Abstract
Mitochondrial shape rapidly changes by dynamic balance of fusion and fission to adjust to constantly changing energy demands of cancer cells. Mitochondrial dynamics balance is exactly regulated by molecular motor consisted of myosin and actin cytoskeleton proteins. Thus, targeting myosin–actin molecular motor is considered as a promising strategy for anti-cancer. In this study, we performed a proof-of-concept study with a natural-derived small-molecule J13 to test the feasibility of anti-cancer therapeutics via pharmacologically targeting molecular motor. Here, we found J13 could directly target myosin-9 (MYH9)–actin molecular motor to promote mitochondrial fission progression, and markedly inhibited cancer cells survival, proliferation and migration. Mechanism study revealed that J13 impaired MYH9–actin interaction to inactivate molecular motor, and caused a cytoskeleton-dependent mitochondrial dynamics imbalance. Moreover, stable isotope labeling with amino acids in cell culture (SILAC) technology-coupled with pulldown analysis identified HSPA9 as a crucial adaptor protein connecting MYH9–actin molecular motor to mitochondrial fission. Taken together, we reported the first natural small-molecule directly targeting MYH9–actin molecular motor for anti-cancer translational research. Besides, our study also proved the conceptual practicability of pharmacologically disrupting mitochondrial fission/fusion dynamics in human cancer therapy.
KEY WORDS: Anti-cancer, Liver hepatocellular carcinoma, Mitochondrial fission, Small molecule, Target identification, Molecular motor, MYH9, HSPA9
Abbreviations: CAM, chick embryo chorioallantoic membrane; CETSA, cellular thermal shift assay; Co-IP, co-immunoprecipitation; DAPI, 4′,6-diamidino-2-phenylindole; ER, endoplasmic reticulum; HE, hematoxylin–eosin staining; HSPA9, heat-shock protein A9; HUVEC, human umbilical vein endothelial cells; IHC, immunohistochemistry; LIHC, liver hepatocellular carcinoma; MMP, mitochondrial membrane potential; MYH9, myosin-9; SILAC, stable isotope labeling with amino acids in cell culture; SPR, surface plasmon resonance; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
Graphical abstract
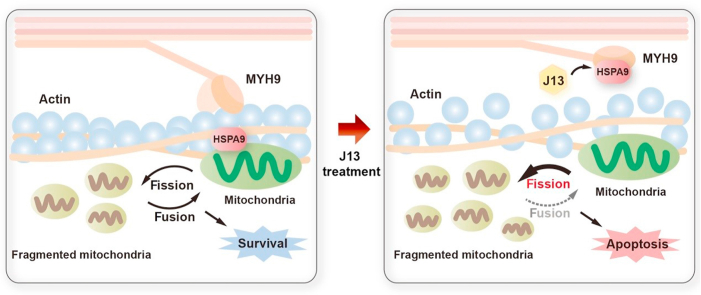
Natural-derived small-molecule J13 restrained the interaction between myosin-9 (MYH9) and actin molecular motor via prompting heat-shock protein A9 (HSPA9) attached to MYH9, resulting in excessive mitochondrial fission with implications for human cancer therapy.
1. Introduction
Mitochondria are crucial organelles to produce energy for efficient maintenance of cell functions1,2. Mitochondria possess capability of changing self-morphology, which is called mitochondrial dynamics, in response to external dangerous stimuli including oxidative stress, nutrient deprivation, UV radiation, and toxic chemicals3,4. Mitochondrial dynamics is regulated by two opposed processes: mitochondrial fission and fusion, which are well conserved between mammals, flies and yeast5. Appropriate fission/fusion dynamics is critical to maintain mitochondrial homeostasis, and is accurately controlled in cells6,7. Mitochondrial fission plays a key role in mitochondrial movement to the regions that lack of energy by forming smaller fragmented mitochondria. Meanwhile, mitochondria tend to fuse together under damaged condition to share nutrients and genetic mtDNA for preserving normal oxidative phosphorylation and ATP energy supply8. Dysregulation of mitochondrial fission/fusion process is widely implicated in various cell stresses, and seriously affects mitochondrial quality control, which has been linked to various human diseases especially for cancers9, 10, 11, 12.
Nowadays, specifically targeting mitochondrial dynamics has become an attractive issue in cancer therapy13. A consensus has emerged that increased mitochondrial fission is a pro-tumorigenic phenotype. Thus, inhibition of mitochondrial fission has been considered as a potential strategy for cancer treatment10,11,14, 15, 16, 17. However, limited attempts have been reported so far in clinical studies, implying a potential high risk and uncertainty of this anti-cancer strategy. Curiously, it has been recently reported that an excess of fission promotes mitochondrial dysfunction, which may play an unfavorable role in cancer cell growth18. Therefore, pharmacological induction of mitochondrial fission could provide novel insight into anti-cancer therapy, but this concept has not yet been adequately validated.
Currently, mitochondrial fission/fusion dynamics modulation mechanism has not been widely clarified19. Accumulating evidence show that molecular motor proteins are master regulators powering mitochondrial dynamics in living organisms20,21. Of note, myosin is a crucial molecular motor protein that slides along actin filaments by utilizing energy from ATP hydrolysis22. Previous report suggests that myosin and actin could form archetypical molecular motor complex to regulate mitochondria fission/fusion dynamics, morphology and cellular localization23,24. Genetic downregulation of myosin markedly suppresses mitochondrial fission to promote elongated mitochondria number25. Intriguingly, it has been also found that mitochondrial fission occurs at mitochondria-endoplasmic reticulum (ER) contact sites with F-actin accumulation, which drives dynamin-related protein DRP1 recruitment to fuel mitochondrial fission26. Therefore, it is significant to explore potential strategy for regulating mitochondrial fission via targeting myosin–actin molecular motor24.
In this study, we discovered a natural-derived small-molecule J13 from medicinal plant Ablizia julibrissin with obvious anti-hepatoma carcinoma effects in vitro and in vivo by directly targeting myosin-9 (MYH9)–actin molecular motor. Further investigation showed that J13 bound to actin-binding motif in MYH9 to impair MYH9 interaction with actin and induced molecular motor dysfunction for mitochondrial fission progression. Moreover, heat-shock protein A9 (HSPA9) was identified as a critical substrate protein of MYH9–actin molecular motor for modulating mitochondrial fission. In addition, high expressions of MYH9 and HSPA9 were associated with poor progression and prognosis in liver hepatocellular carcinoma (LIHC). Collectively, for the first time, our proof-of-concept study revealed that J13 was a novel molecule template for regulating molecular motor function. Moreover, MYH9–actin molecular motor may represent a promising pharmacological therapeutic target of mitochondrial dynamics-related diseases especially for human cancers.
2. Materials and methods
2.1. Chemicals and reagents
Julibraside J13 was previously separated and purified from Ablizia julibrissin by our laboratory27, and the purity is above 98% by HPLC analysis. Fatty acid-free bovine serum albumin (BSA) was purchased from Equitech-Bio (Kerrville, TX, USA). 3-[4,5-Dimethylthiazol-2-yl] 2,5-diphenyltetrazolium bromide (MTT) and recombinant actin and myosin heavy chain were obtained from Sigma Chemical (Santa Clara, CA, USA). TransDetect Annexin V-FITC/PI cell kit was obtained from Transgene (Beijing, China). Hoechst 33258 staining kit and mitochondrial membrane potential assay kit were obtained from Beyotime (Nanjing, China). Fetal bovine serum (FBS) was from PAN-Biotech (Logan, UT, USA). High glucose Dulbecco's modified Eagle's medium (DMEM, 4.5 g/L glucose), antibiotics and trypsin were from Macgene (Beijing, China). Primary antibodies and secondary antibodies were from Cell Signaling Technology (Beverly, MA, USA). Chemiluminescent HRP substrate was purchased from Thermo Scientific (Waltham, MA, USA).
2.2. Methods
2.2.1. Cell culture
Human hepatoma cell lines (HepG2/Hepa1-6/Bel7402/SMMC7721), human umbilical vein endothelial cells (HUVEC), human embryonic kidney 293 cells (HEK293) and mouse hepatoma cell line (H22) were obtained from the State Key Laboratory of National and Biomimetic Drugs, Peking University Health Science Centre (Beijing, China), and cultured in DMEM supplemented with 10% FBS, 100 U/mL streptomycin at 37 °C in a humidified incubator with 95% air and 5% CO2.
2.2.2. Plasmids and siRNA
The plasmids of HA-MYH9, His-HSPA9, MYH9 F (83–764 aa) and MYH9 M (del 654–676 aa) were constructed by Biogot Technology (Nanjing, China). Specific MYH9 siRNAs were designed and synthesized at GenePharma (Shanghai, China) as listed in Supporting Information Table S1. The indicated plasmids or siRNA were transfected into HepG-2 cells using Lipofectamine 2000 (Invitrogen, Waltham, CA, USA) or Lipofectamine RNAiMAX (Thermo Scientific, Waltham, MA, USA) respectively. After 48 h, cells were collected for further experiments.
2.2.3. Cell viability assay
Cell viability was detected via colorimetric MTT assay. After treatment, MTT solutions (5 mg/mL) were added to each well. After incubation for 2 h, supernatants were aspirated and formazan crystals were dissolved in DMSO. The amount of purple formazan was assessed by measuring the absorbance at 570 nm using Infinite F50 Absorbance Microplate Reader (Tecan, Mannedorf, Switzerland).
