Abstract
The microbiome refers to a population of microbes that colonize the skin, nasopharynx, oral cavity, gastrointestinal tract, and urogenital tract. The human microbiome consists of bacteria, archaea, fungi, viruses, and phages. Recent advances in genomic sequencing have catalyzed a deeper understanding of complex microbe-microbe and host-microbe interactions. Dysregulation of these interactions, or dysbiosis of the gastrointestinal tract, has been implicated in a growing list of pathologies including nonalcoholic fatty liver disease, cardiovascular disease, obesity, diabetes, depression, Parkinson’s disease, autism, and various gastrointestinal cancers. Gastric and esophageal cancer, for example, continue to remain as two of the most common causes of cancer-related deaths worldwide, therefore there is an increased emphasis on investigating the role of dysbiosis on these cancers. In this review, we discuss the development and structure of the gut microbiome, its homeostatic and dysbiotic mechanisms, and the key microbes in esophageal and gastric carcinogenesis with a focus on bacterial biology. Further clarification of these pathways and discovery of diagnostic or therapeutic targets could have broad impacts on global subpopulations. It is important to understand the nature of the gastrointestinal tract microbiome and its potentional risk factors for dysbiosis in order to tailor its application to the individual patient and create an era of highly personalized, precision medicine.
Keywords: Microbiome, gut, gastrointestinal tract, cancer, cancer prevention, human
Introduction
The microbiome refers to a population of microbes that colonize the skin, nasopharynx, oral cavity, gastrointestinal tract, and urogenital tract in a ratio of at least one microbe cell to one human cell. Though the above estimate accounts for only bacterial cells, it should be noted that the microbiome includes archaea, fungi, viruses, and phages (1). More important than the actual number of cells or even the number of species inhabiting these microenvironments is the additional genetic diversity, which by some estimates, are orders of magnitude larger than the human genome (2). Though interactions amongst commensal microbiota and between microbiota and the host are complex, the underlying mechanisms are beginning to be elucidated and even manipulated. A healthy gut microbiome confers in humans the ability to digest complex dietary polysaccharides, synthesize essential amino acids, and absorb vitamins, metabolize certain drugs, develop the immune system, and to defend against pathobionts (3-7). Dysregulation of the microbiome, or dysbiosis, has been linked to a growing list of pathologies including nonalcoholic fatty liver disease, cardiovascular disease, obesity, diabetes, depression, Parkinson’s disease, autism, and various cancers (8-12) (Table 1).
Table 1. Glossary of terms.
| Term | Definition | Reference |
|---|---|---|
| Commensalism | A relationship between two organisms in which one organism benefits, whereas the other is not harmed or helped | (13) |
| Dysbiosis | An abnormality—in composition and/or function—of the host symbiotic microbial ecosystem that exceeds its restitutive capacity and has negative effects on the host | (14) |
| Enterotypes | Distinct microbiome configurations depending on composition or metabolic profile of component microbial organisms | (15) |
| Esotypes | Functionally distinct community types found in the esophageal microbiome depending on the relative abundances of microbial organisms | (13) |
| Genetic diversity | The variation in a DNA sequence between distinct individuals of a given species (or population) | – |
| Genetic variability | Genotype differences in a given population | – |
| Genome | The genetic product of an organism | – |
| Human Microbiome Project | A National Institutes of Health project to characterize the human microbial flora launched in 2007 | (16) |
| Microbiome | The complex ecosystem of microbial cells including bacteria, archaea, eukaryotes, and viruses, that colonize the human body. The microbiome has the capability of encoding genetic products that affect health and disease in the human host | – |
| Microenvironment | The immediately local habitat of a microbial cell, usually referring to distinct space within an organ in the host | – |
| Mutualism | A relationship between two organisms in which both organisms benefit | (14) |
| Pathobiont | Normally innocuous microorganisms that can behave like pathogens if their abundance increases and/or their environmental conditions change | (13) |
| Pathogen | A microorganism that can cause disease | (14) |
| Phylum | A taxonomic classification that ranks in between kingdom and class | – |
| Taxon | A group of organisms | – |
| Taxonomic level | A relative level of a group of organisms | – |
| Virulence genes or virulence factors | Molecules expressed by pathogenic microorganisms that help them to gain a growth advantage in a specific ecosystem. These molecules are often responsible for disease manifestation in the host | (13) |
Once regarded as the “forgotten organ”, the gut microbiome is now the subject of renewed fervor. In fact, more than 80% of all microbiome publications were published after 2012 (17,18). Given the dizzying pace of discovery in this field, it may be difficult for clinicians to keep up with the latest insights on gut microbiomes and the implications of dysbiosis in gastrointestinal malignancies. We will briefly discuss perturbations in the gastrointestinal microflora or gut “dysbiosis” focusing on the development and structure of gut microbiota, their reputed role in the protection against cancer, and the proposed mechanisms by which gut dysbiosis may contribute to some types of cancer such as esophageal and gastric cancers.
