Abstract
Background
Viral infections of the respiratory tract represent a major global health concern. Co-infection with bacteria may contribute to severe disease and increased mortality in patients. Nevertheless, viral-bacterial co-infection patterns and their clinical outcomes have not been well characterized to date. This study aimed to evaluate the clinical features and outcomes of patients with viral-bacterial respiratory tract co-infections.
Methods
We included 19,361 patients with respiratory infection due to respiratory viruses [influenza A and B, respiratory syncytial virus (RSV), parainfluenza] and/or bacteria in four tertiary hospitals in Hong Kong from 2013 to 2017 using a large territory-wide healthcare database. All microbiological tests were conducted within 48 h of hospital admission. Four etiological groups were included: (1) viral infection alone; (2) bacterial infection alone; (3) laboratory-confirmed viral-bacterial co-infection and (4) clinically suspected viral-bacterial co-infection who were tested positive for respiratory virus and negative for bacteria but had received at least four days of antibiotics. Clinical features and outcomes were recorded for laboratory-confirmed viral-bacterial co-infection patients compared to other three groups as control. The primary outcome was 30-day mortality. Secondary outcomes were intensive care unit (ICU) admission and length of hospital stay. Propensity score matching estimated by binary logistic regression was used to adjust for the potential bias that may affect the association between outcomes and covariates.
Findings
Among 15,906 patients with respiratory viral infection, there were 8451 (53.1%) clinically suspected and 1,087 (6.8%) laboratory-confirmed viral-bacterial co-infection. Among all the bacterial species, Haemophilus influenzae (226/1,087, 20.8%), Pseudomonas aeruginosa (180/1087, 16.6%) and Streptococcus pneumoniae (123/1087, 11.3%) were the three most common bacterial pathogens in the laboratory-confirmed co-infection group. Respiratory viruses co-infected with non-pneumococcal streptococci or methicillin-resistant Staphylococcus aureus was associated with the highest death rate [9/30 (30%) and 13/48 (27.1%), respectively] in this cohort. Compared with other infection groups, patients with laboratory-confirmed co-infection had higher ICU admission rate (p < 0.001) and mortality rate at 30 days (p = 0.028), and these results persisted after adjustment for potential confounders using propensity score matching. Furthermore, patients with laboratory-confirmed co-infection had significantly higher mortality compared to patients with bacterial infection alone.
Interpretation
In our cohort, bacterial co-infection is common in hospitalized patients with viral respiratory tract infection and is associated with higher ICU admission rate and mortality. Therefore, active surveillance for bacterial co-infection and early antibiotic treatment may be required to improve outcomes in patients with respiratory viral infection.
Keywords: Respiratory, viral-bacterial co-infection, Mortality
Research in context.
Evidence before this study
Respiratory viral-bacterial co-infection can result in severe lower respiratory complications and is associated with higher mortality. Streptococcus pneumoniae and Haemophilus influenzae co-infected with influenza A in the 1957 and 2009 pandemics were common, but bacterial co-infection with other viral infection were rarely reported. PubMed was searched using the keywords [(Respiratory co-infection) OR (bacteria-viral infection)] AND (community-acquired pneumonia) from title/abstract. By the time of November 2020, a total of 150 full-text articles on epidemiology or animal studies about respiratory co-infection were identified. Apart from the studies on animals or in the pediatric field, there were 61 studies focus on adult viral-bacterial co-infection. Clinical outcomes or biomarker prediction were reported. Although the majority of the studies have reported viral-bacterial co-infection has a worse clinical outcome, these studies mainly focus on specific pathogens (Influenza A or Staphylococcus species mainly).
Added value of this study
This study reports on the epidemiology, comorbidity, and clinical outcomes of 19,361 cases with community-acquired, laboratory-confirmed respiratory infection. It contains 1087 patients with laboratory-confirmed viral-bacterial co-infection, the largest retrospective cohort to date. It presents the latest status, common pathogens and outcomes of community-acquired respiratory infection in hospitalized adults in Hong Kong. The mean age of patients was 69 years (standard deviation (SD) 19), and 3655 (18.9%) patients had chronic lung disease. 760 (3.9%) patients required intensive care unit admission, and 1160 (6.0%) died within 30 days. Co-infection with P. aeruginosa had both a relatively high prevalence (180/1087; 16.6%) and death rate (19/180; 10.5%).
Implications of all the available evidence
Respiratory viral-bacterial co-infection is associated with worse mortality and morbidity than viral or bacterial infection alone. More efforts should be made to understand the interactions between viral and bacterial co-infections to improve patient outcomes. Furthermore, patients with respiratory viral infection co-infected with non-pneumococcal streptococci species or methicillin-resistant Staphylococcus aureus resulted in higher mortality rates and require further investigation.
Alt-text: Unlabelled box
1. Introduction
Community-acquired respiratory infection is one of the major causes of morbidity and mortality, especially among the elderly population [1,2]. The prevalence of contributory pathogens have, however, changed over time [3]. Influenza pandemics have demonstrated the harmful effects of viral-bacterial co-infection [4,5]. Secondary bacterial infection was the predominant cause of death among patients infected with the Spanish flu in 1918 [6]. During the pre-antibiotic era, almost all patients who died in the Spanish flu pandemic had at least one bacterial pathogen isolated from the respiratory tract. In the subsequent 1957 Asian influenza and H1N1 pandemics, bacterial pathogens were commonly found in patients who died, albeit less consistently (25–50%) [[7], [8], [9]]. In the latest 2009 H1N1 pandemic, only 17.5% of patients with viral pneumonia had bacterial co-infection [10]. In addition, respiratory syncytial virus (RSV) that commonly infects children and the elderly [11], is associated with Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus [[12], [13], [14]]. Temporal changes in vaccination [15], population structure [16,17], selection pressure from antibiotics [18,19], and availability of highly sensitive detection method with viral polymerase chain reaction (PCR) [20], may account for the changing pattern.
Numerous studies have demonstrated the detrimental interactions between viruses and bacteria that commonly infect the human respiratory tract. Such interactions could produce impaired mucociliary clearance, enhanced bacterial binding, changes in respiratory tract receptor expression, dysregulated immune response and decreased bacterial clearance [[21], [22], [23], [24], [25], [26], [27], [28]]. There is also evidence that bacterial infection or altered microbiome in the respiratory tract may enhance subsequent viral infection [29]. The complex pathogen-host interactions in patients with viral-bacterial co-infection may therefore lead to serious adverse outcomes [16,30,31]. In this respect, a meta-analysis of 31 studies showed an increase in mortality among patients with pneumonia due to viral-bacterial co-infection [32]. Another study focusing on patients admitted to the intensive care unit (ICU) produced similar results [31]. However, recent epidemiological data failed to demonstrate higher mortality of viral-bacterial co-infection [16,33]. Furthermore, apart from influenza and RSV, interactions between other respiratory viruses and bacteria have not been characterized due to the limited sample size. Using a large database, we analyzed the 30-day mortality, intensive care unit (ICU) admission and duration of hospital stay in adult patients hospitalized for viral and/or bacterial respiratory infection. We hypothesized that patients with laboratory-confirmed viral-bacterial co-infection were associated with worse outcome compared with those having bacterial or viral infection alone.
2. Methods
2.1. Study design
In this analysis, we included consecutive adult (>18 years) patients hospitalized for respiratory infection, who had both viral and bacterial respiratory sampling, between 1 January 2013 and 31 December 2017 from four major hospitals in Hong Kong. These patients were identified through the Clinical Data Analysis and Report System (CDARS), which is a computerized clinical database of the Hospital Authority of Hong Kong. As the sole independent public health provider, the Hospital Authority has built a public healthcare infrastructure that covers over 90% acute in-patient services in Hong Kong [34]. Patients were also excluded if they were managed as outpatient, had incomplete clinical records, or had multiple viral or bacterial infections. After excluding these cases, we extracted clinical data including baseline demographics, comorbidities using the International Classification of Diseases, Ninth Revision (ICD9) diagnosis codes (Table S1), medication used, ICU admission, and 30-day mortality. Ethics approval was obtained from the Chinese University of Hong Kong with wavier of informed consent (CREC Ref. No.: 2018.358).
