗P < .001, and ∗∗∗∗P < .0001 (Foz+WD vs all other groups for A, B, and D; the rest are as indicated). IP, intraperitoneal.
Foz+WD Mice Develop NAFL, NASH, and Advanced Fibrosis
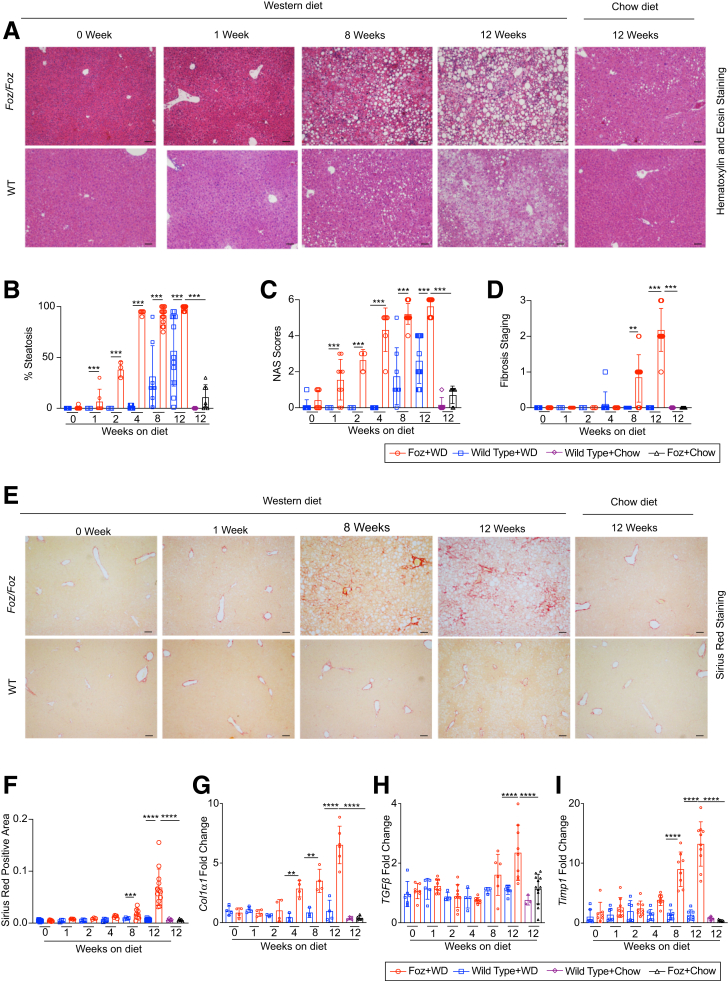
Foz+WD livers have earlier onset of steatosis (by 1 week) and become 100% steatotic by 4 weeks (Figure 3A and B). Based on the NAFLD activity scoring system, Foz+WD mice develop NASH by 4 to 8 weeks (Figure 3C). WT+WD mice on the other hand only developed steatosis at 8 weeks and showed very little inflammation even at 12 weeks (Figure 3B and C). Importantly, none of the WT+WD mice developed fibrosis by 12 weeks (Figure 3D), while Foz+WD mice started to develop fibrosis by 8 weeks (average fibrosis score, 1) and continued to progress to grades 2 and 3 fibrosis by 12 weeks (Figure 3D–F). Activation of hepatic stellate cells (HSCs) and profibrogenic genes, such as Col1α1, TGFβ, and Timp1, occurred within 8 weeks in Foz+WD mice, while WT+WD mice along with WT+chow and Foz+chow controls remained largely unaltered (Figure 3G–I).
Figure 3.
Foz/Foz mice sequentially develop steatosis, steatohepatitis, and advanced fibrosis. (A) Representative liver sections stained with H&E. Pathology scoring for the (B) percentage of steatosis, (C) composite NAFLD activity score (NAS), and (D) fibrosis staging. (E) Representative Sirius red–stained liver sections and (F) quantification (ImageJ). (G–I) qRT-PCR of indicated genes (fold change with respect to WT+WD 0 wk). Data are expressed as the means ± SEM; ordinary 1-way analysis of variance. ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. Scale bars: 200 μm.
Immunohistochemistry (IHC) for HSC activation markers α-smooth muscle actin and desmin confirms HSC activation in Foz+WD livers (Figure 4A–C). Therefore, the classic human NASH signatures including inflammatory infiltrates, pericellular fibrosis, and bridging fibrosis were observed within 8 weeks of WD in Foz/Foz mice and were histologically similar to the patterns found in human NASH (Figure 4D). Even though hepatocyte ballooning-like features have been reported in Foz/Foz11 and other mouse models,5 we believe that rodent hepatocyte ballooning does not recapitulate the cytologic features of their human counterparts. We used a strict definition of hepatocyte ballooning and excluded mimics that are caused mostly by small droplet fat accumulation.
Figure 4.
Foz/Foz mice sequentially develop steatosis, steatohepatitis, and advanced fibrosis. (A–C) Livers were collected at the indicated time points and Formalin-Fixed Paraffin-Embedded (FFPE) sections were subjected to IHC for α-smooth muscle actin (αSMA) and desmin, and the positive area normalized to the nonlipid area was quantified. (D) Representative tissue sections from human NASH patients (I, II) and Foz/Foz NASH mice (III–VI) stained with H&E (I, III, V), Masson's trichrome (II), and Sirius red (IV, VI), showing similarity in key histologic features such as grade 2 steatohepatitis (I, III, V) and bridging fibrosis (II, IV, VI). Inset (I) shows hepatocyte ballooning in human NASH and (V) the corresponding features in Foz/Foz NASH mice that are mostly owing to small droplet fat accumulation. (E–H) Both male and female WT and Foz/Foz mice were fed either chow or WD. (E) Body weight, (F) liver weight, (G) liver weight to body weight ratio, and (H) eWAT weight were measured at the indicated time points and plotted. (I) Representative male and female Foz/Foz liver tissue sections stained with H&E and Sirius red. (J–L) Pathologist scoring. (J) Steatosis percentage, (K) composite NAFLD Activity Score (NAS), (L) fibrosis staging (NASH Clinical Research Network scoring) in the indicated groups. Data are expressed as the means ± SEM; 1-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001. Scale bars: 200 μm.
Because of the conflicting human and rodent data on the relative susceptibility of males and females to NASH,12,13 we also conducted studies on Foz/Foz female mice on a B6 background. Foz+WD females gained less weight than Foz+WD males (Figure 4E–G). Foz+WD females compartmentalized fat in eWAT while Foz+WD males had decreased eWAT weight at 12 weeks (Figures 4H and 2F). Although Foz+WD females developed equivalent steatosis (Figure 4I and J) and had a similar inflammation and total composite NAFLD activity score as the male littermates (Figure 4K), Foz+WD females were resistant to fibrosis development (Figure 4I and L).
Foz+WD Mice Have a Transcriptomic Profile Similar to Human NASH
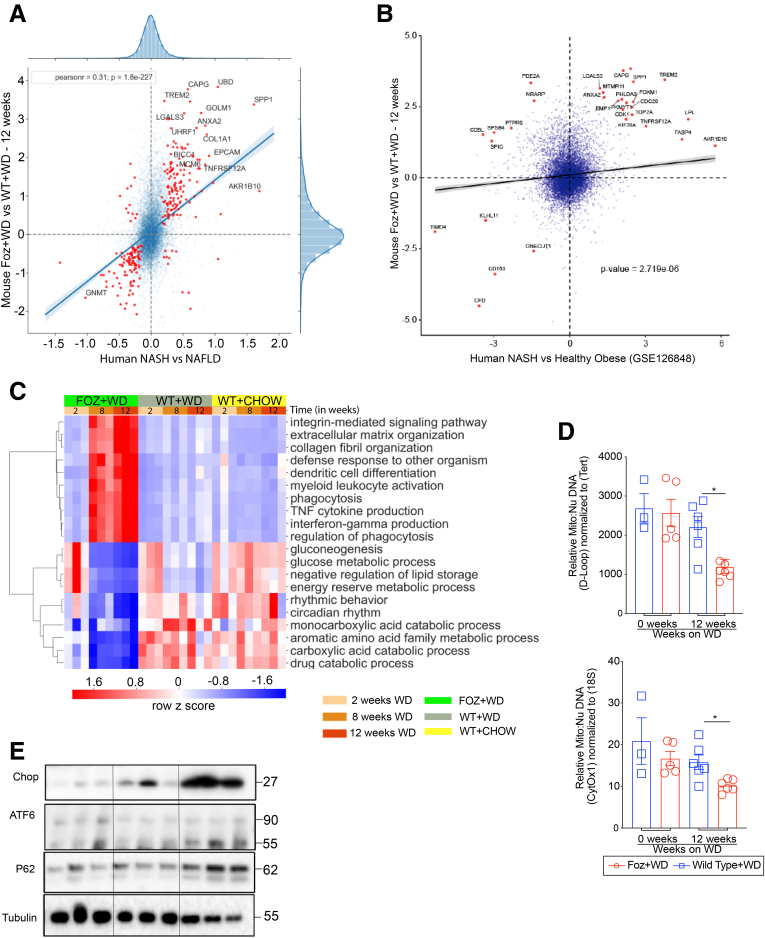
To determine the changes in gene expression that drive NAFL to NASH, we compared the liver transcription profile of Foz+WD at 12 weeks representing NASH plus fibrosis with the WT+WD at 12 weeks representing NAFL. For human liver gene expression, publicly available data (GSE48452)14 were mined and NASH (n = 18) was compared with healthy obese (n = 27) patients. Scatterplot analyses (Figure 5A) show the 257 genes that were dysregulated significantly in both human NASH and in Foz+WD 12-wk NASH (red dots), including key genes such as COL1A1, LGALS3, SPP1, ANXA2, and TREM2. To confirm the robustness of the Foz/Foz model, we performed a similar comparison with a second human data set (GSE126848)15 (Figure 5B), again showing a strong association of Foz+WD NASH to human NASH.
Figure 5.
Foz+WD NASH have a transcriptomic profile similar to human NASH. (A) Scatterplot showing the fold change of all expressed genes in human NASH compared with healthy obese patients (x-axis) (GSE48452), and in Foz+WD compared with WT+WD at 12 week (y-axis) (blue translucent dots). A total of 257 genes were commonly dysregulated significantly in both human NASH and in Foz+WD at 12 week (red dots). Of these, 234 were dysregulated in the same direction in both mice and human beings, where genes are up-regulated or down-regulated in both mice and human beings (upper and lower left quadrant, respectively). The 25 genes with the highest absolute value fold change are labeled. (B) Scatterplot showing the fold change of all expressed genes in both human NASH compared with healthy obese patients (x-axis) (GSE126848), and in Foz+WD compared with WT+WD at 12 weeks (y-axis) (blue translucent dots). The 30 genes with the highest absolute fold changes are labeled as red dots. Included in these labeled genes are key genes TREM2, LGALS3, ANXA2, and SPP1, which are up-regulated in both mice and human beings (upper right quadrant). (C) Heat map showing top dysregulated pathways in the indicated groups. (D) Total DNA was extracted from the liver tissues. Relative mitochondrial DNA amounts were quantified by qRT-PCR, with primers specific for the mitochondrial D-loop and normalized to Tert, which is encoded by the nuclear DNA. Similarly, mitochondrial cytochrome oxidase 1 (Cytox1) was normalized to nuclear 18s. (E) Immunoblot of CHOP, ATF6, cleaved ATF6, and P62, with tubulin as loading control. Data are expressed as means ± SEM; 1-way analysis of variance. ∗P < .05.
We performed a global gene expression analysis of the liver at multiple stages of NAFL and NASH progression to determine the differential kinetics of key molecular pathways. Gene Ontology evaluation showed distinct dysregulation of several signaling pathways in Foz+WD NASH liver compared with age-matched normal (WT+chow) or NAFL (WT+WD) livers. Besides the expected up-regulated pathways, such as extracellular matrix and collagen deposition, the inflammatory pathways were predominated by tumor necrosis factor (TNF) and interferon γ (IFNγ) in the NASH livers (Figure 5C, top half of heat map). Dendritic cell differentiation and myeloid leukocyte activation also were up-regulated in the NASH stages (Figure 5C). Interestingly, although most of the metabolic pathways related to glucose, carbohydrates, and lipids were slightly suppressed in steatotic livers (WT+WD at 8–12 wk, top part of the lower half section), these pathways were suppressed even more in the NASH livers (Foz+WD at 8–12 wk). Mitochondrial biogenesis and gluconeogenesis were suppressed more in NASH vs NAFL livers (Figure 5C). In agreement, the mitochondrial:nuclear DNA ratio also was reduced in Foz+WD 12-weeks NASH livers compared with WT+WD NAFL livers (Figure 5D). Pathways that are suppressed exclusively in NASH livers include disruptions in the circadian rhythm, and amino acid and fatty acid metabolic pathways (Figure 5C). Biological pathways (analysis of nonredundant GO biological process terms) deregulated in both mice and human beings (GSE48452 data set, NASH vs healthy obese) are shown in Supplementary Table 1. Furthermore, NASH vs NAFL was hallmarked by increased endoplasmic reticulum (ER) stress and suppressed autophagy as indicated by the immunoblot analyses of Chop/cleaved ATF6 and P62, respectively (Figure 5E).