Recently, the International Cancer Microbiome Consortium released a consensus statement describing four key features of a healthy gut microbiome: (I) synergizing with the host for immune function; (II) metabolic mutualism; (III) resilience to temporary disturbances and adaptation; and (IV) tumor-suppression (14). The first two of these features is covered in the next section. The remaining two features are covered in the following section.
The development and structure of the gut microbiome
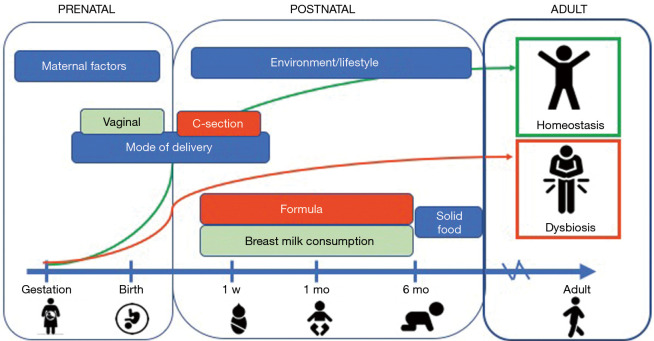
The prenatal intestinal tract was once thought to be sterile, though this notion has recently been challenged. Molecular surveys on infants have shown micro-organism colonization may occur in utero, suggesting that our microbiomes are older than we are (19). Neonatal gut associated lymphoid tissue along with the rest of the immune system is not fully developed upon birth. Mucosal secretion of mucin, IgA, and antimicrobial peptides has not been established. Infants are inoculated with microbes during birth, and the composition of the early microbiome is affected by gestational age and mode of delivery (Figure 1). For example, preterm infants and infants delivered via c-section were found to have reduced diversity and a delayed colonization by commensals as compared to term infants or vaginal deliveries (20). Breastfeeding then provides the infant gut with a “continuous inoculum” of microbes along with human milk oligosaccharides and secretory IgA (21). Breastfed infants have a microbiota composed predominantly of Bifidobacteria spp. and Lactobacilli spp. which consume human milk oligosaccharides and produce short chain fatty acids (SCFAs) (i.e., acetate, propionate, and butyrate). SCFAs are substrates for energy production, downregulate pathogen virulence genes, and promote immunotolerance (21). The microbiome of formula-fed infants is more diverse. This discrepancy of SCFA production in infants has implications in allergic airway inflammation, arthritis, ulcerative colitis, and colic (22-25). Introduction of solid foods at around 6 months of age promotes a shift towards Bacteroides spp. which have are better able to metabolize complex carbohydrates. Diversification of the microbiome increases and adapts to the diet of the child. By age 3, gut microbiota approaches adult-level diversity (26,27).
Figure 1.
Development of the gut microbiome and influencing factors that lead to homeostasis or dysbiosis. Starting from gestation, maternal microbiota influence the developing fetus’s future microbiome. Passage through the vaginal canal confers exposure to mucosal IgA, inoculating the newborns microbiome. At 6 months, the microbiome starts to transition towards adult-like microbiome with the consumption of solid foods. There are many factors present early in life that can tip the scale towards homeostasis vs. dysbiosis.
The adult gut microbiome is composed of intestinal colonization niches in which bacterial microbiota are generally consistent at higher taxonomic levels (18). Firmicutes and Bacteroidetes are the major phyla within the intestinal tract. Stratifying the gut microbiome by just the relative abundance of organisms provides an overly simplistic view because significant variability occurs at lower taxonomic levels. The Human Microbiome Project examined 4,788 specimens from 242 screened and phenotyped healthy adult men and women in the US to characterize the microbial composition of 18 body habitats. The results revealed that microbial compositions were unique to each individual and remained stable over time. Interestingly, no taxa were observed to be universally present among the body habitats or individuals (16). An analysis of monozygotic and dizygotic twins demonstrated that significant gut microbial variability exists even between identical twins (28). Thus, over 80% of individuals can be reliably identified by their “microbial fingerprint” for up to a year after collecting a stool sample (29).