2.2.4. Hoechst 33258 staining assay
Hoechst 33258 staining assay was used to visualize nuclear change and apoptotic body formation. After treatment, cells were washed with PBS and fixed with 4% formaldehyde at 4 °C for 20 min. The fixing solution was removed and cells were washed twice with PBS before staining with Hoechst 33258 (5 μg/mL). After 5 min, cells were washed and observed under a fluorescence microscope (IX73, Olympus, Tokyo, Japan) with excitation at 360 nm and emission at 460 nm.
2.2.5. Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay
TUNEL assay kit (Life, Waltham, MA, USA) was employed to quantify DNA strand breaks in apoptosis. Briefly, after deparaffinization, rehydration, permeabilization and blocking, tumor tissues or HepG2 cells were incubated with TUNEL reaction mixture at 37 °C for 1 h in a humid and dark condition. For staining of nuclei, cells were treated with 4′,6-diamidino-2-phenylindole (DAPI) for 15 min at room temperature. After washing with PBS, tissue or cell samples were imaged by an IX73 fluorescence microscope (Olympus, Tokyo, Japan). DAPI was visualized at 358 nm (excitation) and 460 nm (emission). FITC was visualized at 488 nm (excitation) and 516 nm (emission). Percentage of number of TUNEL-positive cells (green) to total cells (DAPI-positive cells, blue) was calculated as the percentage of apoptotic cells.
2.2.6. Mitochondrial membrane potential (MMP) analysis
After treatment, cells were incubated with 10 μg/mL JC-1 solution for 20 min at 37 °C. Then, cells were washed and observed with the Olympus IX73 fluorescence microscope (Olympus, Tokyo, Japan). Green fluorescence was detected with 488 nm (excitation) and 525 nm (emission) filters, whereas red fluorescence was visualized with 488 nm (excitation) and 590 nm (emission) filters.
2.2.7. Preparation of J13-tagged beads
Epoxy-actived Sepharose 4B beads (GE Healthcare, Chicago, IL, USA) were added into stilled water and left overnight to allow beads completely swelled. Beads were then incubated with J13 or solvent for 12 h under constant rotation at 37 °C. After remove of supernatants through centrifugation at 3000 rpm (6380R, Eppendorf, Hamburg, Germany) for 2 min, residual active sites were quenched using ethanolamine solution (1 mol/L, pH 8.5) for 2 h with agitation. J13-tagged beads were washed with stilled water and stored at 4 °C until further use.
2.2.8. Cellular target identification of J13
Cell lysates were incubated with J13-tagged beads at 4 °C for 12 h. Meanwhile, an excess amount of J13 for competition was added into lysates. Target proteins were specifically bound to J13-tagged beads and then eluted by boiling in SDS-loading buffer for 10 min. Supernatants were obtained and subjected to SDS-PAGE followed by silver staining. The bands with obvious difference upon J13 treatment were separated, trypsin-digested and identified by LC–MS/MS analysis as described in our previous publication28.
2.2.9. Cellular thermal shift assay (CETSA)
Cells were collected and freeze-thawed three times using liquid nitrogen. Cell suspensions were divided into 2 aliquots and treated with or without J13 (5 μmol/L) for 1 h at room temperature. Each aliquot was heated at indicated temperature (38–64 °C) for 3 min followed by cooling for 3 min. Supernatants were isolated and analyzed by Western blot. Additionally, for living cell CETSA experiment, cells were treated with or without J13 (5 μmol/L) for 6 h, harvested, and heated separately at indicated temperature (38–64 °C) followed by 3 min cooling. Cells were collected and freeze-thawed three times using liquid nitrogen. Supernatants were analyzed by Western blot.
2.2.10. Surface plasmon resonance (SPR) assay
SPR measurement was performed on Biocore T200 system (GE Healthcare, Boston, USA) using a carboxymethylated 5 sensor chip. Recombinant MYH9 and β-actin proteins were dissolved in running buffer (Tris–HCl 50 mmol/L, pH 7.2, 100 mmol/L KCl). Protein samples were immobilized on chip via amine coupling chemistry in 10 mmol/L sodium acetate at pH 4.5. Ligand solutions were injected as analytes. Data were analyzed with Biocore evaluation software (T200 Version 2.0) using 1:1 langmuir model to fit kinetic data.
2.2.11. Tryptophan fluorescence quenching assay
Recombinant MYH9 and β-actin proteins were diluted to 500 μmol/L with PBS, and mixed with gradient concentration of J13 (0.39–100 μmol/L). Tryptophan fluorescence was monitored with λex 278 nm and λem 300–500 nm at 1 nm increment on the PerkinElmer fluorescence spectrophotometer (PerkinElmer EnSpire, Waltham, MA, USA). Fluorescence intensities were corrected by buffer contribution.
2.2.12. Co-immunoprecipitation (co-IP) analysis
For co-IP analysis, protein lysates of HepG2 cells were treated with protein A/G agarose for 1 h at 4 °C and centrifuged to obtain supernatants. Samples were incubated with specific primary antibodies at 4 °C overnight and subsequent protein A/G agarose beads for 4 h at 4 °C. Beads were centrifuged and washed five times with IP buffer (0.01% Triton X-100). The immunoprecipitated proteins were eluted by boiling in SDS-loading buffer for 10 min. Final eluate was subjected to SDS-PAGE and analyzed by Western blot.
2.2.13. Molecular docking analysis
Three-dimensional structure of J13 was built in Open Babel software. Since X-ray crystal structures of human β-actin and MYH9 have not been reported, we searched for Protein Data Bank (PDB) (http://www.ncbi.nlm.nih.gow/protein/) to find a proper structural template for homology modeling. We used crystal structure of human α-actin (PDB ID code: 5JLH, chain A) and human MYH14 (PDB ID code: 5JLH, chain F) as input. Homology model was generated in SWISS-MODEL web server as the receptor model in docking stimulation. J13 was placed into the indicated cave which was identified in our study during molecular docking procedure. All calculations were carried out using Autodock Vina software, and Pymol software was used to analyze interaction types of docked proteins with ligand.
2.2.14. Actin polymerization assay
G-actin (200 μg/mL) was mixed with Tris–HCl (pH 8.0), 2 μmol/L FITC-phalloidin methanol solution, 20 mmol/L ATP and different concentrations of J13 (0–0.1 μmol/L). Reaction was started by adding actin polymerization solution (50 mmol/L KCl, 1 mmol/L MgCl2), and continuously detected on the PerkinElmer fluorescence spectrometer (PerkinElmer EnSpire, Waltham, MA, USA) for 15 min (λex 545 nm, λem 570 nm).
2.2.15. Immunofluorescence and morphological staining
For immunofluorescence staining of G-actin, HSPA9 and COX IV, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. After blocking with 1% BSA in PBS for 20 min, cells were incubated with primary antibodies at 4 °C overnight. After incubation, cells were incubated with anti-rabbit IgG Alexa Fluor 488 or anti-mouse IgG Alexa Fluor 594 for 1 h at room temperature. Nuclei were stained with DAPI (50 μg/mL, 358 nm/460 nm) for 15 min at room temperature. For morphological staining of filamentous actin (F-actin), mitochondria and endoplasmic reticulum (ER), cells were permeabilized and blocked as immunofluorescence staining. Then, cells were incubated with actin–tracker Green (5 μmol/L, 496 nm/516 nm), Mito–tracker Green (200 nmol/L, 490 nm/516 nm) or ER–tracker Red (1 μmol/L, 504 nm/511 nm), according to the manufacturers’ instructions. After being stained with DAPI, cells were imaged on Leica TCS-SP5 confocal laser-scanning microscope (Wetzlar, Germany).
2.2.16. Cell migration assay
Migration assay was carried out using a Transwell with an 8 μm pore size. Briefly, HepG2 cells were seeded onto apical side of Transwell membrane in serum-free medium with or without J13. Medium containing 10% serum was added in basolateral side. Following incubation at 37 °C in 5% CO2 for 48 h, noninvasive cells were removed by a cotton swab. Membrane containing the migrated cells was fixed with 4% paraformaldehyde for 20 min and stained with 1% crystal violet for 20 min at room temperature. Images were taken in light field by an IX73 fluorescence microscope (Olympus, Tokyo, Japan) and the migrated cells were quantitated with Image J densitometric software.
2.2.17. Tube formation assay
Matrigel (100 μL, Corning, Kennebunk, ME, USA) was added into 96-well plate, and polymerized for 30 min at room temperature. HUVECs were seeded into each well with or without J13. After 6 h incubation, cells were fixed and tube formation was analyzed by an IX73 fluorescence microscope (Olympus, Tokyo, Japan).
2.2.18. Chick embryo chorioallantoic membrane (CAM) assay
Anti-angiogenesis study was evaluated through CAM assay. Windows were cut on egg shells of 8-day-old chick embryos (Vital River, Beijing, China) and resealed with a transparent film to be incubated at 37 °C for further 5 days. During the incubation, J13 or solvent was dropped onto chick embryo chorioallantoic membranes once a day. Then, membranes were fixed with methanol/acetone at ratio of 1:1 for 20 min. Images of the membranes were obtained by an IX73 inverted microscope (Olympus, Tokyo, Japan) and vessel ramification rate was counted by Image J densitometric software.