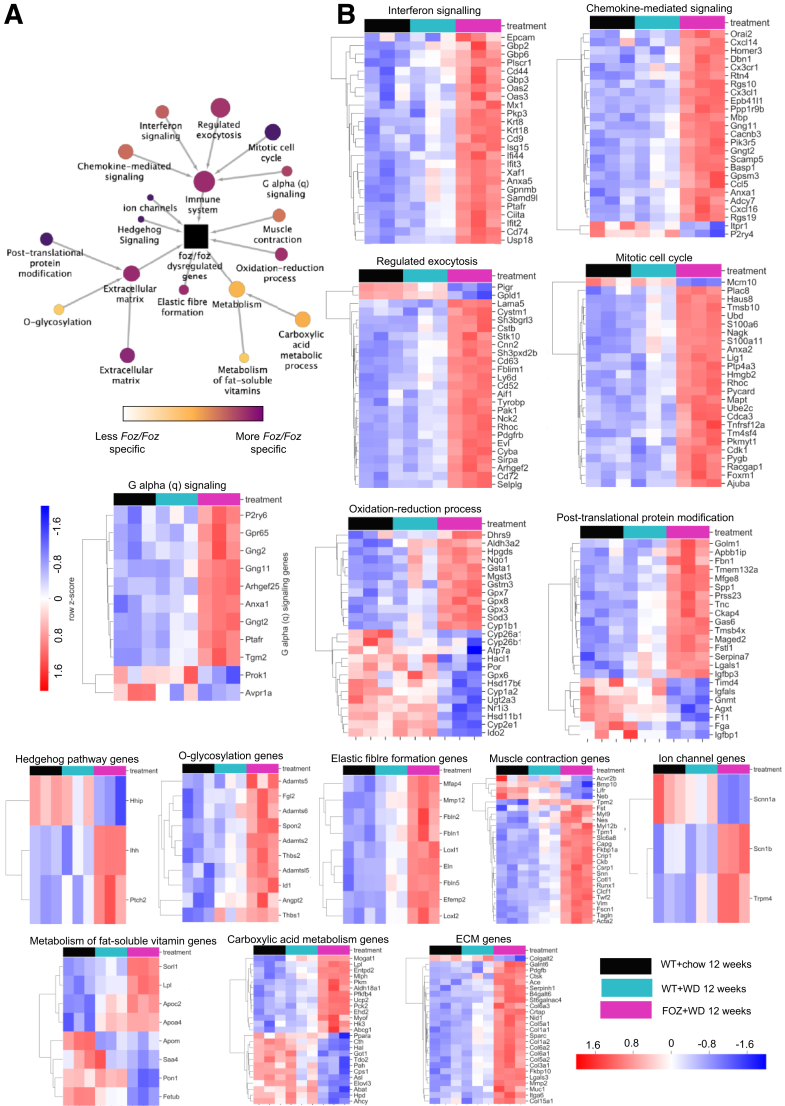
To investigate biological pathways disrupted in Foz+WD NASH livers in greater detail, we performed a 2-step STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, ELIXIR, Wellcome Genome Campus, Cambridgeshire, UK) based hierarchical clustering analyses. First, we integrated genes most significantly dysregulated in Foz+WD at 12 weeks compared with WT+chow at 12 weeks and identified a number of biological pathways underlying the Foz+WD NASH condition (Figure 6). This comparison encompasses all changes based on both diet and disease pathology. In the next step, we investigated these pathways by their specificity to the Foz+WD (NASH) phenotype vs WT+WD (NAFL). Pathways that were highly changed in Foz+WD vs WT+WD comparison (although relatively few genes changed between WT+WD vs WT+chow), include extracellular and elastic fiber formation pathways, chemokine and interferon-mediated signaling, as well as regulated exocytosis, mitotic cell cycle, and G-protein–coupled receptor signaling (Figure 6). On the other hand, pathways such as metabolism of fat-soluble vitamins, the carboxylic acid pathway, and O-glycosylation were notably changed owing to diet, with many genes altered in WT+WD compared with WT+chow, and further dysregulated in Foz+WD compared with WT+WD (Figure 6).
Figure 6.
NASH in WD-fed Foz/Foz mice have a transcriptomic profile similar to human NASH. Results of STRING-based clustering analysis to identify key dysregulated pathways in normal vs NAFL vs NASH liver. (A) The hierarchical clustering analysis of genes (interconnected circles) dysregulated in Foz+WD 12-week livers. Each circle is a system of highly interconnected genes, with the circle size mapped to the number of genes in the system (minimum, 3; maximum, 820). The center system (black square) is the root node, containing all 820 genes significantly differentially expressed in Foz+WD compared with WT+chow. The clusters are ordered hierarchically, with arrows pointing from children to parents. Genes in the child nodes are subsets of the genes in the parent nodes. The pathways are color-coded by Foz+WD specificity, with lighter colors indicating pathways that are dysregulated in Foz+WD (NASH), and to some extent in WT+WD (steatosis), and purple indicating the pathway is more changed in Foz+WD than in WT+WD. (B) The heatmaps show the top 25 genes in the selected clusters, ranked by P value in Foz+WD compared with WT+WD. Row order is determined by hierarchical clustering, and the heatmap shows the relative expression (row-normalized z-score). ECM, extracellular matrix.
NASH in Foz/Foz Mice Is Associated With Distinct Early and Late Inflammatory Signals
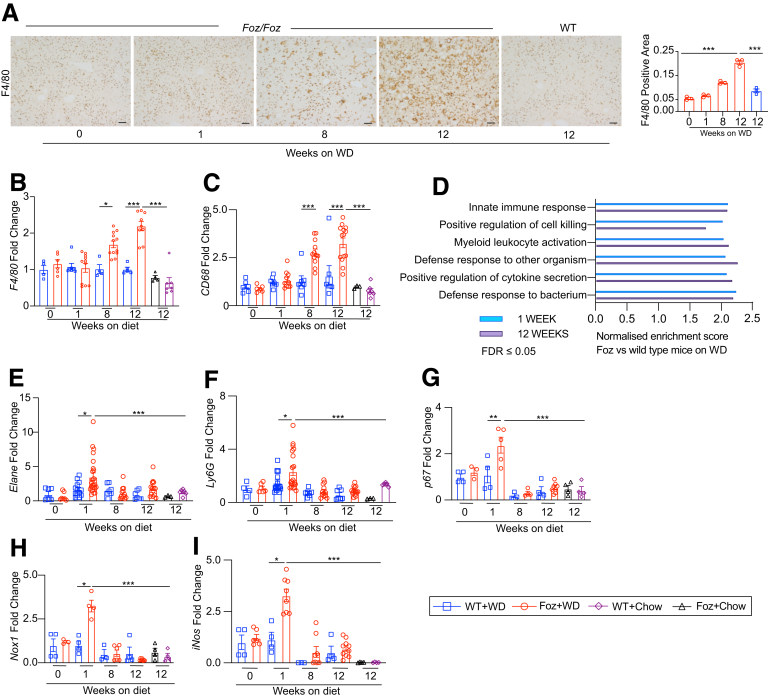
IHC of the NASH-bound Foz+WD livers indicate a progressive increase of F4/80+ macrophages while WT+WD NAFL livers remain unaffected (Figure 7A). This was supported by the F4/80 gene expression analysis of whole-liver RNA (Figure 7B). Circulating monocyte-derived macrophage marker Cd68 also was up-regulated progressively in 8- to 12-wk Foz+WD livers (Figure 7C), which most likely is facilitated by chemokines such as Cxcl14 and Cxcl16 that were up-regulated during this phase in Foz+WD livers (Figure 6, chemokine-mediated signaling). Despite minimal change in the macrophage or monocyte population at acute phases, the liver transcriptome analysis shows innate-immune activation and inflammation at as early as 1 to 2 weeks of WD in Foz/Foz livers (Figure 7D). Gene expression analysis indicated domination of early inflammation in the Foz+WD liver at 1 to 2 weeks by neutrophil infiltration (Figure 7E and F). This early phase response is accompanied by acute oxidonitrosative stress in the liver that subsides over time, as indicated by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analyses of p67, Nox1, and iNos (Figure 7G–I).
Figure 7.
Foz+WD NASH is associated with distinct early and late inflammatory signals. (A) Representative liver sections were stained with anti-F4/80 antibody and the positive area was quantified. (B and C) qRT-PCR of indicated genes (fold change with respect to WT+WD 0 wk). (D) Functional enrichment analysis (WebGestalt 2019) showing the normalized enrichment score of the immune-activation–related pathways that are enriched in Foz+WD compared with WT+WD at 1 week (blue) and at 12 weeks (purple) after diet. (E–I) Total RNA was extracted and qRT-PCR was performed for the indicated genes (fold change with respect to WT+WD 0 wk). Data are expressed as the means ± SEM; 2-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. Scale bars: 200 μm. FDR, False discovery rate.
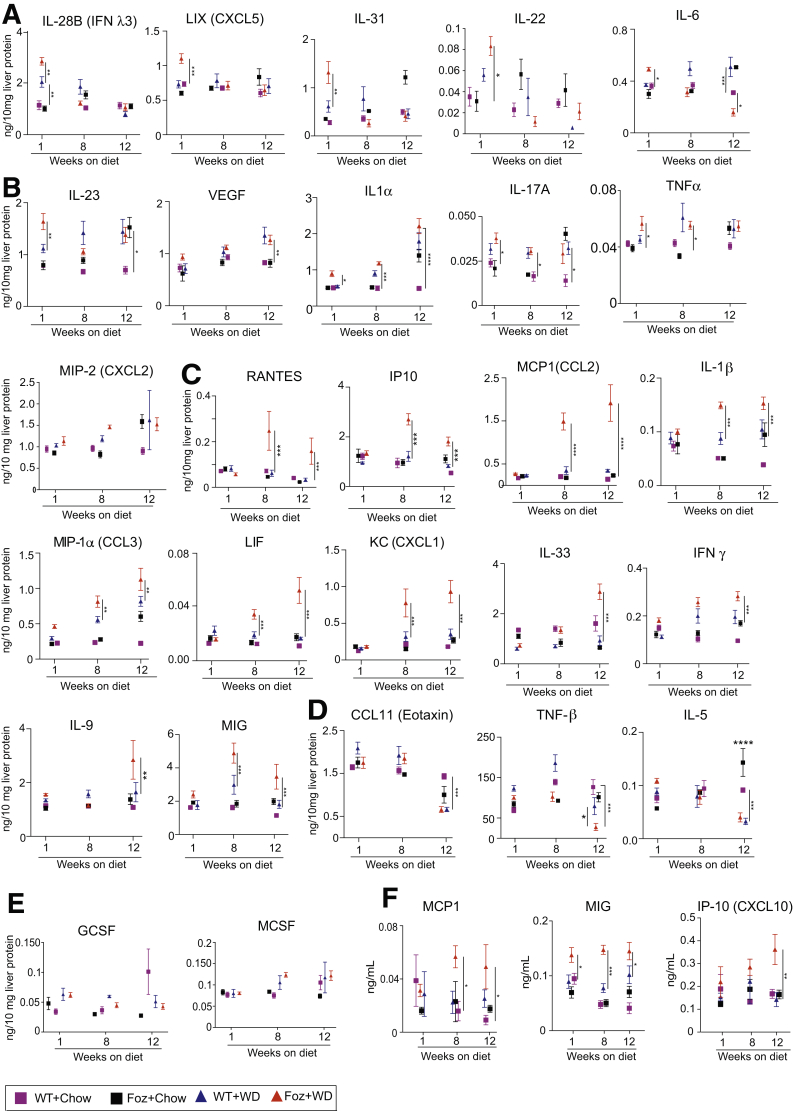
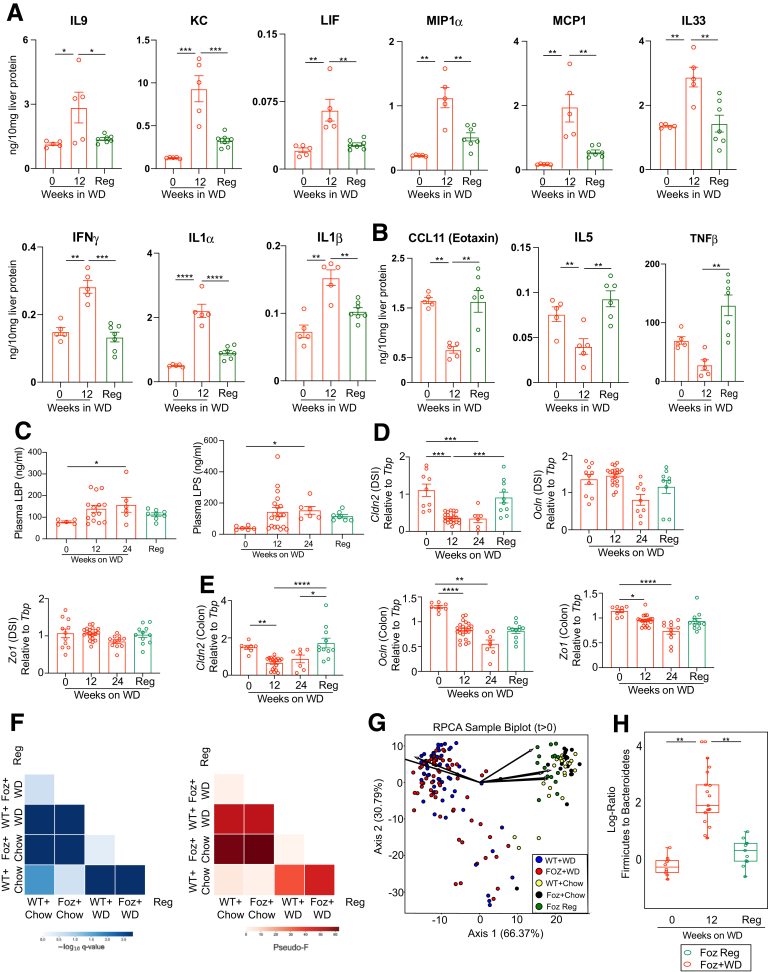
To better understand the kinetics of cytokine and chemokine secretion with respect to NASH progression, we performed Luminex (Luminex Co, Austin, TX) with liver tissue lysates of the Foz+WD and all the controls to account for age, genotype, and diet. Results from the Luminex analysis can be broadly categorized into 3 groups (Table 1). First, cytokines and chemokines that were acutely up-regulated only at early stages in Foz+WD mice and then decreased as NASH progressed, such as interleukin (IL)28B, lipopolysaccharide-induced CXC chemokine (LIX)/CXC Motif Chemokine Ligand 5 (CXCL5), IL31, IL22, and IL6 (Figure 8A). Second, cytokines and chemokines that were up-regulated in both NAFL and NASH, such as IL17A, IL1α, IL23, vascular endothelial growth factor, TNFα, and Macrophage inflammatory protein (MIP2)/CXC Motif Chemokine Ligand 2 (CXCL2) (Figure 8B). Third, cytokines and chemokines that progressively were up-regulated specifically during NASH progression (Figure 8C), including Rantes, Interferon-inducible protein 10 (IP-10)/CXCL10, Monocyte chemoattractant protein-1 (MCP1)/C-C Motif chemokine ligand 2 (CCL2), IL1β, MIP1α, Leukemia inhibitory factor (LIF), keratinocyte chemoattractant (KC), IL33, IFNγ, IL9, and MIG (Monokine induced by IFN-γ/CXCL9). Fourth, in contrast, some cytokines and chemokines C-C Motif chemokine ligand 11 (CCL11), TNF-β, and IL5) were suppressed significantly in NAFL and NASH (Figure 8D). Some cytokines and factors, such as Granulocyte colony-stimulating factor (GCSF) and Macrophage colony-stimulating factor (MCSF), did not show any significant difference among the experimental groups (Figure 8E). Matched serum sample analysis showed that only MCP1 (CCL2), MIG (CXCL9), and IP-10 (CXCL10) increased specifically in NASH (Figure 8F).