Metabolic pathways of microbiota, however, remained stable with respect to the individual and the overall population, regardless of the taxonomic variance and geography, suggesting that a “core microbiome” is organized around the functional gene level and not just at the organismal level (16,28-30). Indeed, clustering models accounting for frequencies of both microbial genes and populations indicate reproducible patterns of variation driven by subsets of three taxa (Prevotella, Bacteroides and Ruminococcaceae) which appear to be mutually negatively correlated (15). These clusters, or “enterotypes”, suggest that the core metagenomic functions and structure of gut microbiomes are conserved. Therefore, it follows that the gut microbiome has the capacity to achieve homeostasis between the host, the gut microbiome, the component microbiota, the host’s environment, the host’s disease state, and pharmacology.
The gut microbiome in homeostasis, dysbiosis, and tumor suppression
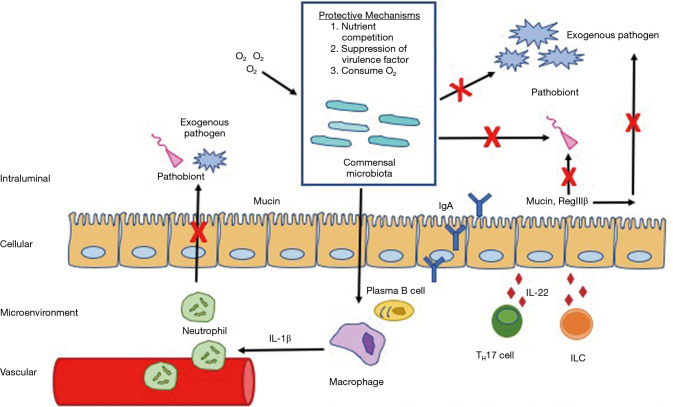
The homeostatic drive of the gut microbiome can be illustrated through its protective role against pathogen colonization (Figure 2). Given that approximately 15% of all cancers have been attributed to infectious agents, the gut microbiome is a crucial shield against infection and the subsequent carcinogenic sequela (31,32). Commensals competitively inhibit pathogenic growth by consuming nutrients and residual oxygen, depleting the resources available to pathobionts. Commensal microbes produce bacteriocins and SCFAs which downregulate virulence gene expression and alter pH. Commensals fortify the intestinal epithelial barrier function by upregulating mucin production, inducing antimicrobial peptides (RegIIIβ), and promoting IgA secretion. Commensal microbiota enhance innate host immunity via the MyD88 pathway by upregulating intestinal macrophage production of pro-IL-1β, which is activated to rapidly recruit neutrophils in enteric infections. Additionally, resident commensals facilitate differentiation of TH17 cells and innate lymphoid cells which release IL-22 and through a MyD88 independent mechanism (18).
Figure 2.
Commensal microbiota inhibit colonization of exogenous pathogens and pathobionts through upregulation of host-immunity mediated resistance such as activating plasma B cells, macrophages, T-helper cells, and innate lymphoid cells. Commensal microbiota also competitively inhibit pathogens and pathobionts by uptake of nutrients, suppressing virulence factors, and consuming O2 needed for pathogen and pathobiont survival.
When the gut microbiome is unable to return to homeostasis, it exists in a state referred to as dysbiosis. It is tempting to define dysbiosis as an anomalous composition of resident microbes (i.e., bloom of pathobionts, loss of commensals, and loss of diversity) (33). Experts, however, define dysbiosis an “abnormality—in composition and/or function—of the host symbiotic microbial ecosystem that exceeds its restitutive capacity and has negative effects on the host” (14). Compositional and functional abnormalities probably coexist and propagate each other. Dysbiosis is not applicable beyond the context of the individual host and specific pathology. A sustained state of dysbiosis may lead to carcinogenesis (14). In fact, dysbiosis could promote carcinogenesis by several potential mechanisms. For example, studies on the vaginal microbiome have found that the human papillomavirus inserts its DNA into the host DNA in a strategy known as genomic integration. In genotoxicity, pathogens or their metabolites damage host DNA structure causing cell death, promoting oncogenes, or disabling tumor suppressor genes. Chronic inflammation by microbial virulence factors can affect host intracellular signaling pathways inducing cellular proliferation and deregulating apoptosis. Pathogens may weaken cancer immunosurveillance by targeting host immune cells. Even commensal metabolism can be manipulated to produce reactive metabolites or convert pro-carcinogens into carcinogens.