2.2.19. Western blot assay
Protein samples were separated by SDS-PAGE and transferred onto PVDF membranes. Then, membranes were incubated with primary antibodies at 4 °C for 12 h, followed by incubation with secondary antibody for 1 h at room temperature. Membranes were washed and developed using enhanced chemiluminescence, and detected with Tanon 5200 Imaging Analysis System (Tanon, Shanghai, China).
2.2.20. In vivo anti-tumor studies
2.2.20.1. Animals
Male ICR mice (5–6 weeks old, weight 20–22 g) were obtained from Vital River Laboratories (Beijing, China), and raised in animal house at 25 ± 1 °C under constant dark and light cycles. ICR mice had free access to standard laboratory food and water. All animal experiments were performed under the guidelines of EIACUC-PKU (Ethical Institutional Animal Care and Use Committee of Peking University).
2.2.20.2. Anti-tumor effects evaluation in vivo
After 5 days of acclimatization, H22 cells (1 × 107/mL) were resuspended in saline (0.2 mL) and injected into the flank of ICR mice to form subcutaneous tumor. Five days later, mice were sorted to give different treatments: control-ip (normal saline, intraperitoneal injection), control-ig (normal saline, intragastric administration), J13-ip (0.5, 1.0 and 2.0 mg/kg per day, intraperitoneal injection), J13-ig (1.0, 2.0 and 4.0 mg/kg per day, intragastric administration), and cyclophosphamide (CTX, 30 mg/kg per day, intraperitoneal injection). Animal body weights were measured every day. After 12 days, tumors were weighed, fixed in 4% paraformaldehyde and embedded in paraffin for HE staining and immunohistochemistry (IHC) analysis.
2.3. Statistical analysis
Data were represented as mean ± standard deviation (SD). Statistical analysis was performed by one-way ANOVA with Tukey's multiple comparison post-test. Statistical significance between two groups was performed using Student's t-test. A probability value of P < 0.05 was considered as statistically significant.
3. Results
3.1. MYH9–actin molecular motor is a direct cellular target of anti-cancer small-molecule J13
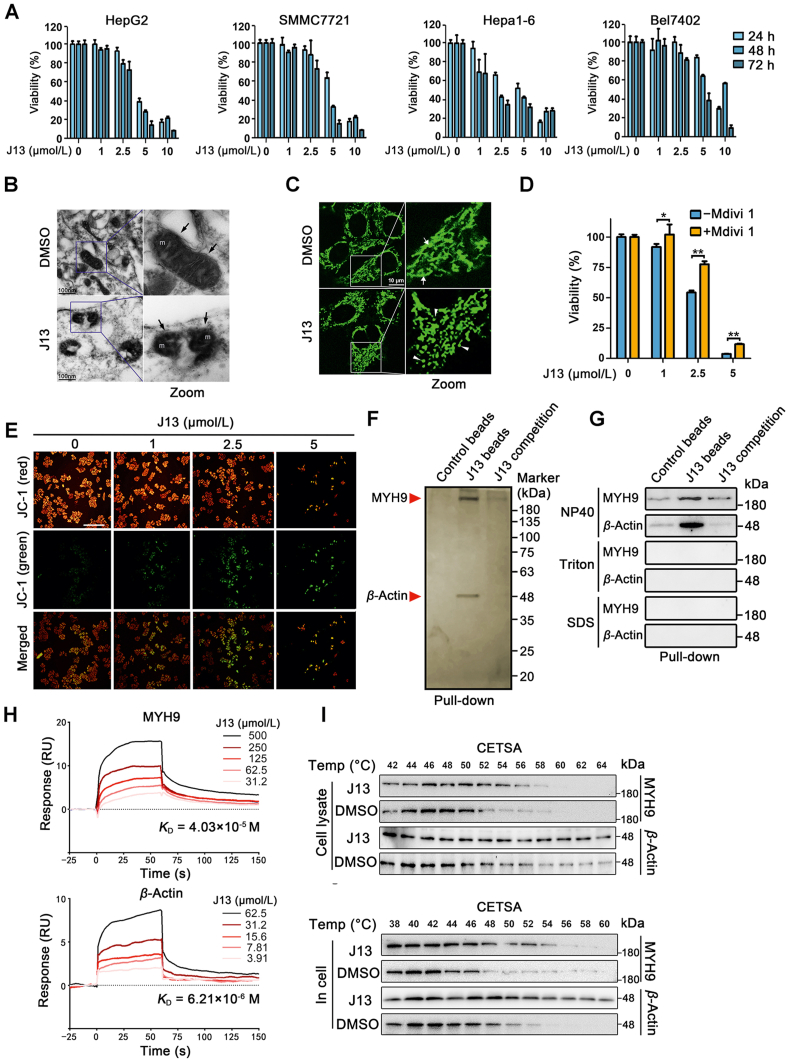
To identify effective anti-cancer drugs that might disrupt mitochondrial dynamics, we previously conducted an activity screen of a natural compound library consisting of around 4500 small molecules. Small-molecule J13 was isolated from medicinal plant Ablizia julibrissin. MTT assay revealed that J13 exerted obvious anti-hepatoma carcinoma effect in multiple cancer cells (Fig. 1A) but with low toxicity on normal hepatocytes (Supporting Information Fig. S1). We then investigated the mitochondrial morphology change upon J13 treatment. Strikingly, mitochondrial ultrastructure analysis illustrated distorted cristae and many fragmented mitochondria in response to J13 (Fig. 1B). Moreover, mitochondrial fission was also determined by Mito-tracker staining. As shown in Fig. 1C, J13 obviously caused an increase in mitochondria stained by Mito-tracker from elongated and tubular mitochondria into dots, implicating that mitochondrial morphology shifted towards fission. Inhibition of mitochondrial fission by Mdivi-1 markedly reversed J13-dependent anti-cancer effects, suggesting that mitochondrial fission is involved in J13-induced hepatoma carcinoma HepG2 cell death (Fig. 1D). Mitochondrial dynamics impairment contributes to mitochondrial membrane potential (MMP) decrease. Our findings show that J13 significantly upregulated JC-1 green fluorescence in HepG2 cells (Fig. 1E), reflecting an increase of depolarized mitochondria. In addition, immunofluorescence analysis indicates that J13 promoted mitochondrial fission protein DRP-1 translocation without affecting its expression (Supporting Information Fig. S2A and S2B). Therefore, these observations indicated that J13 exerted anti-cancer activities by potentially promoting mitochondrial fission.
Figure 1.
MYH9-actin molecular motor is a direct cellular target of anti-cancer small-molecule J13. (A) J13 inhibited cell viability of HepG2, SMMC7721, Hepa1-6 and Bel7402 cells (n = 6). (B) J13 induced mitochondrial fission in HepG2 cells. Mitochondrial morphology of HepG2 after J13 treatment was detected by transmission electron microscopy scanning (scale bar = 100 nm). (C) J13 caused mitochondrial fragmentation in HepG2 cells, which was visualized by Mito-tracker staining (scale bar = 10 μm). (D) Inhibition of mitochondrial fission by Mdivi-1 markedly reversed J13-dependent anti-cancer effects by MTT assay (n = 6). (E) J13 induced mitochondrial depolarization by JC-1 staining (scale bar = 25 μm). (F) MYH9 and β-actin were identified as J13-binding proteins using pull-down assay, followed by LC–MS/MS analysis with silver staining. (G) Non-covalent bindings of J13 with MYH9 or β-actin were confirmed by immunoblotting assay with different surfactants NP40, Triton, and SDS. (H) Dissociation constants (KD) of J13 with MYH9 or β-actin were detected by surface plasmon resonance (SPR) analysis. (I) and (J) Cellular thermal shift assays (CETSA) were performed to confirm the binding specificity of J13 with MYH9 or β-actin in cell lysates and living cells. Data are presented as mean ± SD from independent experiments; ∗P < 0.05, ∗∗P < 0.01.
To explore direct cellular target of J13, we prepared J13-coupled agarose beads to capture target proteins from HepG2 cell lysate. Notably, we identified two obvious protein bands (220 kDa for MYH9 and 48 kDa for β-actin) by MS/MS in SDS-PAGE stained with silver (Fig. 1F), which was confirmed by Western blot. Since MYH9 and β-Actin belong to molecular motor proteins, we then used different surfactants (NP40, Triton and SDS) to explore the binding characteristic of J13 to MYH9 and β-actin. As shown in Fig. 1G, rigorous surfactants Triton and SDS obviously inhibited J13 binding to MYH9 and β-actin, but not for weak surfactant NP40, indicating non-covalent bindings of J13 with MYH9 and β-actin. Then surface plasmon resonance (SPR) analysis was performed to show specific bindings of J13 to MYH9 and β-actin (Fig. 1H). Moreover, cellular thermal shift assay (CETSA) showed that J13 markedly promoted MYH9 and β-actin protein stability in HepG2 lysate and cultured HepG2 cells (Fig. 1I and G). Collectively, these findings suggest that J13 directly targeted molecular motor proteins of MYH9 and β-actin in human hepatoma cells.