Table 1.
Summary of Multiplex Analysis of Hepatic Cytokine Expression (Liver Lysate)
| Up-regulated only at early stages of NASH-bound mice | Expressed in both NAFLD and NASH | Up-regulated only in NASH mice | Moderately reduced during reversal | Reduced to baseline during reversal | Suppressed in NASH and recovered during regression |
|---|---|---|---|---|---|
| IL28b (IFNλ3) | IL17A | KC (CXCL1) | MIG (Cxcl9) | KC (Cxcl1) | CCL11 (eotaxin) |
| LIX (CXCL5) | IL23 | MIG (CXCL9) | MIP-1α (CCL3) | MCP1 (CCL2) | TNF-β |
| IL31 | TNFα | IP-10 (CXCL10) | IL1β | IP-10 (CXCL10) | IL12p40 |
| IL22 | VEGF | MCP1 (CCL2) | IL17A | IL9 | IL5 |
| IL6 | MIP2 (CXCL2) | MIP-1α (CCL3) | IL1α | IL33 | |
| IL1α | IL9 | IFNγ | LIF | ||
| IL33 | IL21 | ||||
| LIF | |||||
| IL1β | |||||
| IFNγ | |||||
| Rantes |
IP-10, Interferon inducible protein 10; LIF, Leukemia inhibitory factor; MIP, Macrophage inflammatory protein; Rantes, Regulated on activation, normal T cell expressed and secreted; VEGF, vascular endothelial growth factor.
Figure 8.
NASH in Foz/Foz mice is associated with distinct early and late inflammatory signals. (A–E) Luminex cytokine analyses of indicated cytokines detected in the liver lysates. Cytokine/chemokines are grouped by the following: (A) early phase cytokines, (B) cytokines up-regulated in both NASH and NAFLD, (C) NASH-specific cytokines, (D) cytokines down-regulated during NAFL and NASH, and (E) cytokines unaltered during NASH progression. (F) Luminex was performed with plasma samples to identify systemic cytokine/chemokine alterations. Only MCP1, MIG, and IP-10 are shown that were found significantly altered in NASH. Data are expressed as the means ± SEM; 1-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. GCSF, Granulocyte colony-stimulating factor; KC, Keratinocyte chemoattractant; LIF, Leukemia inhibitory factor; MCSF, Macrophage Colony-Stimulating Factor; RANTES, Regulated on activation, normal T cell expressed and secreted; VEGF, vascular endothelial growth factor.
Microbial Dysbiosis and Gut Inflammation Precedes NASH Fibrosis in WD-Fed Foz/Foz Mice
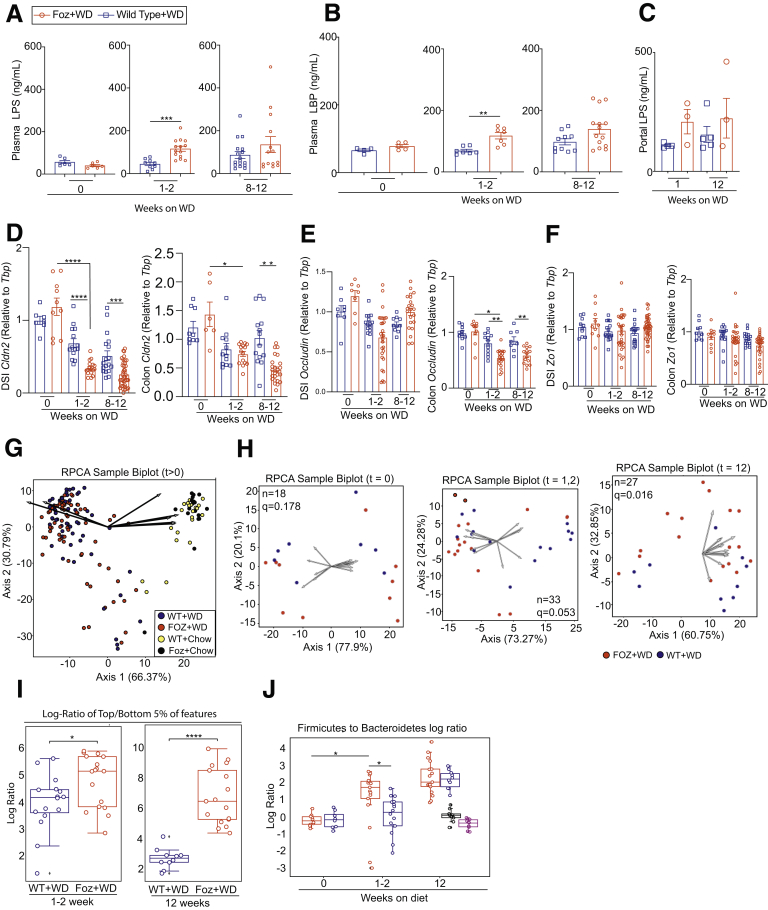
Because we observed an early inflammatory response with increased cytokine expression in the liver preceding NASH development (within 1-2 weeks of WD feeding) (Figures 7 and 8), we hypothesized that inflammation potentially is triggered by translocation of microbial products facilitated by increased intestinal permeability. Plasma lipopolysaccharide (LPS) (Figure 9A) and LPS binding protein (LBP) (Figure 9B) were increased in Foz+WD mice within 1 to 2 weeks and remained high at 12 weeks. LPS levels in the portal blood also showed a trend of increase in Foz+WD mice compared with WT+WD mice (Figure 9C).
Figure 9.
Gut inflammation and microbial dysbiosis precedes NASH-fibrosis in Foz/Foz mice. (A–C) Plasma from systemic blood as well as portal vein blood was isolated and subjected to enzyme-linked immunosorbent assay. Levels of (A) LPS and (B) LBP in systemic circulation and (C) levels of LPS in portal circulation. Total RNA was isolated from DSI and colon and was subjected to qRT-PCR for (D) Cldn2, (E) Occludin, and (F) Zo1 (relative to Tbp). (G–J) DNA was extracted from cecum contents and the microbiome diversity was analyzed by 16s rRNA sequencing. (G) RPCA β-diversity ordination of the indicated samples after WD feeding, showing distinct separation of the WD and NC-fed group. Arrows represent microbes of the highest magnitude contributing to sample differences. (H) RPCA β-diversity ordination of Foz+WD and WT+WD samples at week 0, weeks 1–2, and week 12; n = sample number in each group; q= PERMANOVA (Permuted Analysis Of Variance) statistics. (I) Log ratios of the top and bottom 5% of microbes by Foz+WD differential rank. (J) Log ratios of Firmicutes to Bacteroidetes annotated microbes. Bar graph data are plotted as means ± SEM while (I and J) box-and-whisker data are plotted with median (middle line), 25th–75th percentiles (box), and minimum–maximum percentiles (whiskers); (A and B) unpaired t test, (C, D–F) ordinary 1-way analysis of variance, (I and J) Mann–Whitney test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
In parallel, the tight junction protein Cldn-2 expression was suppressed in the distal small intestine (DSI) of Foz+WD at 1 to 2 weeks and 8 to 12 weeks (Figure 9D) compared with age-matched WT+WD. In colon, Cldn-2 expression decreased within 1 to 2 weeks in both Foz+WD and WT+WD mice, and by 12 weeks it was down-regulated further only in Foz+WD mice (Figure 9D). Occludin, on the other hand, largely was unaffected in DSI, while it was down-regulated significantly in the colon of Foz+WD mice at both 1 to 2 weeks and 12 weeks (Figure 9E). Zo1 remained unaltered in both DSI and colon (Figure 9F).
Intestinal permeability often is associated with microbial dysbiosis.16 Robust Aitchison principal component analysis (RPCA) biplot showed clear separation between the chow and WD-fed groups, indicating that WD induced profound changes in the β-diversity of the intestinal microbes in both WT and Foz/Foz mice (Figure 9G). Although WT+WD and Foz+WD groups clustered together in the RPCA biplot (Figure 9G), a β-diversity ordination between the WD-fed groups only (WT+WD vs Foz+WD) showed significant difference (q = 0.016) between WT and Foz/Foz mice at 12 weeks of WD feeding, but not at the baseline of 0 weeks (q = 0.178) (Figure 9H). At the 1- to 2-week time point this RPCA biplot shows borderline significance (q = 0.053) between WT+WD and Foz+WD (Figure 9H). However, there was a significant difference in the log-ratios of the top and bottom 5% of the microbial features among these samples, even at 1 to 2 weeks and at 12 weeks after WD (Figure 9I, Supplementary Tables 2 and 3). Furthermore, the log ratios of Firmicutes to Bacteroidetes increased in the 1- to 2-week Foz+WD mice, and continued to increase over time (Figure 9J).
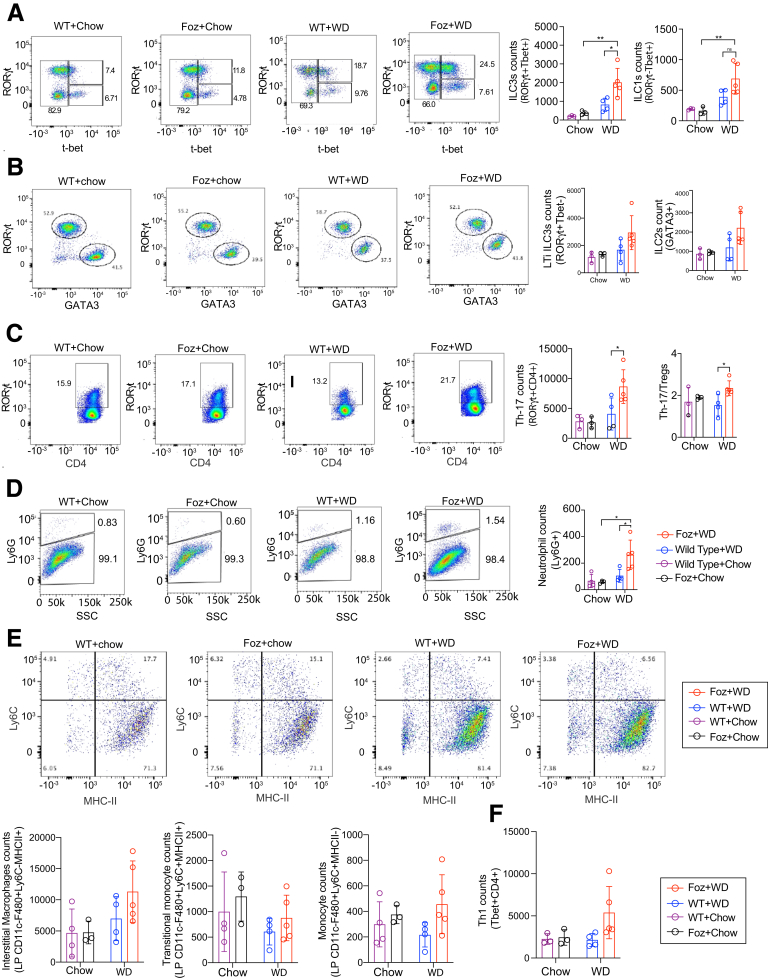
Intestinal homeostasis and defense against pathogens largely are maintained by lymphoid cells expressing the nuclear hormone receptor RORγt (Retineic acid receptor-related orphan nuclear receptor gamma), which includes IL17A-producing T-helper cells and innate lymphoid cells (ILCs) such as lymphoid tissue inducer cells and IL22-producing ILC3s.17,18 To analyze the outcome of gut barrier dysfunction and dysbiosis at early stages of NASH-bound mice, we investigated the frequency of ILCs, myeloid cells, and Th17 cells in the lamina propria of DSI tissues (Figure 10). Flow cytometric analysis by transcription factor labeling of CD45+Lin-CD90+T Cell Receptor (TCR) β-CD127+showed the expansion of ILCs RORγt+Tbet+ in the lamina propria of Foz+WD mice when compared with both WT+WD and chow-fed controls (Figure 10A). No differences were observed in the number of other ILC subsets when Foz+WD were compared with WT+WD mice (Figure 10B). Th17 cells distinguished as RORγt+CD4+ T-cell counts were increased in Foz+WD mice (Figure 10C), and their levels correlated with a concomitant increase of infiltrating neutrophils (Ly6G+) in the DSI of Foz+WD mice (Figure 10D). No differences in the numbers of resident macrophages, inflammatory monocytes, and Th1 cells were found in Foz+WD mice (Figure 10E and F). Altogether, the data suggest that RORγt+Tbet+ ILC, Th17 cells, and neutrophils are recruited with the disruption of the epithelial barrier and dysbiosis.