Esophageal cancer
The esophagus is relatively sterile when compared to the rest of the GI tract, harboring less than one trillionth of the bacterial population in the colon (17). Pei et al. demonstrated that approximately 100 species are endemic to the normal esophagus within six phyla: Firmicutes (e.g., Streptococcus), Bacteroides (e.g., Prevotella), Actinobacteria (e.g., Rothia), Proteobacteria (e.g., Haemophilus), Fusobacteria (e.g., Fusobacterium), and TM7/Saccharibacteria (34). Deshpande et al. later performed cluster analyses on bacteria found in patients with a normal esophagus and described the existence of three clusters or “esotypes”. Esotypes were characterized by dominant organisms and had distinct functional signatures with the first type largely comprised of Streptococcus spp. (enriched pentose phosphate pathway, fructose/mannose metabolism), the second Prevotella spp. [enriched lipopolysaccharide (LPS) biosynthesis], and the third Prevotella, Haemophilus, and Rothia (enriched glycolysis pathway, SCFA metabolism) (35). Notably, the authors found that patients with gastroesophageal reflux disease (GERD) or Barrett’s esophagitis (BE) had a taxonomic shift towards gram-negative organisms including Fusobacterium and Campylobacter spp. with enhanced lactic acid production Tan pathways. Yang et al. also concluded that patients with esophagitis or BE have a propensity for gram-negative bacterial colonization (36) (Table 2).
Table 2. Flora alterations in gut microbiota and proposed dysbiotic mechanisms promoting tumorigenesis.
| Condition | Associated flora | Proposed mechanisms inducing pathology | References |
|---|---|---|---|
| Normal esophagus | Streptococcus, Prevotella, Rothia, Hemophilus, Fusobacterium, TM7/Saccharibacteria | – | (34,35) |
| Barrett’s esophagus, esophageal adenocarcinoma | ↑gram-negative flora, ↓Streptococcus | ↑lactic acid production → chronic inflammation; activation of LPS-TLR-NF-κB pathway → chronic inflammation | (35-41) |
| ↑Lactobacillus | ↑lactic acid production → chronic inflammation | ||
| Esophageal squamous cell cancer | ↓microbial diversity; alterations in adjacent microbiota (oral, gastric corpus) | – | (42-47) |
| ↑Porphyromonas gingivalis | Activation of Jak1/Akt/Stat3 pathway → dysregulation of apoptosis; conversion of ethanol to acetaldehyde → genotoxin formation | ||
| ↑Fusobacterium nucleatum | ↑CCL20 chemokine production → dysregulated immune surveillance | ||
| – | Degradation of epithelial basement membrane via MMP release, induction of DAMPs → chronic inflammation | ||
| Normal stomach | Streptococcus, Lactobacillus, Veillonella, Prevotella, Rothia, Neisseria | – | (48) |
| Gastric carcinoma | Pathogenic strains of Helicobacter pylori | NF-κB pathway mediated cytokine release → chronic inflammation, dysregulation of apoptosis | (47-52) |
| – | CagA oncoprotein activation of Ras-Erk mitogenic pathway → disruption of host cellular signaling; VacA oncoprotein induced host cell destruction → chronic inflammation | ||
| Pathobiont overgrowth promoted by Helicobacter pylori | Urease allows neutralization of acidic environment and reaction with neutrophilic metabolites → genotoxin formation | ||
| ↑Lactobacillus, Lachnospiraceae, Burkholderia, Escheria, Shigella | ↑N-nitroso compound metabolism → genotoxin formation | ||
| ↑Lactobacillus, Lactococcus | ↑lactic acid production → chronic inflammation |
LPS-TLR-NF-κB, lipopolysaccharide-toll-like receptor-nuclear factor-kappa beta; CCL20, C-C chemokine ligand 20; DAMP, danger associated membrane proteins.
The shifts in the GERD/BE microbiome may contribute to malignant transformation into esophageal adenocarcinoma (EAC). Gram-negative bacteria could promote expression of inflammatory signals due to interactions between bacterial metabolites and inflammatory cells resulting in chronic inflammation (37). Toll-like receptors (TLRs) normally mediate host-microbiota interactions and are key players in recognition of pathogen-associated molecular patterns. TLR4 expression is abundant in normal esophageal epithelium and increases in BE/EAC (38). Gram-negative bacteria produce LPSs that bind to TLR4, thereby activating the NF-κB pathway and potentially upregulating oncogenes like COX-2 (39). The enhanced expression of lactic acid producing pathways and dominance of lactate fermenters could indicate EAC is a glycolytic tumor. In these tumors, oncogenic and tumor suppressor mutations result in a synchronized process to produce lactate continuously without regulation (40). The heavy concentration of lactate promotes an acidic microenvironment, potentially inhibiting competitor growth while selecting for other organisms with similar metabolic profiles (41).