2.2. Laboratory confirmed infections and treatment
Respiratory microbiological samples were tested for viral and bacterial pathogens. For viral tests, samples collected from nasal flock swab, nasopharyngeal aspirate, tracheal aspirate, bronchial aspirate and/or bronchoalveolar lavage were analyzed with PCR for influenza A and B, RSV, and parainfluenza types 1, 2, 3, and 4. Patients who were tested positive for adenovirus, enterovirus, rhinovirus, or human metapneumovirus were not included in the study because test availability was inconsistent during the study period. For bacterial tests, samples from sputum, tracheal aspirates, bronchoalveolar lavage, bronchial washing, pleural biopsy and/or blood cultures were sent for bacterial or mycobacterium cultures (Table S2). Matrix-assisted laser desorption/ionization-time of flight mass spectrometry was used for bacteria identification. All included cases had both viral and bacterial tests done. All samples were collected within 48 h of hospital admission and have at least one upper respiratory infection symptoms. We excluded cases with multiple organisms in the viral tests or bacterial cultures to avoid cases with commensal contamination.
During the study period, the clinical management protocols were consistent among all hospitals. Antibiotics were generally prescribed upon hospital admission and usually stopped < 4 days if the patient did not respond clinically or bacterial cultures were found negative. The duration of antibiotics was chosen as the threshold to indicate non-bacterial infection according to the recommended use of antibacterial in adult sepsis event defined by the Centers for Disease Control and Prevention [35]. Patients who had antibiotics but died within four days of admission were considered to have been treated for bacterial infection clinically.
2.3. Group categorization and outcomes
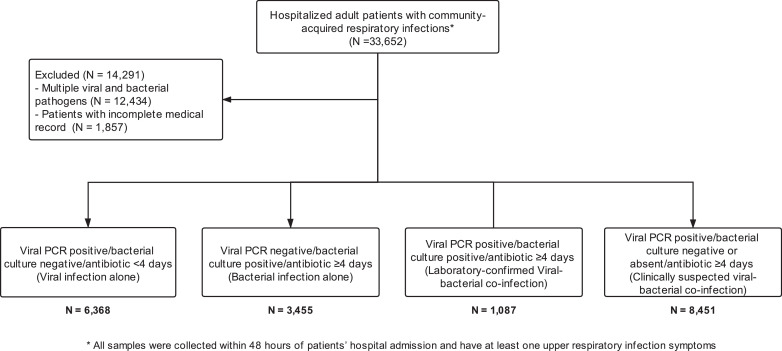
Four groups of patients were defined in this study: (1) Viral infection alone group included patients who were tested positive for any respiratory virus by PCR but negative for bacteria by bacterial or mycobacterial culture, and received antibiotics for < 4 days; (2) Bacterial infection alone group included patients who had positive culture for any respiratory bacterial or mycobacterial species but negative for respiratory virus by PCR, and received a course of antibiotics ≥ 4 days; (3) Laboratory-confirmed viral-bacterial co-infection group included patients with positive laboratory confirmation of viral PCR test and bacterial/mycobacterial culture, and received a course of antibiotics ≥ 4 days; (4) Clinically suspected viral-bacterial co-infection group included patients with a positive viral PCR test, negative bacterial culture and received a course of antibiotics ≥ 4 days (Fig. 1).
Fig. 1.
Flowchart of research methodology, screenig, eligibility and enrollment of patients with respiratory infection.
The primary outcome was all-cause mortality within 30 days of hospital admission. Secondary outcomes were the proportions of patients requiring ICU admission and number of days of hospital length of stay.
2.4. Statistics
Baseline characteristics were compared among four groups using analysis of variance for continuous variables (including age, length of hospital stay and the laboratory data), post-hoc analysis was adjusted by Tukey's test. For categorical data (sex and comorbidities), we used chi square test. Since there were variations in the baseline characteristics among groups, we adjusted outcome comparisons using propensity analysis. Three pairs of between group comparisons were made. In each pair, the laboratory-confirmed viral-bacterial co-infection group was compared with one of the three reference groups, i.e. either viral infection alone, bacterial infection alone, or clinically suspected viral-bacterial co-infection. Propensity score was calculated as the probability of laboratory-confirmed viral-bacterial co-infection versus the respective reference group, adjusted by sex, age, and the baseline risk factors in the Charlson's comorbidity index using binary logistic regression [36]. One-to-one propensity matching was performed with the matchit package in R using the nearest neighbor approach within a caliper distance (i.e. standard deviation of logit of the propensity score) of 0.2 [37,38]. Before propensity matching, several baseline variables showed standardized mean difference >0.1 between the laboratory-confirmed viral-bacterial co-infection group and the respective reference group, including hemiplegia or paraplegia, peripheral vascular disease, rheumatic disease, liver disease and acquired immunodeficiency syndrome (Tables S3–5). After matching, standardized mean difference between groups was reduced to <0.1 in all baseline variables. Cox proportional hazard model was used to calculate the hazard ratio of 30-day mortality, with robust variance estimator to account for the clustering within matched pairs [39]. Kaplan–Meier plot was used to demonstrate the difference in survival probabilities between groups after propensity score matching. Subgroup analysis was performed for patients requiring ICU admission. Relative risk was calculated for the association between co-infection and ICU admission outcome. Crude 30-day mortality and prevalence of different viral-bacterial co-infection combinations were calculated for selected combinations with an incidence ≥ 20 cases. Multiple testing was adjusted by Bonferroni correction. All statistical analysis was conducted in R version 4.0.0 (R Project for Statistical Computing). Nominal p-value less than 0.05 was considered as statistically significant.
2.5. Role of funding
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
3. Results
3.1. Study population and baseline characteristics
We included 19,361 adult hospitalized patients (mean age 69 years (SD: 19)) with positive PCR tests for respiratory virus and/or bacteriological culture over the 5-year period. A total of 6368 patients had viral infection alone (32.9%), 3455 (17.8%) had bacterial infection alone, 1087 (5.6%) had laboratory-confirmed viral-bacterial co-infection, and 8451 (43.6%) had clinically suspected viral-bacterial co-infection (Fig. 1). Table 1 shows significant differences in the distribution of age, sex and chronic illness across the four groups. Patients with laboratory-confirmed viral-bacterial co-infection and bacterial infection alone had higher neutrophil count, neutrophil-to-lymphocyte ratio, C-reactive protein, and platelet count than viral infection alone and clinical suspected viral-bacterial co-infection (Table 1; p values of the post-hoc analysis ranged from <0.001 to 0.005).
Table 1.