Figure 10.
Gut inflammation and microbial dysbiosis precedes NASH fibrosis in Foz+WD mice. Flowcytometric analysis of intestinal lamina propria for (A) ILC3s, ILC1s; (B) lymphoid tissue inducer (LTi) ILC3s, ILC2s; (C) Th-17, regulatory T cells (Tregs); (D) neutrophils, (E) indicated monocyte/macrophages, and (F) Th1 cells. Representative pseudocolor plots illustrate the gating strategy used to detect the indicated cells in the lamina propria of WD and chow-fed Foz/Foz and WT mice, bar diagrams show the changes in the total count of the indicated cells under different dietary conditions. Bar graph data are plotted as means ± SD. Ordinary 1-way analysis of variance. ∗P < .05, ∗∗P < .01. MHC-II, major histocompatibility complex class II.
Dietary Switch to Normal Chow Leads to NASH Regression
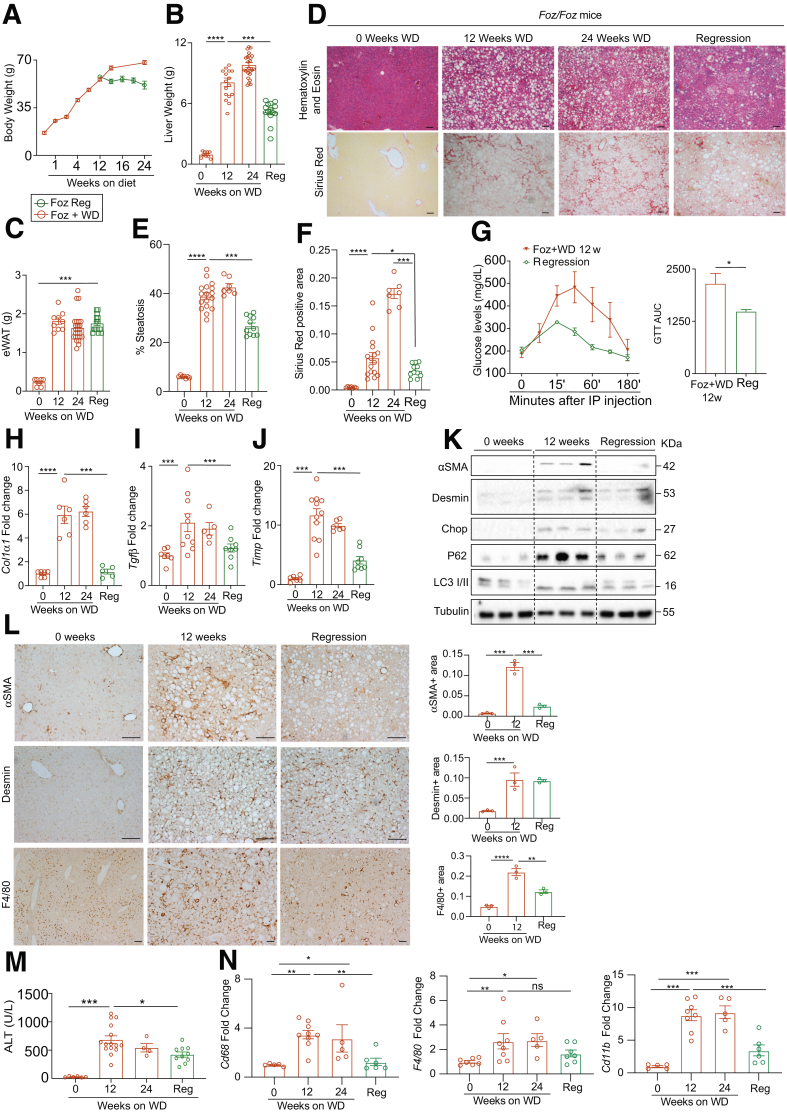
To examine regression of NASH, a cohort of 12-week Foz+WD mice with NASH and fibrosis was switched to chow for an additional 8 to 12 weeks (Foz regression). Another cohort of Foz+WD mice continued on WD for an additional 12 weeks (Foz+WD 24 wk). Although the mice that continued on WD (Foz+WD 24 wk) continued to gain body weight (Figure 11A), after switching to normal chow, the mice maintained a steady weight (Figure 11A). The Foz regression mice had a significant reduction in total liver weight when compared with both 12-week and 24-week Foz+WD groups (Figure 11B). The eWAT weight remained unchanged in the Foz regression mice (Figure 11C).
Figure 11.
Dietary switch to normal chow attenuates fibrosis progression.Foz/Foz mice were fed WD for 12 weeks. One group continued on WD and another group was switched to regular chow for an additional 8–12 weeks. (A) Body weight, (B) liver weight, and (C) eWAT weight were plotted. Representative liver sections (D) stained with H&E and Sirius red and quantified for (E) steatosis and (F) fibrosis using ImageJ software. (G) A glucose tolerance test (GTT) was performed. The data also are represented as the area under the curve (AUC) for the indicated groups. (H–J) qRT-PCR of key fibrogenic genes (fold change with respect to Foz+WD 0 wk). (K) Liver tissues were subjected to immunoblot analyses with the indicated antibodies with tubulin as loading control. (L) FFPE (Formalin-Fixed Paraffin-Embedded) liver sections were stained with α-smooth muscle actin (αSMA), desmin, and F4/80, and quantified. (M) Plasma ALT levels. (N) Total RNA isolated from liver was subjected to qRT-PCR for CD68, F4/80, and CD11b (represented as fold change relative Foz+WD 0 wk). Data are expressed as the means ± SEM; ordinary 1-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. Scale bars: 200 μm. αSMA, α-smooth muscle actin.
Foz regression mice had resolution of hepatic steatosis (Figure 11D and E), consistent with decreased liver weight and improved glucose tolerance (Figure 11G). Moreover, there was a reduction in liver fibrosis (Figure 11D and F), with a reduction in fibrogenic genes (Figure 11H–J), and HSC activation markers (Figure 11K and L). The hepatic levels of ER stress marker proteins, Chop and ATF6, and autophagy markers, P62 and LC3II, also were reduced to normal levels during regression (Figure 11K). Regression was accompanied by a decrease in liver injury (Figure 11M). The hepatic expression of monocyte/macrophage markers Cd68 and Cd11b (Figure 11N) were reduced significantly during regression. The number of F4/80+ macrophages was reduced in the regressed livers (Figure 11L).
Luminex analysis showed that several cytokines and chemokines that were up-regulated in the Foz+WD 12-week NASH group returned toward baseline during regression (Figure 12A) (Table 1). Cytokines/chemokines that were suppressed during NASH, such as Ccl11 (eotaxin), IL5, and TNFβ, recovered during NASH regression (Figure 12B) (Table 1). Although the circulating LBP and LPS levels did not change during regression (Figure 12C), the expression of tight junction protein Cldn-2 improved significantly, with regression in both DSI and colon (Figure 12D and E). Ocln expression also showed a trend of recovery in the regressed mice (Figure 12D and E).
Figure 12.
Dietary switch to normal chow attenuates fibrosis progression.Foz/Foz mice that continued on WD or switched to normal chow after 12 weeks of WD (regression) were analyzed. (A and B) Luminex analyses of liver lysates for the indicated cytokines and chemokines. (C) Plasma LBP and LPS levels in the indicated groups. Total RNA isolated from (D) DSI and (E) colon region of the intestine was subjected to qRT-PCR for Cldn2, Ocln, and Zo1. Data are expressed as the means ± SEM; 1-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. (F) Pairwise PERMANOVA (Permuted Analysis of Variance) of RPCA β-diversity plot by diet-genotype combination. Left: -log10 q-value; right: pseudo–F-statistic. (G) RPCA β-diversity ordination of all samples after 0 weeks. Samples are colored by diet–genotype combination. Arrows represent microbes of the highest magnitude contributing to sample differences. (H) 16s rRNA sequencing of cecal contents was analyzed for the log ratios of Firmicutes to Bacteroidetes annotated microbes in different groups as indicated. Data are shown as box-and-whisker with median (middle line), 25th–75th percentiles (box), and minimum–maximum percentiles (whiskers). Mann–Whitney test. KC, Keratinocyte chemoattractant; Reg, Regression.
Pairwise Permuted analysis of varianc (PERMANOVA) analysis indicated that the gut microbiomes in the regression samples and in the standard chow samples were very similar (Figure 12F). This was confirmed by RPCA biplot showing that the regression samples clustered with the chow-fed samples (Figure 12G). Furthermore, the log-ratio of Firmicutes to Bacteroidetes, which increased during NASH, returned to baseline in the Foz regression group (Figure 12H).
NASH to HCC Progression in Foz+WD Mice
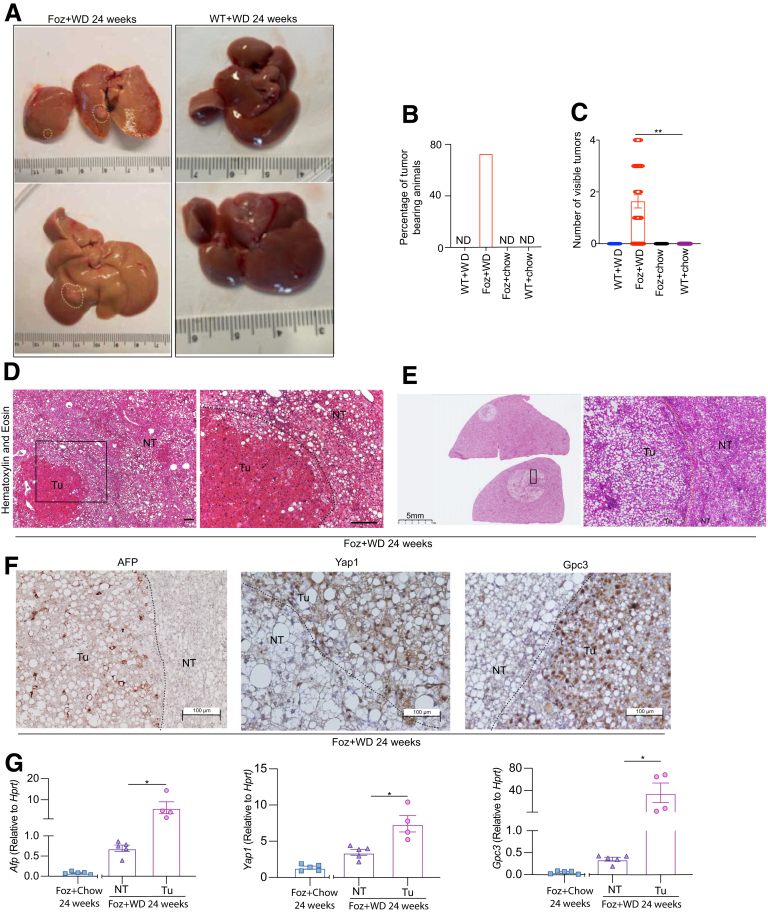
Foz/Foz mice that continued on a WD for 24 weeks developed macroscopically visible tumors (Figure 13A). Approximately 75% of the mice had at least 1 visible tumor (Figure 13B), with an average of 2–3 tumors/liver (Figure 13C). Age-matched WT+WD mice and chow-fed mice did not develop any tumors (Figure 13A–C). The Foz+WD tumors are a mix of conventional (trabecular) HCC (Figure 13D), as well as steatohepatitic HCC (Figure 13E), a HCC variant found in patients with NASH and alcoholic liver cirrhosis.19 IHC evaluation of classic HCC markers such as α-fetoprotein, Yap, and Gpc3 (Figure 13F) and qRT-PCR (Figure 13G) indicated that the tumors are HCC. Although these genes and proteins were most highly increased in the tumors, the nontumor liver in the Foz+WD also had slightly increased levels (Figure 13G), which might reflect microscopic lesions in the nontumor areas of the tumor-bearing mice.
Figure 13.
Foz+WD mice progress from NASH to cirrhosis and HCC by 24 weeks. (A) Representative gross liver images from 24-week WD-fed mice showing macroscopic tumor nodules in Foz+WD livers (white circles). (B) Tumor incidence and (C) visible tumor numbers (>1 mm) were plotted. Representative H&E-stained tissue sections showing (D) trabecular and (E) steatohepatitic HCC in Foz+WD 24-week livers. Classic HCC markers AFP, YAP1, and GPC3 were detected either by (F) IHC or (G) qRT-PCR. RNA from Foz+chow at 24 weeks was used as an age matched control for the gene expression. Data are expressed as the means ± SEM. (C) One-way analysis of variance and (G) unpaired t test. ∗P < .05, ∗∗P < .01. Scale bar: 200 μm, or as indicated. NT, nontumor region from tumor-bearing livers; Tu, tumor tissue.