The link between dysbiosis and esophageal squamous cell cancer has not been investigated as well as EAC. Microbial diversity, especially in neighboring microbiota, has been inversely associated with esophageal squamous dysplasia (42). Patients with squamous dysplasia and squamous cell cancer had higher proportions of Clostridiales and Erysipelotrichales spp. in their gastric microbiomes relative to healthy patients (43). Salivary concentrations of Lautropia, Bulleidia, Catonella, Corynebacterium, Moryella, Peptococcus, and Cardiobacterium spp. were lower in esophageal squamous cell carcinoma compared to healthy controls (53). In studies of the esophageal microbiome, patients with esophageal squamous cell carcinoma were more likely to infected with Porphyromonas gingivalis and presence of the microbe was correlated with severity of disease (44). In patients who underwent esophagectomy for ESCC, tissue analysis revealed that approximately 23% of specimens contained Fusobacterium nucleatum and presence was associated with shorter survival (45).
Gastric cancer
The stomach’s acidic microenvironment, release of antimicrobial enzymes, and its rapid intraluminal flow make it inhabitable to one billionth of the bacterial population in the colon. Taxa commonly found in the stomach include Streptococcus, Lactobacillus, Veillonella, Prevotella, Rothia, and Neisseria spp. representing a more diversified taxonomic profile than the esophagus (48). Fundamentally, colonizers of the healthy stomach must have the ability to resist acid degradation. Tumorigenic organisms possess the additional ability to incite chronic inflammation thereby inducing dysbiosis in susceptible hosts. The International Agency for Research in Cancer (IARC) has designated Helicobacter pylori (H. pylori) is a class I carcinogen with definite links to gastric cancer (54-56). Yet, only 3% of the people infected with H. pylori actually go on to develop gastric cancer (57). Furthermore, a nationwide effort to eradicate H. pylori in patients with chronic gastritis failed to reduce the incidence of gastric cancer in Japan over a 5-year period (58). This implies an indirect, non-causal relationship between H. pylori status and gastric cancer. Indeed, H. pylori could promote a microenvironment that leads to gastric carcinogenesis to varying degrees depending on virulence factors, bacterial and host genotypes, geography, and environmental factors.
H. pylori has coevolved with humans over the past 60,000 years and has only recently disappeared from the West’s microbiome for reasons that are not fully understood (59). The bacteria’s impact on the human gut microbiome is not well understood and is limited by sequencing techniques, interindividual variability, and small sample sizes. One study of 23 healthy subjects found that H. pylori status had no effect on phylotype distribution within the stomach. Clustering analysis did not demonstrate distinct clusters of microbes between positive and negative H. pylori groups (60). A more recent comparison between these groups found that H. pylori did affect microbiome composition. Relative abundance of Proteobacteria (excluding H. pylori) and Acidobacteria was higher in H. pylori infected patients, while relative abundance of Actinobacteria and Firmicutes was higher in H. pylori negative patients. H. pylori has been shown to significantly reduce gastric microbiome diversity, comprising 24–97% of sequences found in samples (60-62). How microbial diversity relates to gastric cancer is unclear, though the relative paucity of H. pylori, and a shift towards nitrosating organisms is associated with carcinogenesis (49,50,63-65).
When involved in antral gastritis, H. pylori induces gastrin secretion and local acid production contributing to peptic ulcer disease but protecting against gastric cancer. Strains causing corpus gastritis stimulate release of inflammatory mediators which degrade gastric glands resulting in deficient acid production and a more hospitable environment for pathobionts (66). Interestingly, H. pylori prefers normal gastric mucosa and the population dwindles in atrophic gastritis, allowing proliferation of tumorigenic bacteria (49,51,61,67,68). These lesions progress to intestinal metaplasia followed by dysplasia, and eventually gastric adenocarcinoma (69).
Additionally, some strains of H. pylori employ several mechanisms to colonize the stomach, destroy gastric epithelium, upregulate inflammatory cascades, disrupt host signaling, alter cell permeability, and promote epigenetic changes. H. pylori gains access to gastric mucosa by producing urease, an enzyme which catalyzes urea into ammonia for neutralization of acid and reaction with neutrophilic metabolites. Carcinogenic strains contain the cytotoxin-associated gene A (CagA) and deliver the oncoprotein into epithelial cell cytoplasm via type IV secretion. CagA is then phosphorylated by tyrosine kinase, binds to SHP2 and activates the Ras-Erk pathway (FGFR2, KRAS, EGFR, ERBB2, and MET), dysregulating downstream cell signaling, promoting cell proliferation, destroying the epithelial barrier, and dismantling the cytoskeleton. Vacuole-forming factor (VacA) induces vacuole formation by the host cell and ultimately compromise cellular membrane integrity. H. pylori adheres to epithelial cells with outer membrane proteins; the interaction triggers release of IL-6, IL-8, IL1-β, TNF-α. Activation of NF-κB prompts cytokine release and inhibits apoptosis. H. pylori also has been implicated in DNA methylation, histone changes, and production of reactive oxygen species (ROS) induced dsDNA breaks (52).