3.2. J13 suppresses MYH9–actin interaction to promote mitochondrial fission
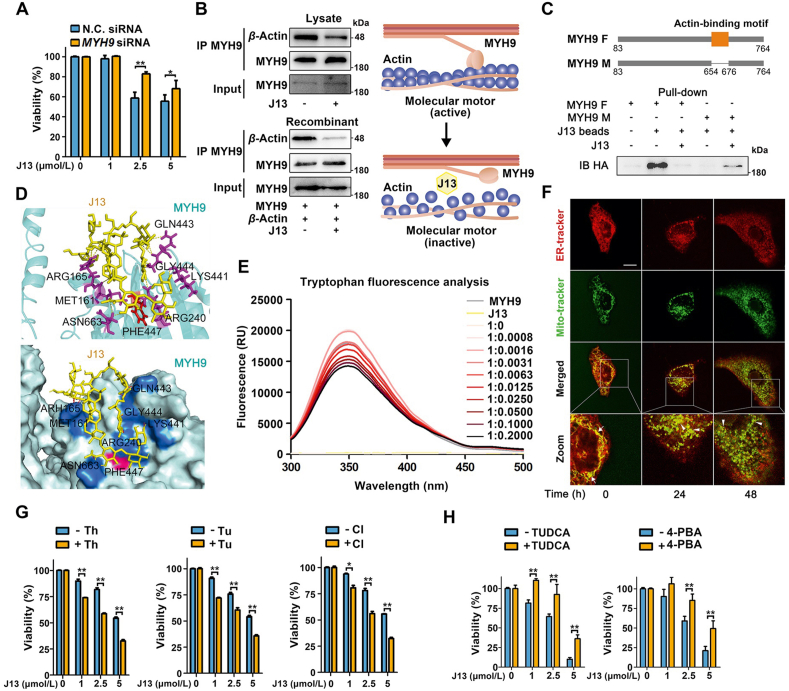
Since molecular motor proteins play crucial roles in mitochondrial morphology and transport, we then hypothesized that J13 may promote mitochondrial fission to exert anti-cancer effect in a MYH9-dependent manner. As shown in Fig. 2A, genetic knockdown of MYH9 significantly reversed J13-mediated cell viability decrease. Moreover, downregulation of MYH9 could significantly blunt J13-induced mitochondrial fission (Fig. S2D), indicating that MYH9 is of importance for J13-induced anti-cancer effects. Since MYH9–actin interaction contributes to mitochondrial dynamics, we thus explored whether J13 regulated MYH9–actin interaction. Here, co-IP assay suggested that J13 significantly inhibited MYH9 interaction with β-actin in cell lysate and recombinant proteins mixture (Fig. 2B), underlining the potential importance of MYH9–β-actin interaction in J13-mediated mitochondrial fission.
Figure 2.
J13 suppresses direct MYH9–actin interaction to promote mitochondrial fission. (A) MYH9 knock-down reversed J13-dependent cell viability decrease in MTT assay. N.C.: knock-down negative control. (B) J13 suppressed the interaction of MYH9 with β-actin by co-IP assay. (C) Pull-down assay revealed that J13 interacted with actin-binding motif of MYH9. MYH9 F and M: full length MYH9 and mutated MYH9 with a deletion of 23 amino acids (654–676). (D) Docking analysis shows the amino acid residues which may form hydrogen bonds with J13 including ARG165, MET161, ARG240, LYS441, GLN443, GLY444, PHE447, and ASN663. (E) J13 induced allosteric effect of MYH9 by tryptophan fluorescence scanning analysis. (F) J13 suppressed the co-location of ER with mitochondria by double fluorescence staining analysis (ER-tracker, red; Mito-tracker, green; scale bar = 10 μm). (G) ER stress inducers including tunicamycin (Tu), thapsigargin (Th), and calcium ionophore III (CI) exacerbated J13-dependent anti-tumor effect by MTT assay. (H) ER stress inhibitor TUDCA and 4-PBA efficiently reversed J13-induced cell viability decrease in HepG2 cells. All data are presented as mean ± SD, n = 6; ∗P < 0.05, ∗∗P < 0.01.
To explore the binding site of J13 to MYH9, we hypothesized that J13 influenced MYH9–actin interaction via actin-binding motif in MYH9. To this end, we compared the difference of J13 binding capacity with MYH9 full-length (MYH9 F) or MYH9 with actin-binding motif deletion (MYH9 M). As shown in Fig. 2C, J13 exhibited an obvious binding to MYH9 F, but a weaker interaction with MYH9 M, demonstrating that actin-binding motif was essential for J13 interaction with MYH9. Then, docking analysis showed that J13 spatially bond around actin-binding motif by forming several hydrogen bonds with arginine (ARG) 165, methionine (MET) 161, arginine (ARG) 240, lysine (LYS) 441, glycine (GLN) 443, glycine (GLY) 444, phenylalanine (PHE) 447 and asparagine (ASN) 663 (Fig. 2D). Thus, we proposed that J13 may engage the actin-binding interface in MYH9 to impair dynamical MYH9–actin interaction. Intriguingly, we observed an obvious allosteric regulation effect of J13 on MYH9 conformation change using fluorescence analysis of tryptophan (Fig. 2E), indicating that J13 may mediate MYH9 binding to β-actin via a putative conformational mechanism.
Since mitochondrial dynamics interferes with mitochondria-ER communication to cause ER stress, we also investigated whether J13-mediated mitochondrial dynamics imbalance impaired ER-mitochondria contact and fueled ER stress. As shown in Fig. 2F, J13 significantly suppressed ER (red)-mitochondria (green) coupling in a time-dependent manner, which was indicated by a decreasing overlap of red/green fluorescence. The reduction of ER-mitochondria contact sites may synergistically enhance J13-induced effects by inducing ER stress. To confirm whether J13-mediated mitochondrial fission contributed to ER stress, we performed MTT assays with ER stress inducers. As shown in Fig. 2G, we observed a notable synergistic inhibition effect of J13 on HepG2 cells with tunicamycin (Tu), thapsigargin (Th) and calcium ionophore III (CI), demonstrating that ER stress may be also a crucial driver for J13-dependent anti-cancer effect. Moreover, we performed MTT assay with two classic ER stress inhibitor TUDCA and 4-PBA, with or without J13, respectively. As shown in Fig. 2H, ER stress inhibitors efficiently reversed J13-induced cell viability decrease in HepG2 cells, further confirming our previous speculation that ER stress indeed is induced by J13 treatment for anti-cancer effect.
3.3. HSPA9 is a MYH9–actin molecular motor substrate protein for controlling mitochondrial dynamics
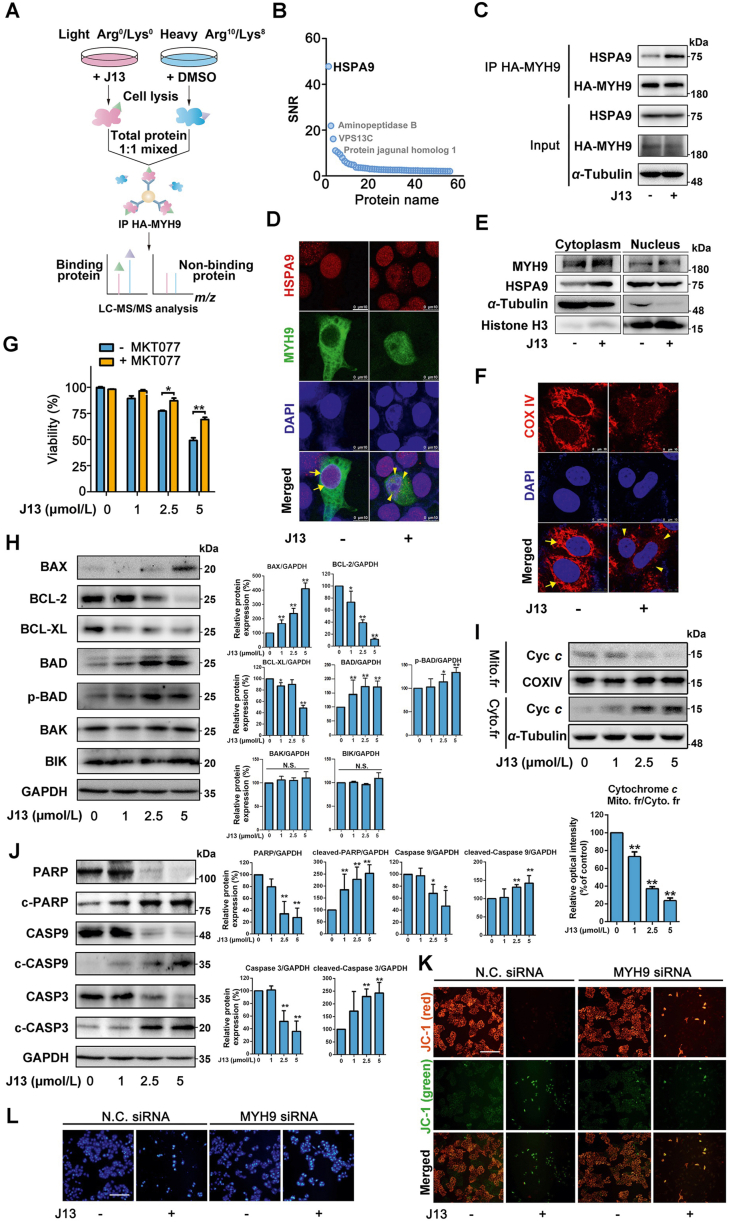
To explore the inter-mediators connecting MYH9–actin molecular motor to mitochondrial dynamics, we performed SILAC-based co-IP experiment to capture potential binding proteins (Fig. 3A). As shown in Fig. 3B, a 74 kDa heat-shock protein A9 (HSPA9) was identified as the most specific interacting protein of MYH9, which was confirmed by co-IP assay with specific antibodies (Fig. 3C). Immunofluorescence analysis also revealed an interesting phenomenon that J13 promoted cytoplasm translocation of HSPA9 to increase its co-location with MYH9 (Fig. 3D). Moreover, we performed cytoplasmic and nuclear fractions separation and found that HSPA9 was preferentially located in the nuclear fraction of control cells. However, upon J13 treatment, HSPA9 was considerably increased in cytosol, and thereby promoting co-location of MYH9 with HSPA9 (Fig. 3E). This observation was supported by previous reports that HSPA9 was a ubiquitously expressed mitochondrial chaperon which facilitated ER membrane interaction with mitochondrial outer membrane29. Then, we observed that J13 significantly decreased fluorescence intensity and disrupted fluorescence distribution of mitochondrial membrane protein cytochrome c oxidase subunit IV (COXIV), indicating an increase of mitochondrial fragmentation (Fig. 3F). To explore whether HSPA9 was involved in J13-meidated decrease of cell viability, HSPA9 inhibitor MKT-077 was used in our study. As shown in Fig. 3G, MKT-077 showed obvious antagonistic effect against J13-mediated decrease of cell viability, suggesting that HSPA9 was a crucial signal hub in regulating MYH9–actin molecular motor-dependent cell survival. Taken together, HSPA9 may serve as a support (scaffold) or “sponge” to provide a platform for protein interactions, more precisely, inactivating the interaction between MYH9 and β-actin.