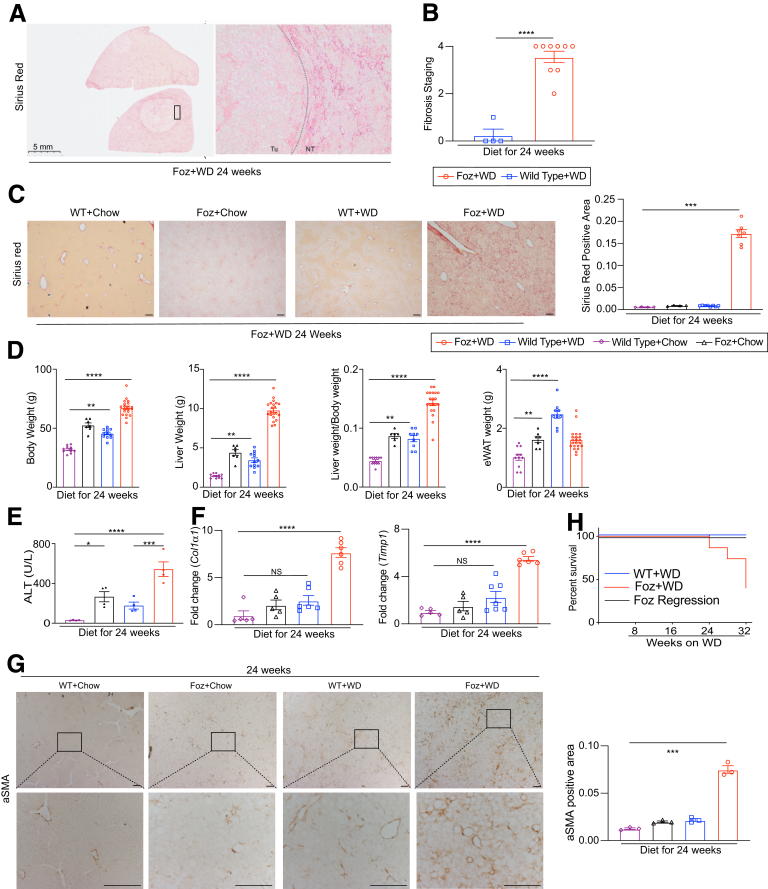
The 24-week Foz+WD livers showed cirrhosis (grade 4 fibrosis) in the nontumor region (Figure 14A–C). Both Foz+chow and WT+WD 24-week mice, despite being obese (Figure 14D), and with increased ALT levels (Figure 14E), did not develop fibrosis (Figure 14C, F, and G) or HCC (Figure 13B and C). Beyond 24 weeks, Foz+WD had reduced survival, with less than 50% of mice surviving by 32 weeks (Figure 14H). WT+WD littermates had no mortality during this study period (Figure 14H). Importantly, Foz regression mice had improved survival, with 100% of mice surviving until 32 weeks (Figure 14H), and they did not develop visible HCC nodules.
Figure 14.
Foz/Foz mice on WD develop cirrhosis and HCC, with increased mortality. (A) Sirius red–stained sections from Foz+WD 24-week livers. (B) Sirius red–stained slides were scored for fibrosis staging (grades, 0–4) (pathologist scoring). (C) Representative Sirius red–stained liver sections and the quantification of Sirius red–positive area (ImageJ). (D) Changes in body weight, liver weight, liver:body weight ratio, and eWAT weight in the indicated groups. (E) Plasma ALT levels. (F) qRT-PCR of Col1α1 and Timp1 (represented as fold change relative WT+chow 24 wk). (G) Representative α-smooth muscle actin (αSMA)-stained liver tissue sections and the quantification of αSMA-positive area. (H) Survival graph indicating survival percentage of WT and Foz/Foz mice under indicated dietary conditions. Data are expressed as the means ± SEM; 1-way analysis of variance. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. Scale bar: 200 μm, or as indicated.
Discussion
A Model of Accelerated NAFL–NASH–HCC Progression
The current rodent models of NASH do not reproduce the complete spectrum of metabolic and histologic NAFLD phenotype.5 Moreover, there is a lack of systemic studies in a single rodent model of NASH that closely recapitulates the human pathology: from insulin resistance–metabolic syndrome, gut barrier disruption–dysbiosis–intestinal inflammation, liver fibrosis, HCC, CKD, and CVD.20 Our systematic analyses of the Foz/Foz model showed that, first, WD feeding in Foz/Foz mice on a C57BL6 background optimally develop NASH with fibrosis. Second, Foz+WD female mice develop NASH but not fibrosis. Third, these mice develop cardiovascular (unpublished data) and kidney dysfunction,10 consistent with the metabolic syndrome. Fourth, a multi-omic analysis of WT+WD with Foz+WD showed factors that differentiate NAFL from NASH. Fifth, switching Foz/Foz mice from WD back to chow results in regression of NASH and fibrosis and improves survival. Sixth, WD increases intestinal permeability and intestinal inflammation. Seventh, NASH progresses in a biphasic wave of cytokines/chemokines that modulate infiltration of neutrophils and monocytes at early and late stages, respectively. Eighth, Foz+WD causes gut microbial dysbiosis, which reverts to normal with a switch back to chow diet. Ninth, prolonged WD results in cirrhosis and HCC, which are prevented by switching back to normal chow.
Foz/Foz NASH Transcriptome Is Similar to Human NASH
Using publicly available data sets for human NASH,14 we normalized the differences in human NASH liver transcriptome to that of healthy obese patients and correlated it to the gene signature from Foz+WD data (NASH) normalized to WT+WD (NAFL). This comparison showed a strong correlation between the human and Foz NASH transcriptome (Figure 5A and B), validating our model to study NASH compared with NAFL.
Global gene expression and STRING-based hierarchical clustering analyses showed several key signaling pathways that are deregulated in NASH compared with NAFL livers. In addition to the expected activation of fibrogenic pathways, the dominating inflammatory pathways were represented by IFN-γ and the TNF superfamily. Pathways that regulate the defense response to other organisms were activated in NASH, consistent with increased gut permeability. However, metabolic processes, including glucose and fatty acids, were suppressed in the NASH liver (Figure 5C). G-protein–coupled receptor signaling, particularly Gαq, is distinctly up-regulated in NASH vs NAFL (Figure 6). Consistent with the metabolic and energy overload in a NASH liver, mitochondrion morphogenesis was extremely down-regulated in NASH (Foz+WD) compared with NAFL livers (WT+WD),21 along with glucose and fatty acid metabolism (Figure 5C and D). Circadian pathways, proposed to regulate NASH development,22 were suppressed specifically in NASH. Furthermore, 2 hallmark pathways of NASH: ER stress and defective autophagy, were exacerbated in Foz+WD NASH livers (Figure 5E). Although steatosis alone can influence a myriad of cellular pathways, it is important to note that this analysis showed key pathways that were altered significantly in NASH but not in NAFL mice that also are obese, diabetic, and steatotic. Studies that compare WD with chow would generate differences that are primarily owing to the diet and mask the subtle differences that distinguish NASH from NAFL, which represents the key window for preventative or therapeutic intervention.
Kinetics of Cytokines and Chemokines During NASH Progression
A detailed timeline analysis of gene expression and multiplex cytokine levels showed distinct acute- and late-phase changes in cytokines/chemokines. The acute phase (1 to 2 weeks after WD initiation) in a NASH-bound mouse was dominated primarily by neutrophil chemoattractants (eg, IL8 and Cxcl5) and lymphocyte chemoattractant (Ccl20) that can be induced by LPS, most likely as a direct consequence of gut barrier disruption and intestinal inflammation (discussed later). This results in an early neutrophil infiltration in these mice. Interestingly, the hepatoprotective cytokine IL2223 also was increased at this stage, and then decreased as NASH progresses. Treatment with IL22 recently was shown to improve neutrophil-driven NASH.24 A number of fibrogenic and inflammatory cytokines/chemokines including chemoattractants for polymorphonuclear leukocytes (neutrophils, basophils, and eosinophils) (eg, Ccl3/Mip1a, Cxcl2/Mip2) were up-regulated only during the NASH phase. Several of these cytokines/chemokines either decreased or returned to baseline levels during NASH regression (Table 1), accentuating the role of immune dysregulation in NASH. Our analysis also showed cytokines commonly up-regulated both during NASH and NAFL, and hence could identify the NASH-driving cytokines as possible targets for disease intervention.
Gut Barrier Disruption, Microbial Dysbiosis, and Intestinal Inflammation
The gut–liver axis plays a pivotal role in NASH pathogenesis.25 Small changes in the gut barrier exposes the liver to endotoxins, microbial-associated molecular patterns, pathogen-associated molecular patterns, and microbial metabolites that elicit liver inflammation. Measurements of gut–barrier dysfunction is technically challenging in human beings, with approximately 40% of NASH patients reported to have increased intestinal permeability.26,27 Moreover, whether intestinal permeability is a cause or consequence of NASH also is debated.27,28 Therefore, it is important to understand the timeline and kinetics of intestinal permeability in the context of both intestinal and liver inflammation, along with associated dysbiosis. The livers of NASH-bound Foz+WD mice have increased innate immune and bacterial defense responses at very early stages (Figure 7D). This potentially is triggered by an increased intestinal permeability, as evidenced by Cldn-2 down-regulation (Figure 9D), resulting in systemic LPS leakage (Figure 9A and B) and local liver inflammation (Figures 7D–F and 8A and B) early in Foz+WD mice.
Metabolic syndrome is associated with a decline in microbial diversity and an increase in the firmicutes/bacteroidetes ratio.29 In our study, WD induced a profound microbial dysbiosis in both WT and Foz/Foz mice (Figure 9G). However, although WD was the determinant trigger for the increase in the firmicutes/bacteroidetes ratio, Foz+WD mice manifest this alteration even at early stages (1–2 weeks post-WD diet) compared with their WT counterparts (Figure 9J). Indeed, comparison of WT+WD with Foz+WD showed subtle differences in microbiome features within 1 to 2 weeks as well as 12 weeks of WD feeding (Figure 9I; Supplementary Tables 2 and 3). Further studies will elaborate the roles of these NASH-specific microbiomial features in the development and progression of NASH fibrosis.
Multiple strong interactions exist between gut microbiota and the immune system to keep homeostasis in the intestine. Microbiota-derived signals can shape the immune system, and the local immune environment also can remodel the microbiota.30 Flow cytometric analyses of the DSI lamina propria showed the biological effect of increased gut permeability on local inflammation. We found increased neutrophil infiltration and expansion of RORyt+T-bet+ ILC3s and Th17 cells in the lamina propria of Foz+WD mice at very early stages (Figure 10). Coordinated activation of these cells is needed to clear invading bacteria and restore gut epithelial integrity. However, under chronic conditions, a constant low-grade inflammation could provoke a systemic response affecting other organs such as the liver.18 Activation of RORγt+Tbet+ ILC3s, in particular, has been shown to occur in response to changes on the microbiota producing type 1 inflammatory immune responses (eg, IFNγ), and also may have pathogenic functions during intestinal inflammation such as inflammatory bowel disease.31 Alterations of the Th17/regulatory T-cell ratio have been reported in chronic inflammatory diseases, associated with dysbiosis and increased permeability.32 In fact, selective recruitment of neutrophils in the intestinal lumen of Foz+WD mice may be triggered by Th17 cell activation in response to acute colonization of segmented filamentous bacteria.33 Taken together, our data suggest that gut leakiness, microbial dysbiosis, and intestinal inflammation occur early in Foz+WD mice, leading to liver inflammation, NASH, and fibrosis.
Modeling NASH Regression
Because human NASH with fibrosis regresses with diet-induced weight loss34 or bariatric surgery,35 we modeled NASH regression in Foz/Foz mice by switching from a WD back to a chow diet. The regressed mice stopped gaining body weight, had improved glucose tolerance, and decreased liver weight with reduced steatosis, fibrosis, and inflammation (Figure 11A–J). The autophagy defect and ER stress improved in the regressed mice (Figure 11K), along with microbial dysbiosis and cytokine dysregulation (Figure 12). Foz regression livers also were devoid of any visible tumor, while the mice that continued on a WD developed HCC with high penetrance (Figure 13A–C). Using the Foz/Foz model of NASH and regression we recently described the heterogeneity of HSCs within NASH.36 We also identified the unique gene signature of inactivated HSCs, which was similar to, but distinct from, the quiescent HSCs. Successful modeling of NASH regression allows for unique opportunities to explore and identify causal relationship among various pathways.
In our experiments, WT+WD mice had mild liver pathology, primarily steatosis with very little NASH and fibrosis, even at 24 weeks. A reason for this outcome might reflect the short duration on the WD. In addition, feeding WT mice with a higher cholesterol diet (2% instead of 0.2%)37 or supplementation of fructose in the drinking water (along with CCl4) for more than 6 months38 increases NASH with fibrosis. The development of CKD and CVD associated with NASH is unknown in these other models.
Taken together, the Foz/Foz model of NASH has a rapid and complete disease progression while mimicking the key transcriptomic and histologic features of human NASH. Using this model, we elucidate important disease modulators that likely are responsible for NAFL to NASH to fibrosis and HCC progression/regression and integrates multi-organ cross-talk for disease manifestation.
Materials and Methods
Animals
Mice with the Alms1 mutation (Foz/Foz mice) on C57BL6/J and a Non-obese Diabetic (NOD) genetic background were kindly provided by Dr Geoffrey C. Farrell (Australian National University Medical School).8 Six- to 7-week-old (time point = 0 w) male and female Foz/Foz along with WT littermates were placed on a WD (AIN-76A; Test Diet, St. Louis, MO, containing 40% fat, 15% protein, 44% carbohydrates based on caloric content, 0.2% cholesterol) or a standard chow diet (12% fat, 23% protein, 65% carbohydrates) for up to 32 weeks. For the regression study, Foz+WD 12-week mice were switched to the standard chow diet for an additional 8–12 weeks. A separate cohort of 6-week-old C57BL6/J Foz/Foz mice was placed on a CD-HFD (D-05010402; Research Diet, New Brunswick, NJ, containing 45% fat, 20% protein, 35% carbohydrates) for 12 weeks. Because Foz/Foz mice are hyperphagic, all mice were fasted for 4 hours before death and tissue collection. All animals were maintained under pathogen-free conditions in filter-topped cages on autoclaved food and water and under a 12/12-hour light/dark cycle, in accordance with National Institutes of Health guidelines for the use and care of live animals and approved by the University of California San Diego Institutional Animal Care and Use Committee (IACUC, Protocol #S07022).
qRT-PCR
Total RNA was isolated using RNeasy columns (Qiagen, Valencia, CA). qRT-PCR was performed using a QuantStudio 5 Real-Time PCR system (Applied Biosystems, Carlsbad, CA ). The expression levels of selected genes were calculated and normalized to housekeeping genes Hprt or Tbp using the ΔΔCT method (Invitrogen). PCR primers were designed either using primerBank (https://pga.mgh.harvard.edu/primerbank/). Primer sequences are shown in Table 2. The data are represented as either fold change with respect to the indicated group, or as relative expression to a housekeeping gene.