Baseline characteristics between patients with different types of infection.
| Viral infection alone N = 6368 | Bacterial infection alone N = 3455 | Laboratory-confirmed viral-bacterial co-infection N = 1087 | Clinically suspected viral-bacterial co-infection N = 8451 | p value | |
|---|---|---|---|---|---|
| Age, Mean (SD) | 0.114 | ||||
| 68.7 (20.1) | 69.2 (17.9) | 70.9 (17.0) | 69.1 (18.8) | ||
| Age group (%) | <0.001* | ||||
| <65 | 2252 (35.4) | 1208(35.0) | 330 (30.4) | 3028 (35.8) | |
| 65–74 | 945 (14.8) | 665 (19.2) | 215 (19.8) | 1358 (16.1) | |
| 75–84 | 1615 (25.4) | 843 (24.4) | 306 (28.2) | 2132 (25.2) | |
| ≥85 | 1556 (24.4) | 739 (21.4) | 236 (21.7) | 1933 (22.9) | |
| Sex (%) | <0.001* | ||||
| Female | 3358 (52.7) | 1502 (43.5) | 511 (47.0) | 4300 (50.9) | |
| Comorbidities (%) | 4019 (63.1) | 2026 (58.6) | 741 (68.2) | 5155 (61.0) | <0.001* |
| Myocardial infarction | 477 (7.5) | 154 (4.5) | 72 (6.6) | 486 (5.8) | <0.001* |
| Congestive heart failure | 858 (13.5) | 287 (8.3) | 128 (11.8) | 983 (11.6) | <0.001* |
| Peripheral vascular disease | 153 (2.4) | 54 (1.6) | 25 (2.3) | 139 (1.6) | 0.002* |
| Cerebrovascular disease | 1032 (16.2) | 274 (7.9) | 144 (13.2) | 1250 (14.8) | <0.001* |
| Chronic pulmonary disease | 1220 (19.2) | 662 (19.2) | 295 (27.1) | 1478 (17.5) | <0.001* |
| Diabetes mellitus | 1196 (18.8) | 574 (16.6) | 181 (16.7) | 1496 (17.7) | 0.036* |
| Rheumatic disease | 95 (1.5) | 46 (1.3) | 27 (2.5) | 144 (1.7) | 0.047* |
| Peptic ulcer disease | 278 (4.4) | 69 (2.0) | 51 (4.7) | 308 (3.6) | <0.001* |
| Hemiplegia/paraplegia | 184 (2.9) | 30 (0.9) | 18 (1.7) | 248 (2.9) | <0.001* |
| Dementia | 418 (6.6) | 116 (3.4) | 61 (5.6) | 541 (6.4) | <0.001* |
| Renal disease | 531 (8.3) | 225 (6.5) | 94 (8.6) | 608 (7.2) | 0.002* |
| Malignancy | 651 (10.2) | 397 (11.5) | 135 (12.4) | 944 (11.2) | 0.064* |
| Liver disease | 396 (6.2) | 191 (5.5) | 73 (6.7) | 519 (6.1) | 0.411* |
| Metastatic solid tumor | 175 (2.7) | 156 (4.5) | 29 (2.7) | 209 (2.5) | <0.001* |
| Acquired immunodeficiency syndrome | 7 (0.1) | 11 (0.3) | 2 (0.2) | 14 (0.2) | 0.131* |
| Laboratory result | |||||
| Neutrophil | <0.001 | ||||
| Mean (SD) | 6.6 (4.0) | 9.9 (5.5) | 7.9 (4.9) | 6.4 (3.9) | |
| Missing (%) | 256 (4.0) | 97 (2.8) | 35 (3.2) | 276 (3.3) | |
| Lymphocyte | 0.057 | ||||
| Mean (SD) | 1.0 (1.8) | 1.1 (2.7) | 0.9 (0.7) | 1.0 (3.4) | |
| Missing (%) | 256 (4.0) | 97 (2.8) | 35 (3.2) | 276 (3.3) | |
| Neutrophil to lymphocyte ratio | <0.001 | ||||
| Mean (SD) | 9.8 ()9.7 | 14.7 (16.2) | 12.6 (14.8) | 9.6 (10.8) | |
| Missing (%) | 256 (4.0) | 97 (2.8) | 35 (3.2) | 276 (3.3) | |
| Platelet | <0.001 | ||||
| Mean (SD) | 191.5 (78.6) | 227.0 (101.8) | 200.7 (92.2) | 187.1 (76.7) | |
| Missing (%) | 164 (2.6) | 49(1.4) | 23 (2.1) | 157 (1.9) | |
| C-reactive protein | |||||
| Mean (SD) | 5.8 (6.3) | 10.9 (9.8) | 10.1 (8.9) | 5.9 (6.3) | <0.001 |
| Missing (%) | 2405 (37.8) | 842 (24.4) | 365 (33.6) | 3136 (37.1) | |
| Hospital length of stay, Mean (SD) | 7.1 (18.3) | 7.3 (9.3) | 9.3 (22.9) | 5.5 (10.9) | <0.001 |
SD = standard deviations.
p values are for analysis of variance or chi square test*.
3.2. Clinical outcomes of different infection groups
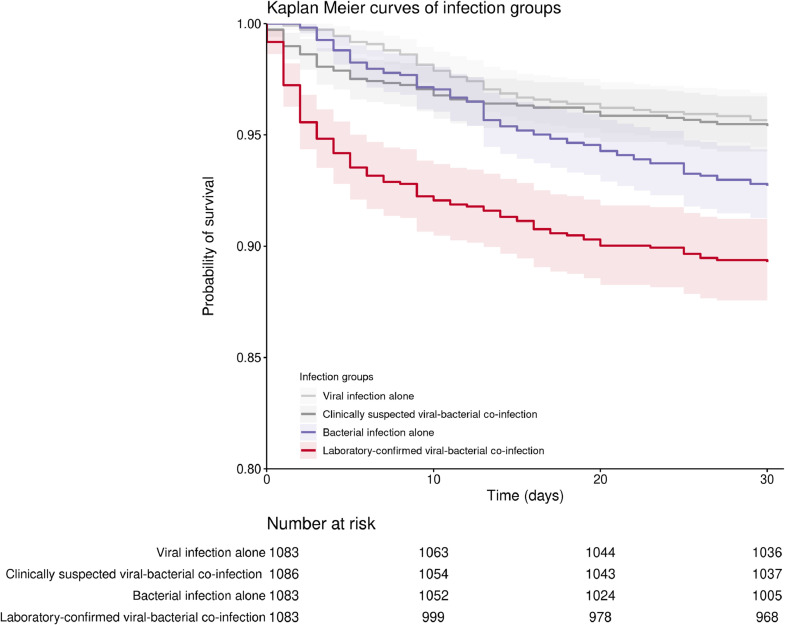
Overall, 30-day mortality in the entire cohort was 6.0% (1160/19,361) with 3.9% (760/19,361) of patients admitted to ICU. Propensity matching for the baseline variables minimized the difference in covariates among different infection groups (Tables S3–5). After propensity matching, laboratory-confirmed viral-bacterial co-infection was significantly higher in 30-day mortality and ICU admission compared with reference group (either viral infection alone, bacterial infection alone, or clinical suspected viral-bacterial co-infection group) (Table 2, Fig. 2). A subgroup analysis among the propensity matched patients requiring ICU admission suggested that laboratory-confirmed co-infection had a significantly higher 30-day mortality than viral infection alone [hazard ratio (HR) = 1.9, 95% confidence level (CI): 1.0–3.5, p = 0.041, N = 98], but not significantly higher than patients with clinically suspected viral-bacterial co-infection (HR = 1.4, 95%CI: 0.8–2.6, p = 0.232, N = 98) or bacterial infection alone (HR = 1.4, 95%CI: 0.8–2.5, p = 0.237, N = 97) (Table S6).
Table 2.
Outcomes of patients with different types of infection.