Conclusions
The “germ theory of disease” or the idea that microscopic pathogens are responsible for disease, has served as a foundational concept of our understanding of the human body for the past two centuries. Robert Koch and Louis Pasteur prompted the departure from miasma theory in which a poisonous vapor from an unhygienic environment was thought to be the culprit for infection. Koch is credited with formalizing criteria to establish the causation of a disease by a microbe: the pathogen must be present in all diseased subjects; the pathogen should be isolated from the diseased subject and grown in culture; when the cultured pathogen is introduced to a healthy experimental host, disease should be observed; the pathogen should be re-isolated from the experimental host and be identical to the originally isolated pathogen (13). In this model of disease, the single pathogen role is overemphasized, and the host factors are unaccounted for. Antoine Bechamp, a vocal rival of Pasteur, believed that “there is an independently living microanatomical element in the cells and fluids of all living organisms” which he termed the “microzyma”. Bechamp posited that the disruption of the host’s inherent “microzymia” was a predisposition to disease (70). Though Bechamp’s “microzymia” was largely ignored by the scientific community then, it has remarkable conceptual similarities to what we now know as the microbiome.
The gut microbiome has been linked to gastric and esophageal cancers, which represent the third and sixth most common causes of cancer-related deaths worldwide (71,72). Both cancers have plausible explanations for carcinogenesis rooted in dysbiosis. Further clarification of these pathways and discovery of diagnostic or therapeutic targets could have broad impacts on global subpopulations (46). As sequencing technology becomes increasingly sophisticated and accessible, researchers will be empowered to elucidate causal, rather than just correlative relationships (47). By reframing the microbiome as a determinant of global health and tailoring its application to the individual patient, we have the power to usher in the era of highly personalized, precision medicine.
Acknowledgments
Funding: Supported by the National Cancer Institute, Division of Cancer Prevention, National Institutes of Health, Bethesda, MD 20892 (NJE).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Provenance and Peer Review: This article was commissioned by the editorial office, Journal of Gastrointestinal Oncology for the series “Global GI Malignancies”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jgo-2019-gi-07). The series “Global GI Malignancies” was commissioned by the editorial office without any funding or sponsorship. JFG served as the unpaid Guest Editor of the series and serves as an unpaid editorial board member of Journal of Gastrointestinal Oncology from Jan 2019 to Dec 2020. The authors have no other conflicts of interest to declare.
References
- 1.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016;164:337-40. 10.1016/j.cell.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 2.Gilbert JA, Blaser MJ, Caporaso JG, et al. Current understanding of the human microbiome. Nat Med 2018;24:392-400. 10.1038/nm.4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Kaoutari A, Armougom F, Gordon JI, et al. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 2013;11:497-504. 10.1038/nrmicro3050 [DOI] [PubMed] [Google Scholar]
- 4.Kau AL, Ahern PP, Griffin NW, et al. Human nutrition, the gut microbiome and the immune system. Nature 2011;474:327-36. 10.1038/nature10213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013;152:39-50. 10.1016/j.cell.2012.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 2010;10:159-69. 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- 7.Chung H, Pamp SJ, Hill JA, et al. 2012 Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012;149:1578-93. 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han R, Ma J, Li H. Mechanistic and therapeutic advances in non-alcoholic fatty liver disease by targeting the gut microbiota. Front Med 2018;12:645-57. 10.1007/s11684-018-0645-9 [DOI] [PubMed] [Google Scholar]
- 9.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012;489:242-49. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 10.Karlsson F, Tremaroli V, Nielsen J, et al. Assessing the human gut microbiota in metabolic diseases. Diabetes 2013;62:3341-49. 10.2337/db13-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang D, Zhao D, Ali Shah SZ, et al. The role of the gut microbiota in the pathogenesis of parkinson's disease. Front Neurol 2019;10:1155. 10.3389/fneur.2019.01155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields CT, Sampson TR, Bruce-Keller AJ, et al. Defining dysbiosis in disorders of movement and motivation. J Neurosci 2018;38:9414-22. 10.1523/JNEUROSCI.1672-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800-12. 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott AJ, Alexander JL, Merrifield CA, et al. International Cancer Microbiome Consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut 2019;68:1624-32. 10.1136/gutjnl-2019-318556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costea PI, Hildebrand F, Arumugam M, et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol 2018;3:8-16. 10.1038/s41564-017-0072-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006;7:688-93. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamada N, Chen GY, Inohara N, et al. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013;14:685-90. 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiménez E, Marín ML, Martín R, et al. Is meconium from healthy newborns actually sterile? Res Microbiol 2008;159:187-93. 10.1016/j.resmic.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 20.Arboleya S, Binetti A, Salazar N, et al. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol 2012;79:763-72. 10.1111/j.1574-6941.2011.01261.x [DOI] [PubMed] [Google Scholar]