Table 2.
List of Primers for qRT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Col1α1 | TAGGCCATTGTGTATGCAGC | ACATGTTCAGCTTTGTGGACC |
| Timp1 | AGGTGGTCTCGTTGATTTCT | GTAAGGCCTGTAGCTGTGCC |
| Tgfβ1 | GTGGAAATCAACGGGATCAG | ACTTCCAACCCAGGTCCTTC |
| sma | GTTCAGTGGTGCCTCTGTCA | ACTGGGACGACATGGAAAAG |
| Gabra3 | ATGTGGCACTTTTATGTGACCA | TCTTGATTCCCCTTGGCTAGT |
| F4/80 | ACCAGAGGAAATTTTCAATAGGC | TGATGCACTTGCAGAAAACA |
| Nox1 | TCGACACACAGGAATCAGGA | TTACACGAGAGAAATTCTTGGG |
| Inos | TTCTGTGCTGTCCCAGTGAG | TGAAGAAAACCCCTTGTGCT |
| P67phox | CCACTCGAGGATTTGCTTCA | ATCTTGGAATGCCTGGGCTC |
| Afp | ACAGGAGGCTATGCATCACC | TGGACATCTTCACCATGTGG |
| Yap1 | TACTGATGCAGGTACTGCGG | TCAGGGATCTCAAAGGAGGAC |
| Gpc3 | CCCTGAATCTCGGAATTGAA | AGTCCCTGGCAGTAAGAGCA |
| Cd68 | CTTCCCACAGGCAGCACAG | AATGATGAGAGGCAGCAAGAG |
| D-loop | AATCTACCATCCTCCGTGAAACC | TCAGTTTAGCTACCCCCAAGTTTAA |
| Tert | CTAGCTCATGTGTCAAGACCCTCTT | GCCAGCACGTTTCTCTCGTT |
| Cytox1 | GCCCCAGATATAGCATTCCC | GTTCATCCTGTTCCTGCTCC |
| 18s | TAGAGGGACAAGTGGCGTTC | CGCTGAGCCAGTCAGTGT |
| Occludin | ACATCTCAGCAACCCACACA | ATTTATGATGAACAGCCCCC |
| Zo-1 | AGCTGTTTCCTCCATTGCTG | GAGATGTTTATGCGGACGG |
| Claudin2 | TTCTTGGATCCGAGCCTCTA | TCCAGAGCTCTTCGAAAGGA |
| Il1 | GGTCAAAGGTTTGGAAGCAG | TGTGAAATGCCACCTTTTGA |
| Tnf | AGGGTCTGGGCCATAGAACT | CCACCACGCTCTTCTGTCTAC |
| Tbp | ACATCTCAGCAACCCACACA | GTGAAGGGTACAAGGGGGTG |
| Cd11b | CCATGACCTTCCAAGAGAATGC | ACCGGCTTGTGCTGTAGTC |
| Hprt | GTTAAGCAGTACAGCCCCAAA | AGGGCATATCCAACAACAAACTT |
Tissue Histology
The left lateral lobe of mice livers was used for histology. Lobes were fixed in 10% neutral-buffered formalin for 48 hours, embedded in paraffin (Formalin-fixed paraffin-embedded [FFPE]), sectioned, and processed for H&E, Picro Sirius red stain, and IHC. Antibody details are provided in Table 3.
Table 3.
List of Antibodies for Immunohistochemistry, Immunoblot, and FACS
| Antibody | Catalog | Vendor | Fluorochrome |
|---|---|---|---|
| Immunohistochemistry | |||
| Desmin | RB-9014-P0 | Thermo Fisher | NA |
| αSMA | ab5694 | Abcam | NA |
| F4/80 | 14-4801-88 | eBioscience, San Diego, CA | NA |
| GPC3 | ab66596 | Abcam | NA |
| AFP | CP028A | Biocare Medical, Pacheco, CA | NA |
| YAP | CS12640 | Cell Signaling | NA |
| Immunoblot | |||
| αSMA | ab5694 | Abcam | NA |
| Desmin | RB-9014-P0 | Thermo Fisher | NA |
| P62 | CS23214 | Cell Signaling | NA |
| LC3 I/II | L7543 | Millipore-Sigma | NA |
| Chop | CS5554 | Cell Signaling | NA |
| ATF6 | alx804381 | Enzo Life Sciences, Farmingdale, NY | NA |
| Tubulin | T5168 | Millipore-Sigma | NA |
| FACS | |||
| ILC/Treg panel | |||
| Marker antibody | |||
| CD45 | 748370 | BD Biosciences, Franklin Lakes, NJ | BUV805 |
| Lineage plus FCeR1a antibody | 133302 | Biolegend, San Diego, CA | FITC |
| CD90 | 564365 | BD Biosciences | BV 785 |
| ST2 | 746115 | BD Biosciences | BB700 |
| GATA3 | 12-9966-42 | Thermo Fisher | PE |
| RORyt | 17-6988-82 | Thermo Fisher | APC |
| CD127 | 135041 | Biolegend | BV605 |
| Tbet | 644816 | Biolegend | BV421 |
| NK1.1 | 47-5941-82 | Thermo Fisher | APCFluor 780 |
| TCRab | 748406 | BD Biosciences | BUV563 |
| Foxp3 | 25-5773-82 | Thermo Fisher | Pecy7 |
| Live dead | 423108 | Biolegend | BUV496 |
| CD25 | 564022 | BD Biosciences | BUV 395 |
| CD4 | 612843 | BD Biosciences | BUV 737 |
| Myeloid cells panel | |||
| CD11b | 612800 | BD Biosciences | BUV 737 |
| CD3 dump channel | 563565 | BD Biosciences | BUV 395 |
| B220 dump channel | 563793 | BD Biosciences | BUV 395 |
| TER119 dump channel | 563827 | BD Biosciences | BUV 395 |
| CD45 | 748370 | BD Biosciences | BUV 805 |
| F4/80 | 749284 | BD Biosciences | BUV 563 |
| CD11c | 563735 | BD Biosciences | BV786 |
| CD206 | 141721 | Biolegend | BV605 |
| MerTK | 47-5751-82 | Thermo Fisher | APCFluor 780 |
| Trem2 | MA5-28223 | Thermo Fisher | FITC |
| IRF5 | 158603 | Biolegend | PE |
| CD80 | 104726 | Biolegend | 421 |
| MHC-II | 25-5321-82 | Thermo Fisher | APC |
| Ly6C | 45-5932-82 | Thermo Fisher | PeCy7 |
| Ly6G | 25-9668-82 | Thermo Fisher | BV480 |
| Live dead | 423108 | Biolegend | BUV496 |
| Siglec F | 740764 | BD Biosciences | BUV 711 |
αSMA, α-smooth muscle actin; MHC-II, major histocompatibility complex class II; TCRαβ, T-cell receptor α/β; Treg, regulatory T cell.
From the stained tissue sections 7 nonoverlapping fields were imaged randomly using an Olympus microscope (Waltham, MA), and quantified using ImageJ (National Institutes of Health, Bethesda, MD) software following a standard protocol. The proportion of nonlipid area quantified from the H&E-stained sections was used to normalize Sirius red and IHC quantifications to account for the degree of steatosis. Whole-tissue section scanning was performed using the Hamamatsu (San Diego, CA) Nanozoomer digital slide scanner (S360).
Pathology Scoring
The H&E-stained tissue sections were scored for steatosis and inflammation, while the Sirius red–stained sections were staged for fibrosis by a blinded trained pathologist (M.H.).39 Evaluation of the degree of parenchymal steatosis was performed by light microscopy at low to medium power and was graded based on Brunt–Kleiner criteria (scale, 0–3), as follows: 0, less than 5%; 1, 5%–33%; 2, more than 33% to 66%; 3, more than 66%.39 The NASH score was calculated from the steatosis grade, inflammation score, and fibrosis stage, using the NASH Clinical Research Network histologic scoring system.
Luminex
Liver tissues were homogenized in 1 mL of the lysis buffer tissue extraction reagent 1 (FNN0071; Thermo Fisher, Waltham, MA) containing EDTA-free protease inhibitor cocktail (04693132001; Roche, Indianapolis, IN). Liver lysates were analyzed with the Luminex mouse cytokine/chemokine magnetic bead-32 plex (MCYTMAG-70K-PX32; Millipore, Burlington, MA) and mouse Th17 magnetic bead kits (MTH17MAG-47K; Millipore Sigma, Burlington, MA). Samples were read on a Luminex MAGPIX Instrument (Luminex, Austin, TX) and MFIs (Mean Fluorescent Intensity) were normalized to absolute values with standard curves generated using the best-fit feature in the Masterplex software (Hitachi Solutions, Irvine, CA). Data normalization was performed using total liver protein levels and concentrations graphed in GraphPad Prism (San Diego, CA) software.
Biochemical Analyses
Peripheral blood was collected from the inferior vena cava and portal blood was collected directly from the portal vein. Plasma was separated using BD microtainer tubes (Franklin Lakes, NJ, cat. 365985). The plasma ALT level was measured using the InfinityTM kit (cat. TR71121; Thermo Fisher Scientific, Inc). Plasma LPS was measured using the LPS Enzyme-Linked Immunosorbent Assay Kit (cat. E91526Ge-1; Lifeome Biolabs, Inc, Oceanside, CA) and LBP was measured using the LBP Enzyme-Linked Immunosorbent Assay kit (ab213876; Abcam, Waltham, MA). Plasma and hepatic levels of cholesterol, triglycerides, and free fatty acids were measured using the Total Cholesterol Assay kit (cat. STA-384; Cell Biolabs, Inc, San Diego, CA), Triglyceride Reagent Set (cat. T7532120; Pointe Scientific, Lincoln Park, MI) and free fatty acids, half micro test Kit (cat. 11383175001; Roche Diagnostics) according to the manufacturer's instructions. The intraperitoneal glucose tolerance test was conducted at 1 week and at 12 weeks after WD initiation. After a 6-hour fast (9 am to 3 pm), mice received a glucose bolus (2 mg/g body weight intraperitoneally), and tail-vein blood samples were taken at 0, 15, 30, 60, and 120 minutes for measurement of glucose using a glucometer (Accu-chek Active; Roche Diagnostics). The area under the blood glucose level curve was determined using Prism software, v6.07 (GraphPad).
RNA Sequencing
RNA sequencing was performed at La Jolla Institute for Immunology and described previously.40 Sequenced reads were aligned to the mouse genome (National Center for Biotechnology Information, MGSCv38, mm10) using STAR (Spliced Transcripts Alignment to a Reference).41 Read quantification was performed with RSEM3 v1.3.0 (University of Wisconsin, Madison, WI) and Ensembl annotation (Gencode v19, Wellcome Genome Campus, Hinxton, Cambridgeshire, UK). The R Bioconductor packages edgeR (The Walter and Eliza Hall Institute of Medical Research, Victoria, Australia)42 and limma (The Walter and Eliza Hall Institute of Medical Research)43 were used to implement the limma-voom43 method for differential expression analysis. Lowly expressed genes were filtered out (genes with counts per million >1 in at least 33% of the total samples were kept). Trimmed mean of M value44 normalization was applied to the filtered data. Functional enrichment analysis of the differential expression results was performed using WebGestalt 2019 (WEB-based Gene SeT AnaLysis Toolkit, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX).45 To determine the similarity in gene expression of Foz/Foz NASH fibrosis to human NASH, the hepatic transcription profile of 12-week Foz+WD (NASH plus fibrosis stage) vs 12-week WT+WD (NAFL stage) was compared with 2 publicly available human NASH vs healthy obese gene expression data sets GSE48452 and GSE126848.15
STRING-based clustering analysis
To identify major dysregulated pathways in NAFL or NASH livers we performed a 2-step analysis. First, significantly changed genes in Foz/Foz WD week 12 compared with WT chow week 12 (adjusted P < .05, absolute value [log fold change], >1.5) were integrated with high confidence interactions from STRING (edge score, >0.7). The resulting network was input to the CDAPS (Community Detection Application and Service) hierarchical clustering tool46 in Cytoscape (Institute for Systems Biology, 2019, https://www.cytoscape.org.)47 to identify biological pathways underlying the Foz/Foz WD phenotype. We used the OSLOM (Order Statistics Local Optimization Method, arXiv:1012.2363 [physics.soc-ph]) clustering algorithm available in CDAPS (Community Detection APplication and Service). The resulting systems of highly interconnected genes were annotated with the KEGG (Kyoto Encyclopedia of Genes and Genomes), GO (Gene Ontology), and REACTOME (https://reactome.org/) pathway databases, using gprofiler (https://biit.cs.ut.ee/gprofiler/gost).48 Network visualization was conducted in Cytoscape, using a spring-embedded layout. Next, we probed these pathways by their specificity to Foz+WD. The specificity of each system to Foz/Foz was assessed by the difference in average absolute value log fold change between Foz+WD 12 weeks and WT+WD 12 weeks, to identify systems in which changes were more or less specific to Foz+WD or WT+WD.