| Before propensity score matching* |
After propensity score matching |
|||||||
|---|---|---|---|---|---|---|---|---|
| Reference group | Laboratory-confirmed viral-bacterial coinfection group | Risk of the coinfection group [95% CI] | p-value | Reference group | Laboratory-confirmed viral-bacterial coinfection group | Risk of the coinfection group [95% CI] | Adjusted p-value | |
| Laboratory-confirmed viral-bacterial co-infection versus viral infection alone | N = 6368 | N = 1087 | N = 1083 | N = 1083 | ||||
| 30-day mortality N (%) | 332 (5.2%) | 118 (10.9%) | HR =2.2 [1.8, 2.7] | <0.001 | 47 (4.3%) | 117 (10.8%) | HR =2.6 [1.9, 3.7] | <0.001 |
| ICU admission N (%) | 207 (3.3%) | 103 (9.5%) | RR = 2.9 [2.3, 3.7] | <0.001 | 35 (3.2%) | 102 (9.42%) | RR =2.9 [2.3, 3.6] | <0.001 |
| Laboratory-confirmed viral-bacterial co-infection versus bacterial infection alone | N = 3455 | N = 1087 | N = 1083 | N = 1083 | ||||
| 30-day mortality N (%) | 310 (9.0%) | 118 (10.9%) | HR =1.3 | |||||
| [1.01, 1.5] | 0.114 | 79 (7.3%) | 116 (10.7%) | HR =1.4 [1.1, 1.9] | 0.028 | |||
| ICU admission N (%) | 196 (5.7%) | 103 (9.5%) | RR= 1.8 | |||||
| [1.3, 2.1] | <0.001 | 44 (4.1%) | 103 (9.5%) | RR =1.6 [1.2, 2.1] | <0.001 | |||
| Laboratory-confirmed viral-bacterial co-infection versus clinically suspected viral-bacterial co-infection | N = 8451 | N = 1087 | N = 1086 | N = 1086 | ||||
| 30-day mortality N (%) | 400 (4.7%) | 118 (10.9%) | HR =2.4 | |||||
| [1.9, 2.9] | <0.001 | 53 (4.9%) | 118 (10.9%) | HR =2.3 [1.7, 3.2] | <0.001 | |||
| ICU admission N (%) | 254 (3.0%) | 103 (9.5%) | RR =3.15 | |||||
| [2.5, 3.9] | <0.001 | 35 (3.2%) | 103 (9.5%) | RR =3.2 [2.5, 3.9] | <0.001 | |||
N: Sample size.
HR: Hazard ratio.
RR: Relative risk.
* The propensity score matching was performed using laboratory-confirmed viral-bacterial co-infection versus respective reference groups (either viral infection alone, bacterial infection alone, or clinical suspected viral-bacterial co-infection group) as dependent variables, the variables listed in Tables S3–5 as independent variables for adjustment.
Fig. 2.
Kaplan–Meier curves for the 30-day mortality of the four types of infection groups. The figure presents the trend of the 30-day survival rate in each group. The log-rank test for reference groups versus laboratory-confirmed co-infection group was done and shown in the figure. Data after propensity score matching were used for the comparison, and the matching result of the laboratory-confirmed viral-bacterial co-infection group versus bacterial infection alone group was used as the co-infection group in this plot.
In general, the distribution of viruses was: influenza A 10,192 (64.1%), influenza B 1968 (12.4%), parainfluenza 2065 (13.0%) and RSV 1681 (10.6%). And patients with parainfluenza (8.0%, 95%CI: 6.9–9.3%) or RSV (8.0%, 95%CI: 6.8–9.4%) infection had higher 30-day mortality than those with influenza A (4.7%, 95%CI: 4.3–5.1%) or influenza B (4.2%, 95%CI: 3.3–5.1%). Regarding ICU admission, patients with parainfluenza (5.0%, 95%CI: 4.1–6.0%) or RSV (3.5%, 95%CI: 2.6–4.4%) also had higher frequency than those from influenza A (3.3%, 95%CI: 3.0–3.8%) or influenza B (3.2%, 95%CI: 2.5–4.1%). The difference in 30-day mortality and ICU admission amongst different viruses remained statistically significant in the viral infection alone group (p < 0.001). The clinically suspected co-infection group also showed the significantly difference for 30-day mortality among different viruses (p < 0.001) but not in the laboratory-confirmed co-infection group (p = 0.752).
Subgroup analysis on patients without chronic pulmonary disease or heart failure showed that patients infected with parainfluenza (8.3%, 95%CI: 6.9–9.9%) or RSV (8.1%, 95%CI: 6.5–9.9%) had higher mortality compared to patients with influenza A (3.8%, 95%CI: 3.4–4.3%) or influenza B (3.3%, 95%CI: 2.5–4.3%) (p < 0.001) (Table 3).
Table 3.
Clinical outcomes amongst different viral infection groups.
| Influenza A | Influenza B | Parainfluenza | RSV | p-value* | |
|---|---|---|---|---|---|
| 30-day mortality | |||||
| Viral infection alone, N (%, 95 CI%) | 181/4042 (4.5, 3.9–5.2) | 31/772 (4.0, 3.0–6.0) | 71/843 (8.4, 6.6–10.5) | 54/711 (7.6, 5.8–9.8) | <0.001 |
| Laboratory-confirmed viral-bacterial co-infection, N (%, 95 CI%) | 64/616 (10.4, 8.1–13.1) | 15/145 (10.3, 6.0–16.5) | 21/185 (11.4, 7.1–16.8) | 19/141 (13.5, 8.3–20.2) | 0.752 |
| Clinically suspected viral-bacterial co-infection, N (%, 95 CI%) | 232/5534 (4.2, 3.7–4.8) | 36/1051 (3.4, 2.4–4.7) | 74/1037 (7.1, 5.6–8.9) | 62/829 (7.5, 5.8–9.5) | <0.001 |
| Total N (%, 95 CI%) | 477/10,192 (4.7, 4.3–5.1) | 82/1968 (4.2, 3.3–5.1) | 166/2065 (8.0, 6.9–9.3) | 135/1681 (8.0, 6.8–9.4) | <0.001 |
| ICU admission | |||||
| Viral infection, N (%, 95 CI%) | 129/4042 (3.2, 2.7–3.8) | 21/772 (2.7, 1.7–4.1) | 42/843 (5.0, 3.6–6.7) | 15/711 (2.1, 1.2–3.5) | 0.008 |
| Laboratory-confirmed viral-bacterial co-infection, N (%, 95 CI%) | 50/616 (8.1, 6.1–10.6) | 20/145 (13.8, 8.6–20.5) | 21/185 (11.4, 7.2–16.8) | 12/141 (8.5, 7.2–16.8) | 0.063 |
| Clinically suspected viral-bacterial co-infection, N (%, 95 CI%) | 161/5284 (3.0, 2.6–3.5) | 22/1051 (2.1, 1.3–3.2) | 40/1037 (3.9, 2.8–5.2) | 31/829 (3.7, 2.6–5.3) | 0.145 |
| Total N (%, 95 CI%) | 340/10,192 (3.3, 3.0–3.8) | 63/1968 (3.2, 2.5–4.1) | 103/2065 (5.0, 4.1–6.0) | 58/1681 (3.5, 2.6–4.4) | 0.002 |
| Subgroup history of patients without chronic pulmonary disease or chronic heart failure | |||||
| Total N (%, 95 CI%) | 292/7594 (3.8, 3.4–4.3) | 51/1537 (3.3, 2.5–4.3) | 111/1342 (8.3,6.9–9.9) | 84/1041 (8.1, 6.5–9.9) | <0.001 |
N: Sample size.
*The p-value refers to the comparisons of the outcomes amongst different viral infection groups using chi-square test.