Figure 3.
HSPA9 is a crucial MYH9–actin molecular motor substrate protein for controlling mitochondrial dynamics. (A) Schematic illustration of MYH9-binding proteins identification using pull-down coupled with SILAC analysis. (B) HSPA9 was identified as a binding protein of MYH9. (C) J13 promoted the interaction of HSPA9 with MYH9 by co-IP assay. (D) J13 promoted MYH9 interaction with HSPA9 in nucleus. (E) J13 treatment promoted cytosol translocation of HSPA9, and thus increased co-location of MYH9 with HSPA9. (F) J13 induced mitochondrial fragmentation by disrupting COXIV fluorescence distribution. (G) HSPA9 inhibitor MKT-077 (0.5 μmol/L for 24 h) reversed J13-induced cell viability decrease by MTT assay. (H) J13 significantly induced BAX and p-BAD expressions, and inhibited BCL-2 and BCL-XL expressions. (I) J13 induced cytochrome c translocation from mitochondria to cytoplasm. (J) J13 activated caspase-9/3-dependent apoptosis pathway. (K) MYH9 knock-down reversed J13-dependent mitochondrial depolarization (scale bar = 25 μm). (L) MYH9 knock-down rescued J13-dependent cell apoptosis by Hoechst 33258 staining assay (scale bar = 25 μm). All data are presented as mean ± SD from independent experiments (G: n = 6, H–J: n = 3; ∗P < 0.05, ∗∗P < 0.01.
Previous reports have revealed that BCL-2 family proteins are regulated by HSPA9 for mitochondrial function control30. STRING analysis suggested that BCL-2 family proteins may be functionally associated with HSPA9 (Supporting Information Fig. S3A). In our study, J13 obviously increased BAX and p-BAD levels, meanwhile decreased BCL-2 and BCL-XL levels in a concentration-dependent manner (Fig. 3H), indicating that BCL-2 family members might be affected by J13 for mitochondrial dysfunction. This finding was also supported by the promotion effect of J13 on cytoplasm translocation of cytochrome c from mitochondria (Fig. 3I). Moreover, J13 induced mitochondria-dependent caspase-9/3 apoptosis pathway activation by up-regulating cleaved caspase-9/-3/PARP expressions (Fig. 3J and Fig. S3B‒S3D). Next, we sought to explore whether MYH9 was a crucial determinant of mitochondrial function. Genetic knockdown of MHY9 markedly reversed JC-1 green fluorescence (Fig. 3K), indicating that MYH9 plays a fundamental role in J13-dependent MMP decline. MYH9 knockdown also markedly suppressed J13-mediated apoptosis in HepG2 cells (Fig. 3L). In summary, these observations suggest that HSPA9–BCL-2 family signal axis is critical to MYH9 mediated-mitochondrial dynamics for apoptosis.
3.4. J13 inhibits actin-dependent cell migration and angiogenesis
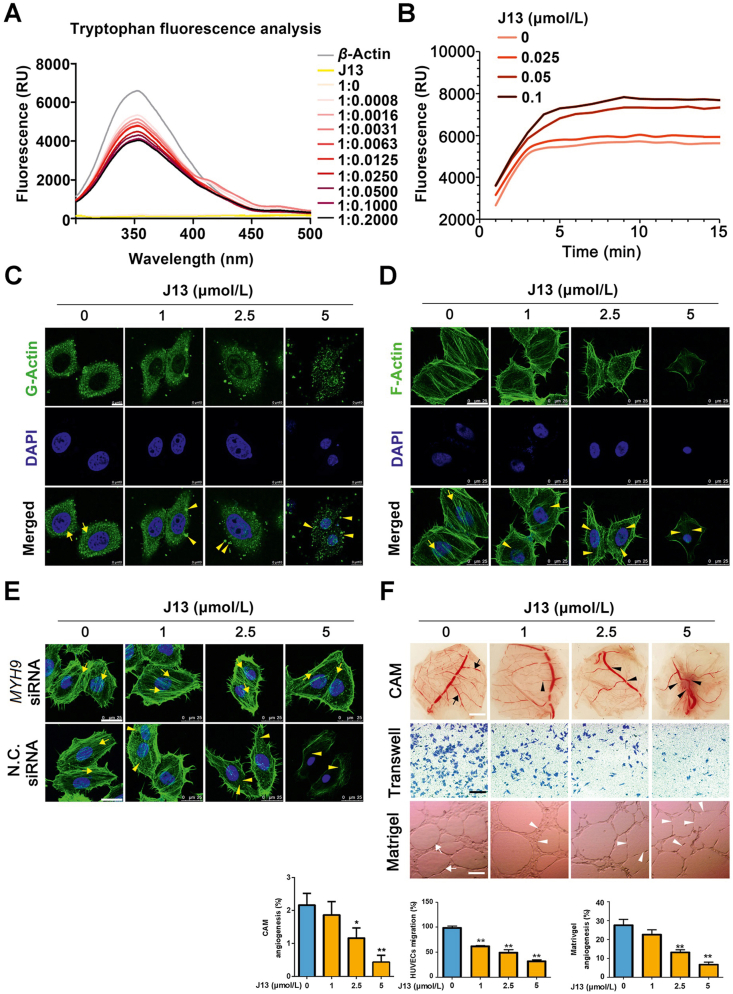
Next, we investigated whether J13 regulated the function of molecular motor protein actin. Fluorescence analysis of tryptophan revealed that the fluorescence intensity of J13–actin complex significantly decreased as J13 concentration increased, suggesting exposure and quenching of more tryptophan residues of proteins, and thus indicating an allosteric regulation effect of J13 on actin (Fig. 4A). Quantitative analysis of actin assembly dynamics showed that polymerized actin filaments rapidly formed as indicated by increasing phalloidin fluorescence after J13 treatment (Fig. 4B). Moreover, actin dynamics measurement in living cells also confirmed that monomeric G-actin was uniformly distributed in cells and formed irregular granulation aggregation upon J13 treatment (Fig. 4C). Actin-tracker for specific F-actin staining was used to explore cytoskeletal structures and result showed that J13 caused significant F-actin cytoskeletal organization collapse (Fig. 4D). Since myosin engages with F-actin to drive the alignment and compaction of F-actin cytoskeleton, we then detected whether MYH9 was involved in J13-mediated F-actin cytoskeletal organization. As shown in Fig. 4E, genetic MYH9 knockdown reversed J13-dependent F-actin cytoskeletal collapse, indicating a crucial role of MYH9 in J13-mediated F-actin cytoskeletal organization.
Figure 4.
J13 inhibits actin-dependent cell migration and angiogenesis. (A) J13 induced allosteric effect of actin in tryptophan fluorescence scanning analysis. (B) J13 induced excessive G-actin assembly which was detected by G-actin polymerization assay. (C) J13 induced G-actin granulation aggregation in cells which was detected by immunofluorescence staining (scale bar = 10 μm). (D) J13 destroyed F-actin cytoskeleton formation which was detected by fluorescent phalloidin staining (scale bar = 25 μm). (E) MYH9 knock-down reversed J13-dependent F-actin cytoskeleton collapse (scale bar = 25 μm). (F) J13 suppressed CAM microvessels formation, cell migration and capillary-like tube formation. All data are presented as mean ± SD from independent experiments performed in triplicate. ∗P < 0.05, ∗∗P < 0.01.
Actin filaments are responsible for diversified cell functions including cell migration and angiogenesis. Therefore, we investigated whether J13 mediated vascular endothelial cells (HUVECs) migration using matrigel-coated Transwell chambers. As shown in Fig. 4F, J13 strikingly suppressed HUVECs migration and impaired the formation of capillary-like tube structures of HUVECs on matrigel. Furthermore, J13 markedly inhibited neovascularization in chicken embryo chorioallantoic membrane (CAM, Fig. 4F). Taken together, these data indicate that J13 affected actin cytoskeletal structures to inhibit cell migration and angiogenesis.