- 21.Wopereis H, Oozeer R, Knipping K, et al. The first thousand days - intestinal microbiology of early life: establishing a symbiosis. Pediatr Allergy Immunol 2014;25:428-38. 10.1111/pai.12232 [DOI] [PubMed] [Google Scholar]
- 22.Duncan SH, Louis P, Flint HJ. Lactate -utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 2004;70:5810-7. 10.1128/AEM.70.10.5810-5817.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Weerth C, Fuentes S, Puylaert P, et al. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics 2013;131:e550-8. 10.1542/peds.2012-1449 [DOI] [PubMed] [Google Scholar]
- 24.Lührs H, Gerke T, Müller JG, et al. Butyrate inhibits NF -kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol 2002;37:458-66. 10.1080/003655202317316105 [DOI] [PubMed] [Google Scholar]
- 25.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009;461:1282-6. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222-7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart CJ, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018;562:583-8. 10.1038/s41586-018-0617-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480-84. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franzosa EA, Huang K, Meadow JF, et al. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci U S A 2015;112:E2930-38. 10.1073/pnas.1423854112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cani PD. Human gut microbiome: hopes, threats and promises. Gut 2018;67:1716-25. 10.1136/gutjnl-2018-316723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet 2012;13:260-70. 10.1038/nrg3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plummer M, de Martel C, Vignat J, et al. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health 2016;4:e609-16. 10.1016/S2214-109X(16)30143-7 [DOI] [PubMed] [Google Scholar]
- 33.Levy M, Kolodziejczyk A, Thaiss C, et al. Dysbiosis and the immune system. Nat Rev Immunol 2017;17:219-32. 10.1038/nri.2017.7 [DOI] [PubMed] [Google Scholar]
- 34.Pei Z, Yang L, Peek RM, Jr, et al. Bacterial biota in reflux esophagitis and Barrett's esophagus. World J Gastroenterol 2005;11:7277-83. 10.3748/wjg.v11.i46.7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deshpande NP, Riordan SM, Castaño-Rodríguez N, et al. Signatures within the esophageal microbiome are associated with host genetics, age, and disease. Microbiome 2018;6:227. 10.1186/s40168-018-0611-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Lu X, Nossa CW, et al. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology 2009;137:588-97. 10.1053/j.gastro.2009.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv J, Guo L, Liu JJ, et al. Alteration of the esophageal microbiota in Barrett's esophagus and esophageal adenocarcinoma. World J Gastroenterol 2019;25:2149-61. 10.3748/wjg.v25.i18.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Francois F, Pei Z. Molecular pathways: pathogenesis and clinical implications of microbiome alteration in esophagitis and Barrett esophagus. Clinical Cancer Research 2012;18:2138-44. 10.1158/1078-0432.CCR-11-0934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaidi AH, Kelly LA, Kreft RE, et al. Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer 2016;16:52. 10.1186/s12885-016-2093-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.San-Millán I, Brooks GA. Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2017;38:119-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott DRF, Walker AW, O'Donovan M, et al. A non-endoscopic device to sample the oesophageal microbiota: a case-control study. Lancet Gastroenterol Hepatol 2017;2:32-42. 10.1016/S2468-1253(16)30086-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Gail MH, Shi J, et al. Association between upper digestive tract microbiota and cancer -predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev 2014;23:735-41. 10.1158/1055-9965.EPI-13-0855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nasrollahzadeh D, Malekzadeh R, Ploner A, et al. Variations of gastric corpus microbiota are associated with early esophageal squamous cell carcinoma and squamous dysplasia. Sci Rep 2015;5:8820. 10.1038/srep08820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao S, Li S, Ma Z, et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer 2016;11:3. 10.1186/s13027-016-0049-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamura K, Baba Y, Nakagawa S, et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res 2016;22:5574-81. 10.1158/1078-0432.CCR-16-1786 [DOI] [PubMed] [Google Scholar]