3.3. Prevalence and outcomes of different co-infections
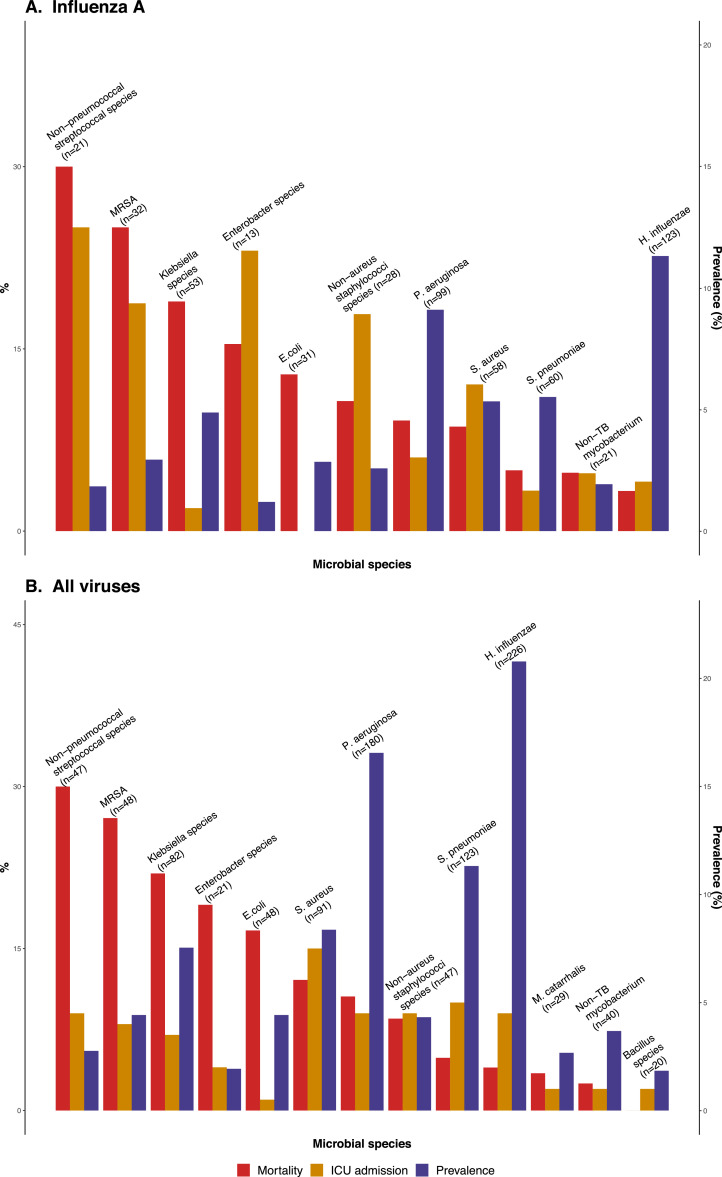
The prevalence of specific pathogen combinations in patients with laboratory-confirmed co-infection and their associated 30-day mortality, ICU admission are shown in Fig. 3. Non-pneumococcal streptococci species, methicillin-resistant S. aureus (MRSA), and Klebsiella species were associated with high mortality ranging from 18.9% to 30.0% in influenza infection group (Fig. 3A) and 22.0% to 30.0% for all viruses (Fig. 3B) but were less common. The prevalence of influenza A and H. influenza co-infection combination was the highest, but mortality was relatively low (4/123, 3.3% of all cases with laboratory-confirmed co-infection) (Fig. 3A). Overall, H. influenzae (226/1087, 20.8%), P. aeruginosa (180/1087,16.6%) and S. pneumoniae (123/1087, 11.3%) were the three most common bacterial pathogens within the laboratory-confirmed co-infection group (Fig. 3B). To determine the contribution of antibiotic resistance to the observed mortality, we carried out a subgroup analysis of 130 patients with co-infection of Klebsiella or Escherichia coli (E. coli). Co-infection with these two bacterial pathogens accounted for high mortality [Klebsiella: 18/82 (22.0%); E. coli: 8/48 (16.7%)]. Within this subgroup, 17 were positive for extended spectrum beta-lactamase (ESBL) isolates [Klebsiella: 3/82 (3.7%); E. coli: 14/48 (13.1%)]. The 30-day mortality was 29.4% (5/17) in this ESBL-positive subgroup versus in 18.6% (21/113) the ESBL-negative subgroup (p = 0.42 by chi-square).
Fig. 3.
Prevalence, 30-day mortality and ICU admission of laboratory confirmed viral-bacterial co-infection combinations, 2013–2017 Crude 30-day mortality and prevalence of different viral-bacterial co-infection combinations were calculated for selected combinations with an incidence ≥ 20 cases. The laboratory confirmed viral-bacterial co-infection combinations were ordered according to the mortality. The percentage of 30-day mortality and ICU admission refereed to the left y-axis and the prevalence refereed to the right one. (A) 30-day mortality, ICU admission and prevalence in influenza A co-infection group (N = 538). (B) A total of 1087 bacterial co-infection cases were identified among a total of 15,906 adults hospitalized for respiratory viral infection.
4. Discussion
In this retrospective cohort study of 19,361 adult patients hospitalized for respiratory infection, 5.6% (1087/19,361) had laboratory-confirmed viral-bacterial co-infection. The 30-day mortality in these patients was significantly higher than those with viral infection alone, bacterial infection alone or clinically suspected co-infection. Patients with laboratory-confirmed co-infection also had a higher rate of ICU admission. Within this co-infection group, the most common bacteria were H. influenzae, P aeruginosa, and S. pneumoniae. Although H. influenzae type b vaccination was recommended by Centers for Disease Control and Prevention in the United States, H. influenzae vaccination is not included in the Childhood Immunization Program in Hong Kong which could be the reason of H. influenzae having such a high prevalence locally.
The rate of laboratory-confirmed viral-bacterial co-infection in this cohort of patients was in the lower range of the previously reported (2–77%) [31,[40], [41], [42], [43], [44], [45]]. This is likely due to heterogeneity of our study population [46], type of respiratory virus [[47], [48], [49]], viral detection methods [20], case definition, community-acquired or nosocomial co-infection, seasonal variation and pandemics [50,51]. Moreover, we showed that laboratory-confirmed co-infection is associated with increased mortality, which is consistent with the findings from a recent meta-analysis on 31 studies consisting of 10, 762 patients with community-acquired pneumonia. Co-infection causes a more complicated course with increased need for mechanical ventilation and vasopressor therapy, that is reflected in our study by the higher rates of ICU admission in patients with laboratory-confirmed viral-bacterial co-infection [10,30,31,[52], [53], [54]]. However, subgroup analysis of ICU patients showed that although the 30-day mortality of the co-infection group was significantly higher than those with viral infection alone, there was no difference to bacterial infection alone group. This may be limited by the small sample size in the bacterial infection alone group.
In our cohort, mortality in patients with clinically suspected viral-bacterial co-infection was similar to those with viral infection alone and lower than those with laboratory-confirmed co-infection. This finding challenges the existence of bacterial infection in the majority of patients with suspected co-infection. In this respect, the neutrophil count, neutrophil-to-lymphocyte ratio and platelet count in patients with clinically suspected co-infection were similar to those with viral infection alone. Clinically, these hematological parameters may be used to differentiate early bacterial infection or viral-bacterial co-infection from viral infection alone.
Another notable finding of this study is that H. influenzae (226/1087, 20.8%), P. aeruginosa (180/1087, 16.8%), and S. pneumoniae (123/1087, 11.3%) were the three most common bacterial pathogens in patients with laboratory-confirmed viral-bacterial co-infection. Given that all co-infection in this cohort was presumably community-acquired (samples collected within 48 h of hospital admission), P. aeruginosa as a more prevalent co-pathogen is surprising. P. aeruginosa had been a rare cause of community-acquired respiratory infection (0.8%−1.9%) [[55], [56], [57]]. However, recent studies reported an increasing rate of P. aeruginosa co-infection with influenza [45]. In our cohort, among the patients who detected with P. aeruginosa, 15.0% (27/180) and 44.4% (80/180) were diagnosed with congestive heart failure and chronic pulmonary disease, respectively. In patients with chronic disease, frequent institutionalized care and recent hospitalization are risk factors for community-acquired P. aeruginosa infection [58,59]. This may explain the higher prevalence of this pathogen in our cohort.
While laboratory-confirmed viral-bacterial co-infection as a nonspecific group had a higher mortality, our results identified specific bacteria that may be associated with increased mortality. Non-pneumococcal streptococcal species and MRSA, as highly virulent species, have high mortality [9/30 (30.0%) and 13/48 (27.1%), respectively]. Infections with these pathogens produce anatomical, functional and immunological changes in the respiratory tract [6,24,[60], [61], [62]], and may predispose and exacerbate subsequent viral or bacterial infections [63], [64], [65], [66]. In addition, co-infection with Klebsiella and E. coli had high mortality [18/82, (22.0%) and 8/48 (16.7%), respectively]. Importantly, ESBL positivity was not associated with a significantly higher mortality, suggesting that the observed high mortality rate in viral-bacterial co-infection with Klebsiella or E. coli was not mainly due to antibiotic resistance.