3.5. J13 showed anti-tumor effects in vivo
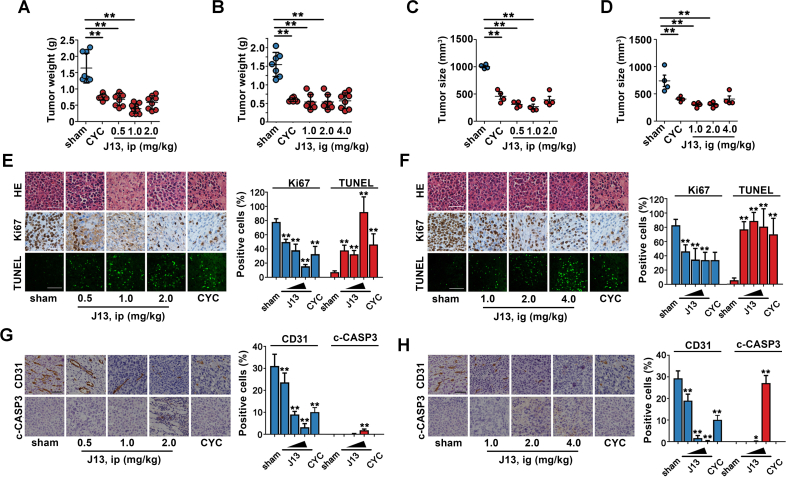
We further investigated the effect of J13 on tumor growth of mouse H22 hepatocarcinoma cells-bearing mice. As shown in Fig. 5A–D, both intraperitoneal injection and intragastric administration of J13 induced significant tumor regression, without obvious immunosuppressive side effects (Supporting Information Fig. S4). Further pathological examinations including HE, Ki67 IHC and TUNEL staining revealed that J13 effectively destroyed cell structure, reduced Ki67 positive cells number, and promoted TUNEL-positive cells increase in tumor tissues (Fig. 5E and F). Moreover, J13 significantly decreased CD31 and increased cleaved caspase-3 (c-CASP3) expressions in tumors of xenograft model, indicating that J13 effectively inhibited tumor angiogenesis and activated tumor apoptosis in vivo (Fig. 5G and H). To sum up, J13 exhibited an obvious anti-tumor effect in vivo through suppression of tumor proliferation and angiogenesis as well as induction of tumor cell apoptosis.
Figure 5.
J13 showed anti-tumor effects in vivo. (A) J13 inhibited liver tumor weight growth in H22-bearing ICR mice in vivo by intraperitoneal injection (ip). (B) J13 inhibited liver tumor weight growth of H22-bearing ICR mice in vivo by intragastric administration (ig). (C) J13 inhibited liver tumor size in H22-bearing ICR mice in vivo by intraperitoneal injection (ip). (D) J13 inhibited liver tumor size of H22-bearing ICR mice in vivo by intragastric administration (ig). (E) J13 destroyed cell structure by HE staining (scale bar = 50 μm), decreased Ki67-positive cells number (scale bar = 50 μm), and promoted TUNEL-positive cell apoptosis (scale bar = 20 μm) by ip. (F) J13 destroyed cell structure by HE staining, decreased Ki67-positive cells number, and promoted TUNEL-positive cell apoptosis (scale bar = 20 μm) by ig. (G) J13 reduced CD31-positive cells number and promoted cleaved-caspase 3 (c-CASP3) expression (scale bar = 50 μm) by ip. (H) J13 reduced CD31-positive cells number and promoted c-CASP3 expression (scale bar = 50 μm) by ig. All data are presented as mean ± SD from independent experiments (A–D: n ≥ 8, E–H: n = 3); ∗P < 0.05, ∗∗P < 0.01.
3.6. MYH9 and HSPA9 expressions are highly associated with poor survival of liver hepatocellular carcinoma (LIHC) patients
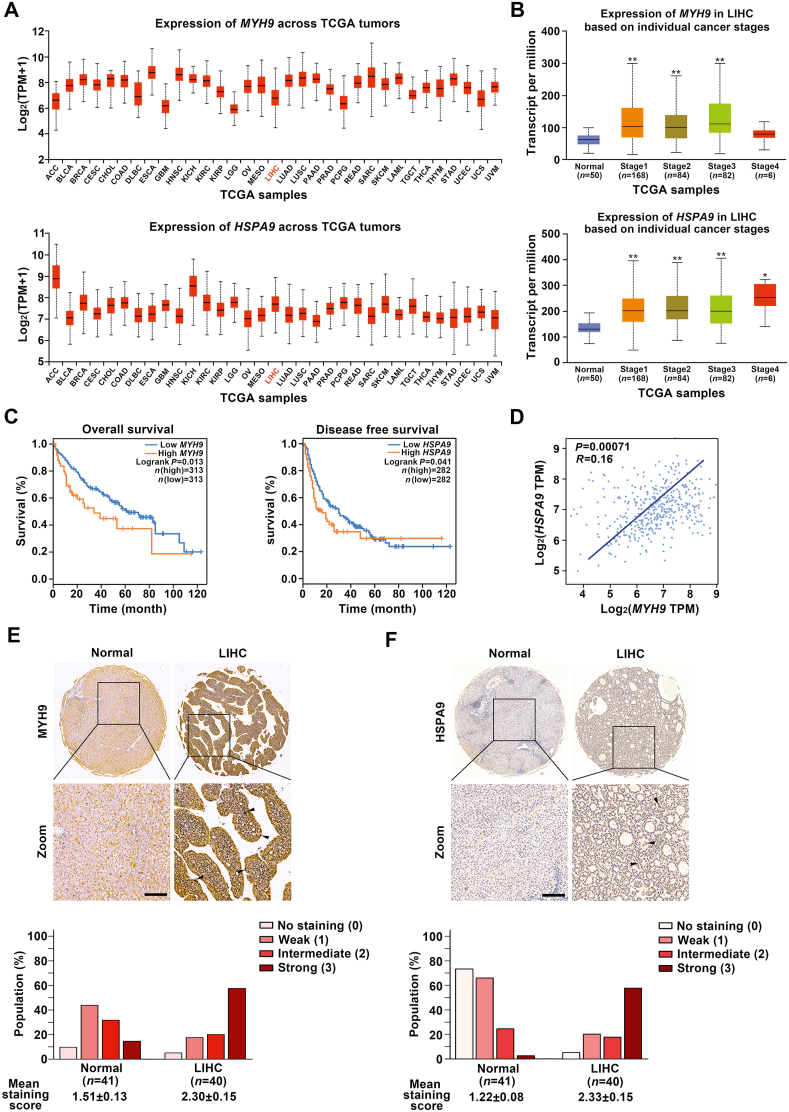
To further clarify the relationship between MYH9/HSPA9 expressions and LIHC, we analyzed clinical samples from TCGA and GTEx databases which are widely used in oncology research31,32. According to TCGA database, MYH9 and HSPA9 showed high expressions in a variety of malignancies, including LIHC [Log2(TPM+1)≥2; Fig. 6A]. Additionally, HSPA9 expression was positively associated with advanced tumor stage, especially for the patients with stage 4. Meanwhile, high expression of MYH9 was significantly related with LIHC patients with stages I to III (Fig. 6B and Supporting Information Fig. S5A). Moreover, MYH9 and HSPA9 were found to be closely associated with the prognosis of LIHC patients. As shown in Fig. 6C, the patients with high MYH9 expression had poor overall survival; meanwhile, those with high HSPA9 expression showed shorter disease free survival. Furthermore, we analyzed the correlation of MYH9 and HSPA9 mRNA expressions, and found MYH9 was positively correlated with HSPA9 (Pearson correlation coefficient, P = 0.00071; Fig. 6D). Patient tissue biopsy specimens of LIHC also supported that MYH9 and HSPA9 had high expression levels in LIHC tissues compared with adjacent normal tissues (Fig. 6E and F). Taken together, these results suggest that MYH9 and HSPA9 are highly associated with LIHC progression, and can be used as prognostic biomarkers for the disease.
Figure 6.
MYH9 and HSPA9 expressions are highly associated with poor survival of liver hepatocellular carcinoma (LIHC) patients. (A) MYH9 and HSPA9 had high expression levels in a variety of malignancies [Log2 (TPM+1) ≥2], including liver hepatocellular carcinoma LIHC from TCGA database. (B) The expression levels of MYH9/HSPA9 in LIHC were positively associated with advanced tumor stage determined by TCGA database. (C) MYH9 expression was positively correlated with LIHC patient overall survival (P = 0.013), while HSPA9 expression was positively related with disease free survival of LIHC patients (P = 0.041) based on GEPIA database. (D) A correlation between MYH9 and HSPA9 mRNA expression levels was shown by scatter plot (P = 0.00071) based on two-tailed Pearson's correlation analysis. (E) and (F) The expression levels of MYH9/HSPA9 were markedly increased in LIHC tissues (n = 40) compared to adjacent normal tissues (n = 41), as shown in representative immunohistochemical images (scale bar = 100 μm) and mean staining scores (P < 0.001, nonparametric ANOVA). ∗P < 0.05, ∗∗P < 0.01.