Gut Microbiome
Cecum contents were used for microbiome analyses. DNA extraction and 16S ribosomal RNA (rRNA) amplicon sequencing were performed using Earth Microbiome Project standard protocols (available: http://www.earthmicrobiome.org/protocols-and-standards/16s).49 16S reads were preprocessed using the default Qiita pipeline (UCSD, San Diego, CA). β-diversity was performed using RPCA with DEICODE (UCSD). To elucidate the association of specific microbial taxa with diet and genotype, multinomial regression was performed using Songbird (UCSD).50
Cecal content collection and 16s rRNA sequencing
Cecal contents were collected aseptically from chow and WD-fed WT and Foz/Foz mice at 0, 1, 2, and 12 weeks after dietary intervention. DNA extraction and 16S rRNA amplicon sequencing were performed using Earth Microbiome Project standard protocols (available: http://www.earthmicrobiome.org/protocols-and-standards/16s).49 Briefly, DNA was extracted with the Qiagen MagAttract PowerSoil DNA kit as previously described.51 Amplicon PCR was performed on the V4 region of the 16S rRNA gene using the primer pair 515f to 806r with Golay (Golay codes) error-correcting barcodes on the reverse primer. Amplicons were barcoded and pooled in equal concentrations for sequencing. The amplicon pool was purified with the MO BIO UltraClean PCR cleanup kit (Qiagen, Valencia, CA) and sequenced on the Illumina MiSeq sequencing platform. Sequence data were demultiplexed and minimally quality filtered using the QIIME 1.9.1 script split_libraries_fastq.py, with a Phred quality threshold of 3 and default parameters to generate per-study FASTA sequence files.
Preprocessing of 16S reads
16S reads were processed using the default Qiita pipeline52 and the feature table output from Deblur (UCSD)53 was used for subsequent analyses. This feature table was filtered to remove experimental blanks. Taxonomic assignment of sOTUs (sub-OTU where OTU stands for Operational Taxonomic Unit) was performed using the pretrained GreenGenes54 classifier available through QIIME2.55
β-diversity
β-diversity was performed using RPCA with DEICODE,56 with 3 components on the full data set. In addition, we performed RPCA on the individual time points of t = 0, t = 12, as well as the pooled samples at t = 1 and 2. We performed PERMANOVA on the results of the RPCA to determine whether the differences between sample groups was statistically significant.57
Multinomial regression
To assess the association of specific microbial taxa with diet and genotype, we performed multinomial regression using Songbird.50 Multinomial regression models were used to generate microbial counts of samples according to the following formula: diet+genotype+diet:genotype, where diet:genotype indicates the interaction term between diet and genotype. All models used were compared with a null model (formula = 1) to verify we were not overfitting. Differentials generated from Songbird were input to Qurro (UCSD)58 to determine microbial taxa more associated with diet and/or genotype relative to all others. The Qarcoal command in Qurro was used to obtain log ratios of microbes according to GreenGenes-assigned taxonomy.
Software
Microbiome analyses were performed using QIIME. Plots were generated using matplotlib and seaborn.
Immunoblot Analysis
Livers were homogenized by dounce homogenizer (Thomas Scientific, Swedesboro, NJ) with 30 strokes. Whole-tissue lysates were made in RIPA buffer with protease and phosphatase inhibitors. Equal amounts of protein (30 μg) were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis on a 4%–12% Bis-Tris Mini Gels (Bio-Rad), then transferred to polyvinylidene difluoride membranes, and incubated with primary antibodies. Membranes were incubated with appropriate horseradish peroxidase–conjugated secondary antibodies (Cell Signaling Technology, Danvers, MA). Bound antibody was visualized using the SuperSignal chemiluminescent kit (Thermo Scientific Pierce Biotechnology, San Diego, CA) and the chemiluminescent signal was detected using the Chemidoc gel imaging system (Bio-Rad). The antibody details are listed in Table 2.
Isolation of Lamina Propria Immune Cells
Small intestine (SI) tissues were dissected from WT and Foz/Foz mice and ileum sections were cut longitudinally, feces were removed, and sections were placed in a petri dish with fresh phosphate-buffered saline (PBS). Tissue sample was cut into 3–5 pieces and transferred to 50 mL conical tubes containing 5 mL complete media (RPMI, 10% fetal bovine serum [FBS], 1× penicillin/streptomycin, 1× L-glutamine, 1 mmol/L sodium pyruvate, and 20 mmol/L HEPES). Complete media was removed, and tissues were washed 3 times in RPMI media supplemented with 2% FBS and 1× penicillin/streptomycin using a 25-mL pipette. SI sections were resuspended in 5 mL epithelial strip buffer (PBS, 5 mmol/L EDTA, 1 mmol/L dithiothreitol, 5% FBS, 1× penicillin/streptomycin), incubated for 10 minutes while shaking (100–200 rpm) at 37°C (in a water bath), and vortexed aggressively every 3 minutes. SI sections were transferred to a new tube containing 15 mL fresh epithelial strip buffer and incubated for 20 minutes while shaking (100–200 rpm) at 37°C. Sections were vortexed and supernatants were removed. Tissues were washed 3 times in RPMI media supplemented with 2% FBS and 1× penicillin/streptomycin using a 25-mL pipette. To isolate lamina propria cells, SI sections were resuspended in 5 mL digestion solution (complete media including 1 mg/mL collagenase VIII [Sigma] and 20 ug/mL DNase I [Sigma]) and incubated for 15 minutes at 37°C while shaking. Ten milliliters digestion was added to the solution and was shaken for 15 extra minutes at 37°C. The entire contents were passed through a cell strainer (100 mmol/L), and intestine pieces were mashed and washed with 10 mL complete media. Cells were centrifuged for 5 minutes at 1500 rpm, supernatants were removed, cells were resuspended in 4 mL of 40% Percoll made in RPMI medium, contents were transferred to a 15-mL tube, with 2 mL of 70% Percoll underlay made in RPMI medium. Samples were centrifuged at 2500 rpm for 20 minutes. Cells present in the interphase were collected and transferred to a new tube with 10 mL complete media.
Flow Cytometry
Two fluorescence-activated cell sorter (FACS) antibody panels were used for the analysis of mouse lamina propria cell suspensions. For ILC/T-cell analysis, the following antibodies were used: BUV805 anti-CD45, fluorescein isothiocyanate (FITC) antilineage (anti-CD3, anti–Gr-1, FITC anti-CD11b, FITC anti-CD45R [B220], and FITC anti–Ter-119, and anti-FCeR1a antibodies) cocktail, FITC–anti-FCeR1a, BV785 anti-CD90, BB700 anti-IL33R (ST2), PE (phycoerythrin) anti-GATA3, APC anti-RORγ(t), BV605 anti-CD127, BV421 anti-Tbet, APC (allophycocyanin)-efluor780 anti-NK1.1, BUV563 anti-TCRβ chain, PE–cyanine 7 anti-Foxp3, BUV395 anti-CD25, and BUV737 anti-CD4 antibodies. For myeloid cell analysis, the antibodies used were the BUV325-conjugate antibody cocktail (anti-CD3, anti-CD45R/B220, and anti–TER-119), BUV737 anti-CD11b, BUV805 anti-CD45, BUV653 anti-F4/80, BV786 anti-CD11c, BV605 anti-CD206, APC-efluor780 anti-MerTK, FITC anti-Trem2, PE anti-IRF5, BV421 anti-CD80, APC anti–MHC-II (major histocompatibility complex), PeCy7 anti-Ly6C, BV480 anti-Ly6G, and BUV711 anti-SiglecF antibodies (Table 2). Lamina propria cell suspensions were first incubated for 30 minutes at 4°C in the dark with the Zombie UV fixable viability kit and purified Fc-block (2 mg/mL, final dilution) in plain, ice-cold PBS to allow discrimination of live cells and to block nonspecific antibody binding, respectively. Samples were transferred to a 0.5-mL deep 96-well plate format and then washed with 500 mL ice-cold FACS buffer (2% FBS), centrifuging cells at 1500 rpm at 4°C for 5 minutes. Cell pellets were labeled with antibody cocktails in 200 mL FACS buffer to extracellular markers for 30 minutes at 4°C in the dark. Samples were washed in 500 mL FACS buffer, centrifuging cells at 1500 rpm at 4°C for 5 minutes, cells were resuspended in 200 μL Foxp3 fixation/permeabilization working solution and incubated at 4°C for 12–16 hours, protected from light. All samples were washed twice with 1× permeabilization buffer, centrifuging 400 × g for 5 minutes at room temperature, and discarding supernatants. Cell pellets were resuspended to be labeled a second time with antibody cocktails in 200 mL permeabilization buffer for 45 minutes at 4°C in the dark. Samples then were washed first with 1× permeabilization buffer and then with FACS buffer. Cell pellets were resuspended with PBS 1× for collection by FACS. Events from all samples were collected immediately within 12 hours after labeling. Samples were read in a FACSymphony A5 Cell analyzer (5 lasers) and analyzed using BD FACS Diva software (BD). FACS files were exported and analyzed using FlowJo 10.7.1 software (Ashland, OR).
Statistical Analysis
Data are represented as means ± SEM unless otherwise mentioned. Differences between groups were compared using analysis of variance, t test, or the Mann–Whitney U test as applicable. A P value less than .05 was considered significant. GraphPad Prism software was used for all statistical analyses.
Acknowledgments
The authors thank Kevin Lam, Naser Khader, Asra Irfan, and Rolando Arreola for their help with experiments, and the University of California San Diego Microscopy Core (NS047101) for Nanozoomer imaging support.
CRediT Authorship Contributions
Souradipta Ganguly, PhD (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead; Investigation: Equal; Methodology: Equal; Writing – original draft: Equal)
German Aleman Muench, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Validation: Supporting; Writing – original draft: Supporting)
Linshan Shang, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Validation: Supporting)
Sara Brin Rosenthal, PhD (Data curation: Supporting; Formal analysis: Supporting; Software: Supporting; Visualization: Supporting)
Gibraan Rahman, BS (Data curation: Supporting; Formal analysis: Supporting; Software: Supporting; Visualization: Supporting)
Ruoyu Wang, BS (Formal analysis: Supporting; Methodology: Supporting)
Yanhan Wang, PhD (Investigation: Supporting; Methodology: Supporting; Validation: Supporting)
Hyeok Choon Kwon, MD, PhD (Investigation: Supporting; Methodology: Supporting)
Anthony M Diomino, BS (Formal analysis: Supporting; Methodology: Supporting; Software: Supporting)
Tatiana Kisseleva, MD, PhD (Investigation: Supporting; Methodology: Supporting; Resources: Supporting)
Pejman Soorosh, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Resources: Supporting)
Mojgan Hosseini, MD (Formal analysis: Supporting; Methodology: Supporting)
Rob Knight, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Software: Supporting)
Bernd Schnabl, MD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting)
David A Brenner, MD (Conceptualization: Equal; Funding acquisition: Equal; Resources: Equal; Supervision: Equal)
Debanjan Dhar, PhD (Conceptualization: Lead; Data curation: Equal; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Methodology: Equal; Project administration: Lead; Resources: Equal; Supervision: Lead; Writing – original draft: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts. The University of California San Diego and D.A.B receives grant support from Janssen.
Funding Supported by the American Liver Foundation (ALF) Liver Scholar award, UC San Diego Altman Clinical and Translational Research Institute (ACTRI)/National Institutes of Health grant KL2TR001444, The San Diego Digestive Diseases Research Center (SDDRC) (National Institutes of Health grant DK120515), and the Southern California Research Center for Alcoholic Liver and Pancreatic Diseases (ALPD) and Cirrhosis funded by the National Institute on Alcohol Abuse and Alcoholism (NIAAA)/National Institutes of Health grant 5P50AA011999 (D.D.); by National Institutes of Health grants U01AA022614, P50AA011999, and AI043477, and an unrestricted research grant from Janssen (D.A.B.); and by National Institutes of Health grants R01DK101737, U01AA022614, R01DK099205, and R01DK111866 (T.K.).
Note: to access the supplementary materials accompanying this article, visit the online version of Cellular and Molecular Gastroenterology and Hepatology at www.cmghjournal.org.
Contributor Information
David A. Brenner, Email: dbrenner@health.ucsd.edu.
Debanjan Dhar, Email: ddhar@health.ucsd.edu.