Among patients with viral infection alone, RSV and parainfluenza infection resulted in lower survival rate than influenza. The mortality difference persisted even in the subgroup of patients without chronic lung disease and congestive heart failure. However, because influenza, predominantly influenza A, was more common, it caused a higher number of deaths. Our results are consistent with previous studies comparing RSV or parainfluenza with influenza [[67], [68], [69]]. Unlike the global initiative for influenza prevention with vaccination among elderly (achieved a coverage of 32.7% to 40.8% in 2013–2017 in those aged >65 population in Hong Kong) [70]. However there is currently no effective vaccination for RSV or parainfluenza, which could cause a higher mortality than influenza in Hong Kong elderly population.
Our study has several limitations. First, this was a retrospective study analyzing electronic health records that was prone to bias from case selection and other processes in the healthcare system. Second, only the first incidence of single viral/bacterial infection was included [71,72]. However, we used a population database to capture all adult hospitalized patients with respiratory viral and bacterial microbiological tests at four major hospitals in Hong Kong over a 5-year period. This resulted in the largest single cohort to date on co-infection in community-acquired respiratory infection. Further studies focusing on multiple bacterial infections or microorganism interactions on metagenomic level are valuable to access the effect of the co-infection combinations on subsequent respiratory infections. Third, because we only included patients who had both viral and bacterial respiratory tests, this excluded patients who did not or could not have concurrent viral and bacterial sampling. Fourth, we did not include adenovirus, enterovirus, rhinovirus, or human metapneumovirus because of inconsistent testing during the study period. This may affect the prevalence of viral-bacterial co-infections. Fifth, unlike viral PCR which has high sensitivity, our study relied on matrix-assisted laser desorption/ionization-time of flight mass spectrometry to identify bacterial pathogens. Furthermore, we were limited by timestamp resolution of antibiotic administration by date and not hours. Therefore some patients may have been given antibiotics prior to bacterial culture which may have reduced yield and biased the reported bacterial pathogen distribution. Similarly, positive sputum culture such as S. pneumoniae or S. aureus could be due to asymptomatic carriage rather than real infection [73]. Nevertheless both of these factors would have reduced rather than exaggerated the mortality difference in patients with co-infection. Another disadvantage of studying co-infection using bacterial culture against viral PCR is the inability to capture pathogens which cannot be cultured. Future research using metagenomics and metatranscriptomics may be useful to develop a complete blueprint of viral-bacterial co-infection of the respiratory tract. Sixth, we were unable to assess the appropriateness of antibiotic therapy according to bacterial sensitivity. However clinicians were trained to adjust antibiotic therapy according to sensitivity, therefore the lack of this assessment should not affect the findings in this study. Furthermore, the rates of resistant organisms (MRSA and ESBL) were low in this cohort (160/4542, 3.5%), therefore this limitation is unlikely to cast major impact in this cohort.
Viral-bacterial co-infection is not uncommon (6.8%) among adult patients hospitalized for respiratory viral infection and is associated with higher mortality and increased need for ICU admission. The most common co-infected bacteria were H. influenzae, P aeruginosa, and S. pneumoniae. Co-infection with non- pneumococcal streptococcal species and MRSA are associated with high mortality.
5. Funding
Commissioned Programmes for Influenza Research, Health and Medical Research Fund (HMRF), FHB (Ref. No.: INF-CUHK-2);
National Natural Science Foundation of China (81873560);
Shenzhen Science and Technology Programme, Shenzhen Science and Technology Innovation Commission (JCYJ20180307150626228);
Health and Medical Research Fund (18170092).
6. Contribution
Lin Zhang, Matthew TV Chan and William KK Wu designed the study, reviewed the data analyses, and approved the final manuscript; Lin Zhang, Lowell Ling and Wai T Wong applied the clinical ethics from Chinese University of Hong Kong; Ying Zhi Liu provided analysis for data interpretation, literature search, writing of the manuscript; Lowell Ling and Sunny H Wong review and design the methodology base on their clinical background, reviewed and drafted the manuscript and data interpretation and verification of submitted data; Maggie HT Wang reviewed the data analyses, and provided statistical comments; J. Ross Fitzgerald, Xuan Zou, Shisong Fang, Xiaodong Liu, and Xiansong Wang helped to oversight of results interpretation and critical review of manuscript; Wei Hu, Hung Chan, Yan Wang, Dan Huang, and Qing Li provided literature search and review, and reviewed the manuscript; Wai T Wong, Gordon Choi, Huachun Zou, David SC Hui, Jun Yu, Gary Tse, and Tony Gin reviewed the manuscript; All authors have read and finally approved the version being submitted. Lin Zhang, Matthew TV Chan, William KK Wu and Lowell Ling had full access to the raw data in the study and accept responsibility to submit for publication.
7. Data sharing statement
Individual participant data that underlie the results reported in this article, after de-identification (text, tables, figures, and appendices), an associated data dictionary is available from the corresponding author. This will be made available to researchers who provide a methodologically sound proposal as well as ethics approval from clinical research ethics committee of Hong Kong Hospital Authority to achieve the aims of the proposal. Please contact: linzhang@cuhk.edu.hk.
Declaration of Competing Interest
Lowell Ling has received consulting fees from Merck Sharp & Dohme. Other authors declared that they have no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.eclinm.2021.100955.
Contributor Information
William KK Wu, Email: wukakei@cuhk.edu.hk.
Matthew TV Chan, Email: mtvchan@cuhk.edu.hk.
Lin Zhang, Email: linzhang@cuhk.edu.hk.
Appendix. Supplementary materials
References
- 1.Bulla A., Hitze K.L. Acute respiratory infections: a review. Bull World Health Organ. 1978;56(3):481–498. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2395579/ [PMC free article] [PubMed] [Google Scholar]
- 2.Millett E.R.C., Quint J.K., Smeeth L., Daniel R.M., Thomas S.L. Incidence of community-acquired lower respiratory tract infections and pneumonia among older adults in the united kingdom: a population-based study. PLoS ONE. 2013;8(9):e75131. doi: 10.1371/journal.pone.0075131. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0075131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garibaldi R.A. Epidemiology of community-acquired respiratory tract infections in adults: incidence, etiology, and impact. Am J Med. 1985;78(6):32–37. doi: 10.1016/0002-9343(85)90361-4. http://www.sciencedirect.com/science/article/pii/0002934385903614 Supplement 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morens D.M., Taubenberger J.K., Fauci A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198(7):962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Metersky M.L., Masterton R.G., Lode H., File T.M., Babinchak T. Epidemiology, microbiology, and treatment considerations for bacterial pneumonia complicating influenza. Int J Infect Dis. 2012;16(5):321. doi: 10.1016/j.ijid.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Morris D.E., Cleary D.W., Clarke S.C. Secondary bacterial infections associated with influenza pandemics. Front Microbiol. 2017;8:1041. doi: 10.3389/fmicb.2017.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson L., Caley J.P., Moore J. Importance of staphylococcus aureus in pneumonia in the 1957 epidemic of influenza a. The Lancet. 1958;272(7040):233–236. doi: 10.1016/S0140-6736(58)90060-6. https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(58)90060-6/abstract [DOI] [PubMed] [Google Scholar]