- 46.Blackett KL, Siddhi SS, Cleary S, et al. Oesophageal bacterial biofilm changes in gastro -oesophageal reflux disease, Barrett's and oesophageal carcinoma: association or causality? Aliment Pharmacol Ther 2013;37:1084-92. 10.1111/apt.12317 [DOI] [PubMed] [Google Scholar]
- 47.Shao D, Vogtmann E, Liu A, et al. Microbial characterization of esophageal squamous cell carcinoma and gastric cardia adenocarcinoma from a high -risk region of China. Cancer 2019;125:3993-4002. 10.1002/cncr.32403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C, Powell SE, Betel D, et al. The gastric microbiome and its influence on gastric carcinogenesis: current knowledge and ongoing research. Hematol Oncol Clin North Am 2017;31:389-408. 10.1016/j.hoc.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 49.Wang L, Zhou J, Xin Y, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol 2016;28:261-6. 10.1097/MEG.0000000000000542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castaño-Rodríguez N, Goh KL, Fock KM, et al. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep 2017;7:15957. 10.1038/s41598-017-16289-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018;67:226-36. 10.1136/gutjnl-2017-314205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yousefi B, Mohammadlou M, Abdollahi M, et al. Epigenetic changes in gastric cancer induction by Helicobacter pylori. J Cell Physiol 2019;234:21770-84. 10.1002/jcp.28925 [DOI] [PubMed] [Google Scholar]
- 53.Chen X, Winckler B, Lu M, et al. Oral microbiota and risk for esophageal squamous cell carcinoma in a high -risk area of China. PLoS One 2015;10:e0143603. 10.1371/journal.pone.0143603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watari J, Chen N, Amenta PS, et al. Helicobacter pylori associated chronic gastritis, clinical syndromes, precancerous lesions, and pathogenesis of gastric cancer development. World J Gastroenterol 2014;20:5461-73. 10.3748/wjg.v20.i18.5461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen XZ, Schottker B, Castro FA, et al. Association of helicobacter pylori infection and chronic atrophic gastritis with risk of colonic, pancreatic and gastric cancer: a ten-year follow-up of the ESTHER cohort study. Oncotarget 2016;7:17182-93. 10.18632/oncotarget.7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.International Agency for Research on Cancer. IARC Monographs of Carginogenic Hazards to Humans and Handbooks of Cancer Prevention. 2018. Available online: https://monographs.iarc.fr/wp-content/uploads/2019/01/OrganSitePoster.PlusHandbooks.17012019.pdf
- 57.Peek RM, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol 2006;208:233-48. 10.1002/path.1868 [DOI] [PubMed] [Google Scholar]
- 58.Uno Y. Prevention of gastric cancer by Helicobacter pylori eradication: a review from Japan. Cancer Med 2019;8:3992-4000. 10.1002/cam4.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atherton JC, Blaser MJ. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest 2009;119:2475-87. 10.1172/JCI38605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 2006;103:732-37. 10.1073/pnas.0506655103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu G, Torres T, Hu N, et al. Molecular characterization of the human stomach microbiota in gastric cancer patients. Front Cell Infect Microbiol 2017;7:302. 10.3389/fcimb.2017.00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andersson AF, Lindberg M, Jakobsson H, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 2008;3:e2836. 10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X, Shao L, Liu X, et al. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine 2019;40:336-48. 10.1016/j.ebiom.2018.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park CH, Lee AR, Lee YR, et al. Evaluation of gastric microbiome and metagenomic function in patients with intestinal metaplasia using 16S rRNA gene sequencing. Helicobacter 2019;24:e12547. 10.1111/hel.12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jo HJ, Kim J, Kim N, et al. Analysis of gastric microbiota by pyrosequencing: minor role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. Helicobacter 2016;21:364-74. 10.1111/hel.12293 [DOI] [PubMed] [Google Scholar]
- 66.Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest 2004;113:321-33. 10.1172/JCI20925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coker OO, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018;67:1024-32. 10.1136/gutjnl-2017-314281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eun CS, Kim BK, Han DS, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 2014;19:407-16. 10.1111/hel.12145 [DOI] [PubMed] [Google Scholar]
- 69.Correa P. Gastric cancer: overview. Gastroenterol Clin North Am 2013;42:211-7. 10.1016/j.gtc.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young RO. Who had their finger on the magic of life - Antoine Bechamp or Louis Pasteur? Int J Vaccines Vaccin 2016;2:00047. 10.15406/ijvv.2016.02.00047 [DOI] [Google Scholar]
- 71.Cook MB, Chow WH, Devesa SS. Oesophageal cancer incidence in the United States by race, sex, and histologic type, 1977-2005. Br J Cancer 2009;101:855-9. 10.1038/sj.bjc.6605246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.International Agency for Research on Cancer. Estimated number of new cases in 2018, worldwide, both sexes, all ages. Available online: http://gco.iarc.fr/today/online-analysis-table 72