Supplementary Material
References
- 1.Anstee Q.M., Reeves H.L., Kotsiliti E., Govaere O., Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019;16:411–428. doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 2.Tan H., Xu L., Liu X., Si S., Sun Y., Liu L., Zhou W., Yang Z. Hepatocellular carcinoma in nonalcoholic fatty liver disease mimicking benign hemangioma: two case reports and literature review. Int J Clin Exp Pathol. 2015;8:15350–15355. [PMC free article] [PubMed] [Google Scholar]
- 3.Wong R.J., Aguilar M., Cheung R., Perumpail R.B., Harrison S.A., Younossi Z.M., Ahmed A. Nonalcoholic steatohepatitis is the second leading etiology of liver disease among adults awaiting liver transplantation in the United States. Gastroenterology. 2015;148:547–555. doi: 10.1053/j.gastro.2014.11.039. [DOI] [PubMed] [Google Scholar]
- 4.Adams L.A., Anstee Q.M., Tilg H., Targher G. Non-alcoholic fatty liver disease and its relationship with cardiovascular disease and other extrahepatic diseases. Gut. 2017;66:1138–1153. doi: 10.1136/gutjnl-2017-313884. [DOI] [PubMed] [Google Scholar]
- 5.Febbraio M.A., Reibe S., Shalapour S., Ooi G.J., Watt M.J., Karin M. Preclinical models for studying NASH-driven HCC: how useful are they? Cell Metab. 2019;29:18–26. doi: 10.1016/j.cmet.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kubota N., Kado S., Kano M., Masuoka N., Nagata Y., Kobayashi T., Miyazaki K., Ishikawa F. A high-fat diet and multiple administration of carbon tetrachloride induces liver injury and pathological features associated with non-alcoholic steatohepatitis in mice. Clin Exp Pharmacol Physiol. 2013;40:422–430. doi: 10.1111/1440-1681.12102. [DOI] [PubMed] [Google Scholar]
- 7.Girard D., Petrovsky N. Alstrom syndrome: insights into the pathogenesis of metabolic disorders. Nat Rev Endocrinol. 2011;7:77–88. doi: 10.1038/nrendo.2010.210. [DOI] [PubMed] [Google Scholar]
- 8.Farrell G.C., Mridha A.R., Yeh M.M., Arsov T., Van Rooyen D.M., Brooling J., Nguyen T., Heydet D., Delghingaro-Augusto V., Nolan C.J., Shackel N.A., McLennan S.V., Teoh N.C., Larter C.Z. Strain dependence of diet-induced NASH and liver fibrosis in obese mice is linked to diabetes and inflammatory phenotype. Liver Int. 2014;34:1084–1093. doi: 10.1111/liv.12335. [DOI] [PubMed] [Google Scholar]
- 9.Arsov T., Silva D.G., O'Bryan M.K., Sainsbury A., Lee N.J., Kennedy C., Manji S.S., Nelms K., Liu C., Vinuesa C.G., de Kretser D.M., Goodnow C.C., Petrovsky N. Fat aussie--a new Alstrom syndrome mouse showing a critical role for ALMS1 in obesity, diabetes, and spermatogenesis. Mol Endocrinol. 2006;20:1610–1622. doi: 10.1210/me.2005-0494. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y.C., Ganguly S., Nespoux J., Freeman B., Zhang H., Brenner D., Dhar D., Vallon V. Western diet promotes renal injury, inflammation, and fibrosis in a murine model of Alstrom syndrome. Nephron. 2020;144:400–412. doi: 10.1159/000508636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larter C.Z., Yeh M.M. Animal models of NASH: getting both pathology and metabolic context right. J Gastroenterol Hepatol. 2008;23:1635–1648. doi: 10.1111/j.1440-1746.2008.05543.x. [DOI] [PubMed] [Google Scholar]
- 12.Giles D.A., Moreno-Fernandez M.E., Stankiewicz T.E., Graspeuntner S., Cappelletti M., Wu D., Mukherjee R., Chan C.C., Lawson M.J., Klarquist J., Sunderhauf A., Softic S., Kahn C.R., Stemmer K., Iwakura Y., Aronow B.J., Karns R., Steinbrecher K.A., Karp C.L., Sheridan R., Shanmukhappa S.K., Reynaud D., Haslam D.B., Sina C., Rupp J., Hogan S.P., Divanovic S. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat Med. 2017;23:829–838. doi: 10.1038/nm.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto E., Tokushige K. Prevalence, gender, ethnic variations, and prognosis of NASH. J Gastroenterol. 2011;46(Suppl 1):63–69. doi: 10.1007/s00535-010-0311-8. [DOI] [PubMed] [Google Scholar]
- 14.Ahrens M., Ammerpohl O., von Schonfels W., Kolarova J., Bens S., Itzel T., Teufel A., Herrmann A., Brosch M., Hinrichsen H., Erhart W., Egberts J., Sipos B., Schreiber S., Hasler R., Stickel F., Becker T., Krawczak M., Rocken C., Siebert R., Schafmayer C., Hampe J. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Suppli M.P., Rigbolt K.T.G., Veidal S.S., Heeboll S., Eriksen P.L., Demant M., Bagger J.I., Nielsen J.C., Oro D., Thrane S.W., Lund A., Strandberg C., Konig M.J., Vilsboll T., Vrang N., Thomsen K.L., Gronbaek H., Jelsing J., Hansen H.H., Knop F.K. Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am J Physiol Gastrointest Liver Physiol. 2019;316:G462–G472. doi: 10.1152/ajpgi.00358.2018. [DOI] [PubMed] [Google Scholar]
- 16.Tripathi A., Debelius J., Brenner D.A., Karin M., Loomba R., Schnabl B., Knight R. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan H., Wang A., Wang Y., Sun Y., Han J., Chen W., Wang S., Wu Y., Lu Y. Innate lymphoid cells: regulators of gut barrier function and immune homeostasis. J Immunol Res. 2019;2019:2525984. doi: 10.1155/2019/2525984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kempski J., Brockmann L., Gagliani N., Huber S. TH17 cell and epithelial cell crosstalk during inflammatory bowel disease and carcinogenesis. Front Immunol. 2017;8:1373. doi: 10.3389/fimmu.2017.01373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olofson A.M., Gonzalo D.H., Chang M., Liu X. Steatohepatitic variant of hepatocellular carcinoma: a focused review. Gastroenterology Res. 2018;11:391–396. doi: 10.14740/gr1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angulo P., Kleiner D.E., Dam-Larsen S., Adams L.A., Bjornsson E.S., Charatcharoenwitthaya P., Mills P.R., Keach J.C., Lafferty H.D., Stahler A., Haflidadottir S., Bendtsen F. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397 e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nassir F., Ibdah J.A. Role of mitochondria in nonalcoholic fatty liver disease. Int J Mol Sci. 2014;15:8713–8742. doi: 10.3390/ijms15058713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saran A.R., Dave S., Zarrinpar A. Circadian rhythms in the pathogenesis and treatment of fatty liver disease. Gastroenterology. 2020;158:1948–1966 e1. doi: 10.1053/j.gastro.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong X., Feng D., Mathews S., Gao B. Hepatoprotective and anti-fibrotic functions of interleukin-22: therapeutic potential for the treatment of alcoholic liver disease. J Gastroenterol Hepatol. 2013;28(Suppl 1):56–60. doi: 10.1111/jgh.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang S., He Y., Xiang X., Seo W., Kim S.J., Ma J., Ren T., Park S.H., Zhou Z., Feng D., Kunos G., Gao B. Interleukin-22 ameliorates neutrophil-driven nonalcoholic steatohepatitis through multiple targets. Hepatology. 2020;72:412–429. doi: 10.1002/hep.31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Federico A., Dallio M., Godos J., Loguercio C., Salomone F. Targeting gut-liver axis for the treatment of nonalcoholic steatohepatitis: translational and clinical evidence. Transl Res. 2016;167:116–124. doi: 10.1016/j.trsl.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Chu H., Duan Y., Yang L., Schnabl B. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut. 2019;68:359–370. doi: 10.1136/gutjnl-2018-316307. [DOI] [PubMed] [Google Scholar]
- 27.Luther J., Garber J.J., Khalili H., Dave M., Bale S.S., Jindal R., Motola D.L., Luther S., Bohr S., Jeoung S.W., Deshpande V., Singh G., Turner J.R., Yarmush M.L., Chung R.T., Patel S.J. Hepatic injury in nonalcoholic steatohepatitis contributes to altered intestinal permeability. Cell Mol Gastroenterol Hepatol. 2015;1:222–232. doi: 10.1016/j.jcmgh.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandl K., Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. 2017;33:128–133. doi: 10.1097/MOG.0000000000000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., Almeida M., Arumugam M., Batto J.M., Kennedy S., Leonard P., Li J., Burgdorf K., Grarup N., Jorgensen T., Brandslund I., Nielsen H.B., Juncker A.S., Bertalan M., Levenez F., Pons N., Rasmussen S., Sunagawa S., Tap J., Tims S., Zoetendal E.G., Brunak S., Clement K., Dore J., Kleerebezem M., Kristiansen K., Renault P., Sicheritz-Ponten T., de Vos W.M., Zucker J.D., Raes J., Hansen T., Meta HITc, Bork P., Wang J., Ehrlich S.D., Pedersen O. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 30.Atarashi K., Umesaki Y., Honda K. Microbiotal influence on T cell subset development. Semin Immunol. 2011;23:146–153. doi: 10.1016/j.smim.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Bernink J.H., Peters C.P., Munneke M., te Velde A.A., Meijer S.L., Weijer K., Hreggvidsdottir H.S., Heinsbroek S.E., Legrand N., Buskens C.J., Bemelman W.A., Mjosberg J.M., Spits H. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 32.Ding S., Lund P.K. Role of intestinal inflammation as an early event in obesity and insulin resistance. Curr Opin Clin Nutr Metab Care. 2011;14:328–333. doi: 10.1097/MCO.0b013e3283478727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flannigan K.L., Ngo V.L., Geem D., Harusato A., Hirota S.A., Parkos C.A., Lukacs N.W., Nusrat A., Gaboriau-Routhiau V., Cerf-Bensussan N., Gewirtz A.T., Denning T.L. IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol. 2017;10:673–684. doi: 10.1038/mi.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Promrat K., Kleiner D.E., Niemeier H.M., Jackvony E., Kearns M., Wands J.R., Fava J.L., Wing R.R. Randomized controlled trial testing the effects of weight loss on nonalcoholic steatohepatitis. Hepatology. 2010;51:121–129. doi: 10.1002/hep.23276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lassailly G., Caiazzo R., Ntandja-Wandji L.C., Gnemmi V., Baud G., Verkindt H., Ningarhari M., Louvet A., Leteurtre E., Raverdy V., Dharancy S., Pattou F., Mathurin P. Bariatric surgery provides long-term resolution of nonalcoholic steatohepatitis and regression of fibrosis. Gastroenterology. 2020;159:1290–1301 e5. doi: 10.1053/j.gastro.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Rosenthal S.B., Liu X., Ganguly S. Heterogeneity of hepatic stellate cells in a mouse model of non-alcoholic steatohepatitis (NASH) Hepatology. 2021 doi: 10.1002/hep.31743. Epub ahead of print. [DOI] [Google Scholar]
- 37.Charlton M., Krishnan A., Viker K., Sanderson S., Cazanave S., McConico A., Masuoko H., Gores G. Fast food diet mouse: novel small animal model of NASH with ballooning, progressive fibrosis, and high physiological fidelity to the human condition. Am J Physiol Gastrointest Liver Physiol. 2011;301:G825–G834. doi: 10.1152/ajpgi.00145.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchida T., Lee Y.A., Fujiwara N., Ybanez M., Allen B., Martins S., Fiel M.I., Goossens N., Chou H.I., Hoshida Y., Friedman S.L. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol. 2018;69:385–395. doi: 10.1016/j.jhep.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Yeh M., McCullough A.J., Sanyal A.J., Nonalcoholic Steatohepatitis Clinical Research Network Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 40.Liu X., Rosenthal S.B., Meshgin N., Baglieri J., Musallam S.G., Diggle K., Lam K., Wu R., Pan S.Q., Chen Y., Dorko K., Presnell S., Benner C., Hosseini M., Tsukamoto H., Brenner D., Kisseleva T. Primary alcohol-activated human and mouse hepatic stellate cells share similarities in gene-expression profiles. Hepatol Commun. 2020;4:606–626. doi: 10.1002/hep4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. e47-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singhal A., Cao S., Churas C., Pratt D., Fortunato S., Zheng F., Ideker T. Multiscale community detection in Cytoscape. PLoS Comput Biol. 2020;16 doi: 10.1371/journal.pcbi.1008239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., Vilo J. g. Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ul-Hasan S., Bowers R.M., Figueroa-Montiel A., Licea-Navarro A.F., Beman J.M., Woyke T., Nobile C.J. Community ecology across bacteria, archaea and microbial eukaryotes in the sediment and seawater of coastal Puerto Nuevo, Baja California. PLoS One. 2019;14 doi: 10.1371/journal.pone.0212355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morton J.T., Marotz C., Washburne A., Silverman J., Zaramela L.S., Edlund A., Zengler K., Knight R. Establishing microbial composition measurement standards with reference frames. Nat Commun. 2019;10:2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marotz C., Amir A., Humphrey G., Gaffney J., Gogul G., Knight R. DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques. 2017;62:290–293. doi: 10.2144/000114559. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez A., Navas-Molina J.A., Kosciolek T., McDonald D., Vazquez-Baeza Y., Ackermann G., DeReus J., Janssen S., Swafford A.D., Orchanian S.B., Sanders J.G., Shorenstein J., Holste H., Petrus S., Robbins-Pianka A., Brislawn C.J., Wang M., Rideout J.R., Bolyen E., Dillon M., Caporaso J.G., Dorrestein P.C., Knight R. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods. 2018;15:796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., Knight R. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2:e00191–e00216. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodríguez A.M., Chase J., Cope E.K., Da Silva R., Diener C., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibbons S.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G.A., Janssen S., Jarmusch A.K., Jiang L., Kaehler B.D., Kang K.B., Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G.I., Lee J., Ley R., Liu Y.-X., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J.T., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S., Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., van der Hooft JJJ, Vargas F., Vázquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y., Wang M., Warren J., Weber K.C., Williamson C.H.D., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y., Zhu Q., Knight R., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martino C., Morton J.T., Marotz C.A., Thompson L.R., Tripathi A., Knight R., Zengler K. A novel sparse compositional technique reveals microbial perturbations. mSystems. 2019;4:e00016–19. doi: 10.1128/mSystems.00016-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 58.Fedarko M.W., Martino C., Morton J.T., González A., Rahman G., Marotz C.A., Minich J.J., Allen E.E., Knight R. Visualizing ’omic feature rankings and log-ratios using Qurro. NAR Genom Bioinform. 2020;2 doi: 10.1093/nargab/lqaa023. lqaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.