- 8.Hers J.F., Masurel N., Mulder J. Bacteriology and histopathology of the respiratory tract and lungs in fatal asian influenza. Lancet. 1958;2(7057):1141–1143. doi: 10.1016/s0140-6736(58)92404-8. [DOI] [PubMed] [Google Scholar]
- 9.Oseasohn R., Adelson L., Kaji M. Clinicopathologic study of thirty-three fatal cases of asian influenza. N Engl J Med. 1959;260(11):509–518. doi: 10.1056/NEJM195903122601101. [DOI] [PubMed] [Google Scholar]
- 10.Martín-Loeches I., Sanchez-Corral A., Diaz E. Community-acquired respiratory co-infection in critically ill patients with pandemic 2009 influenza A(H1N1) virus. Chest. 2011;139(3):555–562. doi: 10.1378/chest.10-1396. [DOI] [PubMed] [Google Scholar]
- 11.Yu J., Xie Z., Zhang T. Comparison of the prevalence of respiratory viruses in patients with acute respiratory infections at different hospital settings in north china, 2012-2015. BMC Infect Dis. 2018;18(1):72. doi: 10.1186/s12879-018-2982-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vikerfors T., Grandien M., Olcen P. Respiratory syncytial virus infections in adults. Am Rev Respir Dis. 1987;136(3):561–564. doi: 10.1164/ajrccm/136.3.561. [DOI] [PubMed] [Google Scholar]
- 13.Morales F., Calder M.A., Inglis J.M., Murdoch P.S., Williamson J. A study of respiratory infections in the elderly to assess the role of respiratory syncytial virus. J Infect. 1983;7(3):236–247. doi: 10.1016/s0163-4453(83)97142-6. [DOI] [PubMed] [Google Scholar]
- 14.Falsey A.R., Cunningham C.K., Barker W.H. Respiratory syncytial virus and influenza A infections in the hospitalized elderly. J Infect Dis. 1995;172(2):389–394. doi: 10.1093/infdis/172.2.389. https://academic.oup.com/jid/article/172/2/389/826766 [DOI] [PubMed] [Google Scholar]
- 15.Lien K.S., Jacobs J., Marcus E.A., Strik R.V. The protective effect of intranasal immunization with inactivated influenza virus vaccine. Postgrad Med J. 1973;49(569):175–179. [PMC free article] [PubMed] [Google Scholar]
- 16.Coughtrie A.L., Morris D.E., Anderson R. Ecology and diversity in upper respiratory tract microbial population structures from a cross-sectional community swabbing study. J Med Microbiol. 2018;67(8):1096–1108. doi: 10.1099/jmm.0.000773. [DOI] [PubMed] [Google Scholar]
- 17.Haq K., McElhaney J.E. Ageing and respiratory infections: the airway of ageing. Immunol Lett. 2014;162(1 Pt B):323–328. doi: 10.1016/j.imlet.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Bell B.G., Schellevis F., Stobberingh E., Goossens H., Pringle M. A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect Dis. 2014;14:13. doi: 10.1186/1471-2334-14-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vila J., Ruiz J., Sanchez F. Increase in quinolone resistance in a haemophilus influenzae strain isolated from a patient with recurrent respiratory infections treated with ofloxacin. Antimicrob Agents Chemother. 1999;43(1):161–162. doi: 10.1128/AAC.43.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahony J.B. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21(4):716–747. doi: 10.1128/CMR.00037-07. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2570148/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levandowski R.A., Gerrity T.R., Garrard C.S. Modifications of lung clearance mechanisms by acute influenza A infection. J Lab Clin Med. 1985;106(4):428–432. [PubMed] [Google Scholar]
- 22.Loosli C.G., Stinson S.F., Ryan D.P., Hertweck M.S., Hardy J.D., Serebrin R. The destruction of type 2 pneumocytes by airborne influenza PR8-A virus; its effect on surfactant and lecithin content of the pneumonic lesions of mice. Chest. 1975;67(2 Suppl):7S–14S. doi: 10.1378/chest.67.2_supplement.7s. 2020. [DOI] [PubMed] [Google Scholar]
- 23.Plotkowski M.C., Puchelle E., Beck G., Jacquot J., Hannoun C. Adherence of type I streptococcus pneumoniae to tracheal epithelium of mice infected with influenza A/PR8 virus. Am Rev Respir Dis. 1986;134(5):1040–1044. doi: 10.1164/arrd.1986.134.5.1040. [DOI] [PubMed] [Google Scholar]
- 24.McCullers J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. 2014;12(4):252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 25.Openshaw P.J., Chiu C. Protective and dysregulated T cell immunity in RSV infection. Curr Opin Virol. 2013;3(4):468–474. doi: 10.1016/j.coviro.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stark J.M., Stark M.A., Colasurdo G.N., LeVine A.M. Decreased bacterial clearance from the lungs of mice following primary respiratory syncytial virus infection. J Med Virol. 2006;78(6):829–838. doi: 10.1002/jmv.20631. https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.20631 [DOI] [PubMed] [Google Scholar]
- 27.Tian X., Xu F., Lung W.Y. Poly I:c enhances susceptibility to secondary pulmonary infections by gram-positive bacteria. PLoS ONE. 2012;7(9):e41879. doi: 10.1371/journal.pone.0041879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Small C., Shaler C.R., McCormick S. Influenza infection leads to increased susceptibility to subsequent bacterial superinfection by impairing NK cell responses in the lung. J Immunol. 2010;184(4):2048–2056. doi: 10.4049/jimmunol.0902772. [DOI] [PubMed] [Google Scholar]
- 29.Bellinghausen C., Rohde G.G.U., Savelkoul P.H.M., Wouters E.F.M., Stassen F.R.M. Viral-bacterial interactions in the respiratory tract. J Gen Virol. 2016;97(12):3089–3102. doi: 10.1099/jgv.0.000627. 2020. [DOI] [PubMed] [Google Scholar]
- 30.Johansson N., Kalin M., Hedlund J. Clinical impact of combined viral and bacterial infection in patients with community-acquired pneumonia. Scand J Infect Dis. 2011;43(8):609–615. doi: 10.3109/00365548.2011.570785. [DOI] [PubMed] [Google Scholar]
- 31.Voiriot G., Visseaux B., Cohen J. Viral-bacterial co-infection affects the presentation and alters the prognosis of severe community-acquired pneumonia. Crit Care. 2016;20(1):375. doi: 10.1186/s13054-016-1517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burk M., El-Kersh K., Saad M., Wiemken T., Ramirez J., Cavallazzi R. Viral infection in community-acquired pneumonia: a systematic review and meta-analysis. Eur Respir Rev. 2016;25(140):178–188. doi: 10.1183/16000617.0076-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi S., Hong S., Ko G. Viral infection in patients with severe pneumonia requiring intensive care unit admission. Am J Respir Crit Care Med. 2012;186:325–332. doi: 10.1164/rccm.201112-2240OC. [DOI] [PubMed] [Google Scholar]
- 34.Ho J., Dai R.Z.W., Kwong T.N.Y. Disease burden of clostridium difficile infections in adults, Hong Kong, China, 2006-2014. Emerg Infect Dis. 2017;23(10):1671–1679. doi: 10.3201/eid2310.170797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hospital toolkit for adult sepsis surveillance. https://www.cdc.gov/sepsis/clinicaltools/index.html. Updated 2020.