4. Discussion
Albizia julibrissin, also called mimosa or Persian silktree, is a member of the family Leguminosae. Albizziae cortex (AC), the stem bark of Albizia julibrissin, has been widely used for the treatment of insomnia in traditional Chinese medicine as a sedative and anti-inflammatory agent33. It has also been used as a vital composition of clinical anti-tumor prescriptions34, 35, 36. Previous studies have reported that the chemical compositions of AC include saponins, flavonoids, alkaloids and polysaccharides. Of note, triterpenoid saponins are considered to be the major anti-tumor components37, 38, 39, 40. Julibroside J13 was one of the most abundant saponin components in AC, and has been identified by our laboratory41. The molecular structure of J13 promotes it to be a suitable lead compound for anti-cancer reagents development. However, the underlying anti-cancer mechanisms still remain to be elucidated.
In this study, we used J13 as a molecule probe to explore the anti-hepatoma carcinoma targets. Particularly, we deeply investigated the regulatory mechanism of J13 on target proteins function and associated molecular signaling pathways. Therefore, J13 may serve as a lead compound for further drug development by activity improvement via structural modification.
Despite accumulating studies on mitochondrial fission/fusion in cancer pathological processes, little is known about potential therapeutic target proteins for mitochondrial dynamics modulation17,42. Myosin is actin-based molecular motor protein using ATP energy to maintain mitochondrial dynamics. Interaction of myosin with actin effectively powers mitochondrial fission/fusion process to drive cancer cells division, proliferation, and motility43. Thus, targeting myosin–actin molecular motor may be a potential strategy for anti-cancer44. However, until nowadays, this hypothesis has not been well explored for its pharmacological application in cancer therapeutics. Thus, we performed a proof-of-concept investigation with natural small-molecule J13, and revealed that myosin–actin molecular motor can be used as a druggable target for anti-cancer effects.
Alterations in mitochondrial dynamics due to chemical regulations may advance the development of novel anti-cancer strategy45. However, there has been lack of precise and reliable chemical agents to effectively modulate mitochondrial fission/fusion dynamics46. Our finding has provided important clues that mitochondrial dynamics can be regulated by targeting myosin–actin molecular motor using a small-molecule J13. Thus, J13 may represent the first small-molecule lead compound targeting myosin–actin molecular motor for further candidate drug structure optimization and structure-guided anti-cancer drug design. Curiously, previous reports have suggested that contacts between mitochondria and ER are vital for ER homeostasis47. Meanwhile, ER wraps around mitochondria and regulates mitochondrial constriction at ER–mitochondria contact sites. In this study, we observed that J13 caused mitochondrial fission by inhibiting MYH9–actin interaction, and disturbed ER–mitochondria contacts which resulted in ER stress. Thus, our observation provides a promising avenue for therapeutic interventions for anti-cancer by synergistically using molecular motor-targeting molecules with ER regulators.
As a highly conserved heat-shock chaperone, HSPA9 possesses several binding partners with multiple biological functions especially for cell stress. HSPA9 is also involved in various human diseases including cancers, Alzheimer's disease and Parkinson's disease48,49. Here, HSPA9 was identified as a substrate protein of MYH9. Further study showed HSPA9 was a crucial functional linker of molecular motor and mitochondrial fission. Meanwhile, HSPA9 has previously been found to regulate BCL-2 family protein expressions to modulate mitochondrial homeostasis50. Therefore, HSPA9–BCL-2 family signal is a key molecular pathway for controlling mitochondrial function. Here, we observed that J13 induced an obvious cytoplasm translocation of HSPA9 and overlapped with MYH9. Although there still lacks enough evidence, we tend to speculate that MYH9 might interact with HSPA9 and impair HSPA9–P53 interaction, further disturbing P53-dependant transcriptional regulation and BCL-2 family gene expressions in cancer cells. Of course, this needs further investigation.
Mitochondrial morphologies are dynamically changed between fission/fusion balances and highly associated with mitochondrial ‘health’ status6. Therefore, mitochondrial fission/fusion dynamics contributes to mitochondrial quality control51,52. We propose that J13 induced mitochondrial fission to cause mitochondrial membrane potential (MMP) decrease and resultant caspase-dependent apoptosis signal activation. Notably, genetic knockdown of MYH9 reversed J13-dependent MMP decrease, indicating MYH9 is a key regulator for controlling mitochondrial quality as well as homeostasis. One possible mechanism may be that J13 impaired mitochondrial dynamics by targeting MYH9, and inhibited mitochondrial self-renewal by excessively promoting its fission process. Meanwhile, J13 may also mediate MYH9–HSPA9–BCL-2 family signal axis to induce BAX/p-BAD increase and BCL-2/BCL-XL downregulation. Since previous reports have revealed that HSPA9 is closely associated with P53, RAF/MEK, and HIF-1α53,54, thus we speculate these interactions can regulate mitochondria-related genes expression for maintaining MMP and mitochondrial function.
We note that J13 also showed direct actin-binding effect to regulate actin dynamics, contributing to a great number of cellular processes including cell polarity, morphogenesis, and motility. This indicates that J13 may regulate myosin–actin molecular motor function by targeting multiple players on molecular motor. Moreover, actin filaments are polarized polymers in conjunction with myosin, and myosin movement along actin filaments correlates with mitochondrial fission/fusion dynamics55. Thus we speculate that J13 may promote mitochondrial fission by targeting actin filaments and impair myosin movement. Meanwhile, mitochondria migrate along protein filaments to different cellular locations with energy demands56. Our data suggest that J13 markedly affected mitochondrial distribution in cells, hinting that mitochondria attachment to protein filaments may be strictly regulated by pharmacological targeting filament proteins57.
Mitochondrial fission/fusion imbalance is well documented in LIHC; however, treatment strategies for LIHC by targeting mitochondrial fission/fusion dynamics still remain unclear. In this study, based on bioinformatics and immunohistochemical analysis of clinical samples, we found that MYH9/HSPA9 expressions were significantly upregulated in LIHC tissues, which could be considered as independent prognostic factors for LIHC patient survival. Unexpectedly, HSPA9 expression was barely related with LIHC patient overall survival. We speculate that there is a loss of LIHC patients to follow-up by 40th month. Thus, we analyzed the relationship between HSPA9 expression and disease free survival, which is widely used as an early surrogate endpoint for overall survival58. Moreover, a positive correlation between MYH9 and HSPA9 levels in LIHC indicated that MYH9/HSPA9 may be regulated in the same signaling pathway. In that scenario, combination therapy of MYH9- and HSPA9-inhibitors may synergistically improve cancer outcomes. Collectively, these data indicate that MYH9 and HSPA9 are positively correlated with LIHC progression, which is of clinical significance for the diagnosis and management of LIHC.
5. Conclusions
Taken together, we identified the first small-molecule J13 which can directly target myosin–actin molecular motor to promote mitochondrial fission for cancer therapy. Therefore, our study puts forward a conceptual framework for novel anti-cancer strategy of pharmacologically regulating mitochondrial dynamics by targeting myosin–actin molecular motor.
Acknowledgments
This study was supported by the National Key Technology R&D Program “New Drug Innovation” of China [Nos. 2019YFC1711000 and 2019YFC1708902] and the National Natural Science Foundation of China [Nos. 81973505 and 81773932].
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.01.011.
Contributor Information
Hong Liang, Email: lianghong@bjmu.edu.cn.
Kewu Zeng, Email: ZKW@bjmu.edu.cn.
Author contributions
Kewu Zeng and Hong Liang designed the project. Yi Qian and Meimei Zhao performed most of the experiments and analyzed data. Qinghua Han performed in vivo experiments. Jingkang Wang, Lixi Liao and Heng Yang performed the molecular biology and cell biology experiments. Dan Liu performed MS analysis. Kewu Zeng, Yi Qian and Meimei Zhao wrote the manuscript. Pengfei Tu contributed to the manuscript editing. All authors contributed and approved the final version of the manuscript.
Conflicts of interest
The authors declare that there are no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chan D.C. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Schapira A.H. Mitochondrial diseases. Lancet. 2012;379:1825–1834. doi: 10.1016/S0140-6736(11)61305-6. [DOI] [PubMed] [Google Scholar]
- 3.Wei M.C., Zong W.X., Cheng E.H., Lindsten T., Panoutsakopoulou V., Ross A.J. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 5.Liesa M., Palacin M., Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 6.Chan D.C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 7.Sabouny R., Shutt T.E. Reciprocal regulation of mitochondrial fission and fusion. Trends Biochem Sci. 2020;4:564–577. doi: 10.1016/j.tibs.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Liesa M., Shirihai O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metabol. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabian S.G., Frederic D. Mitochondrial fission and fusion in human diseases. N Engl J Med. 2014;370:1073–1074. doi: 10.1056/NEJMc1316254. [DOI] [PubMed] [Google Scholar]
- 10.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashatus J.A., Nascimento A., Myers L.J., Sher A., Byrne F.L., Hoehn K.L. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serasinghe M.N., Wieder S.Y., Renault T.T., Elkholi R., Asciolla J.J., Yao J.L. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell. 2015;57:521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H., Chan D.C. Mitochondrial dynamics in regulating the unique phenotypes of cancer and stem cells. Cell Metabol. 2017;26:39–48. doi: 10.1016/j.cmet.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan S., Guha M., Kashina A., Avadhani N.G. Mitochondrial dysfunction and mitochondrial dynamics—the cancer connection. Biochim Biophys Acta Bioenerg. 2017;1858:602–614. doi: 10.1016/j.bbabio.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagdas S., Kashatus J.A., Nascimento A., Hussain S.S., Trainor R.E., Pollock S.R. Drp1 promotes KRas-driven metabolic changes to drive pancreatic tumor growth. Cell Rep. 2019;28:1845–1859. doi: 10.1016/j.celrep.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo J.H., Agarwal E., Chae Y.C., Lee Y.G., Garlick D.S., Storaci A.M. Mitochondrial fission factor is a novel Myc-dependent regulator of mitochondrial permeability in cancer. EbioMedicine. 2019;48:353–363. doi: 10.1016/j.ebiom.2019.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senft D., Ronai Z.A. Regulators of mitochondrial dynamics in cancer. Curr Opin Cell Biol. 2016;39:43–52. doi: 10.1016/j.ceb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao J., Wei R., Yao S. Matrine has pro-apoptotic effects on liver cancer by triggering mitochondrial fission and activating Mst1-JNK signaling pathways. J Physiol Sci. 2019;69:185–198. doi: 10.1007/s12576-018-0634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiku V., Tan M.W., Dikic I. Mitochondrial functions in infection and immunity. Trends Cell Biol. 2020;30:263–275. doi: 10.1016/j.tcb.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Detmer S.A., Chan D.C. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 21.Molloy J.E., Veigel C. Biophysics. Myosin motors walk the walk. Science. 2003;300:2045–2046. doi: 10.1126/science.1087148. [DOI] [PubMed] [Google Scholar]
- 22.Kruppa A.J., Itakura C.K., Masters T.A., Rorbach J.E., Grice G.L., Jones J.K. Myosin VI-dependent actin cages encapsulate parkin-positive damaged mitochondria. Dev Cell. 2018;44:484–499. doi: 10.1016/j.devcel.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C., Svitkina T.M. Ultrastructure and dynamics of the actin–myosin II cytoskeleton during mitochondrial fission. Nat Cell Biol. 2019;21:603–613. doi: 10.1038/s41556-019-0313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korobova F., Gauvin T.J., Higgs H.N. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korobova F., Ramabhadran V., Higgs H.N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honrath B., Metz I., Bendridi N., Rieusset J., Culmsee C., Dolga A.M. Glucose-regulated protein 75 determines ER–mitochondrial coupling and sensitivity to oxidative stress in neuronal cells. Cell Death Discovery. 2017;3 doi: 10.1038/cddiscovery.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Q., Qian Y., Wang X., Zhang Q., Cui J., Tu P. Cytotoxic oleanane triterpenoid saponins from Albizia julibrissin. Fitoterapia. 2017;121:183–193. doi: 10.1016/j.fitote.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Liao L.X., Song X.M., Wang L.C., Lv H.N., Chen J.F., Liu D. Highly selective inhibition of IMPDH2 provides the basis of antineuroinflammation therapy. Proc Natl Acad Sci U S A. 2017;114:5986–5994. doi: 10.1073/pnas.1706778114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starenki D., Hong S.K., Lloyd R.V., Park J.I. Mortalin (GRP75/HSPA9) upregulation promotes survival and proliferation of medullary thyroid carcinoma cells. Oncogene. 2015;34:4624–4634. doi: 10.1038/onc.2014.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:556–560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium G.T. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson G.R., Wardell S.E., Cakir M., Yip C., Ahn Y.R., Ali M. Dysregulation of mitochondrial dynamics proteins are a targetable feature of human tumors. Nat Commun. 2018;9:e1677. doi: 10.1038/s41467-018-04033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y., Wang Q., Ye Y., Liu Z., Sun H. The ethnopharmacology, phytochemistry, pharmacology and toxicology of genus Albizia: a review. J Ethnopharmacol. 2020;257 doi: 10.1016/j.jep.2020.112677. [DOI] [PubMed] [Google Scholar]
- 34.Feng L., Tan Y., Qiu L.Y., Gu X.T., Zhou X.Z. Optimizing extraction process for Yiyuan Yiliu decoction by orthogonal design using multi-index. Chin J Hosp Pharm. 2013;33:601–604. [Google Scholar]
- 35.Tan Y., Qiu L.Y., Mao H.Y., Zhou X.Z., Feng L. Study on formation process of “Yiyuan Yiliu Tang” granules. Chin Tradit Pat Med. 2013;35:2050–2053. [Google Scholar]
- 36.Yi Q.Q., Tan Y., Li R., Du B., Feng L., Qiu L.Y. Investigation of immunity capability of “Yiyuan Yiliu Tang” in H 22 tumor-burdened mice. Chin J Immunol. 2014;30:205–208. [Google Scholar]
- 37.Zhang L., Zheng J., Wu L.J., Zhao Y.Y. Julibroside J8-induced HeLa cell apoptosis through caspase pathway. J Asian Nat Prod Res. 2006;8:457–465. doi: 10.1080/10286020500173309. [DOI] [PubMed] [Google Scholar]
- 38.Qian Y., Han Q.H., Wang L.C., Guo Q., Wang X.D., Tu P.F. Total saponins of Albiziae Cortex show anti-hepatoma carcinoma effects by inducing S phase arrest and mitochondrial apoptosis pathway activation. J Ethnopharmacol. 2018;221:20–29. doi: 10.1016/j.jep.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 39.Yu D.H., Qiao S.Y., Zhao Y.M. Advances in study on bark of Albizzia julibrissin. China J Chin Mater Med. 2004;29:619–624. [PubMed] [Google Scholar]
- 40.Hua H., Feng L., Zhang X.P., Zhang L.F., Jin J. Anti-angiogenic activity of julibroside J8, a natural product isolated from Albizia julibrissin. Phytomedicine. 2009;16:703–711. doi: 10.1016/j.phymed.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Zou K., Tong W.Y., Liang H., Cui J.R., Tu G.Z., Zhao Y.Y. Diastereoisomeric saponins from Albizia julibrissin. Carbohydr Res. 2005;340:1329–1334. doi: 10.1016/j.carres.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 42.Muller M., Diensthuber R.P., Chizhov I., Claus P., Heissler S.M., Preller M. Distinct functional interactions between actin isoforms and nonsarcomeric myosins. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makowska K.A., Hughes R.E., White K.J., Wells C.M., Peckham M. Specific myosins control actin organization, cell morphology, and migration in prostate cancer cells. Cell Rep. 2015;13:2118–2125. doi: 10.1016/j.celrep.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X., Dong Q.F., Li L.W., Huo J.L., Li P.Q., Fei Z. The cap-translation inhibitor 4EGI-1 induces mitochondrial dysfunction via regulation of mitochondrial dynamic proteins in human glioma U251 cells. Neurochem Int. 2015;90:98–106. doi: 10.1016/j.neuint.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 45.Franco A., Kitsis R.N., Fleischer J.A., Gavathiotis E., Kornfeld O.S., Gong G. Correcting mitochondrial fusion by manipulating mitofusin conformations. Nature. 2016;540:74–79. doi: 10.1038/nature20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erpapazoglou Z., Liger F.M., Corti O. From dysfunctional endoplasmic reticulum–mitochondria coupling to neurodegeneration. Neurochem Int. 2017;109:171–183. doi: 10.1016/j.neuint.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 47.Wu P.K., Hong S.K., Park J.I. Steady-state levels of phosphorylated mitogen-activated protein kinase kinase 1/2 determined by mortalin/HSPA9 and protein phosphatase 1 alpha in KRAS and BRAF tumor cells. Mol Cell Biol. 2017;37 doi: 10.1128/MCB.00061-17. e00061-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park S.J., Shin J.H., Jeong J.I., Sone J.H., Jo Y.K., Kim E.S. Down-regulation of mortalin exacerbates Abeta-mediated mitochondrial fragmentation and dysfunction. J Biol Chem. 2014;289:2195–2204. doi: 10.1074/jbc.M113.492587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vander Heiden M.G., Thompson C.B. Bcl-2 proteins: regulators of apoptosis or of mitochondrial homeostasis?. Nat Cell Biol. 1999;1:209–216. doi: 10.1038/70237. [DOI] [PubMed] [Google Scholar]
- 50.Mishra P., Chan D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 52.Wu P.K., Hong S.K., Veeranki S., Karkhanis M., Starenki D., Plaza J.A. A mortalin/HSPA9-mediated switch in tumor-suppressive signaling of Raf/MEK/extracellular signal-regulated kinase. Mol Cell Biol. 2013;33:4051–4067. doi: 10.1128/MCB.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sane S., Abdullah A., Boudreau D.A., Autenried R.K., Gupta B.K., Wang X. Ubiquitin-like (UBX)-domain-containing protein, UBXN2A, promotes cell death by interfering with the p53-Mortalin interactions in colon cancer cells. Cell Death Dis. 2014;5:e1118. doi: 10.1038/cddis.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altmann K., Frank M., Neumann D., Jakobs S., Westermann B. The class V myosin motor protein, Myo2, plays a major role in mitochondrial motility in Saccharomyces cerevisiae. J Cell Biol. 2008;181:119–130. doi: 10.1083/jcb.200709099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hatch A.L., Gurel P.S., Higgs H.N. Novel roles for actin in mitochondrial fission. J Cell Sci. 2014;127:4549–4560. doi: 10.1242/jcs.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boldogh I.R., Pon L.A. Mitochondria on the move. Trends Cell Biol. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 57.Boldogh I.R., Pon L.A. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 58.Kemp R., Prasad V. Surrogate endpoints in oncology: when are they acceptable for regulatory and clinical decisions, and are they currently overused?. BMC Med. 2017;15:e134. doi: 10.1186/s12916-017-0902-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.