- 36.Charlson M.E., Pompei P., Ales K.L., MacKenzie C.R. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. https://search.datacite.org/works/10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- 37.Austin P.C. Using the standardized difference to compare the prevalence of a binary variable between two groups in observational research. Commun Stat Simul Comput. 2009;38(6):1228–1234. doi: 10.1080/03610910902859574. [DOI] [Google Scholar]
- 38.Ho D.E., Imai K., King G., Stuart E.A. MatchIt: nonparametric preprocessing for parametric causal inference. J Stat Softw. 2011;42(8):1–28. [Google Scholar]
- 39.Austin P.C. The use of propensity score methods with survival or time-to-event outcomes: reporting measures of effect similar to those used in randomized experiments. Stat Med. 2014;33(7):1242–1258. doi: 10.1002/sim.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruuskanen O., Lahti E., Jennings L.C., Murdoch D.R. Viral pneumonia. The Lancet. 2011;377(9773):1264–1275. doi: 10.1016/S0140-6736(10)61459-6. https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(10)61459-6/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein E.Y., Monteforte B., Gupta A. The frequency of influenza and bacterial co-infection: a systematic review and meta-analysis. Influenza Other Respir Viruses. 2016;10(5):394–403. doi: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brealey J.C., Sly P.D., Young P.R., Chappell K.J. Viral bacterial co-infection of the respiratory tract during early childhood. FEMS Microbiol Lett. 2015;362(10) doi: 10.1093/femsle/fnv062. [DOI] [PubMed] [Google Scholar]
- 43.Crotty M.P., Meyers S., Hampton N. Epidemiology, co-infections, and outcomes of viral pneumonia in adults: an observational cohort study. Medicine. 2015;94(50):e2332. doi: 10.1097/MD.0000000000002332. (Baltimore) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ignacio M.L., Marcus J.S., Jean V. Increased incidence of co-infection in critically ill patients with influenza. Intensiv Care Med. 2016;43(1):48–58. doi: 10.1007/s00134-016-4578-y. https://europepmc.org/article/med/27709265 2020. [DOI] [PubMed] [Google Scholar]
- 45.Falsey A.R., Becker K.L., Swinburne A.J. Bacterial complications of respiratory tract viral illness: a comprehensive evaluation. J Infect Dis. 2013;208(3):432–441. doi: 10.1093/infdis/jit190. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.To K.K.W., Chan K.-., Ho J. Respiratory virus infection among hospitalized adult patients with or without clinically apparent respiratory infection: a prospective cohort study. Clin Microbiol Infect. 2019;25(12):1539–1545. doi: 10.1016/j.cmi.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch S.V. Viruses and microbiome alterations. Ann ATS. 2014;11(Supplement 1):S57–S60. doi: 10.1513/AnnalsATS.201306-158MG. https://www.atsjournals.org/doi/10.1513/AnnalsATS.201306-158MG [DOI] [PubMed] [Google Scholar]
- 48.Smith C.M., Sandrini S., Datta S. Respiratory syncytial virus increases the virulence of streptococcus pneumoniae by binding to penicillin binding protein 1a. A new paradigm in respiratory infection. Am J Respir Crit Care Med. 2014;190(2):196–207. doi: 10.1164/rccm.201311-2110OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egorov A. The problem of bacterial complications post respiratory viral infections. Microbiol Indep Res J. 2018;5(1):12–21. doi: 10.18527/2500-2236-2018-5-1-12-21. https://www.mir-journal.org/issues/5/1/ [DOI] [Google Scholar]
- 50.MacIntyre C.R., Chughtai A.A., Barnes M. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a(H1N1)pdm09. BMC Infect Dis. 2018;18(1):637. doi: 10.1186/s12879-018-3548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice T.W., Rubinson L., Uyeki T.M. Critical illness from 2009 pandemic influenza A virus and bacterial co-infection in the united states. Crit Care Med. 2012;40(5):1487–1498. doi: 10.1097/CCM.0b013e3182416f23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah N.S., Greenberg J.A., McNulty M.C. Bacterial and viral co-infections complicating severe influenza: incidence and impact among 507 U.S. patients, 2013-14. J Clin Virol. 2016;80:12–19. doi: 10.1016/j.jcv.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nin N., Soto L., Hurtado J. Clinical characteristics and outcomes of patients with 2009 influenza A(H1N1) virus infection with respiratory failure requiring mechanical ventilation. J Crit Care. 2011;26(2):186–192. doi: 10.1016/j.jcrc.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 54.Loubet P., Voiriot G., Houhou-Fidouh N. Impact of respiratory viruses in hospital-acquired pneumonia in the intensive care unit: a single-center retrospective study. J Clin Virol. 2017;91:52–57. doi: 10.1016/j.jcv.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neill A.M., Martin I.R., Weir R. Community acquired pneumonia: aetiology and usefulness of severity criteria on admission. Thorax. 1996;51(10):1010–1016. doi: 10.1136/thx.51.10.1010. https://thorax.bmj.com/content/51/10/1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang G.D, Fine M., Orloff J. New and emerging etiologies for community-acquired pneumonia with implications for therapy. A prospective multicenter study of 359 cases. Medicine. 1990;69(5):307–316. doi: 10.1097/00005792-199009000-00004. https://europepmc.org/article/med/2205784 Baltimore. [DOI] [PubMed] [Google Scholar]
- 57.Blanquer J., Blanquer R., Borrás R. Aetiology of community acquired pneumonia in valencia, spain: a multicentre prospective study. Thorax. 1991;46(7):508–511. doi: 10.1136/thx.46.7.508. https://thorax.bmj.com/content/46/7/508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Restrepo M.I., Babu B.L., Reyes L.F. Burden and risk factors for pseudomonas aeruginosa community-acquired pneumonia: a multinational point prevalence study of hospitalised patients. Eur Respir J. 2018;52(2) doi: 10.1183/13993003.01190-2017. [DOI] [PubMed] [Google Scholar]
- 59.Arancibia F., Bauer T.T., Ewig S. Community-acquired pneumonia due to gram-negative bacteria and pseudomonas aeruginosa: incidence, risk, and prognosis. Arch Intern Med. 2002;162(16):1849–1858. doi: 10.1001/archinte.162.16.1849. [DOI] [PubMed] [Google Scholar]
- 60.McCullers J.A. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19(3):571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun K., Yajjala V.K., Bauer C. Nox2-derived oxidative stress results in inefficacy of antibiotics against post-influenza S. aureus pneumonia. J Exp Med. 2016;213(9):1851–1864. doi: 10.1084/jem.20150514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goncheva M.I., Conceicao C., Tuffs S.W. Staphylococcus aureus lipase 1 enhances influenza A virus replication. mBio. 2020 doi: 10.1128/mBio.00975-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen D.T., Louwen R., Elberse K. Streptococcus pneumoniae enhances human respiratory syncytial virus infection in vitro and in vivo. PLoS ONE. 2015;10(5) doi: 10.1371/journal.pone.0127098. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4430531/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoffmann J., Machado D., Terrier O. Viral and bacterial co-infection in severe pneumonia triggers innate immune responses and specifically enhances IP-10: a translational study. Sci Rep. 2016;6(1):38532. doi: 10.1038/srep38532. https://www.ncbi.nlm.nih.gov/pubmed/27922126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J., Li F., Sun R., Gao X., Wei H., Tian Z. Klebsiella pneumoniae alleviates influenza-induced acute lung injury via limiting NK cell expansion. J Immunol. 2014;193(3):1133–1141. doi: 10.4049/jimmunol.1303303. [DOI] [PubMed] [Google Scholar]
- 66.Zhu X., Ge Y., Wu T. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hung I., Li J., To K., Tam A., Chan J. High mortality associated with parainfluenza virus infection in hospitalized adults. Open Forum Infect Dis. 2017;4 doi: 10.1093/ofid/ofx163.1484. S568–. [DOI] [Google Scholar]
- 68.Ackerson B., Tseng H.F., Sy L.S. Severe morbidity and mortality associated with respiratory syncytial virus versus influenza infection in hospitalized older adults. Clin Infect Dis. 2019;69(2):197–203. doi: 10.1093/cid/ciy991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwon Y.S., Park S.H., Kim M. Risk of mortality associated with respiratory syncytial virus and influenza infection in adults. BMC Infect Dis. 2017;17(1):785. doi: 10.1186/s12879-017-2897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Legislative Council of the Hong Kong Special Administrative Region. Seasonal influenza vaccination. https://www.legco.gov.hk/research-publications/english/essentials-1718ise06-seasonal-influenza-vaccination.htm. Updated 2018.
- 71.Agniel D., Kohane I.S., Weber G.M. Biases in electronic health record data due to processes within the healthcare system: retrospective observational study. BMJ. 2018;361:k1479. doi: 10.1136/bmj.k1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pivovarov R., Albers D.J., Sepulveda J.L., Elhadad N. Identifying and mitigating biases in EHR laboratory tests. J Biomed Inform. 2014;51:24–34. doi: 10.1016/j.jbi.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Esposito S., Terranova L., Ruggiero L. Streptococcus pneumoniae and staphylococcus aureus carriage in healthy school-age children and adolescents. J Med Microbiol. 2015;64(Pt 4):427–431. doi: 10.1099/jmm.0.000029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.